Abstract
The selection, acquisition and use of high quality small molecule libraries for screening is an essential aspect of drug discovery and chemical biology programs. Screening libraries continue to evolve as researchers gain a greater appreciation of the suitability of small molecules for specific biological targets, processes and environments. The decisions surrounding the make-up of any given small molecule library is informed by a multitude of variables and opinions vary on best-practices. The fitness of any collection relies upon upfront filtering to avoiding problematic compounds, assess appropriate physicochemical properties, install the ideal level of structural uniqueness and determine the desired extent of molecular complexity. These criteria are under constant evaluation and revision as academic and industrial organizations seek out collections that yield ever improving results from their screening portfolios. Practical questions including cost, compound management, screening sophistication and assay objective also play a significant role in the choice of library composition. This overview attempts to offer advice to all organizations engaged in small molecule screening based upon current best practices and theoretical considerations in library selection and acquisition.
High through-put screening as a means to discover bioactive agents
High through-put screening (HTS) is a widely utilized enabling technology for drug discovery and biomedical research.1,2 Although there are multiple lead-generating technologies, HTS is universally used as a practical method to query large compound collections in search of novel starting points for the development of biologically active compounds.3,4 HTS involves the integration of automation and biological assay screening technologies to evaluate thousands and even millions of compounds. Screening technologies have rapidly evolved over the past twenty years and most often involve assays that measure how compounds modulate phenotypes, targets or pathways. The main advantage of HTS is the ability to accelerate primary screening. Prior to the HTS revolution, screening was significantly more costly and time-consuming which curbed the need for large compound collections. In the past decade, there has been a dramatic increase in the number of new, screenable biological targets and phenotypes as a result of improved assay design and impressive advances in bioinformatics, genomics, and proteomics.5–7 The sudden influx of new therapeutic targets was one driver of the development of automated, miniaturized assays to increase the speed and lower the cost of screening.
Historical context of screening libraries
The 19th century produced numerous scientific advances including progress in the ability to culture microbes. As a result, scientist such as Paul Ehrlich and Sahachirō Hata were among the first to conduct screens to find agents that killed various parasites in culture. The existing assembly of screening libraries of that day was primarily natural products (NPs) and synthetic dyes. The results of these efforts informed library screening for the next century and both classes of agents remain important components of screening collections. NPs, in particular, remain a consistently successful sources of drug leads.8,9 Traditional NP screening is done using crude or roughly fractionated extracts, followed by the isolation of the active compound from the hit extracts which require systematic fractionation guided by assay activity. Many advances in screening formats, reagent production, robotics, and data management were required to support screening via bioassay-guided fractionation of NPs. Despite impressive procedural advances this technique does not compete in terms of throughput with modern HTS efforts. The use of pure, pre-fractionated NPs has emerged as a means to continue screening NPs as part of HTS libraries.
As HTS platforms improved it was a logical conceptual step to replace NP fermentation broths and extracts with synthetically derived chemical compounds. At pharmaceutical firms the archival collections of synthesized compounds became the first collections available for HTS. Merrifield’s publication on solid-phase peptide synthesis brought high-throughput synthesis into the mainstream and Ellman’s publication of the synthesis of a library of 1,4-benzodiazepines demonstrated the feasibility of synthesizing small molecules in a combinatorial fashion.10,11 These studies laid the groundwork for an immense increase in chemical samples and the initial role for combinatorial chemistry at many pharmaceutical companies was to rapidly expand the size of their corporate compound collections. Many early combinatorial libraries were method driven and focused on production resulting in collections with poor physicochemical properties.12,13 The widespread failure of these libraries in generating translatable leads resulted in an increased awareness that each library member should possess lead-like qualities. The broader awareness of lead-likeness resulted in predominately smaller, focused libraries with increased optimization potential.12,14
These strategic revisions led to a higher degree of successful outcomes from HTS campaigns in industry and academia. However, expansion beyond the traditionally defined ‘druggable genome’ have prompted a desire to again revise the dogma of library design.15 Although the number of drug targets is debatable16, it is likely that many traditional compound libraries are inadequate for targeting macromolecule interactions such as protein-protein interactions and nucleic acid-protein interactions. Efforts to find leads within this region of biological space include a renewed investment in NPs, NP inspired libraries, diversity oriented synthesis (DOS) libraries and fragment based approaches.17–20
The need for high quality screening libraries
Small molecules for HTS come from a variety of sources.21,22 In pharma and academia internal programs for creating compound libraries are often complimented with purchased compounds from compound vendors and contract research organizations (CROs). Purified natural products, natural product extracts, purified metabolites, and synthetic/semi-synthetic analogues of both classes still comprise large fractions of many screening libraries. When the totality of sources for small molecules are considered it is clear that there are many millions of potentially available compounds resulting in difficult decisions when selecting appropriate libraries for HTS.
Planning, selecting and acquiring useful libraries requires upfront choices that are informed by both practical considerations and choices that are unavoidably speculative (Figure 1). Several characteristics are universal including the need for a collection that is free of problematic functionalities, soluble in a carrier solvent (typically DMSO) at relevant concentrations, soluble in the assay environment (water) at relevant concentrations, and directly available or easily reacquired for rapid evaluation in secondary confirmatory assays. Organizations with specific research programs and limited target classes may be better served with focused libraries composed of ‘privileged’ scaffold classes (for kinases or GPCR’s, for instance). The clustering density (degree of structural similarity of library members) is an additional library characteristic that can affect the successful outcome of various screening campaigns. Organizations that repetitively screen similar targets may be served by libraries with a higher clustering density while organizations screening diverse targets may opt for lower density in order to assure the maximum degree of diversity in their collection. There are also numerous theoretical concepts regarding library composition. Investigators are rapidly exploring theories on molecular complexity, three-dimensionality and the inclusion of chirality. There are also a myriad of opinions on screening pure, discrete compounds versus mixtures and agents tethered to beads or plates. Library novelty (originality) is an often debated topic with varied opinions. For organizations with intellectual property considerations, library novelty plays a more significant role than it does within organizations whose aims do not involve commercial outcomes. Many CRO’s offer libraries with increasing novelty and exclusivity that are often paralleled by increasing costs. As the field of library development has evolved it has become clear that all libraries benefit from both empirical and predictive judgments of the lead-likeness of the compounds being considered.23 It is the burden of each organization to decide on what types of libraries will best serve their specific needs taking into account molecular complexity, diversity, clustering density, and the inclusion of a variety of debated chemotypes. These topics elicit strong opinions and can have vast consequences on the ultimate utility of any collection of small molecules.
Figure 1.
Upfront Considerations on library composition.
Another significant question surrounds library size and data quality. Practitioners of HTS will universally argue that the fewer the agents needed to screen in order to find useful leads the better; however, there is often disagreement over how to arrive at this outcome. The majority of HTS campaigns are conducted by screening each agent at a single concentration followed by a variety of cheminformatics methodologies to analyze the putative hits to focus on which agents to advance into confirmatory dose-response studies. Several organizations have chosen to perform dose-response primary screening (referred to as quantitative HTS or qHTS) to improve confidence in the primary data and off-set downstream costs.24 The trade-off between library size and data quality is due to the added well-space required for assaying the same compound at multiple doses this technique is limiting in terms of library size. The comparative utility of these approaches remains a greatly debated topic.
Using cheminformatics tools/filters to craft a library
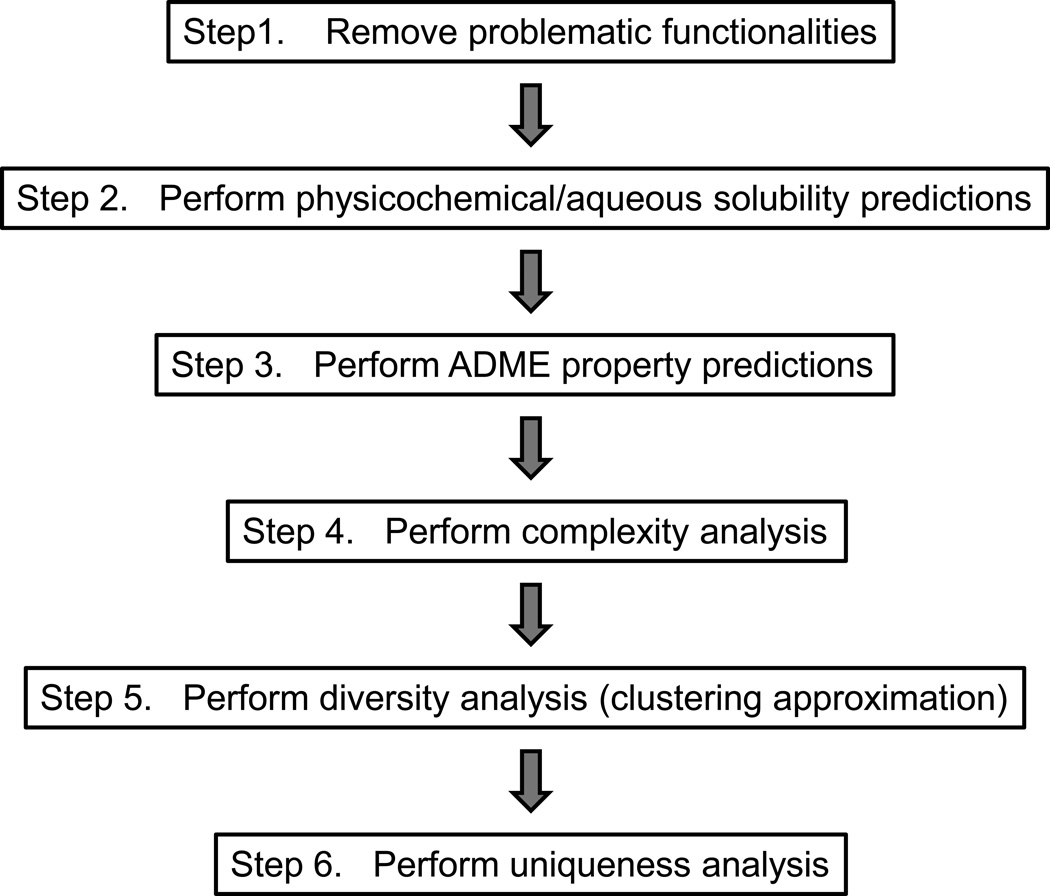
Once an organization has determined their general needs and their theoretical approach to library design there are numerous cheminformatics software tools available to aid researchers in vetting potential libraries to assure that selected constraints will be applied.21,25,26 The use of these tools is nontrivial and experience in cheminformatics staff is an important aspect of any organization wishing to engage in this arena of research. A prerequisite for using cheminformatics tools is the ability to calculate a variety of descriptors for proposed library members using one of the various data formatting systems used to store and sort compounds including SMILES (simplified molecular input line entry specification format) or SDF (structure-data file format)(generally provided by vendors). Many software packages have the ability to perform structural, physicochemical, ADME, complexity and diversity filtering, including software from ACD Labs, Openeye, Tripos, Accelrys, MOE, Pipeline Pilot and Schrodinger. While each organization must choose their specific priorities, a general strategy for filtering is presented in Figure 2.
Figure 2.
A general strategy for cheminformatics filtering.
Step1. Remove problematic functionalities
Among the first considerations for filtering is the elimination of agents that may promiscuously interfere with assay outputs and be confused with authentic assay activity.27 Numerous publications offer advice on compound functionality that is associated with artifactual results. Well known studies include the Pan Assay Interference Compounds (PAINS) listing and the rapid elimination of swill (REOS) filter.28,29 Experience should also inform this aspect of library design and numerous compound functionalities are known to confound HTS results including 2- and 4-halopyridines, sulfonyl halides, aldehydes, alkyl halides, acid halides, iso(thio)cyanates, epoxides, aziridines, thioepoxides, michael acceptors, dihydroxyarenes, disulfides, anhydrides, aromatic azides, trihydroxylarenes, 1,2-dicarbonyls, aminothiazoles, acyl hydrazides, peroxides, perhalo arenes, and anthracenes. Each of these functional groups has a representative SMILES or SMARTS code that can be utilized for batch sorting and elimination (see reference 26 for a comprehensive listing).
Another common source of problematic library members are redox cycling compounds (RCC’s).30 RCC’s are capable of producing hydrogen peroxide in significant enough concentrations to alter many assay outcomes through a variety of non-specific mechanisms including direct oxidation of protein targets (cysteine proteases, phosphatases and metalloenzymes are particularly susceptible), modulation of key assay components (such as DTT, GSH and NADPH) and alteration of cellular systems that respond to changing cellular oxidation states (such as mitochondrial permeabilization and signaling perturbation).31–34 In a recent review on this topic, Johnston summarized several of the molecular functionalities that are common RCC’s including pyrimidotriazinediones, quinones, arylsulfonamides, tolyl-hydrazides and nitrophene-2-carboxylates.35
An interesting province of ‘problematic functionalities’ remains small molecules capable of covalently modifying biochemical targets such as proteins and nucleic acids. Opinions vary on the suitability of these types of agents for inclusion within screening libraries. Undoubtedly, covalent modifiers can alter assay outcomes and small molecules with widely promiscuous covalent labeling of biological constructs have no role as drugs or molecular probes. However, numerous drugs and probes do, in fact, covalently modify specific biological targets resulting in powerful molecular probes and potential clinical agents. A resurgence of efforts to find highly specific covalent modifiers of proteases and subclasses of kinases has challenged existing dogma of removing these agents completely from screening libraries.36,37
Step 2. Perform physicochemical/aqueous solubility predictions
The inability of many small organic molecules to dissolve in aqueous media and/or engage biological targets in a specific, stoichiometric manner was among the primary developmental failures associated with early combinatorial libraries. Outcomes from screening these libraries were plagued with false positives (often attributed to aggregation events) or overly lipophilic leads with limited optimization potential. Lipinski and coworkers were among the first to perform an extensive cheminformatics assessment of solubility and permeability.38 This analysis showed that the 90th percentile of a set of >2000 orally bioavailable agents with acceptable solubility and permeability had a MW under 500, fewer than 5 H-bond donors and 10 H-bond acceptors and a cLogP value less than 5. In the time since this seminal publication a greater appreciation of the physicochemical properties of small molecules has significantly influenced molecular design principles within both library generation and optimization efforts. In addition to Lipinski’s ‘rule of 5’ there are tremendously insightful filters put forth by Ghose, Egan, Oprea, Walters and many others.39–42 These analyses universally put limitations on selected, calculable properties including MW, # of heavy atoms, # of rotatable bonds, # of ring systems, # of hydrogen bond donors, # of hydrogen bond acceptors, LogP and LogD values (see reference 21 for a tabular overview of suggested values).
Filters based upon predictive solubility values are fundamentally associated with analyses of physicochemical properties. However, profiles of experimentally determined solubility values (kinetic and thermodynamic) suggest that solubility cannot be wholly predicted via trend assessments of LogP, PSA, # of rotatable bonds, etc.43–45 There are a number of computational methods for solubility prediction and filtering potential library members utilizing one of these methods may offer an important, additional annotation step.46–50
Step 3. Perform ADME/Tox property predictions
The sophistication in profiling small molecules for selected ADME (absorption, distribution, metabolism and excretion) properties has advanced greatly.51 However, not all organizations engaged in HTS aim to find new drugs.52 As such, filtering potential library members to remove small molecules that possess potential ADME liabilities or toxicity is not universally required. Furthermore, filtering for positive ADME characteristics is achieved, to a degree, during the aforementioned physicochemical filtering step (for instance, small molecules with LogP values between 0.5 and 3 are predicted to have good passive membrane absorption). This not-withstanding, for organizations whose aim is to develop drugs the use of more specific ADME/Tox filters can improve the quality of hits generated by HTS. In recent years, strong predictive models have been developed for intestinal absorption (a proxy of oral bioavailablity), inhibition of selected cytochromes (CYPs)(a predictor of hepatic toxicities and potential drug-drug interactions), binding to the hERG potassium channel (a predictor of potential cardiac associated adverse events), and potential as a substrate for various ATP-binding cassette (ABC) transporters (an indication that a drug may be susceptible to unwanted cellular efflux).53–56 Further, more specific predictive models can aid organizations with targeted drug discovery programs (for instance, blood-brain barrier models exist for organizations with neurological based discovery programs).57 Many of the aforementioned software tools have ADME/Tox predictive capabilities. It should be noted that many ADME/Tox liabilities can be addressed during lead development (post-screen) mitigating the need for stringent filtering during library development.
Step 4. Perform complexity analysis
The complexity of biologically relevant molecules ranges from simple (ethanol) to extraordinarily intricate (actinomycin). The role that molecular complexity plays in the success and failure of HTS is intensely debated.58,59 There are multiple examples of successful drugs and probes with varying degrees of molecular complexity that can be used to justify both highly intricate small molecules and agents with less molecular complexity. Performing a meaningful analysis of the complexity of small molecules is often influenced by personal bias. While computational methods to probe molecular complexities exist60, they are not as widely utilized as those that examine physicochemical and ADME/Tox properties. Principle component analyses that define three-dimensional orientation using dihedral angles and distances and take into account structural and physical properties including potential energy, volume, shape, water-accessible surface area, and conformation dependent charge descriptors such as dipole moment offer the best approximation of overall library complexity. More straightforward assessments can be achieved via the summation of chiral centers per each library member and direct measurement of the sp2:sp3 hybridization ratio for the collection.61 Ultimately, each organization must consider their own needs and biases prior to defining the complexity range of their screening libraries.
Step 5. Perform diversity analysis (clustering approximation)
Choosing the level of diversity (interrelatedness) within a screening collection is inherently tied to the types of screens an organization plans to conduct. Organizations frequently screening kinases, for instance, may choose targeted libraries of privileged scaffolds (agents known to target the ATP-binding domain, for instance) with a higher clustering density. However, even organizations with diverse interests benefit from a degree of structural clustering. Screening libraries embedded with structural similitude will benefit during the triage process in detecting actives amidst the random noise associated with HTS due to the likelihood that active agents with related library members will show uniformity in their activity. As a correlate, a single active compound from a clustering of highly related library members that are inactive should be viewed with skepticism. Further, structurally related actives offer potential insight into structure-activity relationships (SAR) from the primary assay. Achieving the proper balance in clustering density will have a major impact in screening success.
Methods for performing diversity analyses vary and many are based upon structural descriptors and fingerprints contained in SMILES and SMARTs line notation descriptors.62–65 Others utilize chemical scaffolding analyses to gain an appreciation of the similarity of small molecules in three-dimensions.66 Most filters based upon two dimensional descriptors rely upon the Tanimoto coefficient to calculate similarity and a recent analysis by Willett suggested that this construct remains the most reliable method.67 Utilizing the Tanimoto coefficient (or an related similarity scoring construct) within a clustering algorithm (such as Jarvis-Patrick) to define cluster size is a common method for diversity filtering. Organizations will have to define the coefficient values that will define clustering bins and their level of clustering density (a minimum and maximum cluster size).
Step 6. Perform uniqueness analysis
For organizations whose goals include commercialization the uniqueness of their products plays a significant role in all aspects of the drug discovery process. Any small molecule that does not possess freedom to operate as a unique entity will be of limited use to for-profit organizations. Pharma trained medicinal chemists working with patent attorneys are adroit at incorporating freedom to operate assessments into their SAR explorations when necessary. However, screening libraries with high levels of uniqueness can provide leads with no intellectual property-based encumbrances during optimization. As such, performing a uniqueness analysis on small molecule libraries can be worthwhile. Unfortunately, commercial filters for uniqueness are not as commonly used by scientists as filters for physicochemical and ADME properties. Intellectual property attorneys often use searches (known as Markush searches) to perform freedom to operate assessments during patent prosecutions using databases such as the CAS MARPAT and INPI Merged Markush Service databases. Another widely used database is the Derwent World Patents Index which offers fragment codes which can be employed in similar analyses to the aforementioned fingerprint analyses that can be done in batch mode to highlight small molecules with broad patent coverage and limited freedom to operate during optimization. It should be noted that freedom to operate can often be achieved during lead optimization limiting the need for stringent filtering based upon uniqueness. Further, screening only novel, untested libraries may be excessively risky and rigid filtering based upon uniqueness could ultimately disappoint.
Specialty libraries/methodologies
In addition to unbiased compound collections there are numerous classes of specialty libraries available for HTS.68 Specialty libraries can come in a variety of forms however we will highlight five illustrative types based on the intended application or on the source of compounds. These include libraries for specific targets classes, libraries of drugs and bio-actives, libraries for fragment based screening and libraries of complex synthetic compounds.
Target class libraries
Screening collections that are biased towards certain classes of biological targets are increasingly popular. Libraries enriched with privileged chemotypes that target kinases, G-protein coupled receptors (GPCRs), nuclear hormone receptors, ion channels and CNS targets are available from many vendors. The basis for the ‘privileged’ nature of these libraries and the design principles for each vary based upon target class.69 For instance, kinase directed libraries take advantage of the homology associated with the ATP-binding site of kinases. Several chemotypes have been identified that bind to this domain and analogs of these privileged chemotypes are typically present in kinase-biased libraries. GPCRs are characterized by a seven transmembrane bundle. Years of intense focus on GPCRs for the development of drugs has allowed for the identification of compound classes that demonstrate marked affinity to the modestly homologous transmembrane domains. In a similar way, libraries of compounds are available for nuclear hormone receptors and ion channels based on the previously established inhibitors for these classes of targets. The compounds in CNS-biased libraries are typically biased by physical properties typical of compounds that penetrate the blood-brain barrier, a feature necessary for drugs for CNS targets. Collectively, these targeted libraries might offer a higher likelihood of identifying starting leads when the collection is properly applied to target classes. A potential downside to targeted libraries involves the repetitive use of related compound collections across multiple organizations resulting in freedom to operate issues. In some of these instances, the optimization of common, well-known leads must then take into account patentability space in addition to optimizing for SAR and structure property relationships (SPR).
Known drugs/pharmacologically active collections
Approved drug collections can be valuable for many applications including repurposing efforts and validating targets and pathways. Screening libraries of existing drugs that operate by different mechanisms can also lead to new treatments. Researchers such a Jun Liu have reported numerous, surprising findings associating clinically approved agents for one indication with an unrelated efficacy within a separate indication (for instance, the antifungal drug itraconazole as a potential anti-cancer agent).70 A thorough list of approved drugs for human use was recently reported by the National Institutes of Health Chemical Genomics Center (NCGC). Details associated with this collection are freely available online at http://tripod.nih.gov/npc/. This set is publicly available for use in HTS, and other versions of approved drug libraries are available from various commercial vendors.71 In addition to known drug libraries, many organizations make regular use of libraries of optimized agents with well defined pharmacological activities. Popular libraries like Sigma-Aldrich’s LOPAC (Library of Pharmacologically Active Compounds) and the Prestwick Chemical Library collections are often used for validating assays for HTS.
Fragment based screening (FBS) collections
FBS is now routinely carried out in many pharmaceutical companies and academic laboratories as an alternate means to identifying small molecule binders to proteins.72,73 The requirements for small molecules used in FBS are unique.74 A primary difference involves library size. While traditional HTS can be run with tens of thousands or even millions of compounds, FBS is typically run on thousands of agents. A second primary difference is the concentration at which fragment libraries are screened. The functional concentration ranges for traditional HTS range from the nanomolar to micromolar. In contrast, FBS is typically run in the millimolar range. This requirement puts additional pressure on selecting small molecules with aqueous solubility. Fragments, by definition, typically have lower molecular weights and a general cut-off is routinely set at 300 Daltons. Additional criteria include a hydrogen bond donor count less than or equal to 3, hydrogen bond acceptors less than or equal to 3, cLogP less than or equal to 3, and number of rotatable bonds less than or equal to 3. There are a number of methodologies by which fragments are screened including NMR, crystallography and MS based methods.75 An important aspect of the supporting optimization efforts involves the need for linker functionality (masked or unmasked) within the screened agents. Without such functional groups the ability to rapidly optimize ‘hits’ is limited. Libraries of compounds suitable for FBS are available from a number of commercial organizations.
Diversity-oriented synthesis (DOS) collections
The failure of existing compound collections to routinely deliver optimizable hits for ‘undruggable’ targets has inspired many synthetic chemists to explore novel chemical space in the form of DOS. The goal of DOS is to produce compounds with complex architectures that are designed to explore new areas of biological space.76 The structures of such compounds are intended to be highly dissimilar to traditional, commercially available libraries with a focus on three-dimensionality and the unabashed inclusion of chiral centers.19,77,78 As a result, these agents are generally only accessible to organizations willing to make their own investment in library generation. To a limited extent, selected academic labs have contributed DOS libraries to the National Institute of Health Molecular Libraries Small Molecule Repository (NIH-MLSMR). Currently there are about 17,000 compounds (annotated as ‘non-commercial’) in the MLSMR collection that best represents this class of compounds in a publicly available screening collection (see, http://mli.nih.gov/mli/compound-repository/mlsmr-compounds/). Additionally, the Broad Institute has synthesized approximately 100,000 DOS compounds with complex structures that are available for collaborative programs.
In addition to these types of specialty libraries the use of peptides and peptidomimetics is still widely employed in HTS.79,80 Novel classes of small molecule constructs that mimic the structure of biologically relevant molecules are also being screened on a regular basis. These include peptoids, peptide nucleic acids (PNA) and locked nucleic acids (LNA).81–83 Many of these structural classes have found utility versus difficult target classes. For instance, the Kodadek lab has utilized peptoid libraries to find and optimize small molecules with high affinity to antigens that demonstrate utility as biomarkers for Alzheimer’s disease.84 In addition to these library types, many scientific teams have expanded the use of small molecule libraries bound to solid supports for screening. Solid-phase screening on beads and plates is common and other solid supports such as cellulous have proven to be lucrative by innovated groups like the Blackwell lab.85 Ellman and coworkers have also introduced novel libraries that integrate fluorophore reporters directly into library members to facilitate screening.86 This approach, referred to as substrate activity screening, has tremendous relevancy toward finding inhibitors of enzymes that rely upon active site nucleophilic events and has already yielded several novel protease inhibitors.87
Compound sourcing
Once an organization has defined their goals and established a methodology for assembling a set of libraries for screening they must begin the task of compound sourcing. Nontraditional libraries (DOS, peptoids, PNA, LNA) or methodologies (bead-based screening, SAS) are not supported by commercial vendors. As a result, organizations that pursue these types methodologies have to generate their libraries through in-house synthetic efforts or partner with organizations with experience in these areas.
There are numerous CRO’s that provide more conventional libraries for screening efforts. These include fragment libraries, targeted libraries, drug collections, pharmacologically defined collections and unbiased diversity collections. Several online databases offer insight into the composition of these commercial libraries. Chemspider (http://www.chemspider.com/) offers a database of twenty six million small molecules (as of October 2011) searchable by structure, various naming constructs and by descriptors such as reference, physical properties and supplier. The Shoichet lab has made available the ZINC database (http://zinc.docking.org/index.shtml) which has compiled a listing of thirteen million purchasable compounds (as of October 2011). A major advantage of this database is the use of numerous formats that support virtual screening efforts. Another database is eMolecules (http://www.emolecules.com/) which consists of eight million unique structures (as of October 2011) and allows researchers to down-load compound collections. This database will also curate, purchase and format libraries on a fee-for-service basis. Discovery Gate (www.discoverygate.com/) offers a searchable database of over twenty five million structures (as of October 2011) and includes synthetic insights into many of these agents as well as property evaluations and sourcing information. ChemExper (http://www.chemexper.com/) offers a free search engine that allows quires via molecular formula, IUPAC name, common name, CAS number and even catalog numbers. The Pubchem database (http://pubchem.ncbi.nlm.nih.gov/) is the NIH’s repository of both compound collections available for screening through the molecular libraries program (MLI) but also the related biological activity of each library member (updated as activities are defined). DrugBank (http://drugbank.ca/) is an online repository of over six thousand clinical agents and associated data (including structures, activities and targets). This database, along with the previously mentioned NPC database, offers tremendous insight into the existing pharmacopeia. In addition to these databases there are many CRO’s whose non-proprietary libraries are available online for down-loading. A listing of some of the well-known CRO’s who offer libraries is provided in Table 1. ChemNavigator (http://www.chemnavigator.com/) and eMolecules serve as good aggregator by handling sourcing of compounds from multiple vendors.
Table 1.
Selected Vendors for Diversity, Targeted and Specialty Libraries
| ACB Blocks | ACC Corp. | ActiMol | AKOS |
| Albany Mol. Res. | Alchemia Ltd. | Alinda Chem. | AllLab |
| Analyticon Discovery | ApiChem. Corp. | Aronis | ASDI |
| Asinex | Aurora | Axon MedChem | Biofocus |
| Bionet | Biotech Corp. Am. | BOC Sciences | Cerep |
| Chem TI | Chembridge | ChemDiv | ChemRoutes |
| ChemStar | Chemical Block | ChemOvation | Comgenex |
| EMC Microcollection | Enamine | Enzo Life Sciences | EvoBlocks |
| Exclusive Chem. | FCHC | Innovapharm | InterBioScreen |
| InterMed | Key Organics | Labotest | Life Chemicals |
| Librophyt | Maybridge | MDPI | Menai Organics |
| MicroCombiChem e.K. | NanoSyn | Otava | Peakdale |
| PepTech | PharmaBlock | Pharmeks | Princeton BioMol. Res. |
| Pyxis discovery | Scientific exchange | Selleck Chemicals | Sequoia Res. Prod. |
| Sigma-Aldrich | Specs | Spectrum Info | SynKinase |
| Synthon-Lab | Szintekon | TimTec | Tocris Biosciences |
| TOSLab | Tripos | Vitas Labs | Zelinsky Institute |
Specialty Libraries
Integrating virtual compound collections
While different organizations have assorted screening capacities, even the largest pharmaceutical firms are not capable of screening the entirety of available small molecules. As such, virtual screening is a popular method to compliment HTS, specifically when detailed structural knowledge of the target is available.88–90 In addition to complimenting HTS efforts, virtual docking of a library of commercially available compounds provides a facile mechanism to enrich an organizations screening collection with chemotypes predicted to have binding affinity to a target of interest. The cost of virtual screening efforts is comparatively low and negates the need for HTS instrumentation making virtual screening an attractive discovery methodology for some smaller organizations and academic groups. While the effectiveness of virtual screening continues to be debated91, there are an increasing number of reports detailing the successful use of this technique to yield lead agents for optimization.92 Virtual screening methods have been reviewed elsewhere.93 Modeling software is a prerequisite for in house virtual screens and programs from Tripos, Chemical Computing Group, Accelrys and Schrödinger are popular and widely used. There are several online virtual screening platforms including the previously mentioned ZINC database (http://zinc.docking.org/index.shtml)(see reference 88 for a comprehensive listing of available software).
Compound management
Following acquisition the utility of a compound library will be determined by the quality and efficiency of an organizations compound management capability.94–96 Compound management encompasses the receipt, preparation, storage, registration, QC and tracking of all small molecules in a manner that protects the integrity of each sample. Proper receipt, preparation and storage of small organic compounds are critical to their stability and a properly partitioned and stored library plates can dramatically influence the usable lifespan of a library. Registration and tracking of individual compounds is critical and improper documentation of structure or well-location can have drastic effects in screening follow-up efforts.
The totality of steps, variables and potential problems associated with compound management encompass a protocol within its own right and several reviews have been published on the subject that offer glimpses into the complexities of this field.97–100 Receipt of agents involves both receiving of the physical samples [typically sent as neat (or dry) powders/liquids or as stock solutions of known concentrations in a carrier solvent (typically DMSO)] and registration of structures into a usable and flexible compound management software system. A multitude of problems can arise in both steps and seemingly trivial issues such as arriving at a coding system, choice of stock concentration and the number of daughter plates to generate should not be ignored. The conversion of powders/liquids to stock solutions requires an investment in automated weighing/liquid handling systems.101 The solubility of organic compounds in DMSO at typical concentrations (5–30 mM) is not guaranteed and efforts should be made to test the concentration of random samples to assure that automated processes are producing accurate stocks. HPLC systems connected to a chemiluminescent nitrogen detector (CLND) are useful and commonly used system for determining the concentrations of most solutions used in HTS. The level of miniaturization is accounted for during the preparation of testable compound plates and common screening platforms have been uniformly engineered to accommodate 96-, 384-, and 1536-well plates. From the primary plate (mother plate) each organization must decide how many daughter plates to build and store. The plurality of organic molecules stored in DMSO and routinely sourced for HTS will require replacement over time due to degradation.102 While the creation of daughter plates is costly and time consuming they allow for facile replacement of plates that are no longer viable due to compound degradation. A flexible and integrated software system is required to assure that all registered structures are properly associated with plate location. The compound management team typically takes responsibility for the sourcing of samples for follow-up efforts. Most follow-up plates involve the generation of complete response curves to show the dose dependency of activity for each compound. The dilution of each library member to provide a dose response is another non-trivial aspect of compound management and details such as mixing strategies to assure proper concentrations are achieved should be taken into account. Without doubt, managing the complexities of compound management is a vital aspect to HTS efforts.
Library QC and profiling
Once a library is plated a rigorous quality control schedule must be put in place to determine that the library is correct and pure. Traditional LCMS and UPLC systems do not possess the level of throughput to QC libraries of even modest size in a timely manner. LC based judgments of purity (based upon UV, MS and ELSD signatures) are useful for conducting random sampling of compounds and many organizations allow these random assessments to inform on the entirety of the collection. Each organization must set a cutoff for compound purity and many have settled on 90% based upon UV peak integration. QC methods to assess the integrity of an entire library are limited. A recent effort by Auld and coworkers utilized data integrity [as judged by the assays minimum significant ratio (MSR) variation] within a screen of selected Cyps assayed at 7 times over a 37 week time period to QC a library.103 This study resulted in a data set that suggested replacing plates every four months. Profiling libraries for solubility and for native activity within certain assay systems and outputs can also prevent costly and time-consuming mistakes during follow-up efforts. Brenk et al reported a facile proxy assay that informs on solubility by dry spotting compounds into unoccupied plates followed by the addition of PBS (pH 7.4).26 After a 1 hour incubation sample wells were judged for absorption at 620 nm and compared to a 30 µM pyrene control to gain an appreciation of agents that may have solubility issues. As many assay outputs involve the generation or absorption of fluorescence it is useful to know which agents in a collection have fluorescence potential. Inglese and coworkers recently reported the spectroscopic profiling of a large compound library versus six common spectral regions associated with commonly used fluorophores in HTS.104 The results suggested that many compound libraries will have significant interference potential within the blue region of UV/visible spectrum. Another suggested profile involves the examination of the binding/inhibition potential of a library for common reporter constructs like luciferase or beta-lactamase. As these constructs are enzymes there exists the potential for small molecules to modify their behavior which would mimic desired assay outputs within screens designed to utilize these common assay components. Auld et al recently reported a profile that demonstrated that a large fraction of library members are capable of inhibiting luciferase directly.105 Interestingly, small molecule inhibitors of luciferase can paradoxically present as activators in selected, cell based assay systems as well.106 One of the best characterized mechanisms by which small molecules can produce artifactual assay outcomes is through the well-characterized phenomenon of aggregation.107 Austin, Shoichet and coworkers have reported several profiles of aggregation events demonstrating the prevalence of assay artifacts caused by aggregation.108–110 A recent profile of kinetic solubility demonstrated that small molecule aggregation is related to aqueous solubility but exceptions do occur and there is general agreement that aggregation is highly dependent on assay environment.45
Conclusion
The central aim of screening collections of chemical compounds is to provide ligands that perturb biological processes of interest and to identify novel and robust starting points for drug/probe development. The optimal composition and assembly of a compound collection is of vital interest, however there is no single correct methodology for assembling an ideal library. Herein, we have attempted to provide an overview of the myriad of considerations that inform scientist during the assembly, procurement and handling of small molecules intended for HTS. We have highlighted library design strategies that are considered traditional and those emerging strategies that will undoubtedly gain appreciation as new results are generated. As different organizations have unique goals each will ultimately have divergent requirements with only a few conventions (such as aqueous solubility) being universally applied. This notwithstanding, best practices within in silico predictive filtering, compound management and library QC will improve the likelihood that all libraries designs are ultimately useful. These principles are under constant reevaluation and to a very large degree the future of drug discovery hinges on the innovation of the scientists who pursue library design.
Acknowledgements
S.D., N.S. and C.J.T. would like to thank the National Institutes of Health Molecular Libraries Probe Production Centers Network (NIH-MLPCN) at the Broad Institute (U54 HG005032-1) and the NIH Chemical Genomics Center (U54 MH084681) and the Intramural Research Program of the National Human Genome Research Institute, National Institutes of Health for research support.
References
- 1.Mayr LM, Bojanic D. Novel trends in high-throughput screening. Curr. Opin. Pharm. 2009;9:580–588. doi: 10.1016/j.coph.2009.08.004. [DOI] [PubMed] [Google Scholar]
- 2.Inglese J, Johnson RL, Simeonov A, Xia M, Austin CP, Auld DS. High-throughput screening assays for the identification of chemical probes. Nat. Chem. Bio. 2007;3:466–479. doi: 10.1038/nchembio.2007.17. [DOI] [PubMed] [Google Scholar]
- 3.Macarron R, Banks MN, Bojanic D, Burns DJ, Cirovic DA, Garyantes T, Green DVS, Hertzberg RP, Janzen WP, Paslay JW, Schopfer U, Sittampalam GS. Impact of high-throughput screening in biomedical research. Nat. Rev. Drug Disc. 2011;10:188–195. doi: 10.1038/nrd3368. [DOI] [PubMed] [Google Scholar]
- 4.Bleicher KH, Böhm H-J, Müller K, Alanine AI. Hit and lead generation: beyond high-throughput screening. Nat. Rev. Drug Disc. 2003;2:369–378. doi: 10.1038/nrd1086. [DOI] [PubMed] [Google Scholar]
- 5.Ambesi-Impiombato A, di Bernardo D. Computational biology and drug discovery: from single-target to network drugs. Curr. Bioinform. 2006;1:3–13. [Google Scholar]
- 6.Kramer R, Cohen D. Functional genomics to new drug targets. Nat. Rev. Drug Disc. 2004;3:965–972. doi: 10.1038/nrd1552. [DOI] [PubMed] [Google Scholar]
- 7.Cox J, Mann M. Is proteomics the new genomics? Cell. 2007;130:395–398. doi: 10.1016/j.cell.2007.07.032. [DOI] [PubMed] [Google Scholar]
- 8.Butler MS. The role of natural product chemistry in drug discovery. J. Nat. Prod. 2004;67:2141–2153. doi: 10.1021/np040106y. [DOI] [PubMed] [Google Scholar]
- 9.Newman DJ, Cragg GM. Natural Products as sources of new drugs over the last 25 years. J. Nat. Prod. 2007;70:461–477. doi: 10.1021/np068054v. [DOI] [PubMed] [Google Scholar]
- 10.Merrifield RB. Solid phase peptide synthesis. I. The synthesis of a tetrapeptide. J. Am. Chem. Soc. 1963;85:2149–2154. [Google Scholar]
- 11.Bunin BA, Plunkett MJ, Ellman JA. The combinatorial synthesis and chemical and biological evaluation of a 1,4-benzodiazepine library. Proc. Natl. Acad. Sci. USA. 1994;91:4708–4712. doi: 10.1073/pnas.91.11.4708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Myers PL. Will combinatorial chemistry deliver real medicines? Curr. Opin. Biotech. 1997;8:701–707. doi: 10.1016/s0958-1669(97)80123-1. [DOI] [PubMed] [Google Scholar]
- 13.Kennedy JP, Williams L, Bridges TM, Daniels RN, Weaver D, Lindsley CW. Application of combinatorial chemistry science on modern drug discovery. J. Comb. Chem. 2008;10:345–354. doi: 10.1021/cc700187t. [DOI] [PubMed] [Google Scholar]
- 14.Mayr LM, Fuerst P. The future of high-throughput screening. J. Biomol. Screen. 2008;13:443–448. doi: 10.1177/1087057108319644. [DOI] [PubMed] [Google Scholar]
- 15.Drewry DH, Macarron R. Enhancements of screening collections to address areas of unmet medical need: an industry perspective. Curr. Opin. Chem. Bio. 2010;14:289–298. doi: 10.1016/j.cbpa.2010.03.024. [DOI] [PubMed] [Google Scholar]
- 16.Overington JP, Al-Lazikani B, Hopkins AL. How many drug targets are there? Nat. Rev. Drug Discov. 2006;5:993–996. doi: 10.1038/nrd2199. [DOI] [PubMed] [Google Scholar]
- 17.Koehn FE, Carter GT. The evolving role of natural products in drug discovery. Nat. Rev. Drug Discov. 2005;4:206–220. doi: 10.1038/nrd1657. [DOI] [PubMed] [Google Scholar]
- 18.Kumar K, Waldmann H. Synthesis of natural product inspired compound collections. Angew. Chem. Int. Ed. 2009;48:3224–3242. doi: 10.1002/anie.200803437. [DOI] [PubMed] [Google Scholar]
- 19.Tan DS. Diversity-oriented synthesis: exploring the intersections between chemistry and biology. Nat. Chem. Biol. 2005;1:74–84. doi: 10.1038/nchembio0705-74. [DOI] [PubMed] [Google Scholar]
- 20.Hajduk PJ, Greer J. A decade of fragment-based drug design: strategic advances and lessons learned. Nat. Rev. Drug Discov. 2007;6:211–219. doi: 10.1038/nrd2220. [DOI] [PubMed] [Google Scholar]
- 21.Huggins DJ, Venkitaraman AR, Spring DR. Rational Methods for the Selection of Diverse Screening Compounds. ACS Chem. Biol. 2011;6:208–217. doi: 10.1021/cb100420r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hergenrother PJ. Obtaining and screening compound collections: a user’s guide and a call to chemists. Curr. Opin. Chem. Bio. 2006;10:213–218. doi: 10.1016/j.cbpa.2006.04.005. [DOI] [PubMed] [Google Scholar]
- 23.Rishton GM. Nonleadlikeness and leadlikeness in biochemical screening. Drug Discov. Today. 2003;8:86–96. doi: 10.1016/s1359644602025722. [DOI] [PubMed] [Google Scholar]
- 24.Inglese J, Auld DS, Jadhav A, Johnson RL, Simeonov A, Yasgar A, Zheng W, Austin CP. Quantitative high-throughput screening: A titration-based approach that efficiently identifies biological activities in large chemical libraries. Proc. Natl. Acad. Sci. USA. 2006;103:11473–11478. doi: 10.1073/pnas.0604348103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Walters WP, Namchuck M. Designing Screens: How to make your hits and hit. Nat. Rev. Drug Discov. 2003;2:259–266. doi: 10.1038/nrd1063. [DOI] [PubMed] [Google Scholar]
- 26.Brenk R, Schipani A, James D, Krasowski A, Gilbert IH, Frearson J, Wyatt PG. Lessons learnt from assembling screening libraries for drug discovery for neglected diseases. Chem Med Chem. 2008;3:435–444. doi: 10.1002/cmdc.200700139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Thorne N, Auld DS, Inglese J. Curr. Opin. Chem. Bio. 2010;14:315–324. doi: 10.1016/j.cbpa.2010.03.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Baell JB, Holloway GA. New substructure filters for removal of pan assay interference compounds (PAINS) from screening libraries for their exclusion in bioassays. J. Med. Chem. 2010;53:2719–2740. doi: 10.1021/jm901137j. [DOI] [PubMed] [Google Scholar]
- 29.Walters WP, Ajay, Murcko MA. Recognizing molecules with drug-like properties. Curr. Opin. Chem. Bio. 1999;3:384–387. doi: 10.1016/s1367-5931(99)80058-1. [DOI] [PubMed] [Google Scholar]
- 30.Soares K, Blackmon N, Shun TY, Shinde SN, Takyi HK, Wipf P, Lazo JS, Johnston PA. Profiling the NIH small molecule repository for compounds that generate H2O2 by redox cycling in reducing environments. Assay Drug Dev. Technol. 2010;8:152–174. doi: 10.1089/adt.2009.0247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bova MP, Mattson MN, Vasile S, Tam D, Holsinger L, Bremer M, Hui T, McMahon G, Rice A, Fukuto JM. The oxidative mechanism of action of ortho-quinone inhibitors of protein-tyrosine phosphatase (alpha) is mediated by hydrogen peroxide. Arch. Biochem. Biophys. 2004;429:30–41. doi: 10.1016/j.abb.2004.05.010. [DOI] [PubMed] [Google Scholar]
- 32.Lal M, Rao R, Fang X, Schuchmann HP, Sonntag CV. Radical-induced oxidation of dithiothreitol in acidic oxygenated solution: a chain reaction. J. Am. Chem. Soc. 1997;119:5735–5739. [Google Scholar]
- 33.Forman H, Maiorino M, Ursini F. Signaling functions of reactive oxygen species. Biochemistry. 2010;49:835–842. doi: 10.1021/bi9020378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kruglov AG, Subbotina KB, Saris N-EL. Redox-cycling compounds can cause the permeabilization of mitochondrial membranes by mechanisms other than ROS production. Free Radical Bio. Med. 2008;44:646–656. doi: 10.1016/j.freeradbiomed.2007.10.049. [DOI] [PubMed] [Google Scholar]
- 35.Johnston PA. Redox cycling compounds generate H2O2 in HTS buffers containing strong reducing reagents – real hits or promiscuous artifacts. Curr. Opin. Chem. Bio. 2011;15:174–182. doi: 10.1016/j.cbpa.2010.10.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Singh J, Petter RC, Kluge A. Targeted covalent drugs of the kinase family. Curr. Opin. Chem. Bio. 2010;14:475–480. doi: 10.1016/j.cbpa.2010.06.168. [DOI] [PubMed] [Google Scholar]
- 37.Zhou W, Ercan D, Chen L, Yun C-H, Li D, Capelletti M, Cortot AB, Chirieac L, Iacob RE, Padera R, Engen JR, Wong K-K, Eck MJ, Gray NS, Jänne PA. Novel mutant-selective EGFR kinase inhibitors against EFGR T790M. Nature. 2009;462:1070–1074. doi: 10.1038/nature08622. 24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lipinski CA, Lombardo F, Dominy BW, Feeney PJ. Experimental and computational approaches to estimate solubility and permeability in drug discovery and development settings. Adv. Drug Del. Rev. 2001;46:3–26. doi: 10.1016/s0169-409x(00)00129-0. [DOI] [PubMed] [Google Scholar]
- 39.Ghose AK, Viswanadhan VN, Wendoloski JJ. A knowledge-based approach in designing combinatorial or medicinal chemistry libraries for drug discovery. 1. A qualitative and quantitative characterization of known drug databases. J. Comb. Chem. 1999;1:55–68. doi: 10.1021/cc9800071. [DOI] [PubMed] [Google Scholar]
- 40.Egan WJ, Merz KM, Baldwin JJ. Prediction of drug absorption using multivariate statistics. J. Med. Chem. 2000;43:3867–3877. doi: 10.1021/jm000292e. [DOI] [PubMed] [Google Scholar]
- 41.Oprea TI. Property distribution of drug-related chemical databases. J. Comput-Aided Mol. Des. 2000;14:251–264. doi: 10.1023/a:1008130001697. [DOI] [PubMed] [Google Scholar]
- 42.Walters WP, Murcko MA. Methods and Principles in Medicinal Chemistry. Wiley-VCH; Library filtering systems and prediction of drug-like properties. In Virtual Screening for Bioactive Molecules; pp. 15–30. [Google Scholar]
- 43.Kramer C, Heinisch T, Fligge T, Beck B, Clark T. A consistent dataset of kinetic solubilities for early-phase drug discovery. Chem Med Chem. 2009;4:1529–1536. doi: 10.1002/cmdc.200900205. [DOI] [PubMed] [Google Scholar]
- 44.Hill AP, Young RJ. Getting physical in drug discovery: a contemporary perspective on solubility and hydrophobicity. Drug Dis. Today. 2010;15:648–655. doi: 10.1016/j.drudis.2010.05.016. [DOI] [PubMed] [Google Scholar]
- 45.Guha R, Dexheimer T, Kestranek AN, Jadhav A, Chervenak AM, Ford MG, Simeonov A, Roth GP, Thomas CJ. Exploratory analysis of kinetic solubility measurements of a small molecule library. Bioorg. Med. Chem. 2011;19:4127–4134. doi: 10.1016/j.bmc.2011.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Delaney JS. ESOL: Estimating aqueous solubility directly from molecular structure. J. Chem. Inf. Comp. Sci. 2004;44:100–1005. doi: 10.1021/ci034243x. [DOI] [PubMed] [Google Scholar]
- 47.Balakin KV, Savchuk NP, Tetko IV. In silico approaches to prediction of aqueous and DMSO solubility of drug-like compounds: Trends, problems and solutions. Curr. Med. Chem. 2006;13:223–241. doi: 10.2174/092986706775197917. [DOI] [PubMed] [Google Scholar]
- 48.Johnson SR, Chen X-Q, Murphey D, Gudmundsson O. A computational model for the prediction of aqueous solubility that includes crystal packing, intrinsic solubility, and ionization effects. Mol. Pharm. 2007;4:513–523. doi: 10.1021/mp070030+. [DOI] [PubMed] [Google Scholar]
- 49.Hewitt M, Cronin MTD, Enoch SJ, Madden JC, Roberts DW, Dearden JC. In silico prediction of aqueous solubility: the solublity challenge. J. Chem. Inf. Comp. Sci. 2009;49:2572–2587. doi: 10.1021/ci900286s. [DOI] [PubMed] [Google Scholar]
- 50.Lüder K, Lindfors L, Westergren J, Nordholm S, Persson R, Pedersen M. In silico prediction of drug solubility: 4. Will simple potentials suffice? J. Comp. Chem. 2009;30:1859–1871. doi: 10.1002/jcc.21173. [DOI] [PubMed] [Google Scholar]
- 51.van de Waterbeemd H, Gifford E. ADMET in silico modeling: towards prediction paradise? Nat. Rev. Drug Discov. 2003;2:192–204. doi: 10.1038/nrd1032. [DOI] [PubMed] [Google Scholar]
- 52.Austin CP, Brady LS, Insel TR, Collins FS. NIH molecular libraries initiative. Science. 2004;306:1138–1139. doi: 10.1126/science.1105511. [DOI] [PubMed] [Google Scholar]
- 53.Hou T, Li Y, Zhang W, Wang J. Recent developments of in silico predictions of intestinal absorption and oral bioavailability. Comb. Chem. High. T. Scr. 2009;12:497–506. doi: 10.2174/138620709788489082. [DOI] [PubMed] [Google Scholar]
- 54.Refsgaard HHF, Jensen BF, Christensen IT, Hagen N, Brockhoff PB. In silico prediction of cytochrome 450 inhibitors. Drug. Dev. Res. 2006;67:417–429. [Google Scholar]
- 55.Aronov AM. Predictive in silico modeling for hERG channel blockers. Drug Discov. Today. 2005;10:149–155. doi: 10.1016/S1359-6446(04)03278-7. [DOI] [PubMed] [Google Scholar]
- 56.Demel MA, Schwaha R, Krämer O, Ettmayer P, Haaksma EEJ, Ecker GF. In silico prediction of substrate properties for ABC-multidrug transporters. Expert Opin. Drug Metab. Toxicol. 2008;4:1167–1180. doi: 10.1517/17425255.4.9.1167. [DOI] [PubMed] [Google Scholar]
- 57.Clark DE. In silico prediction of blood-brain barrier permeation. Drug Discov. Today. 2003;8:927–933. doi: 10.1016/s1359-6446(03)02827-7. [DOI] [PubMed] [Google Scholar]
- 58.Schreiber SL. Target-oriented and diversity oriented organic synthesis in drug discovery. Science. 2000;287:1964–1969. doi: 10.1126/science.287.5460.1964. [DOI] [PubMed] [Google Scholar]
- 59.Nielsen TE, Schreiber SL. Towards the optimal screening collection: a synthesis strategy. Agnew. Chem. Int. Ed. 2008;47:48–56. doi: 10.1002/anie.200703073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Sauer WHB, Schwarz MK. Molecular shape diversity of combinatorial libraries: a prerequisite for broad bioactivity? J. Chem. Inf. Comput. Sci. 2003;43:987–1003. doi: 10.1021/ci025599w. [DOI] [PubMed] [Google Scholar]
- 61.Lovering F, Bikker J, Humblet C. Escape from flatland: increasing saturation as an approach to improving clinical success. J. Med. Chem. 2009;52:6752–6756. doi: 10.1021/jm901241e. [DOI] [PubMed] [Google Scholar]
- 62.Hert J, Willett P, Wilton DJ. Comparison of fingerprint-based methods for virtual screening using multiple bioactive reference structures. J. Chem. Inf. Comput. Sci. 2004;44:1177–1185. doi: 10.1021/ci034231b. [DOI] [PubMed] [Google Scholar]
- 63.Duan JX, Dixon SL, Lowrie JF, Sherman W. Analysis and comparison of 2D fingerprints: insights into database screening performance using eight fingerprint methods. J. Mol. Graphics Model. 2010;29:157–170. doi: 10.1016/j.jmgm.2010.05.008. [DOI] [PubMed] [Google Scholar]
- 64.Steffen A, Kogej T, Tyrchan C, Engkvist O. Comparison of molecular fingerprint methods on the basis of biological profile data. J. Chem. Inf. Model. 2009;49:338–347. doi: 10.1021/ci800326z. [DOI] [PubMed] [Google Scholar]
- 65.Bender A, Jenkins JL, Scheiber J, Sukuru SCK, Glick M, Davies JW. How similar are similarity searching methods? A principal component analysis of molecular descriptor space. J. Chem. Inf. Model. 2009;49:108–119. doi: 10.1021/ci800249s. [DOI] [PubMed] [Google Scholar]
- 66.Xu J. A new approach to finding natural chemical structure classes. J. Med. Chem. 2002;45:5311–5320. doi: 10.1021/jm010520k. [DOI] [PubMed] [Google Scholar]
- 67.Willett P. Similarity-based virtual screening using 2D fingerprints. Drug Discov. Today. 2006;11:1046–1053. doi: 10.1016/j.drudis.2006.10.005. [DOI] [PubMed] [Google Scholar]
- 68.Djuric SW, Akritopoulou-Zanze I, Cox PB, Galasinski G. Compound collection enhancement and paradigms for high-throughput screening – an update. Ann. Rep. Med. Chem. 2010;45:409–428. [Google Scholar]
- 69.Harris CJ, Hill RD, Sheppard DW, Slater MJ, Stouten PFW. The design and application of target-focused compound libraries. Comb. Chem. High T. Scr. 2011;14:521–531. doi: 10.2174/138620711795767802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Kim J, Tang JY, Gong R, Kim J, Lee JJ, Clemons KV, Chong CR, Chang KS, Fereshteh M, Gardner D, Reya T, Liu JO, Epstein EH, Stevens DA, Beachy PA. Itraconazole, a commonly used antifungal that inhibits hedgehog pathway activity and cancer growth. Cancer Cell. 2010;17:388–399. doi: 10.1016/j.ccr.2010.02.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Huang R, Southall N, Wang Y, Yasgar A, Shinn P, Jadhav A, Nguyen DT, Austin CP. The NCGC Pharmaceutical Collection: A Comprehensive Resource of Clinically Approved Drugs Enabling Repurposing and Chemical Genomics. Sci. Transl. Med. 2011;3:16. doi: 10.1126/scitranslmed.3001862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Hajduk PJ, Greer J. A decade of fragment-based drug design: strategic advances and lessons learned. Nat. Rev. Drug Disc. 2007;6:211–219. doi: 10.1038/nrd2220. [DOI] [PubMed] [Google Scholar]
- 73.Congreve M, Chessari G, Tisi D, Woodhead AJ. Recent developments in fragment-based drug discovery. J. Med. Chem. 2008;51:3661–3680. doi: 10.1021/jm8000373. [DOI] [PubMed] [Google Scholar]
- 74.Schuffenhauer A, Ruedisser S, Marzinzik AL, Jahnke W, Blommers M, Selzer P, Jacoby E. Library design for fragment based screening. Curr. Top. Med. Chem. 2005;5:751–762. doi: 10.2174/1568026054637700. [DOI] [PubMed] [Google Scholar]
- 75.Erlanson DA, McDowell RS, O’Brian T. Fragment-based drug discovery. J. Med. Chem. 2004;47:3463–3482. doi: 10.1021/jm040031v. [DOI] [PubMed] [Google Scholar]
- 76.Schreiber SL. Organic synthesis toward small-molecule probes and drugs. Proc. Natl. Acad. Sci. 2011;108:6699–6702. doi: 10.1073/pnas.1103205108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Clemons PA, Wilson JA, Dancik V, Muller S, Carrinski HA, Wagner BK, Koehler AN, Schreiber SL. Quantifying structure and performance diversity for sets of small molecules comprising small-molecule screening collections. Proc. Natl. Acad. Sci. 2011;108:6817–6822. doi: 10.1073/pnas.1015024108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Burke MD, Schreiber SL. A planning strategy for diversity-oriented synthesis. Angew. Chem. Int. Ed. 2004;43:46–58. doi: 10.1002/anie.200300626. [DOI] [PubMed] [Google Scholar]
- 79.Houghten RA, Pinilla C, Blondelle SE, Appel JR, Dooley CT, Cuervo JH. Generation and use of synthetic peptide combinatorial libraries for basic research and drug discovery. doi: 10.1038/354084a0. [DOI] [PubMed] [Google Scholar]
- 80.Yin H, Lee G-I, Hamilton AD. Alpha-helix mimetics in drug discovery. In: Huang Z, editor. Drug Discovery Research in the Post Genomics Era. John Wiley and Sons; 2007. pp. 280–298. [Google Scholar]
- 81.Yoo B, Kirshenbaum K. Peptoid architechtures: elaboration, actuation, and application. Curr. Opin. Chem. Biol. 2008;12:714–721. doi: 10.1016/j.cbpa.2008.08.015. [DOI] [PubMed] [Google Scholar]
- 82.Shakeel S, Karim S, Ali A. Peptide nucleic acid (PNA) – a review. J. Chem. Technol. Biotechnol. 2006;81:892–899. [Google Scholar]
- 83.Vester B, Wengel J. LNA (locked nucleic acid): high-affinity targeting of complementary RNA and DNA. Biochemistry. 2004;43:13233–13241. doi: 10.1021/bi0485732. [DOI] [PubMed] [Google Scholar]
- 84.Reddy MM, Wilson R, Wilson J, Connell S, Gocke A, Hynan L, German D, Kodadek T. Identification of candidate IgG biomarkers for Alzheimer’s disease via combinatorial library screening. Cell. 2011;144:132–142. doi: 10.1016/j.cell.2010.11.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Stringer JR, Bowman MD, Weisblum B, Blackwell HE. Improved small-molecule macroarray platform for the rapid synthesis and discovery of antibacterial chalcones. ACS Comb. Sci. 2011;13:175–180. doi: 10.1021/co100053p. [DOI] [PubMed] [Google Scholar]
- 86.Patterson AW, Wood WJL, Ellman JA. Substrate activity screening (SAS): a general procedure for the preparation and screening of a fragment-based non-peptidic protease substrate library for inhibitor discovery. Nat. Protocols. 2007;2:424–433. doi: 10.1038/nprot.2007.28. [DOI] [PubMed] [Google Scholar]
- 87.Leyva MJ, DeGiacomo F, Kaltenbach LS, Holcomb J, Zhang N, Gafni J, Park H, Lo DC, Salvesen GS, Ellerby LM, Ellman JA. Identification and evaluation of small molecule pan-caspase inhibitors in Huntington’s disease models. Chem. Biol. 2010;17:1189–1200. doi: 10.1016/j.chembiol.2010.08.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Kalyaanamoorthy S, Chen Y-PP. Structure-based drug design to augment hit discovery. Drug Discov. Today. 2011;16:831–839. doi: 10.1016/j.drudis.2011.07.006. [DOI] [PubMed] [Google Scholar]
- 89.Stahura FL, Bajorath J. Virtual Screening methods that complement HTS. Comb. Chem. High. T. Scr. 2004;7:259–269. doi: 10.2174/1386207043328706. [DOI] [PubMed] [Google Scholar]
- 90.Bajorath J. Integration of virtual and high-throughput screening. Nat. Rev. Drug Disc. 2002;1:882–892. doi: 10.1038/nrd941. [DOI] [PubMed] [Google Scholar]
- 91.Schneider G. Virtual screening: an endless staircase? Nat. Rev. Drug Disc. 2010;9:273–276. doi: 10.1038/nrd3139. [DOI] [PubMed] [Google Scholar]
- 92.Mahasenan KV, Pavlovicz RE, Henderson BJ, González-Cestari TF, Yi B, McKay DB, Li C. Discovery of Novel α4β2 Neuronal Nicotinic Receptor Modulators through Structure-Based Virtual Screening. ACS Med. Chem. Lett. doi: 10.1021/ml2001714. ASAP article online. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Kitchen DB, Decornez H, Furr JR, Bajorath J. Docking and scoring in virtual screening for drug discovery: methods and applications. Nat. Rev. Drug Disc. 2004;3:935–949. doi: 10.1038/nrd1549. [DOI] [PubMed] [Google Scholar]
- 94.Holland-Crimmin S, Gosnell P, Quinn C. Compound management: Guildlines for compound storage, provision, and quality control. Curr. Prot. Chem. Biol. 2011;3:141–152. doi: 10.1002/9780470559277.ch110095. [DOI] [PubMed] [Google Scholar]
- 95.Ray B. Value your compound management team! Drug Discov. Today. 2001;6:563. doi: 10.1016/s1359-6446(01)01816-5. [DOI] [PubMed] [Google Scholar]
- 96.Holden K. The significance of effective compound management. Curr. Drug. Discov. 2003;9:9–10. [Google Scholar]
- 97.Archer JR. History, evolution, and trends in compound management for high throughput screening. Assay Drug Dev. Tech. 2004;2:675–681. doi: 10.1089/adt.2004.2.675. [DOI] [PubMed] [Google Scholar]
- 98.Janzen WP, Popa-Burke IG. Advances in improving the quality and flexibility of compound management. J. Biomol. Screen. 2009;14:444–451. doi: 10.1177/1087057109335262. [DOI] [PubMed] [Google Scholar]
- 99.Yasgar A, Shinn P, Jadhav A, Auld D, Michael S, Zheng W, Austin CP, Inglese J, Simeonov A. Compound management for quantitative high-throughput screening. J. Assoc. Lab. Auto. 2008;13:79–89. doi: 10.1016/j.jala.2007.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Keighley WW, Wood TP. Compound library management: an overview of an automated system. In: Janzen WP, editor. Methods in Molecular Biology. Humana; 2002. [DOI] [PubMed] [Google Scholar]
- 101.Holland-Crimmin S, Gosnell P, Quinn C. Compound management: guidelines for compound storage, provision, and quality control. Curr. Protoc. Chem. Biol. 2011;3:141–152. doi: 10.1002/9780470559277.ch110095. [DOI] [PubMed] [Google Scholar]; Di L, Kerns EH. Biological assay challenges from compound solubility: strategies for bioassay optimization. Drug Discov. Today. 2006;11:446–451. doi: 10.1016/j.drudis.2006.03.004. [DOI] [PubMed] [Google Scholar]
- 102.Kozikowski BA, Burt TM, Tirey DA, Williams LE, Kuzmak BR, Stanton DT, Morand KL, Nelson SL. The effect of freeze/thaw cycles on the stability of compounds in DMSO. J. Biomol. Screen. 2003;8:210–215. doi: 10.1177/1087057103252618. [DOI] [PubMed] [Google Scholar]
- 103.MacArthur R, Leister W, Veith H, Shinn P, Southall N, Austin CP, Inglese J, Auld DS. Monitoring compound integrity with cytochrome P450 assays and qHTS. J. Biomol. Screen. 2009;14:538–546. doi: 10.1177/1087057109336954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Simeonov A, Jadhav A, Thomas CJ, Wang Y, Huang R, Southall NT, Shinn P, Smith J, Austin CP, Auld DS, Inglese J. Fluorescence spectroscopic profiling of compound libraries. J. Med. Chem. 2008;51:2362–2371. doi: 10.1021/jm701301m. [DOI] [PubMed] [Google Scholar]
- 105.Auld DS, Zhang Y-Q, Southall NT, Rai G, Landsman M, MacLure J, Langevin D, Thomas CJ, Austin CP, Inglese J. Characterization of chemical libraries for luciferase inhibitory activity. J. Med. Chem. 2008;51:2372–2386. doi: 10.1021/jm701302v. [DOI] [PubMed] [Google Scholar]
- 106.Auld DS, Thorne N, Nguyen DT, Inglese J. A specific mechanism for nonspecific activation in reporter-gene assays. ACS Chem. Biol. 2008;3:463–470. doi: 10.1021/cb8000793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.McGovern SL, Caselli E, Grigorieff N, Shoichet BK. A common mechanism underlying promiscuous inhibitors from virtual and high-throughput screening. J. Med. Chem. 2002;45:1712–1722. doi: 10.1021/jm010533y. [DOI] [PubMed] [Google Scholar]
- 108.Feng BY, Simeonov A, Jadhav A, Babaoglu K, Inglese J, Shoichet BK, Austin CP. A high-throughput screen for aggregation-based inhibition in a large compound library. J. Med. Chem. 2007;50:2385–2390. doi: 10.1021/jm061317y. [DOI] [PubMed] [Google Scholar]
- 109.Jadhav A, Ferreira RS, Klumpp C, Mott BT, Austin CP, Inglese J, Thomas CJ, Maloney DJ, Shoichet BK, Simeonov A. Quantitative analyses of aggregation, autofluorescence, and reactivity artifacts in a screen for inhibitors of a thiol protease. J. Med. Chem. 2010;53:37–51. doi: 10.1021/jm901070c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Babaoglu K, Simeonov A, Irwin JJ, Nelson ME, Feng B, Thomas CJ, Canian L, Costi P, Maltby DA, Jadhav A, Inglese J, Austin CP, Shoichet BK. Comprehensive mechanistic analysis of hits from high-throughput and docking screens against β-lactamase. J. Med. Chem. 2008;51:2502–2511. doi: 10.1021/jm701500e. [DOI] [PMC free article] [PubMed] [Google Scholar]