Single-molecule resolution particle tracking reveals that mRNAs in S. cerevisiae scan the nuclear periphery before being exported to the cytoplasm and that this process is mediated by both components of the nuclear basket and the mRNP.
Abstract
After synthesis and transit through the nucleus, messenger RNAs (mRNAs) are exported to the cytoplasm through the nuclear pore complex (NPC). At the NPC, messenger ribonucleoproteins (mRNPs) first encounter the nuclear basket where mRNP rearrangements are thought to allow access to the transport channel. Here, we use single mRNA resolution live cell microscopy and subdiffraction particle tracking to follow individual mRNAs on their path toward the cytoplasm. We show that when reaching the nuclear periphery, RNAs are not immediately exported but scan along the nuclear periphery, likely to find a nuclear pore allowing export. Deletion or mutation of the nuclear basket proteins MLP1/2 or the mRNA binding protein Nab2 changes the scanning behavior of mRNPs at the nuclear periphery, shortens residency time at nuclear pores, and results in frequent release of mRNAs back into the nucleoplasm. These observations suggest a role for the nuclear basket in providing an interaction platform that keeps RNAs at the periphery, possibly to allow mRNP rearrangements before export.
Introduction
The export of mRNAs from the nucleus to the cytoplasm is one of many steps along the gene expression pathway and is fundamental for mRNAs to meet with ribosomes for translation in the cytoplasm (Oeffinger and Zenklusen, 2012). Export to the cytoplasm occurs through the nuclear pore complex (NPC), a large macromolecular complex embedded in the nuclear membrane (Aitchison and Rout, 2012). On the nuclear side of the NPC, eight protein filaments protrude from the central scaffold into the nucleoplasm and converge in a distal ring to form the nuclear basket (Beck et al., 2004). The nuclear basket is therefore the first structure messenger RNPs (mRNPs) encounter when reaching the nuclear periphery. Furthermore, mRNA quality control steps are suggested to occur at the nuclear basket, pointing toward a function of the basket as both a gatekeeper and physical barrier (Galy et al., 2004; Vinciguerra et al., 2005). These steps are thought to involve structural rearrangements of mRNPs induced by the local modification of RNA binding proteins and their release from the mRNA before export (Daneholt, 2001; Müller-McNicoll and Neugebauer, 2013). Although exhibiting a weak RNA export phenotype, deletion of nuclear basket proteins does not abolish mRNA export but leads to the increased leakage of partially processed mRNAs to the cytoplasm, suggesting that at least some of these rearrangements are not essential for the transport process per se, but for ensuing proper mRNP maturation, its regulation, and possibly influencing export kinetics (Kosova et al., 2000; Galy et al., 2004; Vinciguerra et al., 2005; Fasken et al., 2008; Powrie et al., 2011).
The myosin-like proteins Mlp1p and Mlp2p are structural components of the basket and are essential for basket integrity; the deletion of Mlps results in basketless pores (Strambio-de-Castillia et al., 1999; Kosova et al., 2000; Niepel et al., 2013). Mlps are composed of a long N-terminal coiled-coil domain and a C-terminal unstructured head domain. The C-terminal region of Mlp1p interacts with the nuclear polyA RNA-binding protein Nab2, a known adapter protein for the mRNA export receptor Mex67, possibly providing an interaction platform for mRNPs at the nuclear basket (Green et al., 2003; Fasken et al., 2008; Grant et al., 2008). In addition to Nab2, Mex67 binds other adapter proteins mediating the interaction between the export receptor and mRNA, including the RNA binding protein Yra1 (Strässer and Hurt, 2000; Stutz et al., 2000; Iglesias et al., 2010). In contrast to Nab2, which accompanies mRNAs to the cytoplasmic side of the NPC, Yra1 is released from the mRNA before translocation to the cytoplasm, a process stimulated by the E3 ubiquitin ligase Tom1 (Duncan et al., 2000; Iglesias et al., 2010). Whether these steps require a potential scaffolding function of the nuclear basket and how the nuclear basket influences the export process has not yet been studied in vivo, largely the result of the lack of experimental tools allowing us to analyze such events. Here, we use single-molecule resolution microscopy to investigate the role of the nuclear basket in mRNA export and reveal general features of mRNA export in budding yeast.
Results and discussion
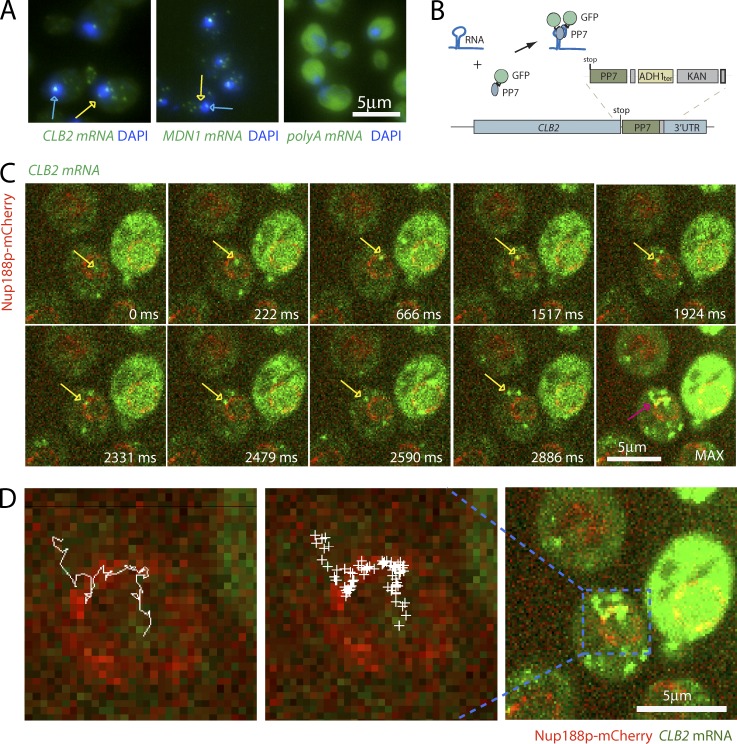
Nuclear mRNPs in mammalian cells reach the nuclear periphery by diffusion (Shav-Tal et al., 2004; Grünwald and Singer, 2010; Mor et al., 2010). Because of the large size of mammalian cell nuclei, mRNPs spend a significant amount of time in the nucleus. Yeast nuclei are much smaller (∼1.5–2 µm in diameter), and, if diffusing freely in the yeast nucleus, mRNPs will reach the periphery quickly, suggesting that mRNA export in yeast is a fast process (Oeffinger and Zenklusen, 2012). Supporting this notion, FISH, using either probes recognizing polyA RNA or probes to specific mRNAs, shows that mRNAs are rarely observed in the nucleus apart from the site of transcription (Fig. 1 A). Interestingly, when observed in the nucleoplasm, mRNAs are generally excluded from the nucleolus and are most frequently found at the nuclear periphery (Fig. 1 A), suggesting a rate-limiting step at the NPC.
Figure 1.
mRNAs scan the nuclear periphery before export to the cytoplasm. (A) Localization of CLB2, MDN1, and polyA mRNA by FISH suggests a rate-limiting step at the nuclear periphery. Blue arrows show sites of transcription. Yellow arrows show single mRNAs. DNA is stained with DAPI (blue). (B) Cartoon illustrating the mRNA labeling strategy using PP7 stem loops. (C) Live cell imaging of CLB2 mRNA. Individual frames from video acquired in 37-ms intervals. MAX shows the maximum intensity projection of all frames. mRNA is shown in green and NPC in red. The yellow arrows show single tracked mRNA in each frame. The purple arrow outlines the same RNA path in the MAX projected image. (D) Track of nuclear CLB2 mRNA from C and Video 1. The RNA position in each frame was determined using 2D Gaussian fitting and was superimposed onto a single frame of Video 1 (middle). Left panel shows connected positions to visualize the path from the nucleus to the cytoplasm.
Insertion of PP7 RNA step loops to the 3′ UTR of MDN1 mRNA allows the detection and tracking of individual mRNAs in yeast (Hocine et al., 2013). To investigate nuclear mRNP movement and to follow the path of single mRNAs toward the cytoplasm, we labeled MDN1 and the cell cycle–regulated CLB2 mRNAs with 12xPP7 stem loops in a strain in which the nuclear pore protein Nup188 was labeled with mCherry (Fig. 1, B and C). Insertion of the PP7 stem loops neither affected growth nor altered mRNA or proteins expression levels, suggesting that PP7 tagging does not affect general mRNA metabolism (Fig. S1). To optimize the signal to noise ratio for single mRNA detection, the PP7-GFP fusion was expressed from an ADE3 promoter, resulting in low expression levels of free PP7-GFP. This setup allows the detection of individual mRNAs in real time using spinning disk confocal microscopy (Fig. 1, C and D). mRNAs were imaged in a single imaging plane in 37-ms intervals, allowing image acquisition for ∼500 frames (18 s) before signals were bleached. Fig. 1 C, Fig. S2, and Video 1 show that, consistent with what was previously observed by FISH, CLB2 and MDN1 mRNAs are mainly cytoplasmic, move rapidly within the cytoplasm, and are frequently lost from the imaging plane. As CLB2 and MDN1 are transcribed at low frequency, nuclear mRNAs are rare, and if observed, only a single CLB2 and MDN1 mRNA is present in the nucleus at most times. When detecting nuclear mRNPs, they spend little time in the nuclear interior and quickly reach the nuclear periphery, consistent with FISH data showing only few RNAs in the nuclear interior. At the periphery, mRNAs are often not immediately exported, but slide along the nuclear envelope (Fig. 1 C). Infrequently, mRNAs lose their association with the periphery and are released back into the nucleoplasm, but then quickly reassociate with the periphery. Export events are difficult to observe, possibly often occurring outside of the imaging plane. mRNPs are also generally excluded from the nucleolus, although infrequently residence in the nucleolus can be observed (Fig. S2, A and B; and Videos 2 and 3). Nucleolar localization is more frequent for MDN1 mRNA, possibly because of the gene’s localization close to the ribosomal DNA repeats. Interestingly, MDN1 mRNPs can get trapped in the nucleolus and exit through pores adjacent to the nucleolus (Fig. S2 C and Video 4). To better visualize the path of nuclear mRNAs, we applied a spot detection and tracking software and plotted all nuclear positions, as well as the mRNP track in a single image. As illustrated for the CLB2 mRNA in Fig. 1 D, mRNPs can scan a large region of the nuclear periphery before being exported.
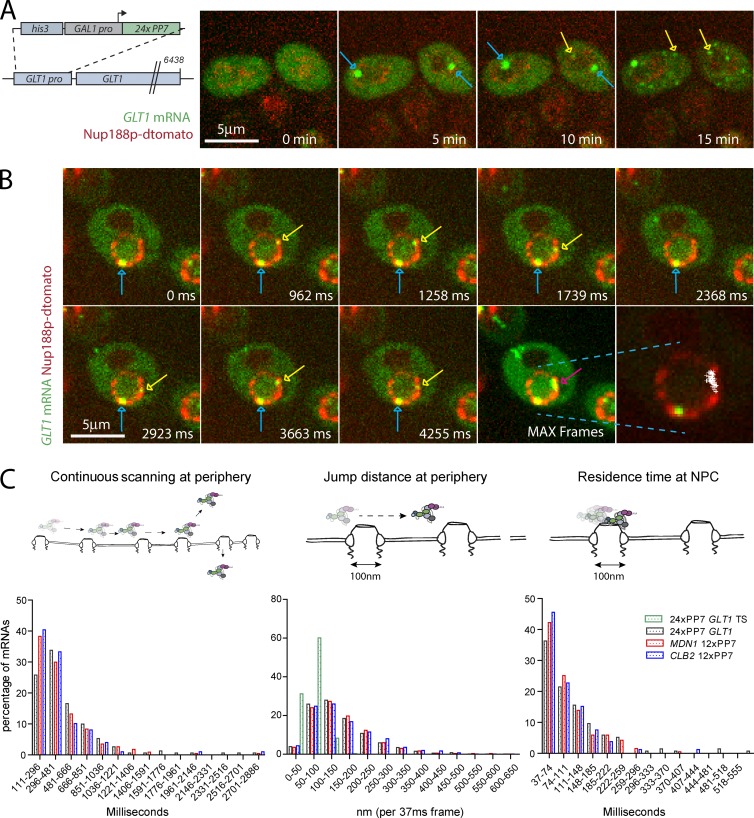
Obtaining nuclear tracks for mRNAs expressed at low levels is challenging. To facilitate nuclear mRNA detection, we constructed a reporter strain where the GLT1 gene is regulated by the inducible GAL1 promoter and labeled with 24xPP7 stem loops in its 5′ UTR (Fig. 2 A). This allows for visualizing RNAs while being synthetized, released into the nucleoplasm, and then exported to the cytoplasm (Fig. 2 A). Images were acquired at early times of induction before RNAs accumulate in high numbers in the cytoplasm. When RNAs are released from the site of transcription into the nucleoplasm, we observe a similar mRNA behavior for GLT1 mRNAs as for the CBL2 and MDN1 mRNAs, spending most of their time in the nucleus scanning the periphery before being exported to the cytoplasm (Fig. 2 B and Video 5).
Figure 2.
Jump distances of scanning RNAs suggest movement between pores. (A) Kinetics of 24PP7-GLT1 mRNA expression upon induction by galactose. Cartoon outlining the GAL1pro-24PP7-GLT1 reporter (left). Single images of 24PP7-GLT1 mRNA (green) and NPC (red) for indicated time points after the addition of galactose (right). Blue arrows show sites of transcription where multiple nascent mRNAs are associated with the GAL1pro-24PP7-GLT1 locus. Yellow arrows show single mRNAs. (B) Live cell tracking of scanning 24PP7-GLT1 mRNAs. Individual frames from a video acquired in 37-ms intervals. Lower right panel shows all nuclear positions of a single RNA superimposed onto a single frame. Blue arrows show sites of transcription, and yellow and purple arrows show the tracked mRNA in individual frames and MAX projected image, respectively. (C) Characterization of perinuclear mRNA scanning for CLB2-12xPP7, MDN1-12xPP7, and GAL1pro-24PP7-GLT1 mRNAs. Timescales of continuous mRNP scanning (left), jump distance at the periphery (middle), and timescales of restricted movements (right) are shown. Jump distances for the GAL1pro-24PP7-GLT1 transcription sites are shown in green. 156 (GLT1), 171 (MDN1), and 104 (CLB2) tracks were analyzed. See text for details.
To quantify mRNA behavior at the periphery, we measured the duration of continuous mRNA movement at the nuclear periphery before mRNAs were either released back into the nucleoplasm, lost from the imaging plane, or exported to the cytoplasm. Perinuclear localization was scored by the overlap of the mRNA signal with the nuclear pore labeled with Nup188-mCherry/dTomato signal. The three mRNAs showed a similar behavior, with approximately two thirds of the mRNAs continuously moving along the nuclear periphery for <500 ms, whereas the remaining RNAs could be observed at the periphery for up to a second, with few RNAs showing longer continuous scanning. We refer to continuous movement at the periphery as scanning that could represent mRNPs moving between pores where transient interactions with NPCs result in increased residence time at the periphery. To better characterize mRNP scanning, we next measured the distances covered by mRNAs during scanning.
Localizing single molecules by Gaussian fitting allows the localization of mRNAs with subdiffraction resolution (Thompson et al., 2002). We first measured the movement of the GLT1 locus marked by nascent GLT1 RNAs (Fig. 2 A, blue arrows). The locus is often found at the nuclear periphery, consistent with observations that the endogenous GAL1 locus associates with the NPC (Casolari et al., 2004; Cabal et al., 2006). Measuring the distance of the center of the nascent RNA spot between two consecutive frames (jump distance) showed that the locus moved <100 nm between frames (Fig. 2 C, middle). This also suggests that if a single mRNA is bound to an NPC, we expect mRNA jump distances similar to that of nascent RNAs. We will therefore consider NPC binding interchangeable with restricted movement similar to the movement of a perinuclear locus. Analyzing the behavior of the peripheral MDN1, CLB2, and GLT1 mRNAs showed that the mRNAs moved a range of distances within 37 ms, varying from 50 to 500 nm. Comparing mRNP movement to the movement of transcription sites shows that only a small portion of mRNPs exhibit jump distances consistent with mRNPs bound to NPCs between 37-ms frames (Fig. 2 C, middle).
Electron microscopy has shown that although neither evenly distributed nor regularly spaced from each other, NPCs do not cluster in close proximity to each other (Winey et al., 1997). Furthermore, a peak density in the distance distribution between the center of two pores of ∼240 nm was observed, with a large fraction of pores even further apart from each other. This suggests that spacing between individual baskets is in most cases farther than 120 nm (Winey et al., 1997). Furthermore, with an NPC diameter of ∼100 nm, the mean distance between two pores is therefore greater than the pore diameter. To determine if scanning mRNPs stay associated with individual pores for multiple consecutive frames, we measured the time span in which individual mRNAs move <100 nm between two consecutive frames. Fig. 2 (C, right) shows that a significant fraction of mRNAs show restricted movement for up to 250 ms, consistent with possible interactions of mRNPs with the NPC during scanning. Such interactions might require the interaction of mRNP components and the NPC and might be responsible for the retention of mRNPs at the periphery during scanning.
The nuclear basket components Mlp1/2 are required for scanning
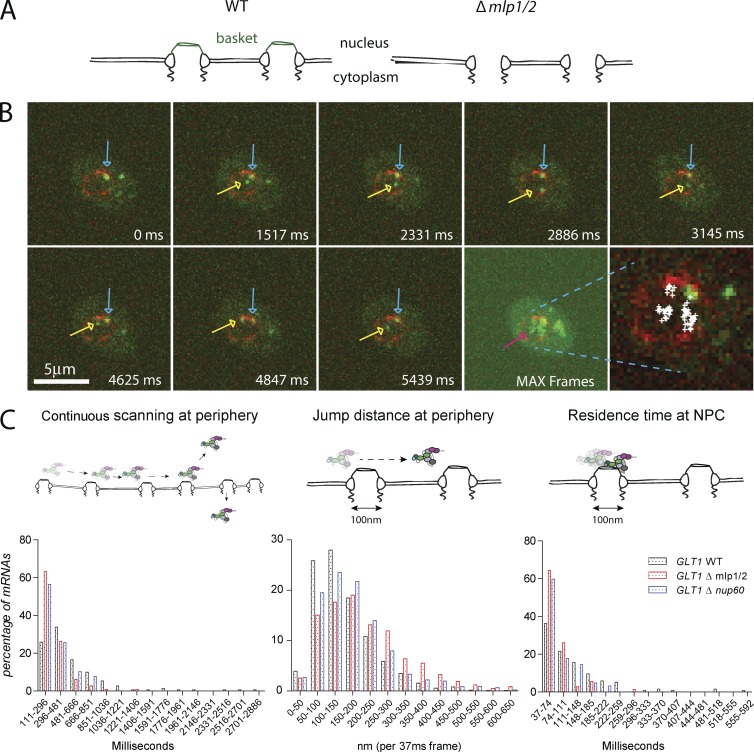
The nuclear basket is the first structure mRNPs encounter when reaching an NPC. The filamentous Mlp1p and Mlp2p proteins are structural components of the basket, and EM studies showed that their deletion leads to basketless NPCs (Krull et al., 2010; Niepel et al., 2013). Surprisingly, MLPs are not essential for viability, and their deletion only leads to a mild growth and mRNA export defect, which is more pronounced in diploid strains (Fig. S3; Strambio-de-Castillia et al., 1999; Kosova et al., 2000; Green et al., 2003; Powrie et al., 2011). Their presence at the NPC, however, is required for quality control processes that ensure that only mature mRNPs are exported to the cytoplasm, and they could therefore serve as an interaction platform for different processes to occur at the nuclear periphery. To determine whether the basket is required for perinuclear mRNP scanning, we tracked mRNPs in a strain where MLP1 and MLP2 genes were deleted. Fig. 3 B and Video 6 show a GLT1 mRNA being released from the site of transcription and diffusing to the nuclear periphery. At the periphery, however, the mRNA does not scan for a prolonged period of time, but is quickly released back into the nucleoplasm. Plotting all positions of the nuclear mRNA while in the imaging plane (5.5-s acquisition period) shows that the mRNA spends more time in the nuclear interior than at the periphery, different than mRNAs in a wild-type (WT) background. Moreover, >60% of mRNPs are released back into the nucleoplasm after a <300-ms residence time at the periphery and do not scan for prolonged amounts of time as observed in WT cells (Fig. 3 C), suggesting a role for the basket in maintaining mRNPs at the periphery. Measuring the jump distance at the periphery shows that mRNPs in basketless cells move longer distances within 37 ms compared with WT, further underlying a role for the basket in restricting the movement of mRNPs at the periphery. Similarly, lack of the basket significantly reduces the number of static frames at the periphery. Deletion of NUP60, a nucleoporin involved in the anchoring of Mlp’s at the pore, causes similar mRNP behavior to MLP1/2 deletion. These data suggest that the nuclear basket is required to restrict mRNP movement at the periphery as well as to retain mRNPs at the periphery. The weak mRNA export phenotype of these mutants might result in part from the more frequent release of mRNAs from the nuclear periphery back into the nucleoplasm. However, most mRNAs are likely exported, probably with some delay, as RNAs accumulate in the cytoplasm (Fig. S3, A and B; Fischer et al., 2002; Powrie et al., 2011). Alternatively, a part of mRNAs released from the periphery might get degraded in the nucleus.
Figure 3.
The nuclear basket is required for perinuclear scanning. (A) Cartoon illustrating the phenotype of mlp1/2 deletion leading to basketless pores. (B) Live cell RNA imaging of GAL1pro-24PP7-GLT1 mRNA shows reduced residence time at the nuclear periphery. Individual frames from a video acquired in 37-ms intervals. Lower right panel shows all nuclear positions of a single RNA superimposed onto a single frame. Blue arrows show sites of transcription, and yellow and purple arrows show the tracked mRNA in individual frames and MAX projected image, respectively. (C) Characterization of perinuclear mRNA scanning for GAL1pro-24PP7-GLT1 mRNAs in WT Δmlp1/2 and Δnup60. Timescales of continuous mRNP scanning (left), jump distance at the periphery (middle), and timescales of restricted movements (right) are shown. 156 (WT), 105 (ΔMLP1/2), and 76 (ΔNUP60) tracks were analyzed. See text for details. P < 0.05, comparing WT versus mutants using a randomized ANOVA followed by posthoc tests.
mRNP interactions with the basket stimulate scanning
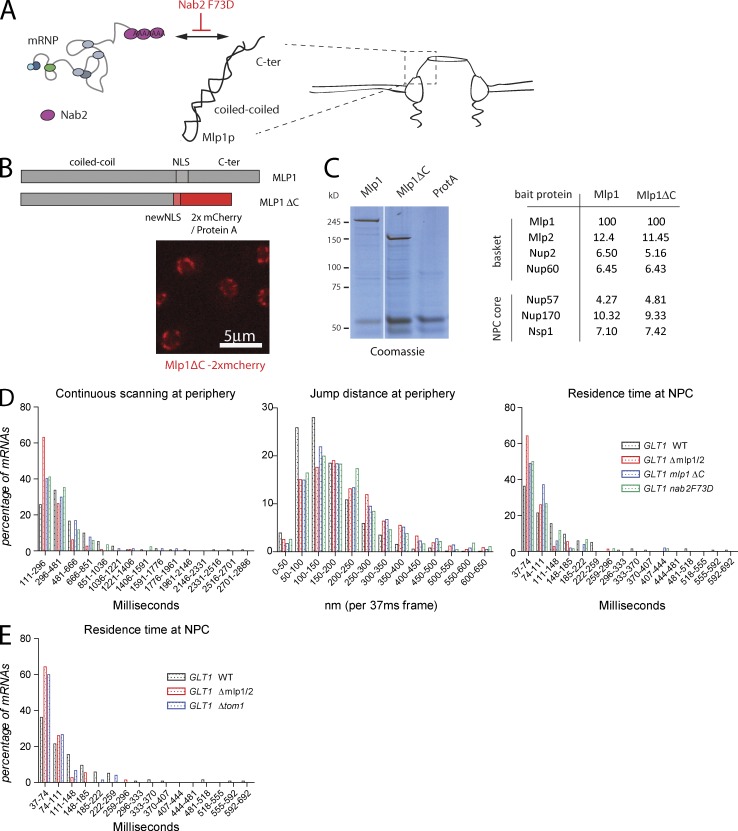
Loss of the nuclear basket has been shown to change the chromatin environment at the nuclear periphery; regions close to the NPC, usually chromatin free, show dense chromatin compared with a WT strain (Krull et al., 2010; Niepel et al., 2013). The phenotypes observed in a ΔMLP1/2 background could therefore, at least in part, be the result of a change in physical properties of the nuclear periphery. Furthermore, lack of the basket could allow mRNPs to diffuse more freely at the periphery, resulting in the larger jump distances observed. Alternatively, these phenotypes could be caused by a change in specific interactions between mRNP components and the basket. The nuclear polyA binding protein Nab2 directly interacts with the C-terminal region of Mlp1 (Green et al., 2003; Fasken et al., 2008; Grant et al., 2008). Nab2 also interacts with the mRNA export receptor Mex67, and the interaction between Nab2 and Mlp1 is likely among the first between the mRNP and the nuclear basket. Furthermore, a single amino acid substitution in Nab2 (Nab2 F73D) was shown to affect the interaction of Nab2p with the C terminus of Mlp1p (Fasken et al., 2008; Grant et al., 2008). To determine whether a Nab2–Mlp1 interaction is required for mRNP scanning at the periphery, we determined mRNP movement in a strain where the C-terminal region of Mlp1 was deleted. In addition to providing an interaction surface for Nab2, the C-terminal region also contains the NLS required to target it to the nucleus. To assure that this C-terminal deletion is targeted to the NPC, we replaced the C-terminal domain of Mlp1p starting at position 4,470 by an exogenous NLS and two copies of mCherry (Fig. 4 B). Furthermore, to test whether the phenotypes observed in the C-terminal deletion are not caused by a change in the overall composition of the basket, we purified Mlp1-associated proteins from an Mlp1–protein A–tagged strain or a strain where the Mlp1 C terminus (Mlp1-ΔC) was replaced by NLS–protein A (Oeffinger et al., 2007b). Fig. 4 C and Table S2 show that Mlp1 and Mlp1-ΔC both copurify basket and NPC components with the same efficiency, suggesting that basket integrity is not disrupted.
Figure 4.
mRNP–NPC interactions mediate perinuclear scanning. (A) Cartoon describing the relationship between the nuclear polyA RNA binding protein Nab2 and the C-terminal domain of Mlp1p. (B) Localization of the Mlp1ΔC-2xmCherry fusion protein to the nuclear periphery. See text for details. (C) Deletion of the C terminus of Mlp1 does not affect basket integrity. Coomassie-stained gel separating protein complexes isolated by single-step affinity purification using Mlp1-ProtA, Mlp1ΔC-ProtA, or ProtA as baits. White line indicates that intervening lanes have been spliced out. Table with normalized peptide counts of copurified proteins as determined by mass spectrometry. Only selected NPC components are shown; for full list, see Table S2. (D) Quantification of GAL1pro-24PP7-GLT1 mRNP scanning behavior in mlp1ΔC and nab2F73D. 156 (WT), 75 (Mlp1-ΔC), and 85 (Nab2 F73D) tracks were analyzed. (E) Frequency of static frames at the periphery for GAL1pro-24PP7-GLT1 mRNPs in Δtom1 strain. P < 0.05, comparing WT versus mutants using a randomized ANOVA followed by posthoc tests, except WT versus Mlp1ΔC for scanning.
Investigation of mRNP movement in an Mlp1-ΔC strain showed reduced continuous mRNP scanning at the periphery, although to a lesser extent than an MLP1/2 deletion. Furthermore, jump distances at the periphery are similar to the deletion strain, suggesting that the interaction between the mRNP and the basket, dependent on the C terminus of Mlp1, is required to restrict mRNP movement (Fig. 4 D). Deleting the C terminus also led to a reduction in extended NPC interactions. In addition, the Nab2 F73D mutant showed a similar phenotype to the Mlp1-ΔC, suggesting that the Nab2-dependent interaction between the mRNP and the C terminus of Mlp1 is required for perinuclear scanning and that the phenotypes observed in MLP1/2 deletions are not the result of the physical absence of the basket at the nuclear periphery.
Tom1 stimulates mRNP binding to the NPC
Although mRNP composition likely varies for different mRNAs, various RNA-binding proteins are thought to be present on most, if not all, mRNAs (Oeffinger and Zenklusen, 2012). We tested whether different nonessential mRNP-associated factors affect perinuclear scanning, including the nuclear cap binding complex (CBC), Pml1 and Pml39, proteins involved in mRNA quality control at the NPC, and the Tom1 E3 ubiquitin ligase (Palancade et al., 2005; Iglesias et al., 2010; Müller-McNicoll and Neugebauer, 2014). Tom1 modifies the mRNP component Yra1, inducing its release from the mRNP before mRNA translocation to the cytoplasm. Neither deletion of the CBC component CBP80 nor deletions of either PML1 or PML39 affected GLT1 mRNP behavior at the periphery (unpublished data). However, deletion of TOM1 led to a strong reduction in the number of static mRNPs at the periphery, similar to an MLP1/2 deletion, whereas scanning times and jump distances were only marginally affected (Fig. 4 E and Fig. S3, C and D). This suggests that Tom1-mediated removal of Yra1 from the mRNP might facilitate binding of mRNPs to the NPC.
The single-molecule tracking data presented here suggest an additional role for the nuclear basket in providing a platform that facilitates mRNPs to stay at the nuclear periphery in the case that they are not immediately exported. mRNP scanning has previously been observed in higher eukaryotes, and here we show that specific interactions of the mRNP with the C-terminal domain of Mlp1 is implicated in maintaining RNAs at the periphery (Grünwald and Singer, 2010). One apparent question is why mRNPs show such a behavior and are not directly exported. One possibility is that rearrangements on the mRNP have to occur before translocation, possibly as part of a quality control mechanism, and that these processes do not take place during the first contact with the periphery. It is also possible that not all pores are available for export, either because of specialized pores or, more likely, because pores might be occupied transporting other molecules; in rapidly growing cells, nucleocytoplasmic transport might get saturated and pore access might be limited. It will be interesting to determine whether different scanning and export rates could be observed for different mRNAs, or under different environmental conditions where nucleocytoplasmic transport is less active, such as the GFA1 mRNA, which is induced upon cell wall stress and does not show extensive scanning (see Smith et al. in this issue).
Materials and methods
Strains and plasmids
Yeast strains were constructed using standard genetic techniques (Amberg et al., 2005) and are listed in Table S1. GAL1p-24xPP7-GLT1–containing strains were constructed by replacing the endogenous GLT1 promoter with a DNA fragment containing a histidine selectable marker, the GAL1 promoter followed by 24xPP7 stem loops using homologous recombination CLB2-12xPP7, and MDN1-12xPP7 strains were constructed by inserting 12xPP7 stem loops into the 3′ UTR of CLB2 and MDN1, respectively, by homologous recombination using a PCR product amplified from pDZ617 (pKAN 12xPP7 V4-ADH1 terminator) and pDZ645 (pKAN mCherry-12xPP7 V4-ADH1 terminator). CLB2 and MDN1 3′ UTRs were reconstituted by removing the ADH1 using CRE recombinase. Nup188 was C-terminal tagged with dTomato or 2xmCherry by homologous recombination using a PCR fragment amplified from pDZ264 (pKAN tdTomato) and from pDZ585 (pKan 2xmCherry), respectively. Haploid ΔMLP1/2 deletion strains were obtained from M. Rout (The Rockefeller University, New York, NY; Niepel et al., 2013). CBC80, TOM1, PML1, and PML39 knockout strains were constructed by homologous recombination using a PCR fragment amplified from plasmid pFA6-hphNT1. nab2F73D strains were constructed by homologous recombination using a PCR fragment inserting the mutant allele and a selection marker replacing the WT allele. Integration of the mutation was confirmed by sequencing. PP7-PS-2xeGFP fusion protein was expressed from either the MET25 (pDZ514 or pDZ529) or ADE3 (pDZ536) promoter. Mlp1 C-terminal deletion was obtained by a C-terminal in-frame integration of a PCR-derived fragment encoding NLS–protein A or NLS-2xmCherry. Correct integration was verified by PCR, Western blot (rabbit anti–protein A antibody; P1291; Sigma-Aldrich), and/or fluorescence microscopy.
Cell synchronization
Cell synchronization to enrich for Clb2 was performed as previously described (Oeffinger et al., 2007a). In brief, cells were grown in synthetic media, arrested in 10-µM α-factor, released into synthetic media, and collected 70 min after release. A harvested cell pellet equivalent to 20 cell ODs was taken up in 6× Laemmli buffer and glass beads, lysed by vortexing and heating cycles, and separated on an SDS-PAGE gel, followed by Western blotting using a rabbit anti-mCherry (PA534974; Invitrogen) and mouse anti-GAPDH antibody (125247; Abcam).
Single-molecule FISH
Single-molecule FISH (smFISH) was essentially performed as described in Rahman and Zenklusen (2013). MDN1 and CLB2 probes were described previously and are listed in Table S3 (mix of 48 for MDN1, 40 for CLB2 20-nt-long oligos containing 3′ amine synthetized by Biosearch Technologies, Inc., and labeled postsynthesis cy5 and cy3, respectively; Castelnuovo et al., 2013; Messier et al., 2013). polyA RNA was detected using a 35-nt dT DNA probe containing 10 locked nucleic acid nucleosides (synthetized by Exiqon) labeled postsynthesis with cy5 (Table S3). Cells were grown in synthetic defined minimal medium (SD)–uracil and 2% glucose at 30°C overnight to mid-log phase (OD600 = 0.6–0.8) and fixed with 4% paraformaldehyde (Electron Microscopy Science) for 30 min at room temperature. Cells were subsequently washed three times with Buffer B (1.2-M sorbitol and 100-mM KHPO4, pH 7.5) and stored overnight at 4°C in Buffer B. Cells were then digested with lyticase (dissolved in 1× PBS to 25,000 U/ml and stored at −20°C; Sigma-Aldrich). Digested cells were plated on poly-l-lysine–treated coverslips and stored in 70% ethanol at −20°C. For hybridization, cells were removed from 70% ethanol, washed twice with 2× saline sodium citrate (SSC), and hydrated in 10% formamide/2× SSC. 10 ng of labeled probe per hybridization (MDN1 cy5, CLB2 cy3, and dT LNA cy5) was resuspended in 10% (vol/vol) formamide, 2× SSC, 1 mg ml−1 BSA, 10-mM ribonucleoside vanadyl complex (New England Biolabs, Inc.), 5-mM NaHPO4, pH 7.5, 0.5 mg ml−1 Escherichia coli tRNA, and 0.5 mg ml−1 single-stranded DNA and denatured at 95°C for 3 min. Cells were then hybridized for 3 h in the dark at 37°C (20-ng probe per sample). Cells were then washed in 10% formamide/2× SSC at 37°C twice for 30 min, followed by a quick wash in 1× PBS at room temperature. The coverslips were then mounted on glass slides using Prolong gold with DAPI mounting medium (Invitrogen). Images were acquired with a 100× NA 1.3 oil objective on a microscope (Axio Imager Z2; Carl Zeiss) equipped with a charge-coupled device camera (AxioCam mRm; Carl Zeiss) and the following filter sets: 488050–9901-000 (Cy5; Carl Zeiss), SP102 v1 (Cy3; Chroma Technology Corp.), SP103 v2 (Cy3.5; Chroma Technology Corp.), and 488049–9901-000 (DAPI; Carl Zeiss). 3D datasets were generated by acquiring multiple 200-nm z stacks spanning the entire volume of cells using acquisition software (Zen; Carl Zeiss). For mRNA counting, 3D datasets were reduced to 2D datasets by applying a maximum projection function in Fiji. RNA signals were detected and quantified using a spot localization algorithm based on 2D Gaussian fitting that was implemented with custom-made software for the interactive data language platform (ITT Visual Information Solutions) as previously described (Zenklusen et al., 2008). Cellular segmentation was performed manually in Fiji.
Preparing cells for live cell imaging
Yeast were grown at 30°C in SD with 3% raffinose or SD with 2% glucose to an OD600 of 0.4–0.6. For galactose induction, galactose was added to a final concentration of 3%. For imaging, 100-µl cell suspension was added to a 96-well glass-bottom plate (MGB096-1-2-LG-L; Brooks Life Science Systems) previously coated with concanavalin A (Con A) and concentrated on the bottom of the well by centrifugation. Wells were coated by adding 100 µl of 1 mg/ml Con A (Sigma-Aldrich) for 10 min before unbound Con A is removed and the Con A activated by adding 100 µl of 50-mM CaCl2/50-mM MnSO4 for 10 min. The solution was then removed, washed once with 100 µl ddH2O, and air dried.
Image acquisition and analysis
Images were acquired on a spinning disk confocal microscope (Observer SD; Carl Zeiss) using a 100×/1.43 NA objective (Carl Zeiss), 488-nm (100 mW) and 561-nm (40 mW) excitation laser lines, and Semrock single bandpass filters for GFP (525 nm/50 nm) and RFPs (617 nm/73 nm). Images were captured using an electron-multiplying charge-coupled device camera (Evolve 512; Photometrics) using Zen blue software. Image sequences were performed by first acquiring a single image of a red fluorescent signal, followed by 500 37-ms frames of GFP channel acquisition. Composite images of NPC and mRNA signals (single NPC image was used for the entire length of the video) were assembled in ImageJ. Only videos with nuclear RNAs were analyzed, and mRNAs showing cytoplasmic scanning were excluded from the analysis. For measuring times of continuous scanning at the periphery, jump distances, and number of static frames, only tracks where RNAs colocalize with the nuclear periphery were used. For each strain, perinuclear mRNAs from 20 cells were tracked. Most cells showed mRNAs associate with the periphery multiple times, either because scanning mRNAs moved RNAs out of the imaging plane and then back in, the tracking algorithm missed more than two frames, or mRNAs were released from the periphery to the nuclear interior, resulting in multiple tracks originating from the same mRNA. Only tracks where mRNAs are observed for at least three frames were used in the analysis, and short associations of mRNAs with the periphery in mutant strains (less than three frames) did not lead to scored tracks, resulting in less tracks observed for mutants compared with WT strains. Plots represent the frequency distribution from data of all perinuclear tracks. The total number of tracks analyzed are as follows: GAL-GLT1 (156), MDN1 (171), and CLB2 (104) in WT (Fig. 2 C), GAL-GLT1 in ΔMLP1/2 (105), and ΔNUP60 (76; Fig. 3 C), Mlp1-ΔC (75), Nab2 F73D (85; Fig. 4 D), and ΔTOM1 (95; Figs. 4 E and S3).
Spot detection using a 2D Gaussian mask fitting algorithm and particle tracking was performed using “localize” as previously described (Coulon et al., 2014). Statistical analysis was performed using Excel (Microsoft), Prism (GraphPad Software), and R software (The R Foundation). Jump distances were calculated using coordinates from the spot detection algorithm using the equation from Excel:
where x1 and y1 are the coordinates at t = 0, and x2 and y2 are the coordinates of the same mRNA at t = 0 + 1 (37 ms). Superimposing all mRNA positions observed in a single cell was done in MATLAB using the coordinates obtained from the tracking software. Statistical significance comparing the distributions of mRNA behavior was performed using randomized ANOVA, performed in R (Horthorn et al., 2008). Where appropriate, posthoc tests were subsequently performed using randomized t tests. P-values were corrected for multiple comparisons using the Holm-Bonferroni method; adjusted p-values <0.05 were considered statistically significant.
NPC purification and mass spectrometry
Affinity purification was performed as previously described (Oeffinger et al., 2007b). In brief, cells were grown to late log phase, frozen by immersion in liquid nitrogen, and mechanically ground using a planetary ball mill (Retsch). 1 g of cell powder was thawed in 9 ml of extraction buffer (1× tributyltin, 50-mM NaCl, 1-mM DTT, 0.5% Triton X-100, 0.5% of solution P, and 0.02% Antifoam), homogenized with a Polytron for 25 s, and cleared by centrifugation at 4,000 g for 5 min. Each lysate was incubated for 30 min with magnetic beads coated with IgG (Dynabeads M-280 sheep anti–rabbit IgG), washed extensively, and resuspended in 50 µl of 20-mM Tris-HCl, pH 8.0. Proteins were digested on beads at 37°C using 1 µg trypsin (Pierce Trypsin Protease, MS Grade) for 16 h and blocked by the addition of 2 µl of 50% formic acid and peptides analyzed by mass spectrometry. Prey proteins were semiquantitatively analyzed by spectral counting, normalized against the bait counts, and compared.
Online supplemental material
Fig. S1 shows that the insertion of PP7 stem loops does not alter MDN1 and CLB2 mRNA and protein expression levels. Fig. S2 shows MDN1 and CLB2 mRNAs scanning the nuclear periphery outside of the nucleolus (A and B) or an MDN1 mRNA trapped in the nucleolus and getting exported to the cytoplasm though NPCs in the nucleolus. Fig. S3 illustrates MLP1/2 deletion growth and mRNA export phenotype and mRNP behavior at the nuclear periphery in a TOM1 deletion strain. Video 1 shows CLB2 mRNA scanning the nuclear periphery from Fig. 1. Video 2 shows that CLB2 mRNA scanning the nuclear periphery occurs outside of the nucleolus as shown in Fig. S2 A. Videos 3 and 4 show MDN1 mRNA scanning the nuclear periphery outside of the nucleolus or being trapped in the nucleolus as shown in Fig. S2 (B and C, respectively). Videos 5–7 show galactose-induced nuclear GLT1 mRNAs in WT (Fig. 3 A), Δmlp1/2 (Fig. 3 B), and Δtom1 (Fig. S3). Table S1 shows yeast strains used in this study. Table S2 is provided as an Excel spreadsheet and summarizes the proteins identified by mass spectrometry purified using Mlp1-ProtA or Mlp1ΔC-ProtA as baits (Fig. 4, B and C). Table S3 is provided as an Excel spreadsheet and lists the smFISH probes. Online supplemental material is available at http://www.jcb.org/cgi/content/full/jcb.201503070/DC1.
Supplementary Material
Acknowledgments
We thank Seckin Sinan Isik for help with MATLAB and Mike Rout for the MLP1/2 deletion strain.
This work is supported by a discovery grant from the Natural Sciences and Engineering Research Council, the Canadian Institute for Health Research (MOP-232642), and the Canadian Foundation for Innovation (awarded to D. Zenklusen). D. Zenklusen holds a Fonds de Recherche du Quebec (FRSQ) Chercheur Boursier Junior I. M. Oeffinger holds a Canadian Institute for Health Research New Investigator Award and an FRSQ Chercheur Boursier Junior I and is supported by a grant from the Canadian Institute for Health Research (MOP-106628).
The authors declare no competing financial interests.
Footnotes
Abbreviations used in this paper:
- mRNP
- messenger RNP
- NPC
- nuclear pore complex
- SD
- synthetic defined minimal medium
- smFISH
- single-molecule FISH
- SSC
- saline sodium citrate
- WT
- wild type
References
- Aitchison J.D., and Rout M.P.. 2012. The yeast nuclear pore complex and transport through it. Genetics. 190:855–883. 10.1534/genetics.111.127803 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amberg D., Burke D., and Strathern J.. 2005. Methods in Yeast Genetics: A Cold Spring Harbor Laboratory Course Manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY. 230 pp. [Google Scholar]
- Beck M., Förster F., Ecke M., Plitzko J.M., Melchior F., Gerisch G., Baumeister W., and Medalia O.. 2004. Nuclear pore complex structure and dynamics revealed by cryoelectron tomography. Science. 306:1387–1390. 10.1126/science.1104808 [DOI] [PubMed] [Google Scholar]
- Cabal G.G., Genovesio A., Rodriguez-Navarro S., Zimmer C., Gadal O., Lesne A., Buc H., Feuerbach-Fournier F., Olivo-Marin J.C., Hurt E.C., and Nehrbass U.. 2006. SAGA interacting factors confine sub-diffusion of transcribed genes to the nuclear envelope. Nature. 441:770–773. 10.1038/nature04752 [DOI] [PubMed] [Google Scholar]
- Casolari J.M., Brown C.R., Komili S., West J., Hieronymus H., and Silver P.A.. 2004. Genome-wide localization of the nuclear transport machinery couples transcriptional status and nuclear organization. Cell. 117:427–439. 10.1016/S0092-8674(04)00448-9 [DOI] [PubMed] [Google Scholar]
- Castelnuovo M., Rahman S., Guffanti E., Infantino V., Stutz F., and Zenklusen D.. 2013. Bimodal expression of PHO84 is modulated by early termination of antisense transcription. Nat. Struct. Mol. Biol. 20:851–858. 10.1038/nsmb.2598 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coulon A., Ferguson M.L., de Turris V., Palangat M., Chow C.C., and Larson D.R.. 2014. Kinetic competition during the transcription cycle results in stochastic RNA processing. eLife. 3 10.7554/eLife.03939 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daneholt B. 2001. Assembly and transport of a premessenger RNP particle. Proc. Natl. Acad. Sci. USA. 98:7012–7017. 10.1073/pnas.111145498 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duncan K., Umen J.G., and Guthrie C.. 2000. A putative ubiquitin ligase required for efficient mRNA export differentially affects hnRNP transport. Curr. Biol. 10:687–696. 10.1016/S0960-9822(00)00527-3 [DOI] [PubMed] [Google Scholar]
- Fasken M.B., Stewart M., and Corbett A.H.. 2008. Functional significance of the interaction between the mRNA-binding protein, Nab2, and the nuclear pore-associated protein, Mlp1, in mRNA export. J. Biol. Chem. 283:27130–27143. 10.1074/jbc.M803649200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischer T., Strässer K., Rácz A., Rodríguez-Navarro S., Oppizzi M., Ihrig P., Lechner J., and Hurt E.. 2002. The mRNA export machinery requires the novel Sac3p–Thp1p complex to dock at the nucleoplasmic entrance of the nuclear pores. EMBO J. 21:5843–5852. 10.1093/emboj/cdf590 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galy V., Gadal O., Fromont-Racine M., Romano A., Jacquier A., and Nehrbass U.. 2004. Nuclear retention of unspliced mRNAs in yeast is mediated by perinuclear Mlp1. Cell. 116:63–73. 10.1016/S0092-8674(03)01026-2 [DOI] [PubMed] [Google Scholar]
- Grant R.P., Marshall N.J., Yang J.-C., Fasken M.B., Kelly S.M., Harreman M.T., Neuhaus D., Corbett A.H., and Stewart M.. 2008. Structure of the N-terminal Mlp1-binding domain of the Saccharomyces cerevisiae mRNA-binding protein, Nab2. J. Mol. Biol. 376:1048–1059. 10.1016/j.jmb.2007.11.087 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Green D.M., Johnson C.P., Hagan H., and Corbett A.H.. 2003. The C-terminal domain of myosin-like protein 1 (Mlp1p) is a docking site for heterogeneous nuclear ribonucleoproteins that are required for mRNA export. Proc. Natl. Acad. Sci. USA. 100:1010–1015. 10.1073/pnas.0336594100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grünwald D., and Singer R.H.. 2010. In vivo imaging of labelled endogenous β-actin mRNA during nucleocytoplasmic transport. Nature. 467:604–607. 10.1038/nature09438 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hocine S., Raymond P., Zenklusen D., Chao J.A., and Singer R.H.. 2013. Single-molecule analysis of gene expression using two-color RNA labeling in live yeast. Nat. Methods. 10:119–121. 10.1038/nmeth.2305 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horthorn T., Hornik K., van de Wiel M.A., and Zeileis A.. 2008. Implementing a class of permutation tests: the coin package. J. Stat. Softw. 28:1–23.27774042 [Google Scholar]
- Iglesias N., Tutucci E., Gwizdek C., Vinciguerra P., Von Dach E., Corbett A.H., Dargemont C., and Stutz F.. 2010. Ubiquitin-mediated mRNP dynamics and surveillance prior to budding yeast mRNA export. Genes Dev. 24:1927–1938. 10.1101/gad.583310 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kosova B., Panté N., Rollenhagen C., Podtelejnikov A., Mann M., Aebi U., and Hurt E.. 2000. Mlp2p, a component of nuclear pore attached intranuclear filaments, associates with nic96p. J. Biol. Chem. 275:343–350. 10.1074/jbc.275.1.343 [DOI] [PubMed] [Google Scholar]
- Krull S., Dörries J., Boysen B., Reidenbach S., Magnius L., Norder H., Thyberg J., and Cordes V.C.. 2010. Protein Tpr is required for establishing nuclear pore-associated zones of heterochromatin exclusion. EMBO J. 29:1659–1673. 10.1038/emboj.2010.54 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Messier V., Zenklusen D., and Michnick S.W.. 2013. A nutrient-responsive pathway that determines M phase timing through control of B-cyclin mRNA stability. Cell. 153:1080–1093. 10.1016/j.cell.2013.04.035 [DOI] [PubMed] [Google Scholar]
- Mor A., Suliman S., Ben-Yishay R., Yunger S., Brody Y., and Shav-Tal Y.. 2010. Dynamics of single mRNP nucleocytoplasmic transport and export through the nuclear pore in living cells. Nat. Cell Biol. 12:543–552. 10.1038/ncb2056 [DOI] [PubMed] [Google Scholar]
- Müller-McNicoll M., and Neugebauer K.M.. 2013. How cells get the message: dynamic assembly and function of mRNA-protein complexes. Nat. Rev. Genet. 14:275–287. 10.1038/nrg3434 [DOI] [PubMed] [Google Scholar]
- Müller-McNicoll M., and Neugebauer K.M.. 2014. Good cap/bad cap: how the cap-binding complex determines RNA fate. Nat. Struct. Mol. Biol. 21:9–12. 10.1038/nsmb.2751 [DOI] [PubMed] [Google Scholar]
- Niepel M., Molloy K.R., Williams R., Farr J.C., Meinema A.C., Vecchietti N., Cristea I.M., Chait B.T., Rout M.P., and Strambio-De-Castillia C.. 2013. The nuclear basket proteins Mlp1p and Mlp2p are part of a dynamic interactome including Esc1p and the proteasome. Mol. Biol. Cell. 24:3920–3938. 10.1091/mbc.E13-07-0412 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oeffinger M., and Zenklusen D.. 2012. To the pore and through the pore: a story of mRNA export kinetics. Biochim. Biophys. Acta. 1819:494–506. 10.1016/j.bbagrm.2012.02.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oeffinger M., Fatica A., Rout M.P., and Tollervey D.. 2007a Yeast Rrp14p is required for ribosomal subunit synthesis and for correct positioning of the mitotic spindle during mitosis. Nucleic Acids Res. 35:1354–1366. 10.1093/nar/gkl824 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oeffinger M., Wei K.E., Rogers R., DeGrasse J.A., Chait B.T., Aitchison J.D., and Rout M.P.. 2007b Comprehensive analysis of diverse ribonucleoprotein complexes. Nat. Methods. 4:951–956. 10.1038/nmeth1101 [DOI] [PubMed] [Google Scholar]
- Palancade B., Zuccolo M., Loeillet S., Nicolas A., and Doye V.. 2005. Pml39, a novel protein of the nuclear periphery required for nuclear retention of improper messenger ribonucleoparticles. Mol. Biol. Cell. 16:5258–5268. 10.1091/mbc.E05-06-0527 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Powrie E.A., Zenklusen D., and Singer R.H.. 2011. A nucleoporin, Nup60p, affects the nuclear and cytoplasmic localization of ASH1 mRNA in S. cerevisiae. RNA. 17:134–144. 10.1261/rna.1210411 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rahman S., and Zenklusen D.. 2013. Single-molecule resolution fluorescent in situ hybridization (smFISH) in the yeast S. cerevisiae. Methods Mol. Biol. 1042:33–46. 10.1007/978-1-62703-526-2_3 [DOI] [PubMed] [Google Scholar]
- Shav-Tal Y., Darzacq X., Shenoy S.M., Fusco D., Janicki S.M., Spector D.L., and Singer R.H.. 2004. Dynamics of single mRNPs in nuclei of living cells. Science. 304:1797–1800. 10.1126/science.1099754 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith C., Lari A., Derrer C.P., Ouwehand A., Rossouw A., Huisman M., Dange T., Hopman M., Joseph A., Zenklusen D., et al. . 2015. In vivo single-particle imaging of nuclear mRNA export in budding yeast. J. Cell Biol. 10.1083/jcb.201503135 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strambio-de-Castillia C., Blobel G., and Rout M.P.. 1999. Proteins connecting the nuclear pore complex with the nuclear interior. J. Cell Biol. 144:839–855. 10.1083/jcb.144.5.839 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strässer K., and Hurt E.. 2000. Yra1p, a conserved nuclear RNA-binding protein, interacts directly with Mex67p and is required for mRNA export. EMBO J. 19:410–420. 10.1093/emboj/19.3.410 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stutz F., Bachi A., Doerks T., Braun I.C., Séraphin B., Wilm M., Bork P., and Izaurralde E.. 2000. REF, an evolutionary conserved family of hnRNP-like proteins, interacts with TAP/Mex67p and participates in mRNA nuclear export. RNA. 6:638–650. 10.1017/S1355838200000078 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson R.E., Larson D.R., and Webb W.W.. 2002. Precise nanometer localization analysis for individual fluorescent probes. Biophys. J. 82:2775–2783. 10.1016/S0006-3495(02)75618-X [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vinciguerra P., Iglesias N., Camblong J., Zenklusen D., and Stutz F.. 2005. Perinuclear Mlp proteins downregulate gene expression in response to a defect in mRNA export. EMBO J. 24:813–823. 10.1038/sj.emboj.7600527 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winey M., Yarar D., Giddings T.H. Jr., and Mastronarde D.N.. 1997. Nuclear pore complex number and distribution throughout the Saccharomyces cerevisiae cell cycle by three-dimensional reconstruction from electron micrographs of nuclear envelopes. Mol. Biol. Cell. 8:2119–2132. 10.1091/mbc.8.11.2119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zenklusen D., Larson D.R., and Singer R.H.. 2008. Single-RNA counting reveals alternative modes of gene expression in yeast. Nat. Struct. Mol. Biol. 15:1263–1271. 10.1038/nsmb.1514 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.