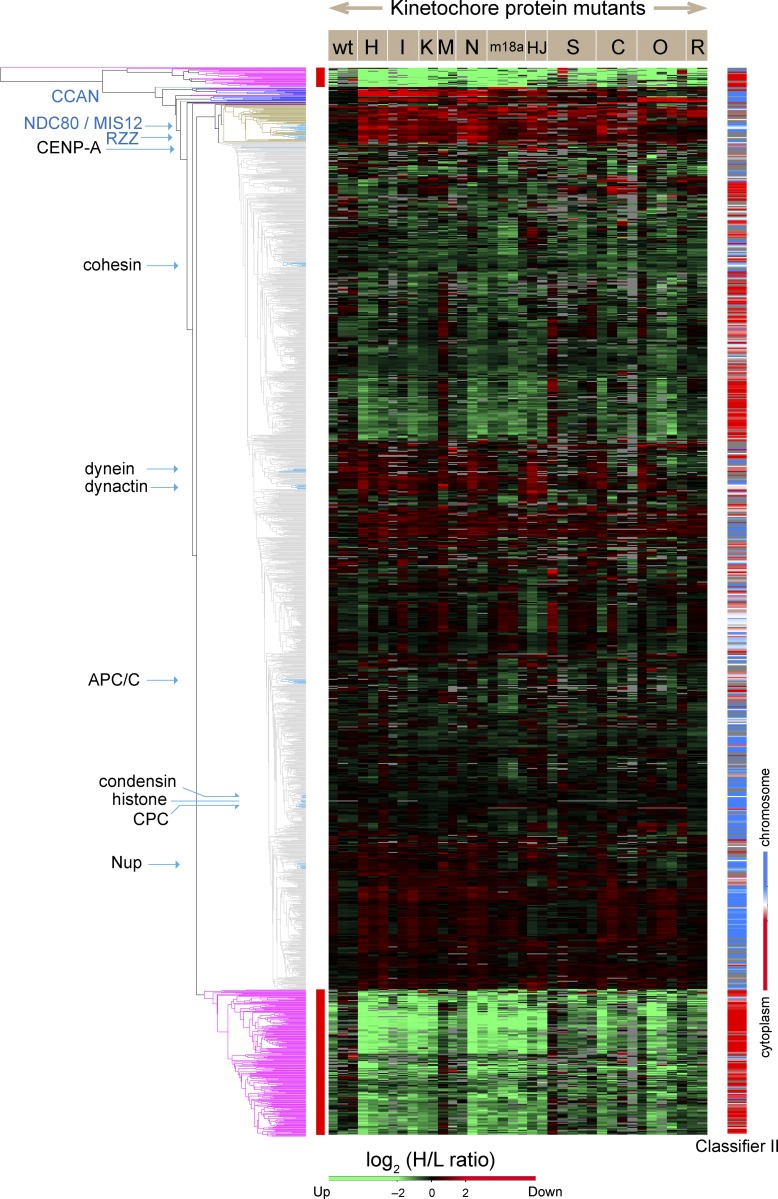
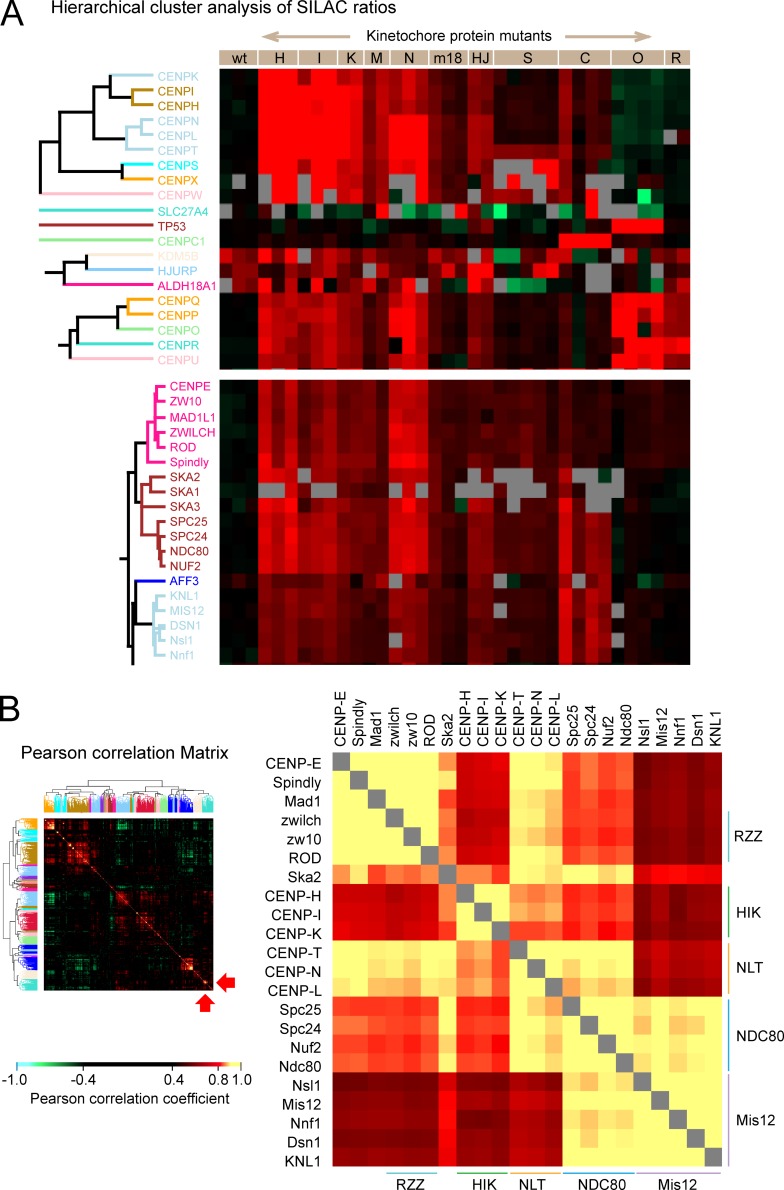
Whole-proteome analysis of isolated mitotic chromosomes from 11 kinetochore structural and assembly mutants is used to develop dependency and correlation maps for protein subcomplexes that confirm many published interactions and also reveal novel dependencies between kinetochore components.
Abstract
Kinetochores orchestrate mitotic chromosome segregation. Here, we use quantitative mass spectrometry of mitotic chromosomes isolated from a comprehensive set of chicken DT40 mutants to examine the dependencies of 93 confirmed and putative kinetochore proteins for stable association with chromosomes. Clustering and network analysis reveal both known and unexpected aspects of coordinated behavior for members of kinetochore protein complexes. Surprisingly, CENP-T depends on CENP-N for chromosome localization. The Ndc80 complex exhibits robust correlations with all other complexes in a “core” kinetochore network. Ndc80 associated with CENP-T interacts with a cohort of Rod, zw10, and zwilch (RZZ)–interacting proteins that includes Spindly, Mad1, and CENP-E. This complex may coordinate microtubule binding with checkpoint signaling. Ndc80 associated with CENP-C forms the KMN (Knl1, Mis12, Ndc80) network and may be the microtubule-binding “workhorse” of the kinetochore. Our data also suggest that CENP-O and CENP-R may regulate the size of the inner kinetochore without influencing the assembly of the outer kinetochore.
Introduction
Genetic and biochemical analyses have led to identification of ∼100 proteins that localize at centromeres and kinetochores (Takahashi et al., 1994; Obuse et al., 2004; Foltz et al., 2006; Okada et al., 2006; Ohta et al., 2011; Perpelescu and Fukagawa, 2011; Biggins, 2013; Tanaka, 2013). A group of 16 proteins known as the CCAN (constitutive centromere-associated network) comprises the inner, chromatin-proximal portion of the kinetochore (Cheeseman and Desai, 2008; Westhorpe and Straight, 2013; Fukagawa and Earnshaw, 2014). CCAN components remain associated with centromeres across the cell cycle and are a foundation for outer kinetochore assembly during mitosis. The CCAN is composed of discrete subcomplexes (De Wulf et al., 2003; Foltz et al., 2006; Okada et al., 2006; Hori et al., 2008b; Nishino et al., 2012). The precise interactions among CCAN proteins and complexes in chromosomes are being studied in many laboratories, and several models have been proposed (DeLuca and Musacchio, 2012; Fachinetti et al., 2013; Cheeseman, 2014; Fukagawa and Earnshaw, 2014; Westhorpe and Straight, 2015; Kim and Yu, 2015).
The mid and outer kinetochore are also characterized by the presence of protein complexes, including CENP-O/-P/-Q/-R/-U (throughout, we will use this shorthand when listing multiple CENP proteins, e.g., CENP-O, CENP-P, and CENP-Q), Ndc80, Mis12, Rod, zw10, and zwilch (RZZ), and the Nup107/Nup160 nucleoporin complexes. The Ndc80 and Mis12 complexes, together with the protein KNL-1, form the KMN network, which is a major site of microtubule binding in the outer kinetochore (Cheeseman et al., 2006).
Some of these complexes have been isolated and characterized structurally. In addition to several CENP-A nucleosome-related structures (Sekulic et al., 2010; Tachiwana et al., 2011; Kato et al., 2013), structures have also been reported for the dimerization domain of CENP-C (Cohen et al., 2008), a bonsai form of the Ncd80 complex (Ciferri et al., 2008), as well as several of the protein domains involved in that complex (Wei et al., 2006, 2007), the CENP-T-W-S-X complex (Nishino et al., 2012), the Knl1/Mis12 complex (Petrovic et al., 2014), the Iml3/Chl4 heterodimer (Hinshaw and Harrison, 2013), and the Ska complex (Jeyaprakash et al., 2012). However, functional analysis of these complexes is complicated, because in chromosomes, they form an insoluble network. Thus, studies of the functions and dependencies of the proteins in their “native environment” (e.g., in situ in mitotic chromosomes) are crucial.
Here, we combined quantitative proteomics using stable isotope labeling by amino acids (SILAC) technology (Ong et al., 2002) with DT40 genetics to map a vertebrate kinetochore by monitoring the behavior of the entire mitotic chromosome proteome after the depletion of defined components. We previously showed that this approach can track the behavior of individual protein complexes in the presence of hundreds or thousands of other proteins (Ohta et al., 2010).
Our studies define dependency pathways for the stable association of a number of protein complexes with mitotic chromosomes. Several unexpected relationships were found. The data suggest that the organization of kinetochore proteins into functional complexes may be more complicated than suggested by previous studies.
Results
Strategy and overview
Analysis of the proteome of DT40 mitotic chromosomes by mass spectrometry (Ohta et al., 2010) enabled us to both make a list of proteins that associate with mitotic chromosomes and also unearth specific protein–protein interactions within and between protein complexes (Ohta et al., 2010; Samejima et al., 2012). Here, we used similar unbiased whole-proteome quantification to define dependency relationships for kinetochore protein assembly.
Several caveats should be kept in mind when comparing our data with other studies. First, because chromosome isolation requires a very high mitotic index (>80%), cultures must be healthy enough to continue to cycle. Therefore, most of our experiments with conditional knockout (KO) cells (with the exception of null alleles of nonessential genes) were performed before the target protein was completely lost. Second, in order to obtain a mitotic index of >80%, cultures were blocked for 12 h with nocodazole. This may induce some alterations in the kinetochore, but all of our experiments compare wild-type and mutant chromosomes subjected to an identical treatment. Third, our analysis detected changes in the entire proteome of mitotic chromosomes, not only at kinetochores. This is potentially significant, as despite the apparent specificity of antibody staining, a recent study reported that ∼70% of CENP-A is located at noncentromeric sites in RPE-1 cells (Bodor et al., 2014). The ratio of centromeric to noncentromeric binding is not known for other kinetochore proteins.
We isolated mitotic chromosomes from 11 DT40 mutant cell lines for whole-proteome analysis. Eight of those lines are conditional knockouts where the target gene is disrupted and cells are kept alive by a rescuing cDNA that is silenced by adding doxycycline (CENP-C, CENP-H, CENP-I, CENP-K, CENP-M, CENP-N, HJURP, and mis18α; we refer to these cells as GENENAMEOFF). Three lines are null strains in which the nonessential genes encoding CENP-O, CENP-R, or CENP-S were deleted (referred to as GENENAMEKO). Mutant and wild-type chromosomes were labeled with stable isotopes in vivo (SILAC; Ong et al., 2002). The relative abundance of proteins in the two proteomes was calculated from ratios of heavy and light peptides derived from the wild type and mutant, respectively (expressed as the H/L ratio). 1,500 to 2,200 proteins were quantified in each experiment.
In a control experiment where inversely labeled wild-type chromosomes were compared with one another, ∼80% of the proteins had a H/L ratio between 0.66 (=2−0. 6) and 1.32 (=20.4) (Fig. S1 A). Typically, >50% of proteins quantified in mutant chromosomes had a H/L ratio in this range. After silencing or loss of a CCAN protein, the target protein and a subset of other kinetochore proteins were typically among the most depleted proteins, whereas the remaining kinetochore proteins showed relatively little change in their abundance (Fig. S1 D). The pattern of effects on other kinetochore proteins varied for different target proteins.
Proteins known to form protein complexes exhibited coordinated behavior and were reproducibly grouped together in the same clusters (Fig. 1 and Table S1). This included components of the Ndc80, Mis12, RZZ, chromosome passenger complex (CPC), anaphase-promoting complex/cyclosome, Nup107/Nup160 nucleoporin, dynein, and dynactin complexes (Fig. 1). Meta-analysis of variations in the kinetochore proteome performed by combining results from the different genetic perturbations also allowed us to identify novel cohorts of kinetochore proteins. We define a “cohort” as a group of proteins that exhibit coordinated behavior in response to diverse genetic perturbations but have not been independently shown to form a protein complex.
Figure 1.
Unbiased cluster analysis reveals coordinated behavior of protein complexes in chromosomes isolated from mutant cell lines. The dendrogram (left) summarizes the relationships between all proteins identified in chromosomes based on the H/L SILAC ratio of each protein in all experiments. The mutant cell lines used for isolation of chromosomes are designated in abbreviated form across the top of the heatmap. The heatmap codes the SILAC ratio of each protein in each experiment (vertical column) in color. Green indicates proteins that increased in mutants or the wild type (wt) grown with complete FBS. Red indicates proteins that decreased in mutants or the wild type grown with complete FBS. Gray blocks correspond to missing values, as some proteins were not identified in every experiment. The color-coded bar on the right shows localization propensity between chromosomes (blue) and cytoplasm (red) according to classifier II, the ratio of the amount of a given protein in chromosomes to that in a corresponding cell equivalent of cytosol (Ohta et al., 2010). The exact values are described in Table S1. Clusters marked with a red side bar next to trees in purple were removed from the analysis as described in Materials and methods. Clusters that contain CCAN and outer kinetochore proteins are extracted, and their details are shown in Fig. 9 A.
Hierarchical clustering analysis of the H/L ratios across all experiments identified two clusters of proteins that appeared to correspond to contaminants and hitchhikers (red lines at left in Fig. 1). Hitchhikers were defined in Ohta et al. (2010) as cytoplasmic proteins that bind to mitotic chromosomes after nuclear envelope breakdown but do not have a role in mitotic chromosome structure or function. These and other proteins whose variation correlated with the tissue culture medium in which cells were grown were excluded from our statistical analysis as explained in Materials and methods.
A subset of CCAN proteins is depleted from CENP-IOFF and CENP-HOFF chromosomes
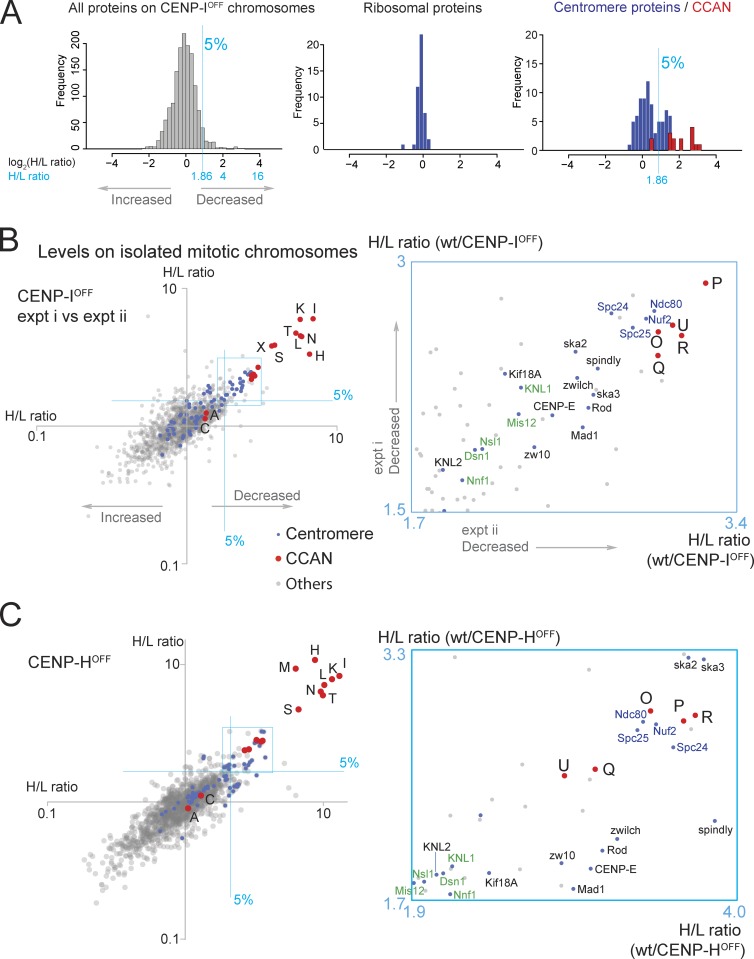
As a first approach to determining the effects of disrupting the CCAN on kinetochore composition, we determined the proteome of chromosomes isolated from CENP-IOFF or CENP-HOFF cells. CENP-I and CENP-H are closely interacting CCAN members (Nishihashi et al., 2002; Okada et al., 2006; Basilico et al., 2014).
In a typical experiment with CENP-IOFF chromosomes, 1,504 proteins were quantified. Depletion of CENP-I was efficient, with ∼12% of the wild-type level remaining on mutant chromosomes (H/L ratio = 8.09). CENP-H/-K/-L/-N/-T were also significantly depleted, with <16% of wild-type levels remaining (Fig. 2, A and B; and Fig. S1 D). In contrast, CENP-A and CENP-C were little affected, with >72% remaining on the chromosomes.
Figure 2.
Behavior of CENP-H,-I,-K,-M,-N,-L,-T in CENP-I and CENP-H KOs. (A) Histograms show the distribution of log2(H/L ratio) of all proteins quantified, ribosomal proteins, and centromere proteins. CCAN proteins (red) are shown as stacked columns. Proteins with a positive score (rightwards on the abscissa) were depleted from CENP-IOFF chromosomes. A blue line designates the top 5%. Those with a negative score (leftward on the abscissa) are accumulated in CENP-IOFF chromosomes. (B) CENP-IOFF: H/L ratios of centromere proteins (blue), CCAN proteins (red), and others (gray) from two replicate experiments are plotted against one another on a log scale in this scatterplot. The blue lines designate the top 5% most depleted proteins. Proteins that decreased in the CENP-IOFF chromosomes map to the top right, and those that increased map to the bottom left. Enlarged plot of the areas boxed in left panel is shown in the blue box on the right. CCAN proteins (red) are designated with the prefix “CENP-“ removed. Members of the Ndc80 complex are in blue, whereas members of the Mis12 complex and KNL1 are in green. (C) CENP-HOFF: H/L ratios from two replicate experiments are plotted against one another. Abbreviations and color-coding are as in B. A magnified plot of the boxed area is shown in a blue box. Note that to appear in scatterplots here and below, proteins must have been identified in both experiments being compared. Very small proteins that give rise to few peptides, such as CENP-M, CENP-S, and CENP-X, were occasionally not detected and are therefore not shown in some of the plots. wt, wild type.
A scatterplot of the H/L ratios of 1,421 proteins from two replicate experiments shows that these observations were reproducible (Fig. 2 B). In these plots of H/L ratios in experiment i versus H/L ratios in experiment ii, good correlation is shown when points cluster along a tight diagonal line. This plot reveals that CENP-H/-I/-K/-L/-N/-T are the proteins most affected by CENP-I depletion, with H/L ratios nearly as large as CENP-I (Fig. 2 B). These proteins form a discrete cluster separate from the rest of the population, suggesting that they comprise one or more protein complexes that are not stably associated with CENP-IOFF chromosomes. CENP-S/-X were also significantly depleted from these chromosomes, and they formed a distinct cluster.
CENP-O/-P/-Q/-R/-U were somewhat less depleted and exhibited a tight cluster of H/L ratios (Fig. 2 B, inset). Outer kinetochore proteins, including members of the Ndc80 complex, clustered with similar H/L ratios, and members of the RZZ and Mis12 complexes were also among the top 5% most reduced proteins (above and to the right of the two orthogonal blue lines).
Similar results were obtained after CENP-H depletion. Fig. 2 C shows a scatterplot of the H/L ratio for 1,515 proteins from two replicate experiments with CENP-HOFF chromosomes.
In summary, depletion of CENP-I or CENP-H from mitotic chromosomes caused a reproducible downstream pattern of depletion of both inner and outer kinetochore proteins from isolated mitotic chromosomes.
Similar proteomes were observed with chromosomes from CENP-HOFF, CENP-IOFF, CENP-KOFF, or CENP-MOFF cells
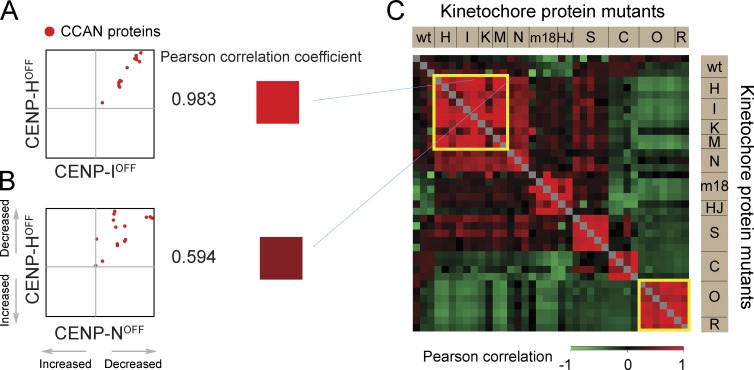
These experiments involved numerous repetitions of proteomics experiments conducted with multiple mutants. A correlation matrix was plotted to compare the results of different experiments (Fig. S2, A and B). The Pearson correlation coefficient was calculated from the distribution of all quantified proteins on chromosomes (gray), all centromere proteins (blue), or CCAN proteins only (red; Fig. S2, C–H). A strong correlation was observed between replicate experiments with the same cell line and also between the CENP-H and CENP-I depletion results (Figs. 3 A and S2), confirming that both mutants have a similar effect on the behavior of centromere proteins, in particular CCAN proteins (Figs. 3 C and S2).
Figure 3.
Pairwise comparisons to identify similar proteomes. (A and B) Scatterplots of CCAN proteins in a pair of experiments. Pearson correlation coefficient between the two experiments is converted into a color according to the scale at the bottom right. (C) Correlation of CCAN proteins in every pair of experiments is presented as a color-coded matrix. wt, wild type.
A similar correlation was obtained for comparisons involving CENP-KOFF and CENP-MOFF. In all, proteomes of chromosomes from CENP-HOFF/-IOFF/-KOFF/-MOFF cells form one closely correlated group (top left yellow box in Fig. 3 C), consistent with them comprising a protein complex (Basilico et al., 2014). Similarly, proteomes of chromosomes from CENP-OKO and CENP-RKO form a distinct group (Fig. 3 C, bottom right yellow box).
Importantly, these correlations were diminished when cross-comparisons were made between proteomes of chromosomes depleted of proteins associated with different kinetochore subcomplexes (Figs. 3 B and S2). For example, proteome comparisons between chromosomes isolated from CENP-COFF, CENP-SKO, CENP-HOFF/-IOFF/-KOFF/-MOFF/-NOFF, and CENP-OKO/-RKO cells showed a significantly reduced correlation. This indicates that these four groups of proteins affect the overall chromosomal association of centromere proteins in different ways.
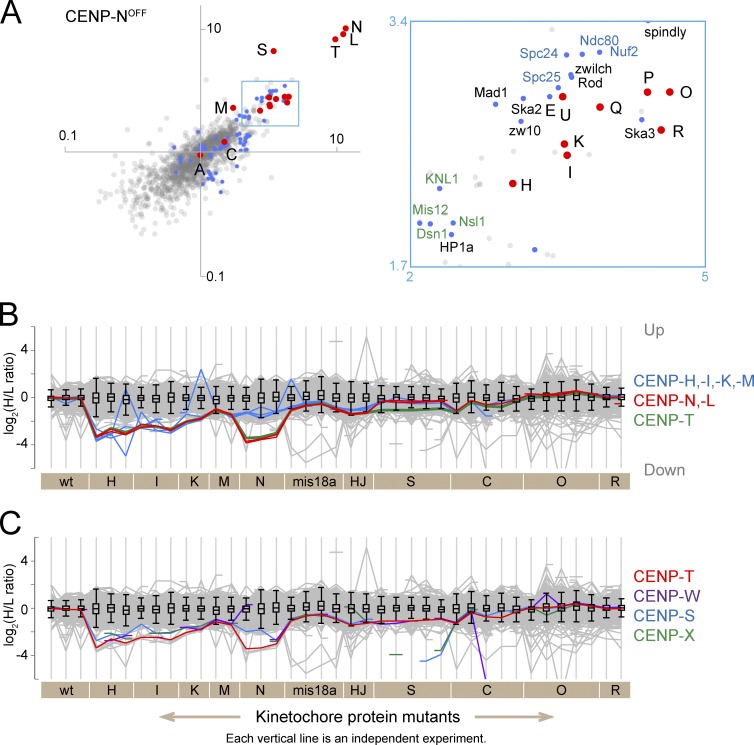
CENP-T behaves coordinately with CENP-N and CENP-L
CENP-N was originally identified as a CCAN member (Foltz et al., 2006; Izuta et al., 2006; Okada et al., 2006) that associates with CENP-A nucleosomes (Carroll et al., 2009; Hellwig et al., 2011). CENP-N, CENP-L, and CENP-T were by far the most depleted proteins in CENP-NOFF chromosomes (Fig. 4 A). CENP-H and CENP-I were also significantly decreased but had H/L ratios in a separate (less depleted) cluster together with members of the CENP-O/-P/-Q/-U complex, the Ndc80 complex, the RZZ complex, Mad1, and CENP-E (Fig. 4 A, inset).
Figure 4.
CENP-N,-L,-T show coordinated behavior in all but the CENP-S KO. (A) For CENP-N, a scatterplot of H/L ratios from two replicate experiments is shown. The boxed area is enlarged to the right. Abbreviations and color-coding are as in Fig. 2. (B) Profile plots show the log2(H/L ratios) of all proteins in all experiments (gray lines). The interquartile population for all proteins detected in each experiment is contained in the box. Whiskers extend to 1.5 times interquartile range away from the edge of the box. The y axis is inversely labeled so that proteins that decrease in mutant chromosomes are plotted downward and those increase are plotted upwards. The behavior of CENP-H,-I,-K,-M is shown in blue with CENP-M as a thin line, CENP-N,-L is in red, and CENP-T is in green. (C) Profile plot showing the behavior of CENP-T (red), CENP-W (purple), CENP-S (blue), and CENP-X (green). Breaks in the lines occur when the small proteins were not detected in individual experiments. wt, wild type.
These data suggest that CENP-N might be part of a protein cohort with CENP-L and CENP-T that interacts with CENP-H/-I/-K/-M to form a larger protein assembly in intact mitotic chromosomes. To reiterate, we use the term cohort to describe a group of proteins whose correlated behavior is consistent with them forming a protein complex that has yet to be independently confirmed.
CENP-H/-I/-K/-M and CENP-N/-L had very similar profiles in virtually all experiments, with the exception of CENP-NOFF cells (Fig. 4 B). (A “profile plot” is an illustrative summary of changes in the abundance of proteins across all experiments [see Materials and methods]. The gray lines plot levels of all 1,409 proteins analyzed in all experiments.) An unexpected result was obtained when comparing the CENP-NOFF proteome against its CENP-HOFF or CENP-IOFF counterparts. Despite CENP-N being part of the CENP-H/-I/-K/-M/-N cluster, when proteomes comprising only CCAN proteins were compared, those derived from CENP-NOFF chromosomes did not exhibit strong correlations with the others. This is seen as scatter away from the diagonal in Fig. 3 B and a lower correlation in the matrix of Fig. 3 C. This suggests that CENP-N is not always associated with CENP-H/-I/-K/-M.
The profile plots for CENP-N and CENP-L are nearly identical, consistent with studies that these proteins form a heterodimeric complex when they are loaded on chromosomes (Carroll et al., 2009; Hinshaw and Harrison, 2013). Both proteins were significantly depleted from chromosomes isolated from CENP-HOFF/-IOFF/-KOFF/-MOFF/-NOFF cells. Their reduction was, however, more modest in chromosomes from other CCAN mutant cells, i.e., CENP-COFF/-SKO/-OKO/-RKO (Fig. 4 B).
Unexpectedly, the profile plot for CENP-T was strikingly similar to CENP-N and CENP-L except in CENP-SKO experiments (Fig. 4 B). CENP-T was one of the most depleted proteins in chromosomes from CENP-HOFF/-IOFF/-KOFF/-MOFF/-NOFF cells (Fig. 4, B and C). This suggests a strong dependency of CENP-T on CENP-H/-I/-K/-M/-N/-L, particularly on CENP-N and CENP-L. Although CENP-T on its own has a DNA binding activity in vitro (Hori et al., 2008a), its centromere association may also require CENP-H/-I/-K/-M/-N. Indeed a reduction of CENP-T levels in CENP-HOFF cells was reported previously (Hori et al., 2008a). CENP-T and CENP-W make a DNA binding complex with their histone fold domains (Hori et al., 2008a), and both proteins show essentially identical profiles in these experiments (Fig. 4 C).
CENP-S and CENP-X may function in part independently of CENP-T
In addition to a CENP-T/-W dimer, a CENP-T/-W/-S/-X tetramer has also been reported at centromeres (Nishino et al., 2012). The profile plot of CENP-S, however, did not always follow that of CENP-T in isolated mitotic chromosomes (Fig. 4 C). For instance, CENP-S was less affected than CENP-T by depletion of CENP-H/-I/-N, suggesting that CENP-S/-X may participate in another complex. Indeed, CENP-S/-X also associates with FANCM (Singh et al., 2010; Yan et al., 2010). For comparison, protein profiles for known complexes, such as the four core histones, the cohesin complex and CPC appear as single lines (Fig. S3).
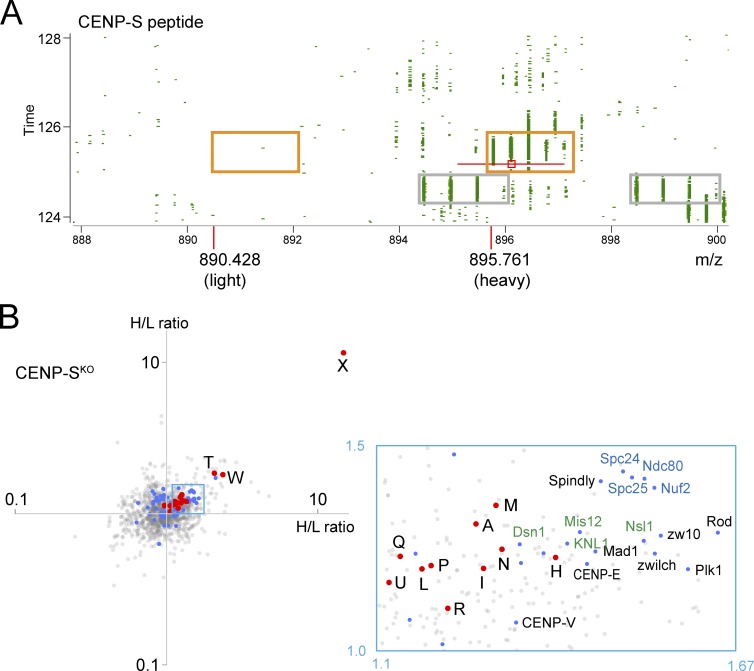
Because CENP-S is dispensable for growth in DT40 cells, we could obtain a complete KO cell line (Amano et al., 2009). CENP-S peptides were not detected on the chromosomes from CENP-SKO cells. This results in an H/L SILAC ratio of zero or infinite value, and the H/L ratios for individual CENP-S peptides were sometimes dealt with as missing values by the Andromeda search engine (i.e., disregarded in calculating the H/L ratio for a given protein). We therefore reviewed the data manually, and the MS1 spectrum of one representative peptide (AAVHYTTGCLCQDVAEDKGVLFSK, charge +4) was identified with heavy-labeled species (i.e., from wild-type cells; Fig. 5 A, orange boxes). We observed no trace of the corresponding light-labeled peptide, confirming the absence of CENP-S from CENP-SKO chromosomes.
Figure 5.
Analysis of chromosomes from CENP-SKO cells. (A) 2D plot of m/z versus retention time. The area around the peptide, AAVHYTTGCLCQDVAEDKGVLFSK, charge +4 is shown. The right orange box contains the isotope pattern for heavy peptide. The left orange box is placed where the light peptide would be expected. MS1 peak marked with a red square was selected for MS/MS fragmentation with the m/z window as indicated by the red line. Monoisotopic m/z values for heavy and light peptides are shown. The gray boxes contain isotope patterns of an irrelevant peptide as an example of a SILAC pair. (B) CENP-SKO scatterplot from two replicate experiments. Note CENP-X alone at the far top right. The boxed area contains outer kinetochore proteins. Abbreviations and color-coding are as in Fig. 2.
By far, the most strongly affected protein in CENP-SKO chromosomes was its binding partner, CENP-X (Amano et al., 2009), with <10% remaining (Fig. 5 B). Inspection of MS1 spectra of a representative CENP-X peptide (QAQAEDLEKVDIEHVEK, charge +4) confirmed that this peptide was virtually absent (data not shown). After CENP-X, the next most depleted centromere proteins were CENP-T and CENP-W, but their depletion was much less extreme. Thus CENP-SKO chromosomes still retain some functional CENP-T/-W dimer. This may explain the viability of CENP-SKO cells.
CENP-N,-L were not reduced as much as CENP-T/-W on CENP-SKO chromosomes (Figs. 4 B and 5 B). Rather, the H/L ratio of CENP-N and CENP-L fell in the same cluster as CENP-H/-I/-K/-M and close to CENP-P/-Q/-R/-U (Fig. 5 B). This suggests that CENP-N and CENP-L are upstream of CENP-T for localization on chromosomes.
Interestingly, outer kinetochore components, including the KMN, RZZ, and Ska complexes plus CENP-E and Mad1, were more depleted than CCAN components on chromosomes lacking CENP-S (Fig. 5 B). Indeed, CENP-T can recruit the Ndc80 complex to chromosomes (Hori et al., 2013; Nishino et al., 2013).
CENP-C is required for association of the KMN network with chromosomes
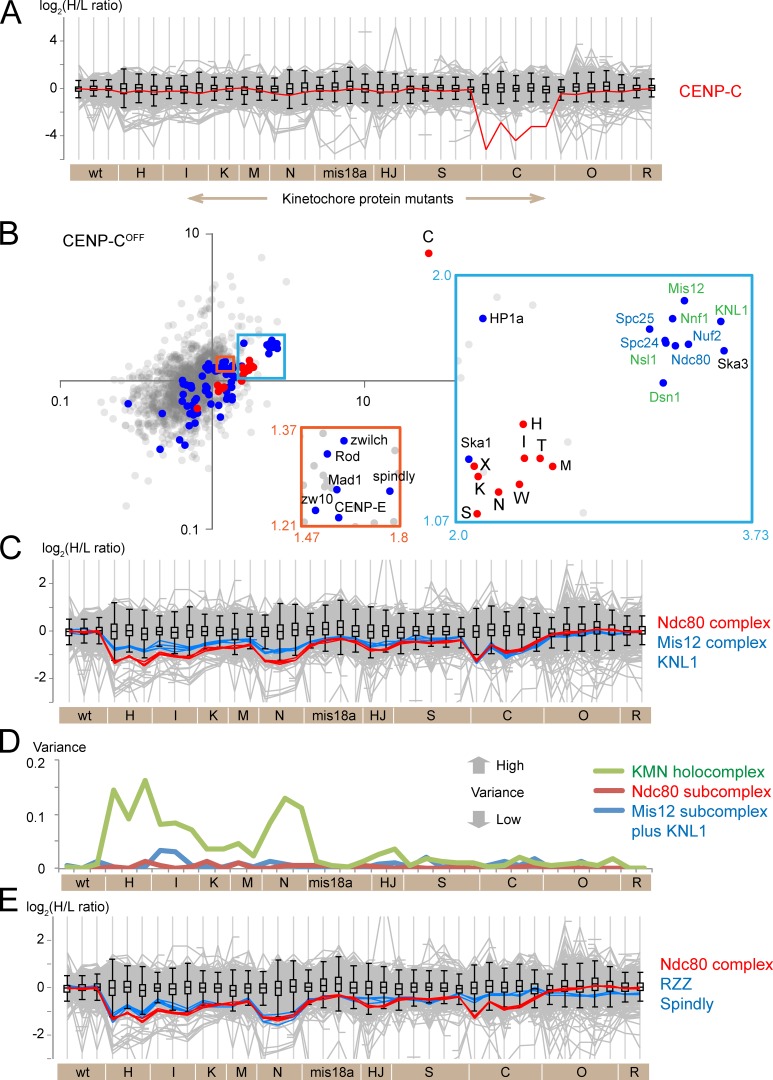
CENP-C links the centromeric chromatin with the outer kinetochore. The C-terminal and central regions of CENP-C interact with CENP-A nucleosomes (Kato et al., 2013), the N terminus binds to Mis12 in the KMN network (Carroll et al., 2010; Przewloka et al., 2011; Screpanti et al., 2011), and the PEST region binds CENP-H/-I/-K/-M (Klare et al., 2015). Profile plots showed only modest changes in CENP-C abundance after depletion of other CCAN proteins (Fig. 6 A). Notably, significant reduction of CENP-A in HJURPOFF or Mis18αOFF chromosomes (see Fig. 7) did not impact on CENP-C levels. We conclude that CENP-C associates with chicken mitotic chromosomes independently of other CCAN proteins.
Figure 6.
Characterization of CENP-C dependencies. (A) Profile plot shows the behavior of CENP-C protein in all experiments. (B) Scatterplot showing the H/L ratios from two replicate experiments. Note CENP-C alone at the far top right. Insets show enlarged views of the boxed areas. Abbreviations and color-coding are as in Fig. 2. (C) Profile plot showing the behavior of Ndc80 complex proteins (red), KNL1 (blue), and Mis12 complex proteins (blue) in all experiments. (D) Plot of the variance of the log2(H/L ratio) among the nine proteins that constitute the entire KMN complex (green), the four proteins of the Ndc80 subcomplex (red), and the four proteins of the Mis12 complex plus KNL1 (blue). (E) Profile plot comparing the behavior of the Ndc80 complex proteins (red) and RZZ complex plus Spindly (blue). wt, wild type.
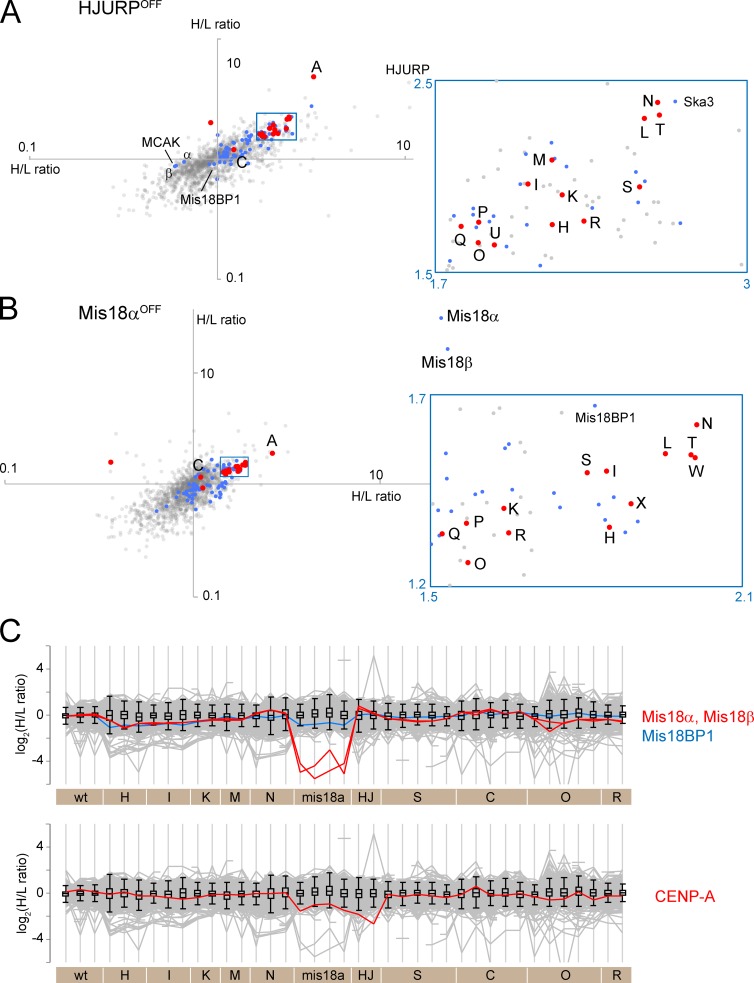
Figure 7.
Strategies to deplete CENP-A from mitotic chromosomes. (A and B) Scatterplots of H/L ratios from two replicate experiments are plotted for HJURPOFF (A) and Mis18αOFF chromosomes. (B). Abbreviations and color-coding are as in Fig. 2. (C) Profile plot shows the behavior of Mis18 complex proteins (top) and CENP-A (bottom).
CENP-C expression was turned off by treatment with doxycycline for 48 h. The H/L ratio for CENP-C reveals that only 14% remained on the chromosomes. No other CCAN protein was similarly affected. After CENP-C, the most depleted proteins in CENP-COFF chromosomes were members of the KMN network (Fig. 6 B). They formed a grouping distinct from the cluster of CENP-H/-I/-K/-M/-N/-L/-T on the scatterplot. Although CENP-H/-I/-K/-M/-N/-L/-T were reduced, their abundance on CENP-COFF chromosomes remained about half of the wild-type level.
CENP-C depletion caused a reduction in Mis12 complex proteins and KNL1 to an extent similar to that of the Ndc80 complex (Fig. 6, B and C). This stoichiometric codepletion is consistent with a direct interaction between CENP-C and the Mis12 complex (Przewloka et al., 2011; Screpanti et al., 2011) and contrasts with depletion of CENP-H/-I/-K/-M/-N, where Ndc80 levels fell with much less effect on the Mis12 complex (Fig. 6 C).
Proteins engaged in a protein complex should show coordinated behavior (i.e., low variance in their SILAC ratios) in our experiments. The variance of the SILAC ratios for the Ndc80 and Mis12 subcomplexes and KNL1 was small in chromosomes from mis18αOFF, CENP-COFF/-SKO/-OKO/-RKO cells (Fig. 6 D). This suggests that components of the KMN network holocomplex behaved together as a single entity in those mutants. In contrast, the variance between the subcomplexes was significantly larger in chromosomes from CENP-HOFF/-IOFF/-KOFF/-MOFF/-NOFF cells. This suggests that not all Ndc80, Mis12, and KNL1 behave as a single holocomplex. Importantly, the Ndc80 and Mis12 complexes, respectively, were apparently intact in chromosomes from those cells because the variance within each subcomplex remained small. Assembly of the KMN network may be defective in chromosomes from CENP-HOFF/-IOFF/-KOFF/-MOFF/-NOFF cells, and this might explain the results of an earlier study (Amaro et al., 2010). Alternatively, a subpopulation of the Ndc80 complex may not be engaged in the KMN network.
The RZZ complex is involved in silencing of spindle checkpoint signaling (Karess, 2005). Spindly bridges RZZ and dynein (Griffis et al., 2007). Profile plots of Ndc80 and RZZ look very similar to each other in most mutant chromosomes (Fig. 6 E). However, RZZ was much less depleted than the Ndc80 complex in CENP-COFF chromosomes, forming a separate cluster in scatterplots of CENP-COFF experiments (note the relative locations of the orange and blue boxes in Fig. 6 B). The reduction of chromosome-associated RZZ complex may be linked to effects on the Ndc80 complex in chromosomes depleted of CENP-H/-I/-K/-M/-N, but this does not occur in CENP-COFF chromosomes (Fig. 6 E).
Partial CENP-A depletion has little effect on CENP-C levels
Available CENP-A conditional KO cells do not die when CENP-A transcription is silenced, suggesting that they have acquired additional mutations (Régnier et al., 2005). We therefore adopted other strategies to deplete centromeres of CENP-A. HJURP is a chaperone that is required for CENP-A loading onto centromeres (Dunleavy et al., 2009; Foltz et al., 2009). We silenced HJURP expression by exposure of KO cells (Perpelescu et al., 2015) to doxycycline for 84 h. This lowered chromosomal levels of HJURP by at least 80%. After this treatment, CENP-A was the most depleted centromere protein, with 18–28% remaining on the chromosomes (Fig. 7 A).
CENP-N/-L/-T/-S were the next most depleted centromere proteins after CENP-A, in HJURPOFF chromosomes. KIF2C (MCAK), Mis18α, and Mis18β accumulated on chromosomes from HJURPOFF cells (Fig. 7 A). Mis18BP1/KNL2 was essentially unaffected (Fig. 7, A and C).
Mis18α works together with Mis18β and Mis18BP1 in telophase to prime sites for CENP-A recruitment on chromosomes (Fujita et al., 2007). Mis18α and Mis18β were the most depleted proteins when Mis18α transcription was silenced (Fig. 7 B). Mis18BP1 was also significantly depleted, though much less so than Mis18α or Mis18β. Profile plots of the three proteins imply that Mis18BP1 may not associate with Mis18α and Mis18β in chromosomes isolated from nocodazole-treated cells (Fig. 7 C). After Mis18α and Mis18β, CENP-A was the most depleted centromere protein in mis18αOFF chromosomes (Fig. 7 B). CENP-N/-L/-T/-W also showed a significant decrease, with CENP-H/-I/-K being less affected. Interestingly, chromosomal levels of CENP-C were little affected by the reduction of CENP-A in HJURPOFF or Mis18αOFF chromosomes despite the fact that CENP-C has been reported to interact with CENP-A (Carroll et al., 2010).
Taken together, reduction in the amount of CENP-A coincided well with the reduction of CENP-N/-L/-T/-W. This observation is reminiscent of the results of CENP-N depletion, where CENP-N/-L/-T were the most affected proteins.
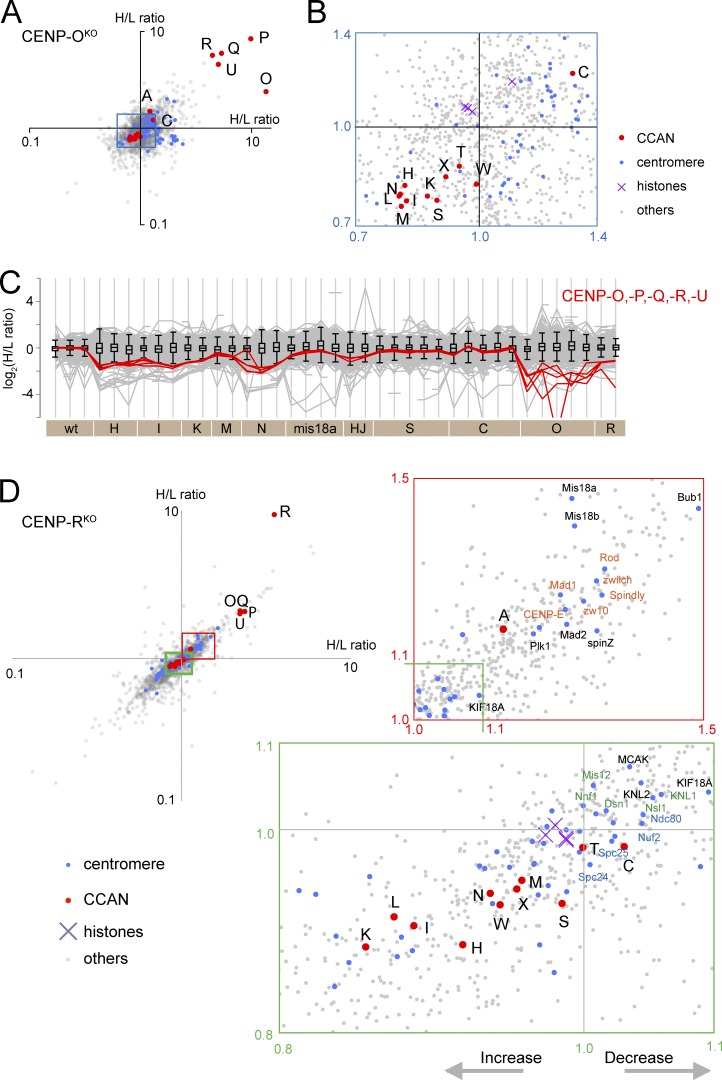
Accumulation of CCAN proteins on CENP-OKO chromosomes
CENP-OKO and CENP-RKO cells are viable (Okada et al., 2006). CENP-O forms a complex with CENP-P/-Q/-U and CENP-R (Hori et al., 2008b), and these proteins were significantly depleted from CENP-OKO chromosomes (Fig. 8, A and C). CENP-O/-P/-Q/-U were depleted to a somewhat lesser extent from CENP-RKO chromosomes (H/L ratio = ∼2; Fig. 8, C and D). The tight cluster suggests that CENP-O/-P/-Q/-U were lost as a single entity in the absence of CENP-R (though their depletion was less severe than seen with loss of CENP-O). Interestingly, CENP-H/-I/-K/-N/-L/-T/-S accumulated on chromosomes isolated from CENP-OKO and CENP-RKO cells (Fig. 8, B and D). This could reflect an enlargement of the inner kinetochore, as cells adapt to continued growth without CENP-O/-P/-Q/-R/-U, either as a direct effect or possibly by modulating the expression or stability of other proteins. Importantly, this accumulation of CCAN proteins was not accompanied by an increase in KMN and RZZ complex components. Thus, increasing the content of CCAN proteins does not necessarily increase the size of the outer kinetochore.
Figure 8.
Effects of depleting CENP-O,-P,-Q,-R,-U from chromosomes. (A) Scatterplot of H/L ratios from two replicate experiments with the CENP-OKO. (B) Enlargement of the blue box in A. Levels of most CCAN proteins (red) are increased in CENP-OKO chromosomes. (C) Profile plot shows the behavior of CENP-O group proteins in all experiments. (D) Scatterplot of H/L ratios from two replicate experiments with the CENP-RKO. Enlargement of the green box confirms the increase in levels of most CCAN proteins, whereas the red box reveals a decrease in RZZ-associated proteins. wt, wild type.
Clustering and correlation networks within the kinetochore
The fact that proteins in the same complex consistently show very similar H/L ratios across experiments allowed us to perform clustering analysis and map a correlation network of kinetochore proteins (Fig. 9). The clusters seen in Fig. 9 A confirm previous descriptions of CCAN complexes, e.g., CENP-H/-I/-K, CENP-S/-X, CENP-O/-P/-Q/-R/-U, as well as the Mis12, Ndc80, and Ska complexes in the outer kinetochore. They also reveal the existence of two previously undescribed protein cohorts: CENP-N/-L/-T in the inner kinetochore and Rod/Zw10/Zwilch/Spindly/CENP-E/Mad1 in the outer kinetochore.
Figure 9.
Identification of cohorts among kinetochore proteins. (A) Cluster analysis based on SILAC ratio of kinetochore proteins in all experiments. This is an expanded form of the clusters in Fig. 1 that contain CCAN and outer kinetochore proteins. Gray blocks correspond to missing values, as some proteins were not identified in every experiment. (B) Cluster analysis of the correlation coefficient between pairs of named proteins. The color-coding is shown in the bar. Blowup of a portion of the correlation matrix (right). In the heatmap (left), the location of the magnified portion is shown by red arrows at the bottom right. The Pearson correlation coefficients between pairs of kinetochore proteins are shown using a color-coded scale, with the highest positive correlation (yellow). wt, wild type.
The similarity of H/L ratio profiles between pairs of proteins was gauged by Pearson correlation coefficient (Fig. S4). The correlation between every pair of proteins in the chromosomes was summarized as a matrix shown in Fig. 9 B, with a blowup in Fig. S5. A profile of these correlations against the rest of the proteome was used for hierarchical clustering analysis so that proteins with strong correlations are grouped together. One cluster that was enriched for kinetochore proteins was scrutinized in detail (Fig. 9 B). The Mis12 complex, KNL1, and the Ndc80 complex together comprised a single correlation cluster (yellow). This is the KMN network (Cheeseman and Desai, 2008). The Ndc80 complex also formed part of a second cluster together with CENP-N/-L/-T. Interestingly, CENP-N/-L/-T and CENP-H/-I/-K were grouped separately.
In both clustering analysis of H/L ratios and the correlation meta-analysis, CENP-E and Mad1 made a single cluster with the RZZ complex (Fig. 9, A and B). Correlation analysis revealed a link of this RZZ supercohort with CENP-N/-L/-T (Fig. 9 B). This RZZ cohort also showed a good correlation with the Ndc80 complex, suggesting a significant interaction. On the other hand, the RZZ cohort did not exhibit a high correlation with the Mis12 complex, suggesting that RZZ assembly on chromosomes is linked with Ndc80CENP-T rather than with Ndc80CENP-C.
Discussion
This analysis differs from previous studies of kinetochore assembly in that it exclusively concerns untagged endogenous proteins in situ in native mitotic chromosomes. The results were not constrained by requiring antibodies that may exhibit varying affinities or accessibilities to their target epitopes, tags that might impair protein function in subtle ways, or the need to find experimental conditions where the proteins are soluble.
Dependencies on CENP-A and CENP-C
Neither CENP-A nor CENP-C depends on any other CCAN protein for association with mitotic chromosomes. Depletion of CENP-A and CENP-C was not accompanied by reduction of other proteins to the same extent as themselves, even though CENP-A depletion decreased the chromosomal accumulation of CENP-N/-L/-T and, to a lesser extent, CENP-H/-I/-K/-O/-P/-Q/-R/-U. Thus, CENP-A and CENP-C do not have protein cohorts as defined by this analysis. This is not the case for most kinetochore components. In the case of the depletion of CENP-H, CENP-I, CENP-K, CENP-M, and CENP-N and loss of CENP-O and CENP-S, one or more other proteins were reduced as much as the target protein. Our work supports the view that CENP-A is a marker for the site of kinetochore assembly rather than a structural foundation for the kinetochore (Prendergast et al., 2011).
CENP-N binds to CENP-A (Carroll et al., 2009), and in our experiments, chromosomal association of CENP-N/-L depended on CENP-A. CENP-T/-W/-S/-X has been reported to bind to DNA independently of CENP-A (Hori et al., 2008a). The unexpected dependency that we observe for CENP-T/-W on CENP-N/-L may favor accumulation of CENP-T/-W near CENP-A nucleosomes. Consistent with this, CENP-N and CENP-T coprecipitated from chromatin extracts as part of the CENP-A associated complex (Foltz et al., 2006) and map at a similar radial position by superresolution microscopy (Suzuki et al., 2014).
CENP-C depletion also had a relatively minor effect on assembly of the inner kinetochore in these experiments. This was surprising in light of the recent study that CENP-C interacts in vitro with a complex of CENP-H/-I/-K/-M (Basilico et al., 2014; Klare et al., 2015) and previous work showing that CENP-C is required for kinetochore localization of CENP-N/-L in yeast (Tanaka et al., 2009; Hinshaw and Harrison, 2013). One possibility is that our depletion of CENP-C (14% remaining on chromosomes) was insufficient to phenocopy the effects seen in previous studies. Alternatively, contributions with other components may also promote the centromere localization of CENP-H,-I,-K,-M in DT40 cells.
CENP-C depletion severely disrupted outer kinetochore assembly. Levels of the entire KMN network were severely reduced on CENP-C–depleted chromosomes. This fits with the view of CENP-C as an essential link between the inner and outer kinetochore (Przewloka et al., 2011; Screpanti et al., 2011).
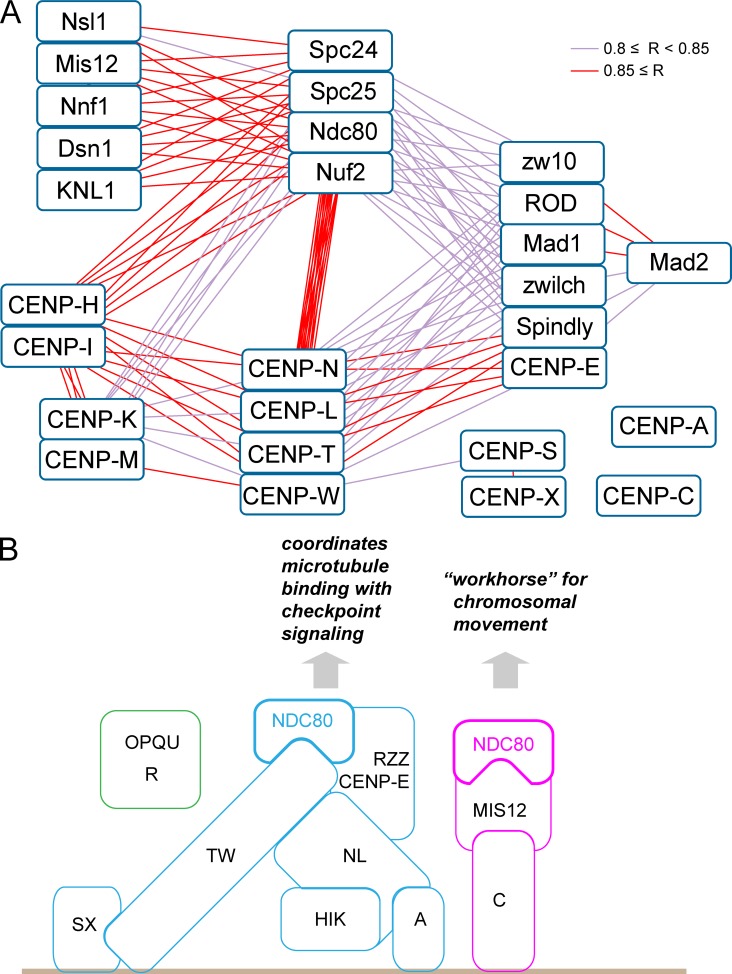
A dependency correlation network for the chicken kinetochore
A network constructed from the correlation matrix based on SILAC ratios for all proteins provides a graphic demonstration of relationships between kinetochore proteins and integrates the diverse experiments reported here (Fig. 10 A). Importantly, no novel proteins clustered among the CCAN members, suggesting that all members of this group of proteins are now known.
Figure 10.
Correlation network and model for the modular organization of the kinetochore. (A) Correlation network of kinetochore proteins constructed using Cytoscape (Shannon et al., 2003). Pairs with a correlation coefficient 0.8 ≤ R < 0.85 (violet lines) and 0.85 ≤ R (red lines) are shown. (B) Model diagram based on dependencies between CCAN proteins observed in these experiments.
The correlation analysis splits “core” CCAN components into the CENP-H/-I/-K/-M complex and CENP-N/-L/-T/-W cohort. These are defined not only by their coordinated behavior when constituent kinetochore proteins are deleted but also functionally. CENP-N/-L/-T/-W exhibit a strong interdependency with members of the RZZ cohort (which includes CENP-E, Mad1, and Spindly [see next section]), whereas CENP-H/-I/-K/-M do not.
CENP-O/-P/-Q/-R/-U and CENP-S/-X are distinct complexes that appear to be “peripheral” in the sense that they do not correlate nearly as tightly with the other CCAN components as they do within themselves. Depletion of CENP-O/-P/-Q/-R/-U actually caused an increase in chromosomal levels of CENP-H/-I/-K/-N/-L/-T/-S. This was not accompanied by an increase in the KMN and RZZ complexes. These results suggest that CENP-O/-P/-Q/-R/-U may have a role in limiting the size of the inner kinetochore and that the size of the outer kinetochore does not scale directly with the content of CCAN proteins.
The Ndc80 complex as a hub of kinetochore dependencies
Remarkably the Ndc80 complex appears to be a “hub” of the kinetochore dependency network. This complex provides a physical link between kinetochores and dynamic spindle microtubules and has roles in microtubule capture, monitoring the state of microtubule binding (Alushin et al., 2010, 2012), error correction (Cheeseman et al., 2006; DeLuca et al., 2006), and subsequent transport of chromosomes into daughter cells upon anaphase onset (Tooley and Stukenberg, 2011). Ndc80 is the only complex that shows robust correlations with all other cohorts and complexes diagrammed here (Fig. 10 A). For example, behavior of the CENP-N/-L/-T/-W cohort does not correlate with the Mis12 complex, and CENP-H/-I/-K/-M complex behavior does not correlate with either the Mis12 or RZZ complexes. The Ndc80 complex also shows robust interactions with the Ska complex, which was not included in Fig. 10 A because its interactions as deduced from correlation analysis are surprisingly complex.
We noted from analysis of the variance across all experiments that the Ndc80 complex does not always covary with the other components of the KMN network (the Mis12 complex and Knl1). Indeed, two different types of Ndc80 supercomplex had previously been suggested (Gascoigne et al., 2011; Schleiffer et al., 2012; Nishino et al., 2013). We will refer to these as Ndc80CENP-C and Ndc80CENP-T. The spatial distribution of CENP-C and CENP-T on centromeres is different (Ribeiro et al., 2010), and it was suggested from reconstitution experiments that those supercomplexes might have distinct functions (Hori et al., 2013). In Ndc80CENP-C, CENP-C binds to the Mis12 complex and KNL1, thereby forming the KMN network. In Ndc80CENP-T, Spc24 and Spc25 bind CENP-T directly upon phosphorylation of the CENP-T N-terminus (Nishino et al., 2013). The Mis12 complex competes with CENP-T for this binding site. Thus, Ndc80CENP-T does not associate with Mis12 and KNL1 and therefore does not form a KMN network.
Our data suggest that Ndc80CENP-C and Ndc80CENP-T may serve different functions in the kinetochore (Fig. 10 B). Ndc80CENP-C exhibits dependency relationships consistent with interactions with the KMN network. It may therefore function primarily in microtubule binding, although a role in the spindle checkpoint has also been suggested (Kim and Yu, 2015). It was previously suggested that the RZZ complex might associate with the KMN network (Varma et al., 2013); however, our dependency analysis suggests a closer functional association between Ndc80CENP-T and the expanded RZZ cohort. We do not see dependency interactions between the RZZ cohort and Mis12 components.
The RZZ cohort revealed by our analysis consists of RZZ components plus Spindly, CENP-E, and Mad1. These proteins exhibit such tight correlations with one another that they are likely to form a physical complex in chromosomes. We propose terming this the RZZ–MES (Rod, Zw10, Zwilch, Mad1, CENP-E, Spindly) supercomplex. This complex has not been observed in solution and may only form when its constituents are associated with chromosomes. RZZ is involved in recruiting Mad1/Mad2 to kinetochores (Karess, 2005), and Mad1 in turn recruits CENP-E (Akera et al., 2015). CENP-E is required to convert kinetochore–microtubule interactions from lateral to end-on (Shrestha and Draviam, 2013). Thus, Ndc80CENP-T may cooperate with the RZZ–MES to coordinate end-on microtubule attachments with spindle checkpoint signaling. It will be interesting in future experiments to determine whether Mps1 kinase preferentially associates with Ndc80CENP-T (Hiruma et al., 2015; Ji et al., 2015).
Perspectives
This study of the entire mitotic chromosome proteome in a comprehensive set of inner kinetochore mutants has led to the elucidation of a network of protein dependencies throughout the kinetochore. This network divides the CCAN into four subcomplexes that appear to be relatively independent of one another for localization to mitotic chromosomes (Fig. 10). Perhaps surprisingly, we identified close functional links between CENP-N/-L and CENP-T/-W consistent with them acting as a complex in the kinetochore. Our data also suggest that the RZZ complex may actually be part of a larger RZZ–MES supercomplex that coordinates microtubule attachments with checkpoint signaling in the outer kinetochore. This analysis will hopefully stimulate future biochemical and cell biology experiments to explore the novel relationships identified here.
Materials and methods
Cell culture and medium
The chicken lymphoma B cell line DT40 was grown in RPMI1640 medium supplemented with 10% FBS and 1% penicillin/streptomycin.
For SILAC, wild type (clone18), CENP-SKO, and CENP-OKO were grown with 10% dialyzed FBS (mol wt cutoff, 10,000). Other cell lines were grown with complete FBS. For SILAC labeling of lysine and arginine, 100 µg/ml 13C6-,15N2-l-lysine:2 HCl and 30 µg/ml 13C6-,15N4-l-arginine:HCl were added to the medium with dialyzed FBS.
1 µg/ml doxycycline was used to silence the rescuing cDNA expression of conditional KO cell lines. We refer to such doxycycline-treated cells as GENENAMEOFF (e.g., as CENP-COFF).
Cell lines
Details of the following KO cell lines are described in the original publications: CENP-CON/OFF (Kwon et al., 2007), CENP-HON/OFF (Fukagawa et al., 2001), CENP-ION/OFF (Nishihashi et al., 2002), CENP-KON/OFF, CENP-MON/OFF and CENP-OKO (Okada et al., 2006), CENP-NON/OFF (Ribeiro et al., 2010), CENP-RKO (Hori et al., 2008b), CENP-SKO (Amano et al., 2009), and HJURPON/OFF and Mis18αON/OFF (Perpelescu et al., 2015).
Cell lines designated GENENAMEON/OFF are conditional KOs where all or a portion of an essential gene has been removed and cells are kept alive by a cDNA whose expression can be silenced by addition of doxycycline. Nonessential genes were absolute KOs, designated GENENAMEKO. Expression of exogenous “rescue” cDNAs encoding CENP-C/-H/-M/-N or Mis18α were turned off by treatment with doxycycline for 48 h before harvest of cells. CENP-ION/OFF and CENP-KON/OFF cells were incubated with doxycycline for 36 h. HJURPON/OFF cells were treated with doxycycline for 84 h.
In all cases, cells were incubated with nocodazole for the last 12 h before harvest.
Preparation of mitotic chromosomes
DT40 cells were incubated with 0.5 µg/ml nocodazole for 12–13 h before harvest. The length of incubation in doxycycline was calibrated such that a minimum mitotic index of >75% was achieved. Longer incubations resulted in increased apoptosis and a decreased mitotic index as cultures became less healthy. Mitotic chromosomes were isolated in the polyamine-EDTA buffer system (Lewis and Laemmli, 1982). The protocol was optimized for chicken DT40 cells with some modification (Ohta et al., 2010).
Mass spectrometry
The DNA content of chromosome isolates was quantified with picoGreen (Invitrogen). Equal amounts of wild-type and mutant chromosomes (based on DNA content) were mixed and resuspended in SDS-PAGE sample buffer (50 mM Tris, pH 6.8, 2% SDS, 10% glycerol, 0.01% bromophenol blue, and 5% 2-mercaptoethanol). Coomassie-stained gel bands were excised and proteins were digested with trypsin, as previously described (Shevchenko et al., 1996). Briefly, proteins were reduced in 10 mM dithiothreitol (Sigma-Aldrich) for 30 min at 37°C and alkylated in 55 mM iodoacetamide (Sigma-Aldrich) for 20 min at ambient temperature in the dark. They were then digested overnight at 37°C with 12.5 ng/µl trypsin (Pierce). After digestion, samples were acidified with orthophosphoric acid and subjected to strong cation exchange fractionation, using a polySULFOETHYL A column (PolyLC) on an Ultimate 3000 Dionex LC system (Thermo Fisher Scientific). Mobile phase A consisted of 5mM KH2PO4, 10% acetonitrile at pH 3; mobile phase B was 5 mM KH2PO4, 1 M KCl, and 10% acetonitrile, pH 3. The peptides were fractionated using the following gradient: 0%–60% buffer B in 18 min, then to 70% in 2 min, and then to 0% in 6 min. The flow rate was constant at 200 µl/min. Fractions were collected at 1-min time slices.
After strong cation exchange chromatography, fractions were diluted with an equal volume of 0.1% TFA and spun onto StageTips as described by Rappsilber et al. (2003). Peptides were eluted in 20 µl 80% acetonitrile in 0.1% TFA and concentrated down to 4 µl by vacuum centrifugation (Concentrator 5301; Eppendorf). The peptide sample was then prepared for liquid chromatography tandem mass spectrometry analysis by diluting it to 5 µl by 0.1% TFA. Mass spectrometry analyses were performed either on a Velos LTQ-Orbitrap mass spectrometer (Thermo Fisher Scientific) or on a QExactive Mass spectrometer (Thermo Fisher Scientific), both coupled on-line to Ultimate 3000 RSLCnano Systems (Dionex; Thermo Fisher Scientific). The analytical column with a self-assembled particle frit (Ishihama et al., 2002) and C18 material (3 µm; ReproSil-Pur C18-AQ; Dr. Maisch GmbH) was packed into a spray emitter (75-µm ID, 8-µm opening, 300-mm length; New Objective) using an air-pressure pump (Proxeon Biosystems). Mobile phase A consisted of water and 0.1% formic acid, and mobile phase B consisted of 80% acetonitrile and 0.1% formic acid. The gradient used was 220 min. In both mass spectrometers, the peptides were loaded onto the column at a flow rate of 0.5 µl/min and eluted at a flow rate of 0.2 µl/min according to the following gradient: 2%–40% buffer B in 180 min, then to 95% in 16 min
For the LTQ Orbitrap Velos, Fourier transform mass spectrometry spectra were recorded at 60,000 resolution and the 20 most intense peaks with charge ≥2 of the MS scan were selected with an isolation window of 1.0 Thomson in the ion trap for MS2 (normal scan, wideband activation, filling 5.0E5 ions for MS scan, 1.0E4 ions for MS2, maximum fill time 100 ms, dynamic exclusion for 120 s). For the Q Exactive, FTMS spectra were recorded at 70,000 resolution and the top 10 most abundant peaks with charge of two or more and isolation window of 2.0 Thomson were selected and fragmented by higher-energy collisional dissociation (Olsen et al., 2007) with normalized collision energy of 25. The maximum ion injection time for the MS and MS2 scans was set to 20 and 60 ms, respectively, and the AGC target was set to 1 E6 for the MS scan and to 5 E4 for the MS2 scan. Dynamic exclusion was set to 60 s.
The MaxQuant software platform (Cox and Mann, 2008) version 1.5.1.2 was used to process the raw files and search was conducted against chicken complete/reference proteome set of UniProt database (released on May 14, 2014) with additional sequences from our in-house database of chicken proteins, using the Andromeda search engine (Cox et al., 2011). The first search peptide tolerance was set to 20 ppm, whereas the main search peptide tolerance was set to 4.5 pm. Isotope mass tolerance was 2 ppm, and maximum charge was 7. A maximum of two missed cleavages was allowed. Carbamidomethylation of cysteine was set as a fixed modification. Oxidation of methionine and acetylation of the N terminus were set as variable modifications. Peptide and protein identifications were filtered to 1% false discovery rate.
Processing of data
The combined data from 38 experiments contained 2,760 protein IDs. To minimize noise, we removed protein entries that occurred in <25 experiments. 1,667 proteins remained after this filtering process (Table S1). Unprocessed data were used to draw scatter plots and histograms.
In cases where discrepancies in protein H/L ratio were found in the replicates, we inspected the MS1 data manually. Where MS1 peaks suffered from interference because of coeluting signals, the corresponding peptide H/L ratio was removed from the analysis. Centromere proteins affected by this data curation were CENP-M, CENP-R, CENP-U, CENP-W, CENP-X, APC4, and APC5.
Filtering out contaminant proteins
To compare the proteomes of mutant and wild-type chromosomes, it was necessary to differentially label cells with stable isotopes (SILAC) in order to distinguish the chromosomal proteins of mutant and wild-type cells from one another. The metabolic labeling with heavy amino acids used for SILAC requires growth of the cells in medium with dialyzed FBS. Unfortunately, many DT-40 conditional KO cell lines do not grow in medium with dialyzed FBS. We have been unable, despite repeated efforts, to determine the explanation, and we thus grew most mutant cell lines in medium with complete FBS.
To test whether the proteome of isolated mitotic chromosomes changed when cells were cultured in conventional medium or medium with dialyzed FBS, we determined the proteome of wild-type mitotic chromosomes isolated from cells grown under the two sets of conditions. The histogram in Fig. S1 A shows the distribution of log2(H/L ratios) for the 1,667 proteins quantified in this experiment. The distribution was unimodal but skewed to the left. Multipeak analysis identified a second minor peak, comprising ∼15% of the total and centered at 2−0.855 = 0.55 (Fig. S1, A and C).
Cluster analysis with our complete dataset identified three subgroups whose H/L ratios fluctuated in a manner different from the rest (Fig. 1). The first subgroup contained CCAN proteins. The second subgroup is highlighted as purple trees at the top of Fig. 1. They were probably contaminants, many of which were not identified in wild-type mitotic chromosomes. They may be derived from interphase or apoptotic cells. The third group, also highlighted as purple trees, clustered at the bottom of Fig. 1. This group contained proteins that accumulated on chromosomes from cells grown with FBS relative to those grown in dialyzed FBS. The distribution of their H/L ratios coincides with the second minor peak (Fig. S1, B and E). These proteins were almost exclusively identified as cytoplasmic in our previous analysis (Fig. S1 F; Ohta et al., 2010). Because the proteins of the second minor peak were predominantly cytoplasmic, we filtered this set of 259 proteins from our subsequent analyses. This filtration was important, as it decreased random scatter in our data and allowed us to more accurately determine the variance of bona fide chromosomal proteins.
Profile plots
A profile plot is an illustrative summary of changes in the abundance of proteins across experiments (Fig. 4 B). The H/L ratio of a protein in one experiment is plotted along a vertical line, in this case with positive H/L ratios below the x axis so that descending lines reflect decreasing values of the protein in question. Members of the same protein complex typically follow a very similar pattern. The superimposed box plots with whiskers provide a convenient criterion to assess the significance of changes in the abundance or H/L ratio of each protein. The interquartile box contains 50% of all H/L ratios measured in each experiment, with the whiskers encompassing more than 95% of all the total ratios (assuming a normal distribution of values).
Online supplemental material
Fig. S1 shows for several representative experiments the distribution of total proteins and centromere proteins as well as showing the distribution of contaminating proteins that were removed from our statistical analysis. Fig. S2 demonstrates a step-by-step correlation analysis between a pair of experiments with a good correlation (CENP-HOFF vs. CENP-IOFF) and a pair of experiments with a poor correlation (CENP-HOFF vs. CENP-NOFF). Fig. S3 shows profile plots for the CPC, core histones, and the cohesin complexes. Fig. S4 is a step-by-step correlation analysis between the RZZ component Rod and the Mis12 component Nsl1 (poor correlation) and CENP-T (good correlation). Fig. S5 is a blowup of Fig. 9 B, with the positions of several key complexes indicated. Table S1 lists the H/L ratios of all proteins in every experiment. Online supplemental material is available at http://www.jcb.org/cgi/content/full/jcb.201508072/DC1.
Supplementary Material
Acknowledgments
This work was funded by The Wellcome Trust, of which W.C. Earnshaw is a Principal Research Fellow (grant 107022) and J. Rappsilber is a Senior Research Fellow (grant 103139). The Wellcome Trust Centre for Cell Biology is supported by core grant numbers 077707 and 092076 and instrument grant 091020.
The authors declare no competing financial interests.
Footnotes
Abbreviations used in this paper:
- CCAN
- constitutive centromere-associated network
- CPC
- chromosome passenger complex
- KO
- knockout
- RZZ
- Rod, zw10, and zwilch
- SILAC
- stable isotope labeling by amino acids
References
- Akera T., Goto Y., Sato M., Yamamoto M., and Watanabe Y.. 2015. Mad1 promotes chromosome congression by anchoring a kinesin motor to the kinetochore. Nat. Cell Biol. 17:1124–1133. [DOI] [PubMed] [Google Scholar]
- Alushin G.M., Ramey V.H., Pasqualato S., Ball D.A., Grigorieff N., Musacchio A., and Nogales E.. 2010. The Ndc80 kinetochore complex forms oligomeric arrays along microtubules. Nature. 467:805–810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alushin G.M., Musinipally V., Matson D., Tooley J., Stukenberg P.T., and Nogales E.. 2012. Multimodal microtubule binding by the Ndc80 kinetochore complex. Nat. Struct. Mol. Biol. 19:1161–1167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amano M., Suzuki A., Hori T., Backer C., Okawa K., Cheeseman I.M., and Fukagawa T.. 2009. The CENP-S complex is essential for the stable assembly of outer kinetochore structure. J. Cell Biol. 186:173–182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amaro A.C., Samora C.P., Holtackers R., Wang E., Kingston I.J., Alonso M., Lampson M., McAinsh A.D., and Meraldi P.. 2010. Molecular control of kinetochore-microtubule dynamics and chromosome oscillations. Nat. Cell Biol. 12:319–329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Basilico F., Maffini S., Weir J.R., Prumbaum D., Rojas A.M., Zimniak T., De Antoni A., Jeganathan S., Voss B., van Gerwen S., et al. . 2014. The pseudo GTPase CENP-M drives human kinetochore assembly. eLife. 3:e02978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biggins S. 2013. The composition, functions, and regulation of the budding yeast kinetochore. Genetics. 194:817–846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bodor D.L., Mata J.F., Sergeev M., David A.F., Salimian K.J., Panchenko T., Cleveland D.W., Black B.E., Shah J.V., and Jansen L.E.. 2014. The quantitative architecture of centromeric chromatin. eLife. 3:e02137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carroll C.W., Silva M.C., Godek K.M., Jansen L.E., and Straight A.F.. 2009. Centromere assembly requires the direct recognition of CENP-A nucleosomes by CENP-N. Nat. Cell Biol. 11:896–902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carroll C.W., Milks K.J., and Straight A.F.. 2010. Dual recognition of CENP-A nucleosomes is required for centromere assembly. J. Cell Biol. 189:1143–1155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheeseman I.M. 2014. The kinetochore. Cold Spring Harb. Perspect. Biol. 6:a015826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheeseman I.M., and Desai A.. 2008. Molecular architecture of the kinetochore-microtubule interface. Nat. Rev. Mol. Cell Biol. 9:33–46. [DOI] [PubMed] [Google Scholar]
- Cheeseman I.M., Chappie J.S., Wilson-Kubalek E.M., and Desai A.. 2006. The conserved KMN network constitutes the core microtubule-binding site of the kinetochore. Cell. 127:983–997. [DOI] [PubMed] [Google Scholar]
- Ciferri C., Pasqualato S., Screpanti E., Varetti G., Santaguida S., Dos Reis G., Maiolica A., Polka J., De Luca J.G., De Wulf P., et al. . 2008. Implications for kinetochore-microtubule attachment from the structure of an engineered Ndc80 complex. Cell. 133:427–439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen R.L., Espelin C.W., De Wulf P., Sorger P.K., Harrison S.C., and Simons K.T.. 2008. Structural and functional dissection of Mif2p, a conserved DNA-binding kinetochore protein. Mol. Biol. Cell. 19:4480–4491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cox J., and Mann M.. 2008. MaxQuant enables high peptide identification rates, individualized p.p.b.-range mass accuracies and proteome-wide protein quantification. Nat. Biotechnol. 26:1367–1372. [DOI] [PubMed] [Google Scholar]
- Cox J., Neuhauser N., Michalski A., Scheltema R.A., Olsen J.V., and Mann M.. 2011. Andromeda: a peptide search engine integrated into the MaxQuant environment. J. Proteome Res. 10:1794–1805. [DOI] [PubMed] [Google Scholar]
- DeLuca J.G., and Musacchio A.. 2012. Structural organization of the kinetochore-microtubule interface. Curr. Opin. Cell Biol. 24:48–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeLuca J.G., Gall W.E., Ciferri C., Cimini D., Musacchio A., and Salmon E.D.. 2006. Kinetochore microtubule dynamics and attachment stability are regulated by Hec1. Cell. 127:969–982. [DOI] [PubMed] [Google Scholar]
- De Wulf P., McAinsh A.D., and Sorger P.K.. 2003. Hierarchical assembly of the budding yeast kinetochore from multiple subcomplexes. Genes Dev. 17:2902–2921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunleavy E.M., Roche D., Tagami H., Lacoste N., Ray-Gallet D., Nakamura Y., Daigo Y., Nakatani Y., and Almouzni-Pettinotti G.. 2009. HJURP is a cell-cycle-dependent maintenance and deposition factor of CENP-A at centromeres. Cell. 137:485–497. [DOI] [PubMed] [Google Scholar]
- Fachinetti D., Folco H.D., Nechemia-Arbely Y., Valente L.P., Nguyen K., Wong A.J., Zhu Q., Holland A.J., Desai A., Jansen L.E., and Cleveland D.W.. 2013. A two-step mechanism for epigenetic specification of centromere identity and function. Nat. Cell Biol. 15:1056–1066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foltz D.R., Jansen L.E., Black B.E., Bailey A.O., Yates J.R. III, and Cleveland D.W.. 2006. The human CENP-A centromeric nucleosome-associated complex. Nat. Cell Biol. 8:458–469. [DOI] [PubMed] [Google Scholar]
- Foltz D.R., Jansen L.E., Bailey A.O., Yates J.R. III, Bassett E.A., Wood S., Black B.E., and Cleveland D.W.. 2009. Centromere-specific assembly of CENP-a nucleosomes is mediated by HJURP. Cell. 137:472–484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujita Y., Hayashi T., Kiyomitsu T., Toyoda Y., Kokubu A., Obuse C., and Yanagida M.. 2007. Priming of centromere for CENP-A recruitment by human hMis18alpha, hMis18beta, and M18BP1. Dev. Cell. 12:17–30. [DOI] [PubMed] [Google Scholar]
- Fukagawa T., and Earnshaw W.C.. 2014. The centromere: chromatin foundation for the kinetochore machinery. Dev. Cell. 30:496–508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukagawa T., Mikami Y., Nishihashi A., Regnier V., Haraguchi T., Hiraoka Y., Sugata N., Todokoro K., Brown W., and Ikemura T.. 2001. CENP-H, a constitutive centromere component, is required for centromere targeting of CENP-C in vertebrate cells. EMBO J. 20:4603–4617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gascoigne K.E., Takeuchi K., Suzuki A., Hori T., Fukagawa T., and Cheeseman I.M.. 2011. Induced ectopic kinetochore assembly bypasses the requirement for CENP-A nucleosomes. Cell. 145:410–422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griffis E.R., Stuurman N., and Vale R.D.. 2007. Spindly, a novel protein essential for silencing the spindle assembly checkpoint, recruits dynein to the kinetochore. J. Cell Biol. 177:1005–1015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hellwig D., Emmerth S., Ulbricht T., Döring V., Hoischen C., Martin R., Samora C.P., McAinsh A.D., Carroll C.W., Straight A.F., et al. . 2011. Dynamics of CENP-N kinetochore binding during the cell cycle. J. Cell Sci. 124:3871–3883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hinshaw S.M., and Harrison S.C.. 2013. An Iml3-Chl4 heterodimer links the core centromere to factors required for accurate chromosome segregation. Cell Reports. 5:29–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hiruma Y., Sacristan C., Pachis S.T., Adamopoulos A., Kuijt T., Ubbink M., von Castelmur E., Perrakis A., and Kops G.J.. 2015. CELL DIVISION CYCLE. Competition between MPS1 and microtubules at kinetochores regulates spindle checkpoint signaling. Science. 348:1264–1267. [DOI] [PubMed] [Google Scholar]
- Hori T., Amano M., Suzuki A., Backer C.B., Welburn J.P., Dong Y., McEwen B.F., Shang W.H., Suzuki E., Okawa K., et al. . 2008a CCAN makes multiple contacts with centromeric DNA to provide distinct pathways to the outer kinetochore. Cell. 135:1039–1052. [DOI] [PubMed] [Google Scholar]
- Hori T., Okada M., Maenaka K., and Fukagawa T.. 2008b CENP-O class proteins form a stable complex and are required for proper kinetochore function. Mol. Biol. Cell. 19:843–854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hori T., Shang W.H., Takeuchi K., and Fukagawa T.. 2013. The CCAN recruits CENP-A to the centromere and forms the structural core for kinetochore assembly. J. Cell Biol. 200:45–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishihama Y., Rappsilber J., Andersen J.S., and Mann M.. 2002. Microcolumns with self-assembled particle frits for proteomics. J. Chromatogr. A. 979:233–239. [DOI] [PubMed] [Google Scholar]
- Izuta H., Ikeno M., Suzuki N., Tomonaga T., Nozaki N., Obuse C., Kisu Y., Goshima N., Nomura F., Nomura N., and Yoda K.. 2006. Comprehensive analysis of the ICEN (Interphase Centromere Complex) components enriched in the CENP-A chromatin of human cells. Genes Cells. 11:673–684. [DOI] [PubMed] [Google Scholar]
- Jeyaprakash A.A., Santamaria A., Jayachandran U., Chan Y.W., Benda C., Nigg E.A., and Conti E.. 2012. Structural and functional organization of the Ska complex, a key component of the kinetochore-microtubule interface. Mol. Cell. 46:274–286. [DOI] [PubMed] [Google Scholar]
- Ji Z., Gao H., and Yu H.. 2015. CELL DIVISION CYCLE. Kinetochore attachment sensed by competitive Mps1 and microtubule binding to Ndc80C. Science. 348:1260–1264. [DOI] [PubMed] [Google Scholar]
- Karess R. 2005. Rod-Zw10-Zwilch: a key player in the spindle checkpoint. Trends Cell Biol. 15:386–392. [DOI] [PubMed] [Google Scholar]
- Kato H., Jiang J., Zhou B.R., Rozendaal M., Feng H., Ghirlando R., Xiao T.S., Straight A.F., and Bai Y.. 2013. A conserved mechanism for centromeric nucleosome recognition by centromere protein CENP-C. Science. 340:1110–1113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim S., and Yu H.. 2015. Multiple assembly mechanisms anchor the KMN spindle checkpoint platform at human mitotic kinetochores. J. Cell Biol. 208:181–196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klare K., Weir J.R., Basilico F., Zimniak T., Massimiliano L., Ludwigs N., Herzog F., and Musacchio A.. 2015. CENP-C is a blueprint for constitutive centromere-associated network assembly within human kinetochores. J. Cell Biol. 210:11–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwon M.S., Hori T., Okada M., and Fukagawa T.. 2007. CENP-C is involved in chromosome segregation, mitotic checkpoint function, and kinetochore assembly. Mol. Biol. Cell. 18:2155–2168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis C.D., and Laemmli U.K.. 1982. Higher order metaphase chromosome structure: evidence for metalloprotein interactions. Cell. 29:171–181. [DOI] [PubMed] [Google Scholar]
- Nishihashi A., Haraguchi T., Hiraoka Y., Ikemura T., Regnier V., Dodson H., Earnshaw W.C., and Fukagawa T.. 2002. CENP-I is essential for centromere function in vertebrate cells. Dev. Cell. 2:463–476. [DOI] [PubMed] [Google Scholar]
- Nishino T., Takeuchi K., Gascoigne K.E., Suzuki A., Hori T., Oyama T., Morikawa K., Cheeseman I.M., and Fukagawa T.. 2012. CENP-T-W-S-X forms a unique centromeric chromatin structure with a histone-like fold. Cell. 148:487–501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishino T., Rago F., Hori T., Tomii K., Cheeseman I.M., and Fukagawa T.. 2013. CENP-T provides a structural platform for outer kinetochore assembly. EMBO J. 32:424–436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Obuse C., Yang H., Nozaki N., Goto S., Okazaki T., and Yoda K.. 2004. Proteomics analysis of the centromere complex from HeLa interphase cells: UV-damaged DNA binding protein 1 (DDB-1) is a component of the CEN-complex, while BMI-1 is transiently co-localized with the centromeric region in interphase. Genes Cells. 9:105–120. [DOI] [PubMed] [Google Scholar]
- Ohta S., Bukowski-Wills J.C., Sanchez-Pulido L., Alves Fde.L., Wood L., Chen Z.A., Platani M., Fischer L., Hudson D.F., Ponting C.P., et al. . 2010. The protein composition of mitotic chromosomes determined using multiclassifier combinatorial proteomics. Cell. 142:810–821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohta S., Wood L., Bukowski-Wills J.C., Rappsilber J., and Earnshaw W.C.. 2011. Building mitotic chromosomes. Curr. Opin. Cell Biol. 23:114–121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okada M., Cheeseman I.M., Hori T., Okawa K., McLeod I.X., Yates J.R. III, Desai A., and Fukagawa T.. 2006. The CENP-H-I complex is required for the efficient incorporation of newly synthesized CENP-A into centromeres. Nat. Cell Biol. 8:446–457. [DOI] [PubMed] [Google Scholar]
- Olsen J.V., Macek B., Lange O., Makarov A., Horning S., and Mann M.. 2007. Higher-energy C-trap dissociation for peptide modification analysis. Nat. Methods. 4:709–712. [DOI] [PubMed] [Google Scholar]
- Ong S.E., Blagoev B., Kratchmarova I., Kristensen D.B., Steen H., Pandey A., and Mann M.. 2002. Stable isotope labeling by amino acids in cell culture, SILAC, as a simple and accurate approach to expression proteomics. Mol. Cell. Proteomics. 1:376–386. [DOI] [PubMed] [Google Scholar]
- Perpelescu M., and Fukagawa T.. 2011. The ABCs of CENPs. Chromosoma. 120:425–446. [DOI] [PubMed] [Google Scholar]
- Perpelescu M., Hori T., Toyoda A., Misu S., Monma N., Ikeo K., Obuse C., Fujiyama A., and Fukagawa T.. 2015. HJURP is involved in the expansion of centromeric chromatin. Mol. Biol. Cell. 26:2742–2754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petrovic A., Mosalaganti S., Keller J., Mattiuzzo M., Overlack K., Krenn V., De Antoni A., Wohlgemuth S., Cecatiello V., Pasqualato S., et al. . 2014. Modular assembly of RWD domains on the Mis12 complex underlies outer kinetochore organization. Mol. Cell. 53:591–605. [DOI] [PubMed] [Google Scholar]
- Prendergast L., van Vuuren C., Kaczmarczyk A., Doering V., Hellwig D., Quinn N., Hoischen C., Diekmann S., and Sullivan K.F.. 2011. Premitotic assembly of human CENPs -T and -W switches centromeric chromatin to a mitotic state. PLoS Biol. 9:e1001082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Przewloka M.R., Venkei Z., Bolanos-Garcia V.M., Debski J., Dadlez M., and Glover D.M.. 2011. CENP-C is a structural platform for kinetochore assembly. Curr. Biol. 21:399–405. [DOI] [PubMed] [Google Scholar]
- Rappsilber J., Ishihama Y., and Mann M.. 2003. Stop and go extraction tips for matrix-assisted laser desorption/ionization, nanoelectrospray, and LC/MS sample pretreatment in proteomics. Anal. Chem. 75:663–670. [DOI] [PubMed] [Google Scholar]
- Régnier V., Vagnarelli P., Fukagawa T., Zerjal T., Burns E., Trouche D., Earnshaw W., and Brown W.. 2005. CENP-A is required for accurate chromosome segregation and sustained kinetochore association of BubR1. Mol. Cell. Biol. 25:3967–3981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ribeiro S.A., Vagnarelli P., Dong Y., Hori T., McEwen B.F., Fukagawa T., Flors C., and Earnshaw W.C.. 2010. A super-resolution map of the vertebrate kinetochore. Proc. Natl. Acad. Sci. USA. 107:10484–10489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samejima K., Samejima I., Vagnarelli P., Ogawa H., Vargiu G., Kelly D.A., de Lima Alves F., Kerr A., Green L.C., Hudson D.F., et al. . 2012. Mitotic chromosomes are compacted laterally by KIF4 and condensin and axially by topoisomerase IIα. J. Cell Biol. 199:755–770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schleiffer A., Maier M., Litos G., Lampert F., Hornung P., Mechtler K., and Westermann S.. 2012. CENP-T proteins are conserved centromere receptors of the Ndc80 complex. Nat. Cell Biol. 14:604–613. [DOI] [PubMed] [Google Scholar]
- Screpanti E., De Antoni A., Alushin G.M., Petrovic A., Melis T., Nogales E., and Musacchio A.. 2011. Direct binding of Cenp-C to the Mis12 complex joins the inner and outer kinetochore. Curr. Biol. 21:391–398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sekulic N., Bassett E.A., Rogers D.J., and Black B.E.. 2010. The structure of (CENP-A-H4)(2) reveals physical features that mark centromeres. Nature. 467:347–351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shannon P., Markiel A., Ozier O., Baliga N.S., Wang J.T., Ramage D., Amin N., Schwikowski B., and Ideker T.. 2003. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 13:2498–2504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shevchenko A., Wilm M., Vorm O., and Mann M.. 1996. Mass spectrometric sequencing of proteins silver-stained polyacrylamide gels. Anal. Chem. 68:850–858. [DOI] [PubMed] [Google Scholar]
- Shrestha R.L., and Draviam V.M.. 2013. Lateral to end-on conversion of chromosome-microtubule attachment requires kinesins CENP-E and MCAK. Curr. Biol. 23:1514–1526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh T.R., Saro D., Ali A.M., Zheng X.F., Du C.H., Killen M.W., Sachpatzidis A., Wahengbam K., Pierce A.J., Xiong Y., et al. . 2010. MHF1-MHF2, a histone-fold-containing protein complex, participates in the Fanconi anemia pathway via FANCM. Mol. Cell. 37:879–886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki A., Badger B.L., Wan X., DeLuca J.G., and Salmon E.D.. 2014. The architecture of CCAN proteins creates a structural integrity to resist spindle forces and achieve proper Intrakinetochore stretch. Dev. Cell. 30:717–730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tachiwana H., Kagawa W., Shiga T., Osakabe A., Miya Y., Saito K., Hayashi-Takanaka Y., Oda T., Sato M., Park S.Y., et al. . 2011. Crystal structure of the human centromeric nucleosome containing CENP-A. Nature. 476:232–235. [DOI] [PubMed] [Google Scholar]
- Takahashi K., Yamada H., and Yanagida M.. 1994. Fission yeast minichromosome loss mutants mis cause lethal aneuploidy and replication abnormality. Mol. Biol. Cell. 5:1145–1158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka K. 2013. Regulatory mechanisms of kinetochore-microtubule interaction in mitosis. Cell. Mol. Life Sci. 70:559–579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka K., Chang H.L., Kagami A., and Watanabe Y.. 2009. CENP-C functions as a scaffold for effectors with essential kinetochore functions in mitosis and meiosis. Dev. Cell. 17:334–343. [DOI] [PubMed] [Google Scholar]
- Tooley J., and Stukenberg P.T.. 2011. The Ndc80 complex: integrating the kinetochore’s many movements. Chromosome Res. 19:377–391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varma D., Wan X., Cheerambathur D., Gassmann R., Suzuki A., Lawrimore J., Desai A., and Salmon E.D.. 2013. Spindle assembly checkpoint proteins are positioned close to core microtubule attachment sites at kinetochores. J. Cell Biol. 202:735–746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei R.R., Schnell J.R., Larsen N.A., Sorger P.K., Chou J.J., and Harrison S.C.. 2006. Structure of a central component of the yeast kinetochore: the Spc24p/Spc25p globular domain. Structure. 14:1003–1009. [DOI] [PubMed] [Google Scholar]
- Wei R.R., Al-Bassam J., and Harrison S.C.. 2007. The Ndc80/HEC1 complex is a contact point for kinetochore-microtubule attachment. Nat. Struct. Mol. Biol. 14:54–59. [DOI] [PubMed] [Google Scholar]
- Westhorpe F.G., and Straight A.F.. 2013. Functions of the centromere and kinetochore in chromosome segregation. Curr. Opin. Cell Biol. 25:334–340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Westhorpe F.G., and Straight A.F.. 2015. The centromere: epigenetic control of chromosome segregation during mitosis. Cold Spring Harb. Perspect. Biol. 7:a015818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan Z., Delannoy M., Ling C., Daee D., Osman F., Muniandy P.A., Shen X., Oostra A.B., Du H., Steltenpool J., et al. . 2010. A histone-fold complex and FANCM form a conserved DNA-remodeling complex to maintain genome stability. Mol. Cell. 37:865–878. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.