Abstract
How do cancer cells escape tightly controlled regulatory circuits that link their proliferation to extracellular nutrient cues? An emerging theme in cancer biology is the hijacking of normal stress response mechanisms to enable growth even when nutrients are limiting. Pancreatic ductal adenocarcinoma (PDA) is the quintessential aggressive malignancy that thrives in nutrient-poor, hypoxic environments. PDAs overcome these limitations through appropriation of unorthodox strategies for fuel source acquisition and utilization. Additionally, interplay between evolving PDA and whole body metabolism contributes to disease pathogenesis. Deciphering how these pathways function and integrate with one another can reveal novel angles of therapeutic attack.
Keywords: Pancreatic cancer, metabolism, autophagy, lysosome, glycolysis, diabetes, obesity
INTRODUCTION
Characteristic features of pancreatic ductal adenocarcinoma
Pancreatic ductal adenocarcinoma (PDA) is among the most lethal of all cancer types with approximately 48,000 new cases and 40,000 deaths annually in the United States (1). It is projected to become the 2nd leading cause of cancer death by 2020 in the US and has a 5-year survival rate of only ~6%, which has changed little over the last 4 decades. Invasive PDA arises through multistage genetic and histological progression from microscopic precursor lesions designated as Pancreatic Intraepithelial Neoplasia (PanIN) that are believed to develop and progress asymptomatically over several decades (1–3) (Figure 1A).
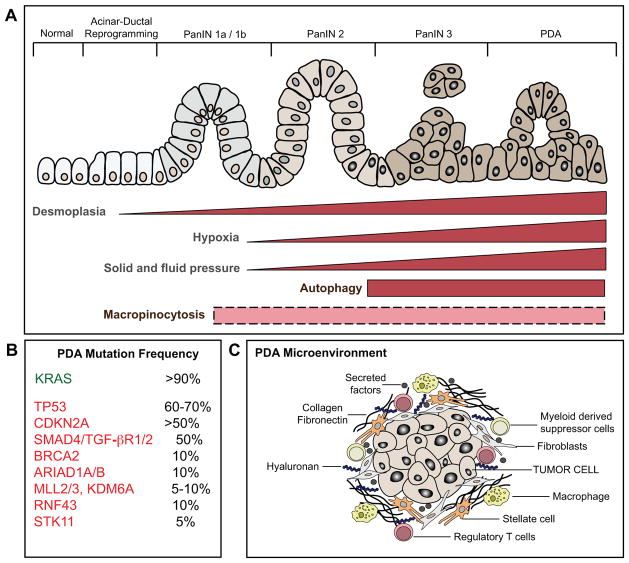
Figure 1. Schematic of the multi-stage progression of pancreatic ductal adenocarcinoma.
A) PDA arises from the multi-stage progression of precursor lesions known as pancreatic intraepithelial neoplasia (PanIN). B) KRAS mutations are an early event in disease pathogenesis, present in the great majority of early stage PanIN lesions. Mutations in a series of tumor suppressors occur as later events, and contribute to disease progression. C) PDA is also associated with evolving alterations in the tumor microenvironment, including increasing fibrosis and extracellular matrix deposition (desmoplasia) and recruitment of immune and inflammatory cells. Increasing desmoplasia accompanies progressive disease (as indicated) and creates intratumoral pressure that compresses the vasculature, resulting in limited blood flow to the tumor and consequent hypoxia and low nutrient delivery. In turn, PDA cells exhibit activation of nutrient scavenging pathways (autophagy and macropinocytosis) that support tumor cell growth. While autophagy activation is a late event in PDA tumorigenesis, the precise temporal dynamics of macropinocytosis is as yet unknown (dotted box).
An early event during malignant transformation is the acquisition of activating mutations in the KRAS oncogene at codons 12, 13, 61, which occurs in >90% of PDA patients. PDAs are highly “addicted” to this oncogene for multiple parameters influencing tumor initiation, progression and maintenance as demonstrated using genetically engineered mouse (GEM) models and human PDA cell lines (4–9). Additionally, inactivating mutations and deletions of tumor suppressor genes, TRP53, CDKN2A, and SMAD4 are also frequently observed and occur later during disease progression (Figure 1B). Metastatic lesions exhibit extensive conservation of genomic alterations with matched primary PDA, although specific mutations in the primary tumor (in SMAD and TRP53) are associated with increased propensity for metastatic dissemination (10–12). GEM models incorporating these genetic alterations have provided functional validation of their roles in progression of PanIN to PDA and in metastasis (4, 6, 13–16).
The identification of these recurrent mutations and additional less common genetic alterations in PDA has not yet pointed to key targets that are readily inactivated by existing drugs (17, 18). While KRAS is clearly a critical driver of tumorigenesis, pharmacologic KRAS inhibitors remain elusive. Thus, the present standard of care involves conventional cytotoxic agents which can yield significant responses but in most patients have limited efficacy (1). These issues highlight the need to further probe the biology of PDA in order to uncover novel vulnerabilities of the cancer cells. Indeed, recent studies have revealed a profound rewiring of metabolic pathways activated downstream of oncogenic KRAS that is essential for PDA growth and holds promise as a source of targets for new therapeutic strategies (19, 20). Activation of these pathways may also be linked to the unique microenvironment of PDA, which is characterized by extensive stromal and matrix deposition (desmoplasia) (21). While other cancer types such as breast, prostate and ovarian cancers also display prominent stromal infiltration, PDA stands out by the remarkable extent of its desmoplastic reaction, which often forms the bulk of the tumor mass. This heterogeneous stromal infiltrate — consisting of activated fibroblasts (pancreatic stellate cells) and diverse inflammatory and immune cells — coevolves with the tumor cells and influences PDA progression and response to therapy (22–28). A prominent consequence of the dense stroma is the generation of high levels of solid stress and fluid pressure in the tumors and compression of the vasculature, which creates a highly hypoxic and nutrient-poor microenvironment (22, 29–31) (Figure 1C). Despite these harsh environmental conditions, PDA cells are able to survive and thrive. How do these cells subsist in the presence of low levels of nutrients derived from the circulation? Which pathways are activated that allow unbridled proliferative capacity? This review will focus on the recently discovered unorthodox strategies employed by PDA cells to acquire nutrients and use them for generation of energy and as building blocks for de novo synthesis of proteins, lipids and nucleic acids. We will also provide an overview of how PDA pathogenesis is influenced by conditions that alter whole body metabolism, such as diabetes and obesity. Finally, we will discuss the translational potential of exploiting knowledge about pancreatic cancer metabolism for improved diagnostics and therapy for this disease.
Uncoupling nutrient sensing in cancer
The adaptive changes in tumor metabolism can broadly be categorized into alterations in the sensing, acquisition and utilization of nutrients, and elimination of toxic byproducts. In non-cancerous cells, the utilization of nutrients is tightly linked to their abundance via the action of multiple nutrient-sensing pathways (32). These sensors are finely tuned to detect drops in cellular nutrient levels before they have deleterious consequences — e.g. Adenosine monophosphate-activated protein kinase (AMPK) (33) — or conversely to respond to signs of plenitude — e.g. Mechanistic Target of Rapamycin Complex 1 (mTORC1) (34). For example, in response to reduced energy charge (ATP:AMP ratio), AMPK triggers two tightly coordinated responses: one is to shut down energy-intensive anabolic processes such as protein and lipid biosynthesis. The second is to increase energy generation both by activating autophagy, a nutrient scavenging/recycling pathway that provides fuel sources by breaking down superfluous cellular components into their constituent building blocks, and by enhancing mitochondrial oxidative phosphorylation. Additional sensors for lipids, amino acids and other key metabolites act to restore homeostasis through similar principles (32). An emerging view is that cancer cells adapt to life under limiting nutrient conditions by breaking these basic rules and removing the dichotomy between states of biosynthesis and catabolism. This bypass endows cancer cells with sustained growth even in challenging environments where nutrients and oxygen are scarce, or following metastasis to distant organ sites. How this occurs has been the focus of extensive study over the last several years (35) and much evidence suggests that cancer cells hijack and modify normal cellular homeostatic response mechanisms to maintain an unrestricted rate of growth.
While relatively little is known to date about how sensing mechanisms themselves may be subverted or appropriated in PDA, significant advances have been made with regards to how these tumors obtain nutrients and channel them into distinct biochemical pathways. PDA and other KRAS-driven cancers thrive in poorly perfused, hypovascular conditions by simultaneously upregulating both nutrient acquisition and utilization pathways (36, 37). This metabolic reprogramming may enable PDA cells to more efficiently maintain adequate intracellular nutrient levels despite limited external supply, providing them with a competitive growth advantage compared to law-abiding normal cells. Thus, severe nutrient and oxygen shortage may function as strong selective pressures favoring survival of aggressive tumor cells able to withstand such harsh environmental conditions. Conversely, the acquired dependence of PDA on these pathways creates new vulnerabilities that can be targeted therapeutically.
GLUCOSE AND GLUTAMINE METABOLISM IN PDA
Anabolic glucose metabolism
To fuel their elevated demand for energy and macromolecular biosynthesis, many cancers show augmented nutrient acquisition that is coupled to increased flux through downstream metabolic pathways. Thus, it is not surprising that mutations in KRAS and other canonical oncogenes (e.g. AKT, MYC and PI3K) and tumor suppressors (e.g. TP53, RB and PTEN) that drive accelerated growth also directly reprogram cellular metabolism by acting at both of these levels (38–40). A common theme associated with these central cancer pathways is the promotion of glucose metabolism, which serves as a major nutrient source for the production of ATP and provides building blocks for anabolic processes. In keeping with their poor perfusion, the overall levels of glucose and its rate of uptake are thought to be modest in PDA compared to other cancer types (29). Measurement of steady state metabolite levels suggests that glucose concentrations are not significantly elevated in most PDAs compared to adjacent pancreatic tissue (29). Nevertheless, among PDAs, higher levels of glucose uptake and expression of the primary glucose transporter, GLUT1 (SLC21A), correlate with worse prognosis (41, 42). Moreover, alterations in glucose delivery and utilization are required for PDA tumorigenesis, and mutant KRAS serves as a major regulator of these processes. Using a GEM model with expression of mutant KRAS under a doxycycline-inducible promoter, it was shown that KRAS silencing markedly reduces glucose uptake in PDA in vivo and in derivative cell lines, associated with downregulation of GLUT1 and of multiple glycolytic enzymes (8, 37) (Figure 2).
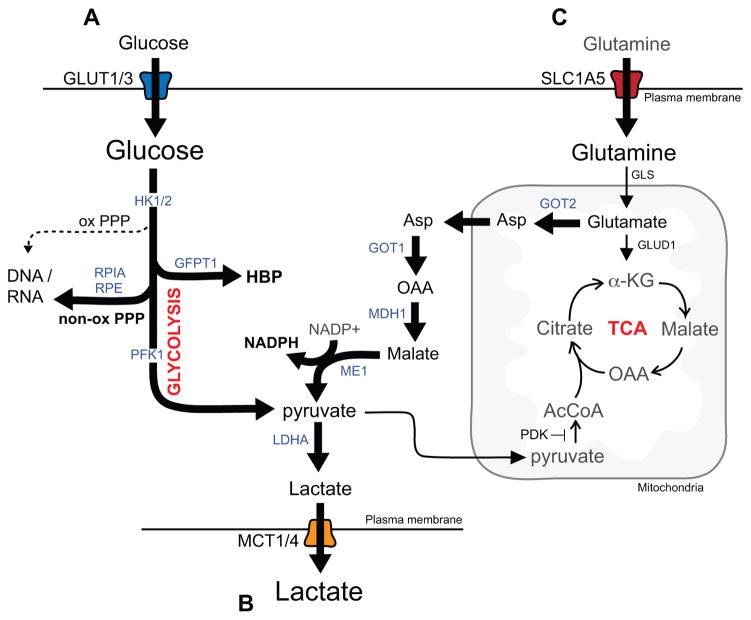
Figure 2. Alterations in metabolite utilization in PDA.
A) KRAS promotes glucose metabolism in PDA cells by upregulating the GLUT1 transporter and driving glycolysis through induction of the expression of multiple glycolytic enzymes. In addition, glycolytic intermediates are shunted toward biosynthetic pathways including the non-oxidative arm of the pentose phosphoate pathway (PPP) for synthesis of DNA and RNA and the hexosamine biosynthesis pathway (HBP), which generates precursors necessary for generation of glycoproteins, glycolipids, proteoglycans and glycosaminoglycans. B) In addition, PDA cells have enhanced activity of monocarboxylate transporters, MCT1 and MCT4 that shuttle lactate in order to prevent intracellular accumulation and subsequent decreases in cytosolic pH. C) KRAS also activates and reprograms glutamine metabolism. A proportion of glutamate is utilized to fuel NAPDH production via the aspartate-malate shunt thus contributing to maintenance of reduced glutathione levels and redox balance. The enzymes whose expression levels are regulated by mutant Kras are indicated in blue.
In PDA cells grown in vitro, as in most cultured cells, glycolysis predominates over mitochondrial oxidative phosphorylation of pyruvate, regardless of oxygen tension – a phenomenon known as the Warburg effect. This is mediated by inhibition of pyruvate dehydrogenase by pyruvate dehydrogenase kinase (PDK) and by increased lactate dehydrogenase (LDH) activity. The decreased fractional utilization of pyruvate for ATP generation in the mitochondria allows for the channeling of glycolytic intermediates into important anabolic pathways including the hexosamine biosynthesis pathway (HBP), which generates substrates for protein and lipid glycosylation, and to the non-oxidative arm of the pentose phosphate pathway (PPP), which generates ribose-5-phosphate for nucleotide biosynthesis. Unlike the well-known oxidative PPP, this latter pathway does not produce NADPH thereby necessitating other mechanisms for redox control (see below). KRAS-mediates these changes by transcriptional induction of genes encoding rate-limiting enzymes in both pathways (8) (Figure 2A). These alterations in glucose metabolism are required for the full tumorigenic growth of PDA cells as demonstrated by the decreased ATP levels and reduced growth of PDA xenografts treated with a small molecule inhibitor of LDHA (FX11, which acts by competing with NADH binding) (43). Similarly, knockdown of key KRAS-regulated enzymes in the non-oxidative PPP or the hexosamine pathway slows the growth of murine PDA cell lines in vitro and suppresses tumorigenicity upon subcutaneous implantation (8, 44).
This dependence on glycolysis also presents additional demands on mobilization and excretion of potentially toxic byproducts. Enhanced shuttling of lactate via the activity of monocarboxylate transporters, MCT1 and MCT4 (encoded by the SLC16A1 and SLC16A3 genes, respectively), was shown to be essential to prevent intracellular accumulation of lactate and decreased cytosolic pH in PDA cells (45, 46) (Figure 2B). These transporters are overexpressed in PDA compared to normal tissue and are required for PDA growth (45), with MCT4 playing a predominant role, supporting the physiologic importance of this detoxification process. PDA cells also show elevated levels of the lactate receptor GPR81, which regulates expression of lactate transporters, and CD147, an essential MCT chaperone protein (47). Thus, in response to increased metabolic demand, PDA cells coordinately enhance glucose utilization and lactate mobilization (44).
The signaling pathways controlling glucose metabolism downstream of KRAS have not been completely resolved, although inhibition of MEK signaling markedly impairs glycolysis of PDA cell lines in vitro (8). Moreover, the regulation of glycolytic enzymes by KRAS is at least in part MEK-dependent and involves transcriptional control by MYC and likely other transcription factors (8). PDA show multiple additional mechanisms for altering glucose metabolism beyond direct KRAS signaling. For example, it appears that hypoxia and the hypoxia-inducible factor (HIF1a) contribute to upregulation of glycolysis and HBP genes in PDA (44). The FOXM1 and KLF4 transcription factors have also been proposed as positive and negative regulators, respectively, of LDHA levels and glycolytic activity (48, 49). In addition to their regulation at the transcriptional level, several glycolytic enzymes are controlled by post-transcriptional mechanisms (50, 51). In PDA, one such mechanism involves removal of inhibitory acetylation on lysine 5 of LDHA by SIRT2, a deacetylase that senses increases in NAD+/NADH ratio (52). The full spectrum of mechanisms regulating glucose metabolism are no doubt complex and likely involve multiple additional levels of KRAS-dependent and –independent control that remain to be deciphered. Moreover, as metabolic pathways may operate differently in vitro and in vivo, the precise utilization of glucose in PDA will require further study.
Glutamine metabolism and redox homeostasis
In addition to glucose, highly proliferative cancer cells rely on glutamine – the most abundant and versatile amino acid (AA) in the cell cytoplasm - as a fuel source for ATP generation and for macromolecular biosynthesis. Glutamine is a non-essential AA that functions as a precursor and amine donor for generation of other AA as well as nucleotides and hexosamine, and as a donor of carbon skeletons for replenishment of TCA cycle intermediates (anaplerosis). While many tissues can synthesize glutamine, cancer cells show addiction to glutamine in culture (53, 54) and thus this AA becomes conditionally essential for growth. The first step in glutamine catabolism involves its conversion to glutamate catalyzed via the glutaminase enzymes (GLS1, GLS2). Glutamate, in turn, is a source of a-ketoglutarate (a-KG) — a TCA cycle intermediate as well as a co-enzyme for DNA and protein modifying dioxygenases — generated via the function of glutamate dehydrogenase (GLUD1) in the mitochondria or by transamination in the cytosol or mitochondria. This latter reaction also produces non-essential AAs. Glutamate is also a precursor of glutathione, the major antioxidant in the cell (55).
As noted above, KRAS mutant PDA cells do not effectively generate NADPH from the PPP, and rather these cells produce NADPH through a non-canonical glutamine-glutamate metabolism pathway (Figure 2C). This pathway involves conversion of glutamate to a-KG and aspartate in the mitochondria catalyzed by aspartate transaminase (GOT2) (56). Aspartate is then trafficked to the cytosol where it is converted sequentially to oxaloacetate, malate and pyruvate, via a GOT1-Malate Dehydrogenase-Malic enzyme cascade that generates NADPH. This pathway is under the control of KRAS, which promotes the transcriptional upregulation of GOT1 and repression of GLUD1, and is necessary for redox balance and growth of PDA cells in vitro and in vivo. In addition, enhanced catalytic activity of GOT2 via lysine acetylation has been reported to be required for redox homeostasis in PDA cells (57). KRAS also mitigates the high levels of ROS generated in rapidly proliferating cells by activating the NRF2 transcription factor that induces an anti-oxidant gene expression program (58).
The diverse roles of glutamine in fueling tumor cell metabolism have spurred the development of inhibitors targeting enzymes along the glutamine metabolism pathway, including GLS inhibitors that are currently being evaluated clinically (see Table 1). However, it should be noted that recent studies have suggested that glutamine may not be a major contributor to anaplerosis in some cancer types in vivo and therefore the dependence of cultured cell lines on exogenous glutamine may not always be conserved in primary tumors (59, 60). Likewise, GLS (and thus glutamine-glutamate conversion) may be dispensable for the growth of some tumors. Nevertheless, this would not undermine the importance of the GOT2-GOT1-ME pathway, which can utilize glutamate regardless of its source. Thus, these downstream components may offer additional therapeutic targets irrespective of the potential utility of GLS inhibitors in PDA.
Table 1.
Clinical trials targeting metabolism in PDA
| Target | Agent | Trial Design | NCT number |
|---|---|---|---|
|
| |||
| Pyruvate dehydrogenase and α-ketoglutarate dehydrogenase | CPI-613 + gemcitibine | Phase I/II | NCT00907166 |
|
| |||
| Lysosome | HCQ + gemcitabine/nab-paclitaxel | Phase I/II | NCT01506973 |
| HCQ + gemcitabine | Phase I/II | NCT01128296 | |
| HCQ + proton beam (neoadjuvant) | Phase II | NCT01494155 | |
| HCQ + gemcitabine/nab-paclitaxel (neoadjuvant) | Phase II | NCT01978184 | |
|
| |||
| Vitamin D receptor | paricalcitol + gemcitabine/abraxane (neoadjuvant) | randomized, pharmacodynamic study | NCT02030860 |
|
| |||
| PPARg | pioglitazone | Phase II | NCT01838317 |
|
| |||
| Mitochondrial Complex I | metformin | Phase I | NCT01954732 |
| metformin + gemcitabine or nab-paclitaxel | Phase I | NCT02336087 | |
| metformin + rapamycin | Phase I/II | NCT02048384 | |
| metformin + gemcitabine | Phase II | NCT02005419 | |
| metformin/gemcitabine | Phase II | NCT01210911 | |
|
| |||
| HMG-CoA reductase. | atorvastatin + metformin | observational | NCT02201381 |
|
| |||
| Glutaminase | CB-839 | Phase I | NCT02071862 |
NUTRIENT SCAVENGING IN PDA
An unusual diet – a pro-tumorigenic role for autophagy in PDA
PDAs employ an intriguing set of scavenging mechanisms that support growth and may mitigate the limited delivery of nutrients from the vasculature that characterizes these tumors (Figure 3A). These include autophagy (also known as macroautophagy), which is a highly conserved cellular catabolic process that mediates degradation of macromolecules as well as whole organelles. Autophagy involves sequestration of cytoplasmic contents within a double membrane vesicle (the autophagosome), which eventually fuses with lysosomes forming autolysosomes where cargo is degraded. Products of autolysosome digestion (amino acids, fatty acids, nucleosides) are recycled back to the cytoplasm to fuel biosynthetic and bioenergetic reactions and ultimately protect the cell during conditions of cellular stress such as nutrient starvation (61). In addition, autophagy functions to remove misfolded proteins, damaged organelles and protein aggregates and therefore provides the cell with an important quality control mechanism. Deregulation of these essential protective functions of basal autophagy has been implicated in the pathogenesis of degenerative and immune disorders as well as in aging (62, 63). Of the numerous stimuli that can activate autophagy above baseline levels, the best characterized and most potent is nutrient starvation, which activates AMPK and turns off mTORC1 (Figure 3A). These kinases phosphorylate key proteins controlling autophagy initiation, namely ULK1/2 and ATG13, to induce (AMPK) or suppress (mTORC1) autophagosome formation. Autophagy can also be activated in response to glucose deprivation in an ULK1-independent manner by increased ammonia levels generated via compensatory amino acid catabolism (64). Thus, decreases in extracellular and intracellular nutrient levels promote autophagy, providing an adaptive response geared toward restoring cellular homeostasis. Extensive studies of the functions of autophagy in cancer reveal context- and stage-specific roles. While its quality control activity serves as a barrier to tumorigenesis through suppression of genomic instability, oxidative stress, and chronic tissue damage, established cancers exploit the macromolecular recycling and detoxifying functions of autophagy to gain a growth advantage and protect the tumor cell (65–74).
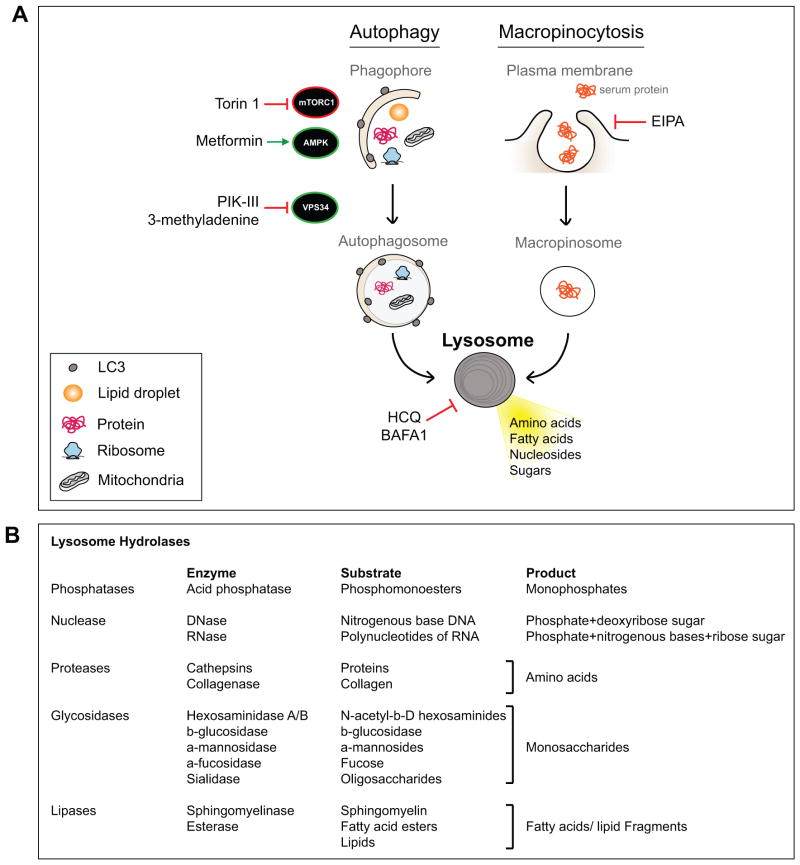
Figure 3. Nutrient scavenging in PDA converges at the lysosome for breakdown of intracellular and extracellular cargo.
A) PDA cells show enhanced autophagy activation and macropinocytosis in vitro and in vivo. Autophagy involves formation of double membrane vesicles that surround a portion of cytoplasm thus encapsulating cargo material (protein, lipid, organelles) that is delivered to lytic organelles (lysosome) for breakdown. Positive (AMPK and VPS34) and negative (mTORC1) kinase regulators of autophagy are indicated. Macropinocytosis, the bulk uptake of extracellular material, occurs via plasma membrane invagination and generation of internalized macropinosomes. These cargo-laden vesicles similarly fuse with lysosomes for efficient degradation of the internalized material. Therefore lysosomes are a key central delivery port for substrates destined for breakdown and serve to recycle the constituent building blocks and support cellular metabolism. Drugs that modulate different aspects of these pathways are shown. B) Resident lysosomal enzymes, their substrates and final products are listed.
Autophagy is constitutively active and required for PDA growth
Autophagy can be gauged by the cleavage and lipidation of the LC3 protein followed by its integration into the autophagosomal membrane (Table 2). By these measures, the great majority of PDA cell lines exhibit high basal autophagy compared to control immortalized pancreatic ductal cells (61). The use of additional assays confirms a true increase in autophagic activity (flux) rather than a block in the pathway. Moreover, autophagy is active in PDA cell lines even when grown in standard tissue culture conditions, suggesting this process is uncoupled from external nutrient availability. This is functionally important since treatment with the anti-malarial drug, chloroquine (CQ)—which inhibits autophagy by increasing lysosomal pH—or knockdown of essential autophagy genes (ATG5 or ATG7), strongly inhibits PDA cell proliferation under full nutrient conditions (75). Correspondingly, treatment with the CQ analogue, hydroxychloroquine (HCQ), suppresses tumorigenic growth in PDA patient-derived xenograft (PDX) and cell line-derived xenograft models, and in the KRASG12D-p53+/− GEM harboring established PDAs or advanced PanIN lesions. Likewise, knockdown of ATG5/ATG7 inhibits the growth of human PDA cell line xenografts.
Table 2.
Assays for monitoring autophagy
| Assay | Visualization | Readout | Interpretation |
|---|---|---|---|
| Autophagy | |||
| Electron microscopy | ultrastructure | éautophagosomes | induction or block in maturation |
| Western blot | LC3 | éLC3-II band/LC3-I band | induction or block in maturation |
| Fluorescence Microscopy | GFP-LC3 | éLC3-GFP puncta | induction or block in maturation |
| Autophagy flux | |||
| Western blot | LC3 ± lysosome inhibitor | éLC3-II band/LC3-I band in the +inhibitor treated sample | increased induction |
| Fluorescence Microscopy | GFP-LC3 ± lysosome inhibitor | éLC3-GFP spots in the +inhibitor treated sample | increased induction |
| Fluorescence Microscopy | mRFP-GFP-LC3 | Yellow fluorescence: autophagosome Red fluorescence: autolysosome |
éYellow/éRed: increased induction éYellow/êRed: block in maturation |
LC3 staining and the use of an autophagy reporter indicate that autophagy is induced as a late event in PDA progression, with elevated levels in the majority of invasive PDA tumors as compared to low grade PanIN (75, 76). Against this backdrop, mouse genetic studies have highlighted the complex, context-specific functions of autophagy in tumorigenesis. Mice with deletion of ATG7 in the pancreas show progressive tissue injury (77, 78), consistent with the important quality control function of basal autophagy in this organ. This inflammatory state promotes the initial formation of PanIN precursor lesions in mice with engineered KRASG12D mutations, although these lesions show significant impairment in full malignant progression to PDA (75, 77, 78). Delayed PDA formation and extended survival are also observed in the KRASG12D-p53+/− model upon deletion of the lysosomal gene, PLAC8, which partially compromises autophagy (76). By contrast, the simultaneous activation of KRAS and homozygous deletion of p53 (incurred during embryogenesis) negates the need for autophagy in PDA pathogenesis (77), although this genetic context is not thought to be representative of the genesis of most human PDAs. Taken as a whole these data strongly suggest that autophagy is required for the full malignant progression of PDA during the typical natural history of these tumors.
The contextual effects of autophagy inhibition bear directly on the potential of targeting this process therapeutically. First, it is important to note that unlike complete deletion of ATG7 (or ATG5) during pancreatic development, CQ treatment does not cause pancreatic injury or cooperation with KRAS in driving PanIN formation. Secondly, in human cell lines and PDX models, CQ inhibits tumor growth irrespective of p53 genotype (78). Therefore, these data provide support for the pharmacological targeting of autophagy as a PDA therapy. While the mechanisms by which autophagy inhibition impairs tumorigenesis are presently under investigation, a number of key observations have been made. Autophagy inhibition in vitro and in vivo is cytostatic rather than cytotoxic. In vitro, this effect is associated with increased ROS and DNA damage as well as a decrease in oxidative phosphorylation. In turn ROS scavengers or supplementation with pyruvate partially rescues growth, indicating that autophagy is required to maintain redox control and supply metabolic intermediates in PDA (65, 75). As discussed below, further examination of the interface of autophagy with cell metabolism will be an important step in the most effective deployment of autophagy inhibition to treat this cancer, potentially providing information regarding metabolic escape pathways and pointing toward combinatorial treatment strategies that may promote cell death rather than cytostasis.
PDAs depend on uptake of extracellular protein and lipid
In addition to intracellular scavenging via autophagy, cells can employ an additional scavenging pathway involving endocytosis-mediated bulk uptake of extracellular material, known as macropinocytosis (Figure 3A). Studies from the Bar-Sagi lab and colleagues recently showed that KRAS mutant cancer cells, including PDAs, upregulate macropinocytosis to import significant quantities of extracellular protein, which is ultimately delivered to lysosomes for proteolysis (79). Macropinocytosis of serum albumin was demonstrated to be a key source of amino acids to fuel multiple metabolic pathways in PDA cells and to support growth upon glutamine restriction. Moreover treatment of PDA cells with inhibitors of endocytosis that block albumin uptake impaired proliferation in vitro and tumor growth in vivo. Importantly, high levels of macropinocytic uptake are observed in PDA GEM models and in human PDA tumors (29). Thus, there is considerable interest in fully understanding the contributions of this pathway to PDA metabolism and in potentially targeting it therapeutically in KRAS mutant cancers.
Other components of the extracellular milieu may also serve as critical sources of nutrients, including lipids. While the overall abundance of lipid species in PDA is limited, with reduced amount of fatty acids, lipids and choline-containing compounds compared to normal pancreatic tissues (31, 80), the tumor cells appear to have efficient means for their retrieval. For example, KRASG12D transformation of immortalized pancreatic ductal epithelial cells (HPNE) induces increased scavenging of extracellular lipids (lysophospholipids) as an alternative source of fatty acids (81). While a role for macropinocytosis is not clear, active uptake of fatty acids contrasts to the common view that cancer cells synthesize the majority of their nonessential fatty acids de novo and suggests a shift in the origin of fatty acid pools in the cell occurs downstream of oncogenic KRAS. Additionally, PDA cells were reported to exhibit increased acquisition of cholesterol, in part through enhanced expression of the low-density lipoprotein receptor (LDLR) (82). Inhibition of LDLR led to alterations in cholesterol distribution in the cell and a decrease in PDA growth both in vitro and in vivo. The detailed assessment of lipid metabolism in PDA growth is an important topic for future investigation. Nevertheless, taken together, these observations show that PDA cells orchestrate multiple nutrient scavenging pathways as sources of additional nutrients. Further discussion of dietary lipids and obesity in PDA pathogenesis is presented in the section on systemic metabolism, below.
ROLE OF LYSOSOMAL CATABOLISM IN PDA
Lysosome activation – a novel hallmark of PDA
The major scavenging pathways in PDA, autophagy and macropinocytosis, converge at the lysosome where cargo is digested by over 40 resident lysosomal acid hydrolases (lipases, proteases, glycosidases, acid phosphatases and sulfatases), which are functional in the acidic environment of the lysosome (Figure 3B). While cancer associated changes in lysosomes have been proposed (83), a recent study showed that the number of lysosomes is markedly increased in PDA specimens from treatment-naïve patients compared to matched normal pancreatic tissue (84). This finding indicates that increased lysosome biogenesis and function may be integral to the nutrient scavenging program, ensuring efficient breakdown and recycling of cellular components and endocytosed material. Moreover, it suggests that there may be coordination between scavenging pathways and lysosome function in cancer.
The MiT/TFE family of basic helix-loop-helix transcription factors (MITF, TFE3, TFEB) have been identified as central regulators of the biogenesis and function of the autophagy-lysosome system in PDA (84). TFEB was first shown by the Ballabio lab to be a master transcriptional regulator of an autophagy-lysosome transcriptional program through direct binding to a consensus sequence present in the regulatory regions of essential autophagy and lysosome genes (85–87). In follow up studies, the MiT/TFE factors were shown to be components of an mTORC1-regulated acute stress response mechanism in normal cells (88–90). Under nutrient-rich conditions, mTOR is activated and localized to the lysosome where it phosphorylates and inactivates the MiT/TFE proteins. Conversely, upon starvation, mTOR is switched off enabling nuclear translocation of unphosphorylated MiT/TFE proteins. Together these studies highlight a lysosome-to-nucleus signaling pathway that monitors the cell’s nutritional status and adjusts catabolic activity accordingly.
In PDA cell lines and patient-derived PDA cultures, the MiT/TFE proteins bypass mTORC1-mediated surveillance and are constitutively localized in the nucleus regardless of external nutrient availability (84). This constitutive nuclear localization is mediated through binding to nuclear import proteins (including Importin 8), which are over-expressed in PDA. Inactivation of MiT/TFE proteins in PDA cells results in downregulation of autophagy and lysosome genes, defective lysosomal function, complete compromise in autophagic flux and degradation of macropinocytosis-derived protein, and consequently, significant impairment in cell proliferation in vitro and in vivo. Collectively, these data show that by governing both autophagic flux and lysosomal catabolism, the MiT/TFE proteins support an integrated cellular clearance program that enables efficient processing of cargo from autophagy as well as macropinocytosis. Thus, PDAs achieve an unusual state which appears to maximize growth processes associated with high mTOR activity while simultaneously benefiting from the metabolic fine-tuning and adaptation to stress afforded by activation of catabolic pathways. Interestingly, the MiT/TFE proteins are established oncogenes that are activated by genomic amplification or translocation in melanomas, renal cell carcinomas and in alveolar soft part sarcoma (91), although contributions of autophagy regulation in these settings have not been explored to date.
What are the products of lysosome degradation?
As noted above, autophagy activation and macropinocytosis represent hardwired programs essential for metabolic adaptation and growth of PDA cell lines and tumors. A precise understanding of which specific metabolite pools are recovered through autolysosome-mediated degradation and how PDA cells utilize these pools will be critical to deciphering the metabolic reprogramming that sustains these tumors. Accordingly, metabolomics studies in cells following knockdown of the MiT/TFE proteins or of ATG7, or treatment with lysosome inhibitors revealed a marked drop in intracellular AA levels, even in full nutrient conditions (84). These differences did not reflect broad changes in the rate of AA import or export and were not seen in non-transformed pancreatic cells. These findings indicate that autolysosome activation has PDA-specific functions in maintaining intracellular AA stores. Similar decreases in intracellular AA levels have also been observed following proteosome inhibition in yeast, Drosophila and various mammalian cell lines despite exposure to full external nutrient conditions (92). Thus, catabolic processes supply a significant fraction of internal AA that is independent of import from the external environment in PDA. These observations raise the intriguing possibility that distinct metabolite pools may fuel different biological processes. If so, how might this segregation occur and what factors dictate this partitioning? Detailed metabolite tracing experiments will provide insight into how these AA pools might be incorporated into different cellular reactions.
Beyond compensating for a paucity of nutrients supplied from the vasculature, the enhanced scavenging capacity of PDA cells may also serve an important quality control mechanism. While initially thought to function as a non-selective method for degradation of cytoplasmic content, recent studies have shown that autophagosomes can sequester and degrade specific cargo (93, 94). This selective breakdown of protein may also be essential for functional maintenance or remodelling of the PDA cellular proteome, a process known as proteostasis. Cancer cells are often characterized by increased rates of protein synthesis, due to activation of oncogenic signaling pathways or extrinsic factors such as hypoxia or nutrient deprivation, which places a heavy burden on the endoplasmic reticulum (ER) for enhanced protein folding capacity. Adaptive responses to ER stress such as autophagy ensure efficient clearance of misfolded protein species that can impair cell function (95). Similarly, removal of damaged organelles, particularly mitochondria, is an important function of autophagy in cancer and has been shown to influence the malignant progression of lung tumors (65–67, 71).
Autophagy has also been linked to resistance to radiotherapy and cytotoxic chemotherapy in several cancer types including PDA (96–98). Both the quality control mechanisms of autophagy and the upregulation of internally generated nutrient sources may cooperate to enhance overall cellular fitness and increase metabolic resilience, thereby sustaining tumor cell survival under these conditions (99, 100). Thus, in addition to treatment of autophagy-addicted tumors, combination strategies incorporating autophagy inhibition may prevent or delay therapy resistance or increase the effectiveness of anticancer drugs in multiple tumor settings.
RELATIONSHIP BETWEEN PDA AND SYSTEMIC METABOLISM
Systemic conditions such as obesity and diabetes have been linked to the onset and progression of PDA, suggesting that alterations in whole body metabolism contribute to the pathogenesis of this cancer. Recent experimental studies support and extend this notion, revealing complex reciprocal interactions between somatic physiologic processes and the tumor cells that at least partially involve modulation of metabolism.
Obesity and diabetes in PDA pathogenesis
Obesity is an established risk factor for PDA in both men and women, increasing risk by an estimated 20%-50%, as observed across multiple large pooled studies and meta-analyses (101). Moreover, the magnitude of risk increases in proportion to body mass index (BMI) in obese individuals. Consistent with an impact on disease initiation, obesity is also associated with increased incidence of PanIN lesions in otherwise normal pancreatic tissue (102). In addition, obesity appears to influence disease progression as well as the behavior of advanced tumors since patients with an elevated BMI pre-diagnosis are more likely to present with advanced stage metastatic PDA at diagnosis compared to healthy weight patients, and these patients show decreased overall survival times (103, 104).
In agreement with these epidemiologic data, administration of a high fat/high calorie diet (HFHCD) in multiple KRAS mutant mouse models accelerates the development of early PanIN lesions and increases their progression to PDA (105, 106). Conversely, calorie-restricted diets have been shown to delay PanIN progression in KRAS mutant mice (107). HFHCD was associated with activation of fibrosis and inflammatory pathways (e.g. COX2, TNFα) and increased immune infiltrate in the pre-malignant pancreatic lesions, although these studies do not establish whether this effect is a cause rather than a consequence of accelerated tumorigenesis. While these models showed some differences regarding the impact of HFHCD on insulin sensitivity and weight gain, they broadly support a connection between increased dietary intake and PDA risk. Based on the biologic alterations observed in mouse models and the aggressive features of obesity-associated PDA in humans, it will be of interest to determine whether PDA arising in this setting has distinct genomic features and differences in metabolic circuitry.
In addition to serving as substrates for anabolic metabolism and energy generation, lipids can act as important signaling molecules (e.g. prostaglandins and leukotrienes) (108). Accordingly, additional studies have explored the role of specific lipid species in PDA pathogenesis using mouse models. Bioactive lipids containing omega-3 polyunsaturated fatty acids (n-3 PUFAs), which have anti-inflammatory properties, were shown to strongly suppress PanIN progression and PDA development in the Ptf1Cre/+-LSL-KrasG12D mouse model. (109, 110). This series of findings on dietary intake and dietary supplementation appears to have implications for PDA prevention, and may be particularly relevant for the management of individuals at high risk for PDA, such as those with hereditary PDA syndromes.
The potential role of diabetes in PDA pathogenesis has long been under debate, however, recent work has provided considerable clarity in this regard, suggesting a ‘bi-directional’ relationship between the two conditions (Figure 4). Firstly, long standing type II diabetes (<2–8 years) correlates with an approximately 1.5–2-fold increased risk of PDA development (111, 112). The specific clinical features of diabetes that contribute to PDA risk have not fully been established. Interestingly, a large prospective case-control study of individuals without diabetes history showed an association between PDA and circulating markers of insulin resistance (e.g. increased pro-insulin levels), but not with islet cell dysfunction or hyperglycemia (113) (Figure 4A). This is in line with the reported increased PDA risk in diabetics treated with insulin or insulin secretagogues and decreased risk in those treated with the insulin sensitizer, metformin (114), although a systematic meta-analysis concluded that additional prospective studies are still needed to support these associations (114). It is not known whether PDA arising in the setting of existing diabetes has distinct genomic profiles. Nevertheless, it is notable that patients with type II diabetes exhibit decreased overall survival compared to non-diabetic PDA patients (115), potentially suggesting differences in tumor biology.
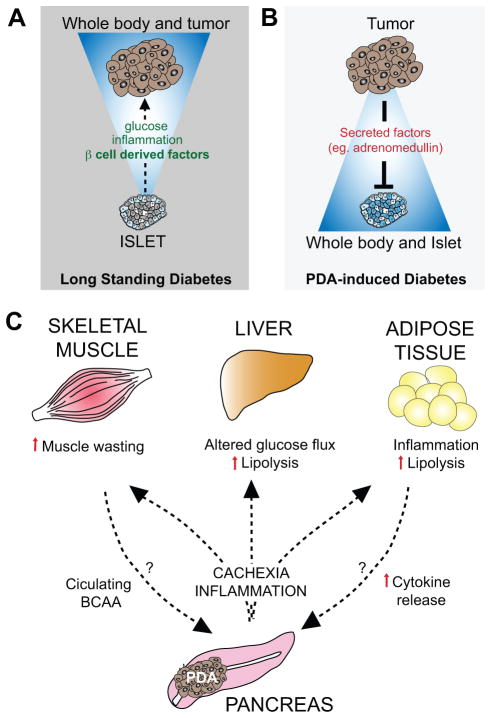
Figure 4. PDA is linked to alterations in whole body metabolism.
A) Conditions associated with altered systemic metabolism—namely long standing diabetes and obesity—are associated with increased PDA risk. In the case of diabetes, the increased secretion of islet-derived factors such as insulin may make particular contributions to PDA development. B) PDAs can reciprocally induce diabetes as a paraneoplastic syndrome (referred to as PDA-induced diabetes or recent-onset diabetes) by secretion of tumor-associated factors (e.g. adrenomedullin) that cause beta cell dysfunction. C) Advanced PDA is associated with cachexia, a condition involving weight loss and altered function of several metabolic tissues (skeletal muscle, liver and adipose tissue). Cachexia is thought to be induced by inflammatory mediators and cytokines produced by the PDA cells themselves as well as components of the PDA microenvironment. In addition, increased pools of circulating branched chain amino acids (BCAA) are an early sign of PDA onset, and may also be liberated from the muscle prior to clinically evident cachexia. These BCAA and the breakdown products of muscle and adipose tissue in cachexia may in turn serve as fuel sources that feed tumor growth.
Importantly, in addition to being a risk factor, diabetes can also signal the onset of PDA (101). In particular, patients newly diagnosed with diabetes have an 8-fold increased risk of developing PDA within the next 36 months over the general population. It is estimated that ~34% of PDA patients have new-onset diabetes (also referred to as pancreatic cancer-induced diabetes) at the time of cancer diagnosis, and that this group represents up to 75% of PDA patients with diabetes. A large retrospective study monitoring blood glucose levels found evidence of diabetes caused by PDA starting 2–3 years prior to diagnosis of the cancer (116). Correspondingly, rather than reflecting destruction of islets and pancreatic parenchyma, this condition appears to be a paraneoplastic syndrome arising due to secretion of factors from the tumor cells, such as adrenomodulin, which inhibits insulin secretion by b-cells (117, 118) (Figure 4B). Consistent with this, new-onset diabetes resolves in some cases following tumor resection, while long standing diabetes patients have persistent disease following surgery (119). Since new-onset diabetes signals subclinical malignancy, it may offer approaches for early cancer diagnosis. However, given that type II diabetes is 100 times more common than pancreatic-cancer-induced diabetes, the potential of utilizing the latter condition for pancreatic cancer screening will require additional biomarkers distinguishing these conditions.
Based on the associations between PDA and whole body metabolism, there is considerable interest in understanding the impact of the widely used anti-diabetic drugs on PDA risk as well as the therapeutic effects of these drugs in established tumors. In particular, there are extensive studies conducted using the biguanide, metformin in this setting. Metformin is a mitochondrial electron transport chain complex I inhibitor, that functions in part via inhibiting ATP synthesis and thereby activating AMPK signaling as well as inhibiting the PKA pathway (120). In addition to decreasing gluconeogenesis in the liver and decreasing blood glucose levels, metformin inhibits anabolic metabolism and increases energy generation in peripheral tissues and cancer cells (121, 122). The basis of the widely observed anti-proliferative effects of metformin observed in cancer cells in vitro and the anti-tumor effects seen in vivo have been attributed variously to AMPK activation, loss of TCA cycle intermediates, and more systemic effects (122, 123).
As noted above, retrospective studies have given inconsistent results regarding association between use of metformin and PDA risk in diabetics. On the other hand, studies conducted in a mutant Kras GEM model of PDA show that preventative treatment with metformin led to a decrease in progression of PanIN to invasive PDA (124). Additionally metformin was found to significantly reduce the growth of human and murine PDA xenografts (125, 126), perhaps relating to reduction of glucose levels, anti-inflammatory effects or direct effects on tumor cell metabolism via mitochondrial complex I inhibition. Evaluation of metformin as an anti-cancer therapeutic is currently ongoing in a number of clinical trials testing combinations of chemotherapy with this drug in metastatic PDA patients (Table 1).
Cachexia in PDA
Given the pronounced alterations in cell metabolism associated with PDA pathogenesis, there is considerable interest in the identification of metabolic biomarkers for early detection. Notably, by conducting a prospective study profiling metabolite changes in pre-diagnostic serum from 4 large cohorts of PDA patients versus matched controls, Wolpin and colleagues found that elevated levels of circulating branched chain amino acids (BCAA) are an independent predictive marker of increased risk (2-fold) of developing future PDA (127). This increase was present early in disease progression (2 to 5 years prior to tumor diagnosis) and preceded clinically evident cachexia, the process of skeletal muscle wasting and loss of body fat. Nevertheless, it is likely the BCAA source is from early stages of tissue breakdown, pointing to a tumor-associated secreted factor that influences whole body metabolic homeostasis during PDA progression (Figure 4C). Whether these BCAA directly nourish the evolving tumor and promote its growth is currently unknown, although these observations suggest an additional mechanism by which PDAs may enhance their acquisition of nutrients. Such utilization of BCAA for tumor growth is suggested by the increases in cell proliferation and tumor volume following administration of the BCAA, leucine, in a subcutaneous tumor model of PDA (128). Along these lines, it is also worth noting that cachexia itself is associated with more aggressive PDA tumors and poor prognosis, perhaps reflecting access of the tumor cells to substrates derived from lipolysis, protein breakdown, and systemic changes in glucose metabolism (127).
The signals inducing cachexia remain under investigation and both components of the tumor stroma as well as the neoplastic cells are thought to contribute to the process. Infiltration of lymphocytes and tumor-associated fibroblasts are detected during early stages of the disease and the cytokines and inflammatory mediators secreted by these cells, including Tumor necrosis factor a and Interleukin 6, have been implicated in promoting cachexia (129). There is also evidence that adrenodullin produced by PDA cells may have lipolytic activity on adipose tissue (130).
Vitamin D
Metabolite levels can also have protective functions in relation to PDA development. Notably, high circulating levels of vitamin D have been associated with reduced risk of PDA in a large prospective study (131). The basis for this effect is not clear. However, it is notable that the activation state of pancreatic stellate cells (PSCs; i.e. PDA-associated fibroblasts) has recently been shown to be under the control of vitamin D receptor (VDR) signaling (26). Activated PSCs promote inflammation and may support PDA growth, whereas a VDR agonist was shown to revert activation of PSCs to a quiescent state. Thus, vitamin D may act in part to reduce tumor-promoting fibrosis and inflammation. The vitamin D analog, paricalcitol, is presently being tested in clinical trials in PDA on the basis of its anti-fibrotic effects and potential to improve delivery of cytotoxic agents administered concurrently (Table 1).
Taken as a whole, it is increasingly apparent that alterations in whole-body metabolism can significantly influence disease pathogenesis and patient outcomes in PDA. With this information comes the promise that the development of suitable assays to detect predictive biomarkers for cancer development, to devise chemopreventative strategies for high-risk individuals, and to assess metabolic features of advanced cancers may allow earlier intervention as well as guide treatment strategies.
THERAPEUTIC TARGETING OF PDA METABOLISM
The superior adaptive capacity and ability to rewire their metabolism allows for sustained growth of PDA cells, but also imparts vulnerabilities that may be targeted therapeutically. Based on this concept, clinical trials aimed at disturbing cancer metabolism are currently ongoing (Table 1). Treatment with HCQ, aims to impair lysosome function and thus block output from autophagy and macropinocytosis, and is currently being tested in combination with a number of chemotherapy regimes. While this drug is well tolerated in patients, the need for micromolar levels for activity and lack of accurate measures of pharmacology have complicated interpretations of the disappointing early clinical results (132). The future development of alternative, more potent, autophagy/lysosome inhibitors [e.g. targeting upstream kinases of the autophagy cascade (ULK1, Vps34)] (133), coupled with intermittent dosing regimens has the potential to have real efficacy in PDA. Moreover, a better understanding of the roles of autophagy in PDA metabolism and a more complete elucidation of metabolic adaptations to autophagy inhibition (such as the roles of increased glycolysis (61)) may help to define combination approaches to change the effects of this treatment from cytostasis to cytotoxicity. Inhibition of parallel catabolic pathways such as proteasome-mediated degradation is one such approach that may have synergistic effects with autophagy inhibition.
As noted above, KRAS is a critical driver of proliferation and a master regulator of metabolic rewiring in PDA. In the context of targeting the metabolic pathways associated with KRAS activation, it is as yet unclear how best to target these enzymes as a cancer therapy, as many have essential functions in non-cancer cells. For example, targeting GLUT1 or other glycolytic enzymes may be associated with severe toxicities due to their near ubiquitous requirement in most normal tissues. By contrast, targeting of LDHA may be a well tolerated therapy strategy, since human syndromes associated with decreased LDHA activity do not present severe abnormalities in organ function in adults (35). In addition, renewed efforts to generate inhibitors of KRAS are currently under-way as part of the National RAS Initiative (134). Anticipating the development of such agents and the potential that resistance mechanisms to KRAS inhibition may eventually arise, the Draetta and DePinho labs developed a GEM model of resistance to genetic inactivation of KRAS in PDA. They found that a series of adaptive metabolic alterations, including elevation in oxidative phosphorylation and potentiation of autophagy were required to mediate survival following KRAS extinction, thus suggesting combinatorial strategies for future KRAS targeted therapy (135). Given the recent emergence of cancer immunotherapy, an added consideration for the deployment of drugs that block tumor cell metabolism is their potential effects on tumor immunity, and on the efficacy of T cell checkpoint inhibition and other approaches of immune activation. As activated immune and stromal cells exhibit a number of metabolic changes that are common with tumor cells (136–140) it will be important to determine whether targeting these metabolic pathways interferes with (or enhances) immune function, thereby informing potential combination therapies.
CONCLUSION
Metabolic rewiring is central to the pathogenesis of PDA and is a critical component of the tumorigenic program driven by KRAS, the signature mutation in this malignancy. A key current challenge is to more fully define how nutrient substrates are generated and utilized in these tumors and to understand how the multiple different cooperating genomic alterations found in PDA influence these processes. Many important areas, such as lipid metabolism, mitochondrial function, and the role of nutrient sensing transcription factors remain to be explored. With the development of more precise techniques for dynamic measurement of metabolic reactions both in vitro and in vivo, coupled with use of faithful cancer models, significant progress in our understanding of the functions of these pathways in disease progression is on the horizon. In the future, information regarding the metabolic dependencies of PDA and the interplay between the tumor, systemic metabolism, and immune function holds promise for highlighting a path towards development of novel cancer diagnostics and therapeutics.
STATEMENT OF SIGNIFICANCE.
Alterations in cell metabolism are a key hallmark of pancreatic ductal adenocarcinoma (PDA) and contribute to tumor cell proliferation and survival. In addition, there is complex interplay between evolving PDA and systemic metabolism that is important for the pathogenesis of this disease. Furthering understanding of these processes can inform new approaches to the detection, prevention and treatment of this deadly cancer
KEY CONCEPT AND RELEVANCE.
Cancers have heightened metabolic requirements for cell growth that need to be coordinated with nutrient supply.
PDAs must contend with further metabolic constraints due to their hypovascular, fibrotic microenvironment and ensuing hypoxia and limited nutrient availability. To support tumor growth, PDAs acquire multiple alterations in metabolic circuitry and activation of nutrient scavenging processes — autophagy and macropinocytosis
PDA development is also influenced by conditions that change whole body metabolism (type 2 diabetes and obesity), and reciprocally PDA incites systemic metabolic alterations (cachexia and PDA-induced diabetes
How these processes are activated, integrated, and regulated has started to come into focus
The recent advances in understanding these metabolic alterations provides new insights into PDA pathogenesis and suggests paths forward for the development of improved therapeutics and diagnostics
Acknowledgments
Grant Support: This work was supported by grants from the NIH (P50CA1270003, P01 CA117969-07, R01 CA133557-05) and the Linda J. Verville Cancer Research Foundation to N.B and a Hirshberg Foundation for Pancreatic Cancer seed grant to R.M.P.
We sincerely apologize to our colleagues for being unable to cite all of the important work in this area due to space constraints. We thank Brian Wolpin for critical reading of the manuscript. This work was supported by grants from the NIH (P50CA1270003, P01 CA117969-07, R01 CA133557-05) and Linda J. Verville Cancer Research Foundation to N.B. N.B holds the Gallagher Endowed Chair in Gastrointestinal Cancer Research. R.M.P holds a Hirshberg Foundation for Pancreatic Cancer seed grant. N.B. and R.M.P. are members of the Andrew Warshaw Institute for Pancreatic Cancer Research.
Abbreviations
- PDA
Pancreatic Ductal Adenocarcinoma
- PanIN
Pancreatic Intraepithelial Neoplasia
- CQ
Chloroquine
- AMPK
Adenosine monophosphate-activated protein kinase
- mTORC1
Mechanistic Target of Rapamycin Complex 1
- GEM
genetically engineered mouse
- PDX
PDA patient-derived xenograft
- AA
Amino acid
- a-KG
a-ketoglutarate
- PDK
Pyruvate Dehydrogenase Kinase
- LDH
Lactate Dehydrogenase
- HBP
Hexosamine biosynthesis pathway
- PPP
Pentose phosphate pathway
- LDLR
low-density lipoprotein receptor
- BMI
body mass index
Footnotes
Conflict of Interest Statement: The authors have no conflicts of interest to declare
References
- 1.Ryan DP, Hong TS, Bardeesy N. Pancreatic adenocarcinoma. N Engl J Med. 2014;371:2140–1. doi: 10.1056/NEJMc1412266. [DOI] [PubMed] [Google Scholar]
- 2.Maitra A, Hruban RH. Pancreatic cancer. Annu Rev Pathol. 2008;3:157–88. doi: 10.1146/annurev.pathmechdis.3.121806.154305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Morris JPt, Wang SC, Hebrok M. KRAS, Hedgehog, Wnt and the twisted developmental biology of pancreatic ductal adenocarcinoma. Nat Rev Cancer. 2010;10:683–95. doi: 10.1038/nrc2899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Aguirre AJ, Bardeesy N, Sinha M, Lopez L, Tuveson DA, Horner J, et al. Activated Kras and Ink4a/Arf deficiency cooperate to produce metastatic pancreatic ductal adenocarcinoma. Genes Dev. 2003;17:3112–26. doi: 10.1101/gad.1158703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Collisson EA, Sadanandam A, Olson P, Gibb WJ, Truitt M, Gu S, et al. Subtypes of pancreatic ductal adenocarcinoma and their differing responses to therapy. Nat Med. 2011;17:500–3. doi: 10.1038/nm.2344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hingorani SR, Petricoin EF, Maitra A, Rajapakse V, King C, Jacobetz MA, et al. Preinvasive and invasive ductal pancreatic cancer and its early detection in the mouse. Cancer cell. 2003;4:437–50. doi: 10.1016/s1535-6108(03)00309-x. [DOI] [PubMed] [Google Scholar]
- 7.Singh A, Greninger P, Rhodes D, Koopman L, Violette S, Bardeesy N, et al. A gene expression signature associated with “K-Ras addiction” reveals regulators of EMT and tumor cell survival. Cancer cell. 2009;15:489–500. doi: 10.1016/j.ccr.2009.03.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ying H, Kimmelman AC, Lyssiotis CA, Hua S, Chu GC, Fletcher-Sananikone E, et al. Oncogenic Kras maintains pancreatic tumors through regulation of anabolic glucose metabolism. Cell. 2012;149:656–70. doi: 10.1016/j.cell.2012.01.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Collins MA, Bednar F, Zhang Y, Brisset JC, Galban S, Galban CJ, et al. Oncogenic Kras is required for both the initiation and maintenance of pancreatic cancer in mice. J Clin Invest. 2012;122:639–53. doi: 10.1172/JCI59227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yachida S, Iacobuzio-Donahue CA. Evolution and dynamics of pancreatic cancer progression. Oncogene. 2013;32:5253–60. doi: 10.1038/onc.2013.29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yachida S, Jones S, Bozic I, Antal T, Leary R, Fu B, et al. Distant metastasis occurs late during the genetic evolution of pancreatic cancer. Nature. 2010;467:1114–7. doi: 10.1038/nature09515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yachida S, White CM, Naito Y, Zhong Y, Brosnan JA, Macgregor-Das AM, et al. Clinical significance of the genetic landscape of pancreatic cancer and implications for identification of potential long-term survivors. Clin Cancer Res. 2012;18:6339–47. doi: 10.1158/1078-0432.CCR-12-1215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bardeesy N, Aguirre AJ, Chu GC, Cheng KH, Lopez LV, Hezel AF, et al. Both p16(Ink4a) and the p19(Arf)-p53 pathway constrain progression of pancreatic adenocarcinoma in the mouse. Proc Natl Acad Sci U S A. 2006;103:5947–52. doi: 10.1073/pnas.0601273103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bardeesy N, Cheng KH, Berger JH, Chu GC, Pahler J, Olson P, et al. Smad4 is dispensable for normal pancreas development yet critical in progression and tumor biology of pancreas cancer. Genes Dev. 2006;20:3130–46. doi: 10.1101/gad.1478706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hingorani SR, Wang L, Multani AS, Combs C, Deramaudt TB, Hruban RH, et al. Trp53R172H and KrasG12D cooperate to promote chromosomal instability and widely metastatic pancreatic ductal adenocarcinoma in mice. Cancer cell. 2005;7:469–83. doi: 10.1016/j.ccr.2005.04.023. [DOI] [PubMed] [Google Scholar]
- 16.Whittle MC, Izeradjene K, Rani PG, Feng L, Carlson MA, DelGiorno KE, et al. RUNX3 Controls a Metastatic Switch in Pancreatic Ductal Adenocarcinoma. Cell. 2015;161:1345–60. doi: 10.1016/j.cell.2015.04.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Biankin AV, Waddell N, Kassahn KS, Gingras MC, Muthuswamy LB, Johns AL, et al. Pancreatic cancer genomes reveal aberrations in axon guidance pathway genes. Nature. 2012;491:399–405. doi: 10.1038/nature11547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Waddell N, Pajic M, Patch AM, Chang DK, Kassahn KS, Bailey P, et al. Whole genomes redefine the mutational landscape of pancreatic cancer. Nature. 2015;518:495–501. doi: 10.1038/nature14169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kimmelman AC. Metabolic Dependencies in RAS-Driven Cancers. Clin Cancer Res. 2015;21:1828–34. doi: 10.1158/1078-0432.CCR-14-2425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.White E. Exploiting the bad eating habits of Ras-driven cancers. Genes Dev. 2013;27:2065–71. doi: 10.1101/gad.228122.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Neesse A, Michl P, Frese KK, Feig C, Cook N, Jacobetz MA, et al. Stromal biology and therapy in pancreatic cancer. Gut. 2010;60:861–8. doi: 10.1136/gut.2010.226092. [DOI] [PubMed] [Google Scholar]
- 22.Hingorani SR. Cellular and molecular conspirators in pancreas cancer. Carcinogenesis. 2014;35:1435. doi: 10.1093/carcin/bgu138. [DOI] [PubMed] [Google Scholar]
- 23.Lee JJ, Perera RM, Wang H, Wu DC, Liu XS, Han S, et al. Stromal response to Hedgehog signaling restrains pancreatic cancer progression. Proc Natl Acad Sci U S A. 2014;111:E3091–100. doi: 10.1073/pnas.1411679111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ozdemir BC, Pentcheva-Hoang T, Carstens JL, Zheng X, Wu CC, Simpson TR, et al. Depletion of carcinoma-associated fibroblasts and fibrosis induces immunosuppression and accelerates pancreas cancer with reduced survival. Cancer cell. 2014;25:719–34. doi: 10.1016/j.ccr.2014.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rhim AD, Oberstein PE, Thomas DH, Mirek ET, Palermo CF, Sastra SA, et al. Stromal elements act to restrain, rather than support, pancreatic ductal adenocarcinoma. Cancer cell. 2014;25:735–47. doi: 10.1016/j.ccr.2014.04.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sherman MH, Yu RT, Engle DD, Ding N, Atkins AR, Tiriac H, et al. Vitamin D receptor-mediated stromal reprogramming suppresses pancreatitis and enhances pancreatic cancer therapy. Cell. 2014;159:80–93. doi: 10.1016/j.cell.2014.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Vonderheide RH, Bayne LJ. Inflammatory networks and immune surveillance of pancreatic carcinoma. Curr Opin Immunol. 2013;25:200–5. doi: 10.1016/j.coi.2013.01.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kraman M, Bambrough PJ, Arnold JN, Roberts EW, Magiera L, Jones JO, et al. Suppression of antitumor immunity by stromal cells expressing fibroblast activation protein-alpha. Science. 2010;330:827–30. doi: 10.1126/science.1195300. [DOI] [PubMed] [Google Scholar]
- 29.Kamphorst JJ, Nofal M, Commisso C, Hackett SR, Lu W, Grabocka E, et al. Human pancreatic cancer tumors are nutrient poor and tumor cells actively scavenge extracellular protein. Cancer Res. 2015;75:544–53. doi: 10.1158/0008-5472.CAN-14-2211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Stylianopoulos T, Martin JD, Chauhan VP, Jain SR, Diop-Frimpong B, Bardeesy N, et al. Causes, consequences, and remedies for growth-induced solid stress in murine and human tumors. Proc Natl Acad Sci U S A. 2012;109:15101–8. doi: 10.1073/pnas.1213353109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ma X, Zhao X, Ouyang H, Sun F, Zhang H, Zhou C, et al. The metabolic features of normal pancreas and pancreatic adenocarcinoma: preliminary result of in vivo proton magnetic resonance spectroscopy at 3. 0 T. J Comput Assist Tomogr. 2011;35:539–43. doi: 10.1097/RCT.0b013e318227a545. [DOI] [PubMed] [Google Scholar]
- 32.Efeyan A, Comb WC, Sabatini DM. Nutrient-sensing mechanisms and pathways. Nature. 2015;517:302–10. doi: 10.1038/nature14190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hardie DG. AMP-activated protein kinase: an energy sensor that regulates all aspects of cell function. Genes Dev. 2011;25:1895–908. doi: 10.1101/gad.17420111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zoncu R, Efeyan A, Sabatini DM. mTOR: from growth signal integration to cancer, diabetes and ageing. Nature reviews Molecular cell biology. 2011;12:21–35. doi: 10.1038/nrm3025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.DeBerardinis RJ, Thompson CB. Cellular metabolism and disease: what do metabolic outliers teach us? Cell. 2012;148:1132–44. doi: 10.1016/j.cell.2012.02.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Guo JY, Xia B, White E. Autophagy-mediated tumor promotion. Cell. 2013;155:1216–9. doi: 10.1016/j.cell.2013.11.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sousa CM, Kimmelman AC. The complex landscape of pancreatic cancer metabolism. Carcinogenesis. 2014;35:1441–50. doi: 10.1093/carcin/bgu097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.DeBerardinis RJ, Lum JJ, Hatzivassiliou G, Thompson CB. The biology of cancer: metabolic reprogramming fuels cell growth and proliferation. Cell Metab. 2008;7:11–20. doi: 10.1016/j.cmet.2007.10.002. [DOI] [PubMed] [Google Scholar]
- 39.Koppenol WH, Bounds PL, Dang CV. Otto Warburg’s contributions to current concepts of cancer metabolism. Nat Rev Cancer. 2011;11:325–37. doi: 10.1038/nrc3038. [DOI] [PubMed] [Google Scholar]
- 40.Weinberg F, Hamanaka R, Wheaton WW, Weinberg S, Joseph J, Lopez M, et al. Mitochondrial metabolism and ROS generation are essential for Kras-mediated tumorigenicity. Proc Natl Acad Sci U S A. 2010;107:8788–93. doi: 10.1073/pnas.1003428107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kitasato Y, Yasunaga M, Okuda K, Kinoshita H, Tanaka H, Okabe Y, et al. Maximum standardized uptake value on 18F-fluoro-2-deoxy-glucose positron emission tomography/computed tomography and glucose transporter-1 expression correlates with survival in invasive ductal carcinoma of the pancreas. Pancreas. 2014;43:1060–5. doi: 10.1097/MPA.0000000000000185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Yamamoto T, Sugiura T, Mizuno T, Okamura Y, Aramaki T, Endo M, et al. Preoperative FDG-PET predicts early recurrence and a poor prognosis after resection of pancreatic adenocarcinoma. Ann Surg Oncol. 2014;22:677–84. doi: 10.1245/s10434-014-4046-2. [DOI] [PubMed] [Google Scholar]
- 43.Le A, Cooper CR, Gouw AM, Dinavahi R, Maitra A, Deck LM, et al. Inhibition of lactate dehydrogenase A induces oxidative stress and inhibits tumor progression. Proc Natl Acad Sci U S A. 2010;107:2037–42. doi: 10.1073/pnas.0914433107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Guillaumond F, Leca J, Olivares O, Lavaut MN, Vidal N, Berthezene P, et al. Strengthened glycolysis under hypoxia supports tumor symbiosis and hexosamine biosynthesis in pancreatic adenocarcinoma. Proc Natl Acad Sci U S A. 2013;110:3919–24. doi: 10.1073/pnas.1219555110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Parks SK, Chiche J, Pouyssegur J. Disrupting proton dynamics and energy metabolism for cancer therapy. Nat Rev Cancer. 2013;13:611–23. doi: 10.1038/nrc3579. [DOI] [PubMed] [Google Scholar]
- 46.Baek G, Tse YF, Hu Z, Cox D, Buboltz N, McCue P, et al. MCT4 defines a glycolytic subtype of pancreatic cancer with poor prognosis and unique metabolic dependencies. Cell Rep. 2014;9:2233–49. doi: 10.1016/j.celrep.2014.11.025. [DOI] [PubMed] [Google Scholar]
- 47.Roland CL, Arumugam T, Deng D, Liu SH, Philip B, Gomez S, et al. Cell surface lactate receptor GPR81 is crucial for cancer cell survival. Cancer Res. 2014;74:5301–10. doi: 10.1158/0008-5472.CAN-14-0319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Cui J, Shi M, Xie D, Wei D, Jia Z, Zheng S, et al. FOXM1 promotes the warburg effect and pancreatic cancer progression via transactivation of LDHA expression. Clin Cancer Res. 2014;20:2595–606. doi: 10.1158/1078-0432.CCR-13-2407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Shi M, Cui J, Du J, Wei D, Jia Z, Zhang J, et al. A novel KLF4/LDHA signaling pathway regulates aerobic glycolysis in and progression of pancreatic cancer. Clin Cancer Res. 2014;20:4370–80. doi: 10.1158/1078-0432.CCR-14-0186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Guan KL, Xiong Y. Regulation of intermediary metabolism by protein acetylation. Trends Biochem Sci. 2010;36:108–16. doi: 10.1016/j.tibs.2010.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Hitosugi T, Chen J. Post-translational modifications and the Warburg effect. Oncogene. 2013;33:4279–85. doi: 10.1038/onc.2013.406. [DOI] [PubMed] [Google Scholar]
- 52.Zhao D, Zou SW, Liu Y, Zhou X, Mo Y, Wang P, et al. Lysine-5 acetylation negatively regulates lactate dehydrogenase A and is decreased in pancreatic cancer. Cancer cell. 2013;23:464–76. doi: 10.1016/j.ccr.2013.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Hensley CT, Wasti AT, DeBerardinis RJ. Glutamine and cancer: cell biology, physiology, and clinical opportunities. J Clin Invest. 2013;123:3678–84. doi: 10.1172/JCI69600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Wise DR, DeBerardinis RJ, Mancuso A, Sayed N, Zhang XY, Pfeiffer HK, et al. Myc regulates a transcriptional program that stimulates mitochondrial glutaminolysis and leads to glutamine addiction. Proc Natl Acad Sci U S A. 2008;105:18782–7. doi: 10.1073/pnas.0810199105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Meister A, Anderson ME. Glutathione. Annu Rev Biochem. 1983;52:711–60. doi: 10.1146/annurev.bi.52.070183.003431. [DOI] [PubMed] [Google Scholar]
- 56.Son J, Lyssiotis CA, Ying H, Wang X, Hua S, Ligorio M, et al. Glutamine supports pancreatic cancer growth through a KRAS-regulated metabolic pathway. Nature. 2013;496:101–5. doi: 10.1038/nature12040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Yang H, Zhou L, Shi Q, Zhao Y, Lin H, Zhang M, et al. SIRT3-dependent GOT2 acetylation status affects the malate-aspartate NADH shuttle activity and pancreatic tumor growth. EMBO J. 2015;34:1110–25. doi: 10.15252/embj.201591041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.DeNicola GM, Karreth FA, Humpton TJ, Gopinathan A, Wei C, Frese K, et al. Oncogene-induced Nrf2 transcription promotes ROS detoxification and tumorigenesis. Nature. 2011;475:106–9. doi: 10.1038/nature10189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Mashimo T, Pichumani K, Vemireddy V, Hatanpaa KJ, Singh DK, Sirasanagandla S, et al. Acetate is a bioenergetic substrate for human glioblastoma and brain metastases. Cell. 2014;159:1603–14. doi: 10.1016/j.cell.2014.11.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Marin-Valencia I, Yang C, Mashimo T, Cho S, Baek H, Yang XL, et al. Analysis of tumor metabolism reveals mitochondrial glucose oxidation in genetically diverse human glioblastomas in the mouse brain in vivo. Cell Metab. 2012;15:827–37. doi: 10.1016/j.cmet.2012.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Rabinowitz JD, White E. Autophagy and metabolism. Science. 2010;330:1344–8. doi: 10.1126/science.1193497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Kroemer G, Marino G, Levine B. Autophagy and the integrated stress response. Molecular cell. 2010;40:280–93. doi: 10.1016/j.molcel.2010.09.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Levine B, Kroemer G. Autophagy in the pathogenesis of disease. Cell. 2008;132:27–42. doi: 10.1016/j.cell.2007.12.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Cheong H, Lindsten T, Wu J, Lu C, Thompson CB. Ammonia-induced autophagy is independent of ULK1/ULK2 kinases. Proc Natl Acad Sci U S A. 2011;108:11121–6. doi: 10.1073/pnas.1107969108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Guo JY, Chen HY, Mathew R, Fan J, Strohecker AM, Karsli-Uzunbas G, et al. Activated Ras requires autophagy to maintain oxidative metabolism and tumorigenesis. Genes Dev. 2011;25:460–70. doi: 10.1101/gad.2016311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Guo JY, Karsli-Uzunbas G, Mathew R, Aisner SC, Kamphorst JJ, Strohecker AM, et al. Autophagy suppresses progression of K-ras-induced lung tumors to oncocytomas and maintains lipid homeostasis. Genes Dev. 2013;27:1447–61. doi: 10.1101/gad.219642.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Karsli-Uzunbas G, Guo JY, Price S, Teng X, Laddha SV, Khor S, et al. Autophagy is required for glucose homeostasis and lung tumor maintenance. Cancer Discov. 2014;4:914–27. doi: 10.1158/2159-8290.CD-14-0363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Lock R, Kenific CM, Leidal AM, Salas E, Debnath J. Autophagy-dependent production of secreted factors facilitates oncogenic RAS-driven invasion. Cancer Discov. 2014;4:466–79. doi: 10.1158/2159-8290.CD-13-0841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Lock R, Roy S, Kenific CM, Su JS, Salas E, Ronen SM, et al. Autophagy facilitates glycolysis during Ras-mediated oncogenic transformation. Mol Biol Cell. 2011;22:165–78. doi: 10.1091/mbc.E10-06-0500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Morgan MJ, Gamez G, Menke C, Hernandez A, Thorburn J, Gidan F, et al. Regulation of autophagy and chloroquine sensitivity by oncogenic RAS in vitro is context-dependent. Autophagy. 2014;10:1814–26. doi: 10.4161/auto.32135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Rao S, Tortola L, Perlot T, Wirnsberger G, Novatchkova M, Nitsch R, et al. A dual role for autophagy in a murine model of lung cancer. Nat Commun. 2014;5:3056. doi: 10.1038/ncomms4056. [DOI] [PubMed] [Google Scholar]
- 72.Strohecker AM, Guo JY, Karsli-Uzunbas G, Price SM, Chen GJ, Mathew R, et al. Autophagy sustains mitochondrial glutamine metabolism and growth of BrafV600E-driven lung tumors. Cancer Discov. 2013;3:1272–85. doi: 10.1158/2159-8290.CD-13-0397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Wei H, Wei S, Gan B, Peng X, Zou W, Guan JL. Suppression of autophagy by FIP200 deletion inhibits mammary tumorigenesis. Genes Dev. 2011;25:1510–27. doi: 10.1101/gad.2051011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Xie X, Koh JY, Price S, White E, Mehnert JM. Atg7 Overcomes Senescence and Promotes Growth of BrafV600E-Driven Melanoma. Cancer Discov. 2015;5:410–23. doi: 10.1158/2159-8290.CD-14-1473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Yang S, Wang X, Contino G, Liesa M, Sahin E, Ying H, et al. Pancreatic cancers require autophagy for tumor growth. Genes Dev. 2011;25:717–29. doi: 10.1101/gad.2016111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Kinsey C, Balakrishnan V, O’Dell MR, Huang JL, Newman L, Whitney-Miller CL, et al. Plac8 links oncogenic mutations to regulation of autophagy and is critical to pancreatic cancer progression. Cell Rep. 2014;7:1143–55. doi: 10.1016/j.celrep.2014.03.061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Rosenfeldt MT, O’Prey J, Morton JP, Nixon C, MacKay G, Mrowinska A, et al. p53 status determines the role of autophagy in pancreatic tumour development. Nature. 2013;504:296–300. doi: 10.1038/nature12865. [DOI] [PubMed] [Google Scholar]
- 78.Yang A, Rajeshkumar NV, Wang X, Yabuuchi S, Alexander BM, Chu GC, et al. Autophagy is critical for pancreatic tumor growth and progression in tumors with p53 alterations. Cancer Discov. 2014;4:905–13. doi: 10.1158/2159-8290.CD-14-0362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Commisso C, Davidson SM, Soydaner-Azeloglu RG, Parker SJ, Kamphorst JJ, Hackett S, et al. Macropinocytosis of protein is an amino acid supply route in Ras-transformed cells. Nature. 2013;497:633–7. doi: 10.1038/nature12138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Yao X, Zeng M, Wang H, Fei S, Rao S, Ji Y. Metabolite detection of pancreatic carcinoma by in vivo proton MR spectroscopy at 3T: initial results. Radiol Med. 2012;117:780–8. doi: 10.1007/s11547-011-0757-7. [DOI] [PubMed] [Google Scholar]
- 81.Kamphorst JJ, Cross JR, Fan J, de Stanchina E, Mathew R, White EP, et al. Hypoxic and Ras-transformed cells support growth by scavenging unsaturated fatty acids from lysophospholipids. Proc Natl Acad Sci U S A. 2013;110:8882–7. doi: 10.1073/pnas.1307237110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Guillaumond F, Bidaut G, Ouaissi M, Servais S, Gouirand V, Olivares O, et al. Cholesterol uptake disruption, in association with chemotherapy, is a promising combined metabolic therapy for pancreatic adenocarcinoma. Proc Natl Acad Sci U S A. 2015;112:2473–8. doi: 10.1073/pnas.1421601112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Kallunki T, Olsen OD, Jaattela M. Cancer-associated lysosomal changes: friends or foes? Oncogene. 2012;32:1995–2004. doi: 10.1038/onc.2012.292. [DOI] [PubMed] [Google Scholar]
- 84.Perera RM, Stoykova S, Nicolay BN, Ross KN, Fitamant J, Boukhali M, et al. Transcriptional control of autophagy-lysosome function drives pancreatic cancer metabolism. Nature. 2015 doi: 10.1038/nature14587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Sardiello M, Palmieri M, di Ronza A, Medina DL, Valenza M, Gennarino VA, et al. A gene network regulating lysosomal biogenesis and function. Science. 2009;325:473–7. doi: 10.1126/science.1174447. [DOI] [PubMed] [Google Scholar]
- 86.Settembre C, Di Malta C, Polito VA, Garcia Arencibia M, Vetrini F, Erdin S, et al. TFEB links autophagy to lysosomal biogenesis. Science. 2011;332:1429–33. doi: 10.1126/science.1204592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Settembre C, Fraldi A, Medina DL, Ballabio A. Signals from the lysosome: a control centre for cellular clearance and energy metabolism. Nature reviews Molecular cell biology. 2013;14:283–96. doi: 10.1038/nrm3565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Martina JA, Diab HI, Lishu L, Jeong AL, Patange S, Raben N, et al. The nutrient-responsive transcription factor TFE3 promotes autophagy, lysosomal biogenesis, and clearance of cellular debris. Science signaling. 2014;7:ra9. doi: 10.1126/scisignal.2004754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Roczniak-Ferguson A, Petit CS, Froehlich F, Qian S, Ky J, Angarola B, et al. The transcription factor TFEB links mTORC1 signaling to transcriptional control of lysosome homeostasis. Science signaling. 2012;5:ra42. doi: 10.1126/scisignal.2002790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Settembre C, Zoncu R, Medina DL, Vetrini F, Erdin S, Huynh T, et al. A lysosome-to-nucleus signalling mechanism senses and regulates the lysosome via mTOR and TFEB. EMBO J. 2012;31:1095–108. doi: 10.1038/emboj.2012.32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Haq R, Fisher DE. Biology and clinical relevance of the micropthalmia family of transcription factors in human cancer. Journal of clinical oncology : official journal of the American Society of Clinical Oncology. 2011;29:3474–82. doi: 10.1200/JCO.2010.32.6223. [DOI] [PubMed] [Google Scholar]
- 92.Suraweera A, Munch C, Hanssum A, Bertolotti A. Failure of amino acid homeostasis causes cell death following proteasome inhibition. Molecular cell. 2012;48:242–53. doi: 10.1016/j.molcel.2012.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Mancias JD, Wang X, Gygi SP, Harper JW, Kimmelman AC. Quantitative proteomics identifies NCOA4 as the cargo receptor mediating ferritinophagy. Nature. 2014;509:105–9. doi: 10.1038/nature13148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Mathew R, Khor S, Hackett SR, Rabinowitz JD, Perlman DH, White E. Functional role of autophagy-mediated proteome remodeling in cell survival signaling and innate immunity. Molecular cell. 2014;55:916–30. doi: 10.1016/j.molcel.2014.07.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Tameire F, Verginadis II, Koumenis C. Cell intrinsic and extrinsic activators of the unfolded protein response in cancer: Mechanisms and targets for therapy. Semin Cancer Biol. 2015;3:3–15. doi: 10.1016/j.semcancer.2015.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Hashimoto D, Blauer M, Hirota M, Ikonen NH, Sand J, Laukkarinen J. Autophagy is needed for the growth of pancreatic adenocarcinoma and has a cytoprotective effect against anticancer drugs. Eur J Cancer. 2014;50:1382–90. doi: 10.1016/j.ejca.2014.01.011. [DOI] [PubMed] [Google Scholar]
- 97.Sui X, Chen R, Wang Z, Huang Z, Kong N, Zhang M, et al. Autophagy and chemotherapy resistance: a promising therapeutic target for cancer treatment. Cell Death Dis. 2013;4:e838. doi: 10.1038/cddis.2013.350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Wang P, Zhang J, Zhang L, Zhu Z, Fan J, Chen L, et al. MicroRNA 23b regulates autophagy associated with radioresistance of pancreatic cancer cells. Gastroenterology. 2013;145:1133–43. e12. doi: 10.1053/j.gastro.2013.07.048. [DOI] [PubMed] [Google Scholar]
- 99.Thorburn A. Autophagy and its effects: making sense of double-edged swords. PLoS Biol. 2014;12:e1001967. doi: 10.1371/journal.pbio.1001967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.White E. Deconvoluting the context-dependent role for autophagy in cancer. Nat Rev Cancer. 2012;12:401–10. doi: 10.1038/nrc3262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Bracci PM. Obesity and pancreatic cancer: overview of epidemiologic evidence and biologic mechanisms. Mol Carcinog. 2012;51:53–63. doi: 10.1002/mc.20778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Rebours V, Gaujoux S, d’Assignies G, Sauvanet A, Ruszniewski P, Levy P, et al. Obesity and fatty pancreatic infiltration are risk factors for pancreatic precancerous lesions (PanIN) Clin Cancer Res. 2015 doi: 10.1016/j.dld. [DOI] [PubMed] [Google Scholar]
- 103.Li D, Morris JS, Liu J, Hassan MM, Day RS, Bondy ML, et al. Body mass index and risk, age of onset, and survival in patients with pancreatic cancer. JAMA. 2009;301:2553–62. doi: 10.1001/jama.2009.886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Yuan C, Bao Y, Wu C, Kraft P, Ogino S, Ng K, et al. Prediagnostic body mass index and pancreatic cancer survival. Journal of clinical oncology : official journal of the American Society of Clinical Oncology. 2013;31:4229–34. doi: 10.1200/JCO.2013.51.7532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Khasawneh J, Schulz MD, Walch A, Rozman J, Hrabe de Angelis M, Klingenspor M, et al. Inflammation and mitochondrial fatty acid beta-oxidation link obesity to early tumor promotion. Proc Natl Acad Sci U S A. 2009;106:3354–9. doi: 10.1073/pnas.0802864106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Philip B, Roland CL, Daniluk J, Liu Y, Chatterjee D, Gomez SB, et al. A high-fat diet activates oncogenic Kras and COX2 to induce development of pancreatic ductal adenocarcinoma in mice. Gastroenterology. 2013;145:1449–58. doi: 10.1053/j.gastro.2013.08.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Lanza-Jacoby S, Yan G, Radice G, LePhong C, Baliff J, Hess R. Calorie restriction delays the progression of lesions to pancreatic cancer in the LSL-KrasG12D; Pdx-1/Cre mouse model of pancreatic cancer. Exp Biol Med (Maywood) 2013;238:787–97. doi: 10.1177/1535370213493727. [DOI] [PubMed] [Google Scholar]
- 108.Wang D, Dubois RN. Eicosanoids and cancer. Nat Rev Cancer. 2010;10:181–93. doi: 10.1038/nrc2809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Mohammed A, Janakiram NB, Brewer M, Duff A, Lightfoot S, Brush RS, et al. Endogenous n-3 polyunsaturated fatty acids delay progression of pancreatic ductal adenocarcinoma in Fat-1-p48(Cre/+)-LSL-Kras(G12D/+) mice. Neoplasia. 2012;14:1249–59. doi: 10.1593/neo.121508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Strouch MJ, Ding Y, Salabat MR, Melstrom LG, Adrian K, Quinn C, et al. A high omega-3 fatty acid diet mitigates murine pancreatic precancer development. J Surg Res. 2011;165:75–81. doi: 10.1016/j.jss.2009.04.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Huxley R, Ansary-Moghaddam A, Berrington de Gonzalez A, Barzi F, Woodward M. Type-II diabetes and pancreatic cancer: a meta-analysis of 36 studies. Br J Cancer. 2005;92:2076–83. doi: 10.1038/sj.bjc.6602619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Muniraj T, Chari ST. Diabetes and pancreatic cancer. Minerva Gastroenterol Dietol. 2012;58:331–45. [PMC free article] [PubMed] [Google Scholar]
- 113.Wolpin BM, Bao Y, Qian ZR, Wu C, Kraft P, Ogino S, et al. Hyperglycemia, insulin resistance, impaired pancreatic beta-cell function, and risk of pancreatic cancer. J Natl Cancer Inst. 2013;105:1027–35. doi: 10.1093/jnci/djt123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Li D, Yeung SC, Hassan MM, Konopleva M, Abbruzzese JL. Antidiabetic therapies affect risk of pancreatic cancer. Gastroenterology. 2009;137:482–8. doi: 10.1053/j.gastro.2009.04.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Yuan C, Rubinson DA, Qian ZR, Wu C, Kraft P, Bao Y, et al. Survival among patients with pancreatic cancer and long-standing or recent-onset diabetes mellitus. Journal of clinical oncology : official journal of the American Society of Clinical Oncology. 2014;33:29–35. doi: 10.1200/JCO.2014.57.5688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Chari ST, Leibson CL, Rabe KG, Timmons LJ, Ransom J, de Andrade M, et al. Pancreatic cancer-associated diabetes mellitus: prevalence and temporal association with diagnosis of cancer. Gastroenterology. 2008;134:95–101. doi: 10.1053/j.gastro.2007.10.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Aggarwal G, Ramachandran V, Javeed N, Arumugam T, Dutta S, Klee GG, et al. Adrenomedullin is up-regulated in patients with pancreatic cancer and causes insulin resistance in beta cells and mice. Gastroenterology. 2012;143:1510–7. e1. doi: 10.1053/j.gastro.2012.08.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Sah RP, Nagpal SJ, Mukhopadhyay D, Chari ST. New insights into pancreatic cancer-induced paraneoplastic diabetes. Nat Rev Gastroenterol Hepatol. 2013;10:423–33. doi: 10.1038/nrgastro.2013.49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Pannala R, Leirness JB, Bamlet WR, Basu A, Petersen GM, Chari ST. Prevalence and clinical profile of pancreatic cancer-associated diabetes mellitus. Gastroenterology. 2008;134:981–7. doi: 10.1053/j.gastro.2008.01.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Baur JA, Birnbaum MJ. Control of gluconeogenesis by metformin: does redox trump energy charge? Cell Metab. 2014;20:197–9. doi: 10.1016/j.cmet.2014.07.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Hardie DG. Molecular Pathways: Is AMPK a Friend or a Foe in Cancer? Clin Cancer Res. 2015 doi: 10.1158/1078-0432.CCR-14-3300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Hardie DG. AMPK: positive and negative regulation, and its role in whole-body energy homeostasis. Curr Opin Cell Biol. 2015;33:1–7. doi: 10.1016/j.ceb.2014.09.004. [DOI] [PubMed] [Google Scholar]
- 123.Janzer A, German NJ, Gonzalez-Herrera KN, Asara JM, Haigis MC, Struhl K. Metformin and phenformin deplete tricarboxylic acid cycle and glycolytic intermediates during cell transformation and NTPs in cancer stem cells. Proc Natl Acad Sci U S A. 2014;111:10574–9. doi: 10.1073/pnas.1409844111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Mohammed A, Janakiram NB, Brewer M, Ritchie RL, Marya A, Lightfoot S, et al. Antidiabetic Drug Metformin Prevents Progression of Pancreatic Cancer by Targeting in Part Cancer Stem Cells and mTOR Signaling. Transl Oncol. 2014;6:649–59. doi: 10.1593/tlo.13556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Cifarelli V, Lashinger LM, Devlin KL, Dunlap SM, Huang J, Kaaks R, et al. Metformin and Rapamycin Reduce Pancreatic Cancer Growth in Obese Prediabetic Mice by Distinct MicroRNA-Regulated Mechanisms. Diabetes. 2015;64:1632–42. doi: 10.2337/db14-1132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Kisfalvi K, Moro A, Sinnett-Smith J, Eibl G, Rozengurt E. Metformin inhibits the growth of human pancreatic cancer xenografts. Pancreas. 2013;42:781–5. doi: 10.1097/MPA.0b013e31827aec40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Mayers JR, Wu C, Clish CB, Kraft P, Torrence ME, Fiske BP, et al. Elevation of circulating branched-chain amino acids is an early event in human pancreatic adenocarcinoma development. Nat Med. 2014;20:1193–8. doi: 10.1038/nm.3686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Liu KA, Lashinger LM, Rasmussen AJ, Hursting SD. Leucine supplementation differentially enhances pancreatic cancer growth in lean and overweight mice. Cancer Metab. 2014;2:6. doi: 10.1186/2049-3002-2-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Fearon KC, Glass DJ, Guttridge DC. Cancer cachexia: mediators, signaling, and metabolic pathways. Cell Metab. 2012;16:153–66. doi: 10.1016/j.cmet.2012.06.011. [DOI] [PubMed] [Google Scholar]
- 130.Sagar G, Sah RP, Javeed N, Dutta SK, Smyrk TC, Lau JS, et al. Pathogenesis of pancreatic cancer exosome-induced lipolysis in adipose tissue. Gut. 2015 doi: 10.1136/gutjnl. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Wolpin BM, Ng K, Bao Y, Kraft P, Stampfer MJ, Michaud DS, et al. Plasma 25-hydroxyvitamin D and risk of pancreatic cancer. Cancer Epidemiol Biomarkers Prev. 2011;21:82–91. doi: 10.1158/1055-9965.EPI-11-0836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Wolpin BM, Rubinson DA, Wang X, Chan JA, Cleary JM, Enzinger PC, et al. Phase II and pharmacodynamic study of autophagy inhibition using hydroxychloroquine in patients with metastatic pancreatic adenocarcinoma. Oncologist. 2014;19:637–8. doi: 10.1634/theoncologist.2014-0086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Ronan B, Flamand O, Vescovi L, Dureuil C, Durand L, Fassy F, et al. A highly potent and selective Vps34 inhibitor alters vesicle trafficking and autophagy. Nat Chem Biol. 2014;10:1013–9. doi: 10.1038/nchembio.1681. [DOI] [PubMed] [Google Scholar]
- 134.McCormick F. KRAS as a Therapeutic Target. Clin Cancer Res. 2015;21:1797–801. doi: 10.1158/1078-0432.CCR-14-2662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Viale A, Pettazzoni P, Lyssiotis CA, Ying H, Sanchez N, Marchesini M, et al. Oncogene ablation-resistant pancreatic cancer cells depend on mitochondrial function. Nature. 2014;517:205–8. doi: 10.1038/nature13611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Balliet RM, Capparelli C, Guido C, Pestell TG, Martinez-Outschoorn UE, Lin Z, et al. Mitochondrial oxidative stress in cancer-associated fibroblasts drives lactate production, promoting breast cancer tumor growth: understanding the aging and cancer connection. Cell Cycle. 2011;10:4065–73. doi: 10.4161/cc.10.23.18254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Ertel A, Tsirigos A, Whitaker-Menezes D, Birbe RC, Pavlides S, Martinez-Outschoorn UE, et al. Is cancer a metabolic rebellion against host aging? In the quest for immortality, tumor cells try to save themselves by boosting mitochondrial metabolism. Cell Cycle. 2012;11:253–63. doi: 10.4161/cc.11.2.19006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.O’Neill LA, Hardie DG. Metabolism of inflammation limited by AMPK and pseudo-starvation. Nature. 2013;493:346–55. doi: 10.1038/nature11862. [DOI] [PubMed] [Google Scholar]
- 139.Rattigan YI, Patel BB, Ackerstaff E, Sukenick G, Koutcher JA, Glod JW, et al. Lactate is a mediator of metabolic cooperation between stromal carcinoma associated fibroblasts and glycolytic tumor cells in the tumor microenvironment. Exp Cell Res. 2011;318:326–35. doi: 10.1016/j.yexcr.2011.11.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Sotgia F, Martinez-Outschoorn UE, Howell A, Pestell RG, Pavlides S, Lisanti MP. Caveolin-1 and cancer metabolism in the tumor microenvironment: markers, models, and mechanisms. Annu Rev Pathol. 2012;7:423–67. doi: 10.1146/annurev-pathol-011811-120856. [DOI] [PubMed] [Google Scholar]