Abstract
Background
There are very limited data on children with pneumonia in Mali. The objective was to assess the etiology and factors associated with community-acquired pneumonia in hospitalized children <5 years of age in Mali.
Methods
A prospective hospital-based case-control study was implemented in the Pediatric department of Gabriel Touré University Hospital at Bamako, Mali, between July 2011-December 2012. Cases were children with radiologically-confirmed pneumonia; Controls were hospitalized children without respiratory features, matched for age and period. Respiratory specimens, were collected to identify 19 viruses and 5 bacteria. Whole blood was collected from cases only. Factors associated with pneumonia were assessed by multivariate logistic regression.
Results
Overall, 118 cases and 98 controls were analyzed; 44.1% were female, median age was 11 months. Among pneumonia cases, 30.5% were hypoxemic at admission, mortality was 4.2%. Pneumonia cases differed from the controls regarding clinical signs and symptoms but not in terms of past medical history. Multivariate analysis of nasal swab findings disclosed that S. pneumoniae (adjusted odds ratio [aOR] = 3.4, 95% confidence interval [95% CI]: 1.6–7.0), human metapneumovirus (aOR = 17.2, 95% CI: 2.0–151.4), respiratory syncytial virus [RSV] (aOR = 7.4, 95% CI: 2.3–23.3), and influenza A virus (aOR = 10.7, 95% CI: 1.0–112.2) were associated with pneumonia, independently of patient age, gender, period, and other pathogens. Distribution of S. pneumoniae and RSV differed by season with higher rates of S. pneumoniae in January-June and of RSV in July-September. Pneumococcal serotypes 1 and 5 were more frequent in pneumonia cases than in the controls (P = 0.009, and P = 0.04, respectively).
Conclusions
In this non-PCV population from Mali, pneumonia in children was mainly attributed to S. pneumoniae, RSV, human metapneumovirus, and influenza A virus. Increased pneumococcal conjugate vaccine coverage in children could significantly reduce the burden of pneumonia in sub-Saharan African countries.
Introduction
Pneumonia is the leading cause of child mortality from infectious diseases, accounting for an estimated 1 million deaths annually, and mainly affecting children in developing countries [1,2]. Mortality attributed to pneumonia has decreased since 2000, but remains a major public health concern [3,4]. The main known causative pathogens reported are Streptococcus pneumoniae, Haemophilus influenzae type B, and respiratory syncytial virus (RSV) [5]. However, their distribution varies by season and location. Data on the etiology and epidemiology of pneumonia in children in developing countries are still insufficient, particularly in sub-Saharan Africa [6].
Mali is one of the poorest countries in the world, with a under-five year mortality rate of 123 per 1,000 live births in 2013 according to the UN Inter-agency Group for Child Mortality Estimation. It has been estimated that among its 14.9 million inhabitants, each year more than 900,000 pneumonia episodes occur in children under 5 years of age, leading to almost 8,000 deaths annually [7]. A previous descriptive study reported that pneumonia was the most frequent cause of admission, representing 18% of total hospital admissions [8]. However, detailed information was not available on clinical presentation and on the etiology of suspected pneumonia cases [8]. Moreover, no control group was included. The presence of a control group of children without pneumonia would allow better interpretation of microbiological findings, particularly with nasal sampling [9]. In March 2011, Mali included pneumococcal vaccination (PCV13) in a routine immunization program; but vaccine coverage is still low [10].
The study objective was to assess the etiology and factors associated with community-acquired pneumonia in hospitalized children in Mali.
Materials and Methods
Setting and Participants
A prospective multicenter case-control study, based on the Global Approach to Biological Research, Infectious diseases and Epidemics in Low-income countries (GABRIEL) network [11], was implemented in the Pediatric department of Gabriel Touré University Hospital at Bamako, Mali, between July 2011 and December 2012. This multicenter study is ongoing, it will include ten study sites, located in 9 countries over 3 continents (Brazil, Cambodia, China, Haiti, India, Madagascar, Mali, Mongolia, and Paraguay). The study protocol has been described in detail elsewhere [12], and pooled results will be analyzed later.
The Gabriel Touré University Hospital is a 447-bed tertiary-care general hospital located in Bamako. It is a primary care hospital for people living in Bamako and a national reference centre for other patients. Various medical and surgical specialities, including pediatrics, are located in the hospital. The pediatrics department, with a capacity of 150 beds, includes a general pediatrics unit and a neonatal/emergency unit. It receives sick children for primary care and severe cases referred from other healthcare settings. On average, 50,000 consultations and 10,000 hospital admissions occur in the pediatrics department annually. Acute respiratory infections represent 34% of admissions in children, and 15% of child hospitalizations.
Case Definition and Enrollment
Pneumonia cases were hospitalized children who fulfilled the following criteria:
- Cough and/or dyspnea, and
- Tachypnea, as characterized by the World Health Organization (WHO) in children between 2 and 12 months of age: breathing rate ≥50 cycles per minute; in children between 12 and 59 months of age: breathing rate ≥40 cycles per minute) [13], and
- Absence of wheezing at auscultation, and
- First symptoms appearing within the last 14 days, and
- Radiological confirmation of pneumonia as per WHO guidelines [14].
The exclusion criterions for cases were presence of wheezing at auscultation, or minors whose parents or legal guardian declined to sign the informed consent statement. Controls were patients hospitalized for surgery or in a routine outpatient practice environment, aged between 2 and 59 months, without any symptoms suggestive of respiratory illness; suspicion of infection of other site was not an exclusion criteria. Cases and controls were matched for age (±1 year) and calendar date of hospital admission (±1 month) to take seasonality into account. Thus, 61% of patients were recruited during the rainy season (May to October) while 39% were recruited during the dry season (November to April).
Biological Samples
Samples were collected in the first 48 hours of patient hospitalization. Nasal swabs were taken from all pneumonia cases and controls. To document antibiotic usage history, urine was sampled from cases for broad spectrum antibiotic detection. Whole blood and pleural effusions (in applicable cases) were collected from cases only. Whole blood was distributed in Ethylenediaminetetraacetic acid (EDTA) vials for preservation and in dry tubes for serum specimens. EDTA-whole blood samples allowed complete blood count, blood culture and real-time multiplex polymerase chain reaction (PCR) assay for the identification of Staphylococcus aureus, Streptococcus pneumoniae and Haemophilus influenzae type b. C reactive protein (CRP) and procalcitonin (PCT) were measured quantitatively in serum.
Respiratory specimens from nasal swabs and pleural effusions were characterized by FTD respiratory pathogens 21 plus (Fast-track Diagnostics, Luxembourg) based on RT-PCR which included a panel of 19 viruses and 5 bacteria. The pathogens identified were: influenza A, influenza A(H1/N1), influenza B, coronavirus 229E, coronavirus OC43, coronavirus NL63, coronavirus HKU1, parainfluenza virus 1, parainfluenza virus 2, parainfluenza virus 3, parainfluenza virus 4, human metapneumoviruses A and B, rhinovirus, RSVs A and B, adenovirus, enterovirus, parechovirus, bocavirus, Mycoplasma pneumoniae, Chlamydia pneumoniae, S. aureus, S. pneumoniae, and H. influenzae type b. S. pneumoniae-positive specimens were serotyped by RT-PCR that detects the 29 main S. pneumoniae serotypes: 1, 3, 4, 5, 6A/B, 7C, 7F, 8, 9V, 10A, 11A, 12F, 14, 15A, 15B/C, 16F, 17F, Sg18C, 19A, 19F, 20, 22F, 23F, 31, 33F, 34, 35B, 35F, 38 and Lyt A.
Statistical Analysis
Demographic characteristics, underlying diseases, medical history, clinical examination at enrollment, radiological findings, vaccinations, and outcome were recorded prospectively for each patient on a standardized form.
Continuous variables were described as median and interquartile range (IQR), categorical variables as number and percentage. Pneumonia cases we compared to the controls by the Mann-Whitney U test for continuous covariates, and by the Chi2 test for categorical variables. Univariate and multivariate logistic regression analyses were performed to assess factors associated with pneumonia. Multivariate analyses were adjusted for gender, age, period per quarter, and other pathogens significantly associated with increased risk of pneumonia. Concordance between serotypes detected in nasopharyngeal samples and blood was tested by Kendall rank correlation.
Crude population attributable fraction (PAF) was calculated after univariate logistic regression analysis to quantify the contribution of the each microorganism to pneumonia occurrence. Conceptuality, PAF permits the estimation of the proportional reduction in pneumonia occurrence that would occur if the pathogen was absent (alternative ideal scenario). The formula was the following:
No patient was excluded because of missing data. P<0.05 was considered significant; all tests were bilateral, and statistical analysis was conducted with Stata version 13.0 (Stata Corp.). With at least 100 cases and 100 controls, analyses had 80% power to detect odds ratios (OR)≥3 with ≥20% prevalence of control exposure, whatever the prevalence of case exposure.
Ethics
The study protocol, informed consent statement, clinical research form, any amendments and all other study documents were submitted to and approved by the institutional ethics committee (Comité National d'Ethique pour la Santé et les Sciences de la Vie). Written informed consent was obtained from all participants.
Results
Overall Population
Among the 119 enrolled cases, 118 conformed with the inclusion criteria and were all sampled. One patient without radiological confirmation was excluded from the analysis. Among the 100 included controls, 2 were excluded because of missing data.
Overall, 216 patient, accounting for 118 (54.6%) pneumonia cases and 98 (45.4%) controls, were analyzed. Among them, 93 (44.1%) were female. The male/female gender ratio was 0.79, and median age was 11 months (IQR: 5–55 months).
Description of Pneumonia Cases
Among the 118 patients with pneumonia, 16 (13.6%) were referred from another health center; median length of hospital stay was 7 days (IQR: 6–10 days) for a total of 909 days of hospitalization (Table 1). The median time period between first symptoms and hospitalization was 6 days (IQR: 4–7 days).
Table 1. Characteristics of children pneumonia cases and controls, Mali, N = 216.
| Characteristics a | Pneumonia cases (n = 118) | Controls (n = 98) | P |
|---|---|---|---|
| Demographics at admission | |||
| Gender, male | 57 (48.3) | 36 (38.7) | 0.16 |
| Age, months, median (IQR) | 12 (5–26) | 11 (5–23) | 0.24 |
| Height, cm, median (IQR) | 73.5 (65–88) | 71 (64–79) | 0.04 |
| Weight, kg, median (IQR) | 7.6 (6–10) | 7 (5.5–9.3) | 0.13 |
| Body mass index, median (IQR) | 13.9 (12.6–15.2) | 14.2 (12.9–15.5) | 0.47 |
| Arm circumference, cm, median (IQR) | 13 (12–14) | 12.5 (11–14) | 0.01 |
| Weight-for-height Z-score ≤2 SD | 52 (44.1) | 41 (44.1) | 0.99 |
| Weight-for-height Z-score ≤3 SD | 32 (27.1) | 22 (23.7) | 0.57 |
| Medical history | |||
| Heart disease | 4 (3.4) | 0 (0) | 0.07 |
| HIV-positive | 3 (2.5) | 0 (0) | 0.12 |
| Contracted a common cold/pharyngitis b | 61 (31.7) | 0 (0) | <0.001 |
| Contracted ILI b | 18 (15.2) | 0 (0) | <0.001 |
| Pneumococcal conjugate vaccine | 0 (0) | 0 (0) | - |
| DPT-Hep. B-Hib vaccine, 1 dose | 102 (86.4) | 80 (86.0) | 0.93 |
| DPT-Hep. B-Hib vaccine, 3 dose | 81 (68.6) | 69 (74.2) | 0.38 |
| Influenza vaccine | 1 (0.8) | 0 (0) | 0.37 |
| Vital signs at admission | |||
| Temperature, °C, median (IQR) | 38.6 (37.8–39.4) | 37 (36.8–37.4) | <0.001 |
| Breathing rate, cycles/min, median (IQR) | 56 (48–67) | 31 (29–35) | <0.001 |
| Cardiac rate, cycles/min, median (IQR) | 151 (132–167) | 122 (112–132) | <0.001 |
| Oxygen saturation, %, median (IQR) | 94.5 (87–96) | 98 (98–99.5) | <0.001 |
| Clinical signs/symptoms at admission | |||
| Dyspnea | 116 (98.3) | 0 (0) | <0.001 |
| Lower chest indrawing | 116 (98.3) | 0 (0) | <0.001 |
| Cough | 114 (96.6) | 0 (0) | <0.001 |
| Pulmonary crackles | 102 (86.4) | 0 (0) | <0.001 |
| Ronchi | 34 (28.8) | 0 (0) | <0.001 |
| Rhinopharyngitis | 32 (27.1) | 0 (0) | <0.001 |
| Prostration or lethargy | 23 (19.5) | 0 (0) | <0.001 |
| Inability to drink | 15 (12.7) | 0 (0) | <0.001 |
| Diarrhea | 12 (10.2) | 20 (21.5) | 0.02 |
| Cyanosis | 11 (9.3) | 0 (0) | 0.002 |
| Vomiting | 9 (7.6) | 13 (14.0) | 0.13 |
| Convulsions | 6 (5.1) | 2 (2.1) | 0.27 |
| Conjunctivitis | 6 (5.1) | 1 (1.1) | 0.11 |
| Diminished breath sounds | 5 (4.2) | 0 (0) | 0.04 |
| Dullness to percussion | 4 (3.4) | 0 (0) | 0.07 |
| Otitis | 3 (2.5) | 1 (1.1) | 0.44 |
| Rasping | 3 (2.5) | 0 (0) | 0.12 |
| Skin rash | 0 (0) | 2 (2.1) | 0.11 |
Abbreviations: IQR, interquartile range; SD, standard deviation; DPT-Hep. B-Hib, Diphtheria, tetanus, Pertussis, hepatitis B, Haemophilus influenza type b; HIV, human immunodeficiency virus; ILI, influenza-like illness
aData are N (%), unless specified otherwise.
bwithin 2 weeks.
Four (3.9%) pneumonia cases presented a ventricular septal defect. None had previous tuberculosis or underlying lung disease. Among cases with pulmonary crackles, 36 (35.3%) were unilateral and 66 (64.7%) were bilateral. Other signs or medical history of pneumonia cases included sickle-cell anaemia (n = 1), dehydration (n = 1), epilepsy (n = 1), goiter (n = 1), edema (n = 1), pallor (n = 1), and trisomy (n = 3).
Among the 47 patients who took antibiotics before hospitalization, 17 (14.4%) received Amoxicillin, 8 (5.6%) Ceftriaxone, 4 (2.8%) Amoxicillin/Clavulanic acid, and 3 (2.1%) Cotrimoxazole. Among the 63 cases tested for antibiotic detection testing in urine, 22 (65.1%) were positive; in this population, sensitivity of declared antibiotic use compared with urine detection was low (sensitivity = 51.2%, 95% confidence interval: 35.1–67.1%).
Median CRP level at admission was 21 mg/l (IQR: 6–63, mean: 56.6 mg/l), median PCT level was 4.6 ng/ml (IQR: 0.4–26.4, mean: 31.5 ng/ml). Median CRP and PCT levels were higher in hypoxemic compared with non-hypoxemic pneumonia cases (P = 0.03 and P = 0.007, respectively).
Median white blood cell count was 15,750*109cells/l (IQR: 10,000–25,700, mean: 20,600*109cells/l), and median neutrophil percentage was 52.5% (IQR: 31–68, mean: 50.3%). X-rays showed generalized, dense, homogeneous opacification in 24 (20.3%) patients, interstitial syndrome in 24 (20.5%), and alveolar infiltrate in 65 (55.1%), without significant differences according to the pattern of infection (viral, bacterial or mixed). No patient had abscess or pneumothorax, but 4 (3.4%) had pleural effusion.
During hospital stay, pneumonia cases received the following antibiotics: Amoxicillin (n = 111, 94.1%), Ceftriaxone (n = 3, 2.6%), Amoxicillin/Clavulanic acid (n = 3, 2.5%), Ciprofloxacin (n = 1, 0.8%), and Vancomycin (n = 2, 1.6%). Fifty-one (43.2%) received oxygen for a median length of 2 days (IQR: 1–3 days), and 3 (2.5%) had blood transfusions. Three patients with pleural effusions were tested for microbial detection. Five patients died (lethality: 4.2%); death was directly related to pneumonia in 4 patients (Table 2).
Table 2. Description of deceased patients with pneumonia, N = 5.
| ID | Gender | Age | Signs | Respiratory rate | Temperature | Oxygen saturation | Delay from admission to death | Treatments | Blood, molecular detection or culture | Nasal aspirate Microbiology, molecular detection | Cause of death |
|---|---|---|---|---|---|---|---|---|---|---|---|
| #1 | F | 4 years | Dyspnea, lower chest indrawing | 77 cycles/min | 38.4°C | 86% | 10 hours | Amoxicillin, paracetamol, oxygen | S. pneumoniae serotype 3 a | S. aureus, H. influenza, Parainfluenzae 2 | Severe pneumonia |
| #2 | F | 3 months | Dyspnea, lower chest indrawing | 47 cycles/min | 39 .7°C | 89% | 2 days | Amoxicillin, paracetamol, oxygen | Negative | Coronavirus HKU1 | Severe pneumonia |
| #3 | F | 2 years | Dyspnea, lower chest indrawing, dehydration, pallor | 82 cycles/min | 40.1°C | 89% | 12 hours | Amoxicillin, gentamycin, rehydration, paracetamol, oxygen | S. pneumoniae serotype 35F a | S. pneumoniae serotypes 19A, 19F, 22F, 35F S. aureus, parainfluenzae 3, enterovirus | Severe, acute malnutrition |
| #4 | M | 8 months | Dyspnea, lower chest indrawing, | 62 cycles/min | 38.8°C | 80% | 12 hours | Amoxicillin, gentamycin, parecetamol, oxygen | Coagulase-negative staphylococci b | S. pneumoniae serotypes 6AB, 15BC, H. influenza, RSV | Severe pneumonia |
| #5 | F | 13 months | Dyspnea, lower chest indrawing, pallor | 60 cycles/min | 38.7°C | 68% | 3 days | Amoxicillin, paracetamol, oxygen | Coagulase-negative staphylococcib | S. pneumoniae serotype 7C, Bocavirus | Pneumonia with moderate, acute malnutrition |
Abbreviations: F, female; M, male; RSV, respiratory syncytial virus.
a By PCR
b Blood culture
Comparison of Characteristics of Pneumonia Cases and Controls
Table 1 compares the clinical signs and symptoms at admission between cases and controls, underlying conditions and biological findings. Pneumonia cases differed from controls regarding clinical signs and symptoms as well as vital signs at admission, but not in terms of demographic factors or past medical history. Pneumonia cases were more frequently hypoxemic (defined as oxygen saturation<90%) at admission than the controls (30.5% vs. 0%, P<0.001). PCV coverage was zero in both groups.
Microbiological Findings
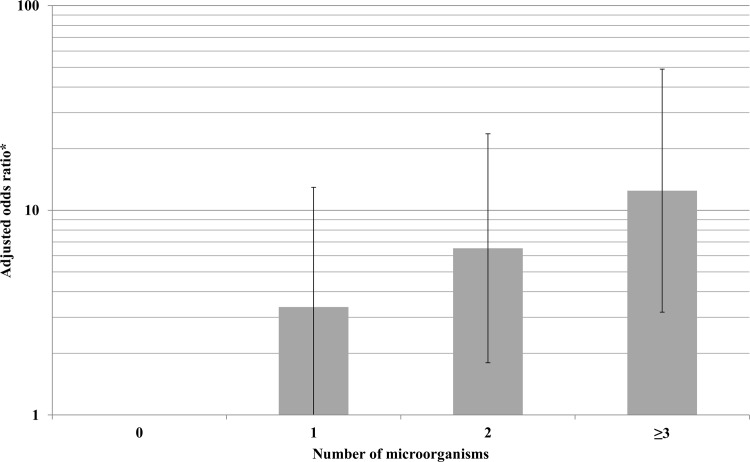
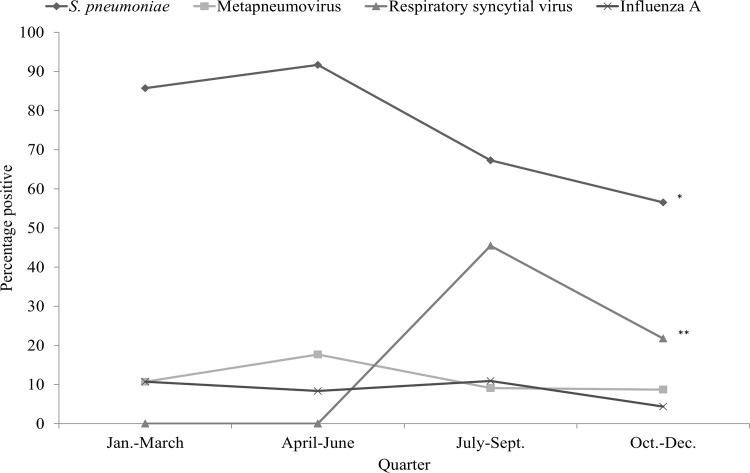
At least 1 microorganism was detected on nasal swabs in 96.6% of cases and 82.3% of controls (crude OR = 6.4, 95% confidence interval [95% CI]: 2.1–19.7, P<0.001). Overall, 78.8% of cases and 54.2% of controls were co-infected or co-colonized (crude OR = 3.3, 95% CI: 1.8–6.0, P<0.001). Co-detection on nasal swab of S. pneumoniae and RSV was more frequent in cases than in controls (respectively, 15.2% [N = 18] vs. 2.0% [N = 2], P = 0.001). Co-detection of S. pneumoniae and rhinovirus was not different in cases and controls (respectively, 16.1% [N = 19] vs. 12.2% [N = 12], P = 0.42; co-detection RSV and rhinovirus was not different between cases and controls (respectively, 5.9% [N = 7] vs. 3.1% [N = 3], P = 0.32). A dose-response relationship was apparent between the number of microorganisms found in nasal swabs and the risk of being a case (Fig 1). Distribution of S. pneumoniae and RSV differed by season with higher rates of S. pneumoniae in January-June and of RSV in July-September (Fig 2).
Fig 1. Relative risk (95% confidence interval) of pneumonia in children according to number of microorganisms from nasal swab, Mali, N = 216.
aAfter multivariate logistic regression, adjusted for gender, age, and period per quarter. bLogarithmic scale.
Fig 2. Temporal trends of pneumonia etiology in childhood cases, Mali, N = 118.
*P < .05, **P < .001.
Univariate analysis revealed that S. pneumoniae, human metapneumovirus, RSV, and influenza A virus detection in nasal swabs were significantly associated with pneumonia in Mali (Table 3). Multivariate analysis reinforced linkage of these pathogens with pneumonia, independently of patient age, gender, period per quarter and the presence of other pathogens significantly coupled with increased risk of pneumonia (Table 3). PAF was the highest for S. pneumoniae (PAF = 46%, 95% CI: 30–59%). Contribution of human metapneumovirus, RSV, and influenza A were lower, with PAFs of 9% (95% CI: 7–11%), 21% (95% CI: 16–25%) and, 8% (95% CI: 6–10%), respectively.
Table 3. Microbiological findings of children pneumonia cases and controls, nasal swabs, Mali, N = 216.
| Microorganisms | Pneumonia cases (n = 118) | Controls (n = 98) | P | Crude odds ratio (95% CI) | Adjusted odds ratio a (95% CI) |
|---|---|---|---|---|---|
| Bacteria | |||||
| Streptococcus pneumoniae | 85 (72.0) | 47 (48.0) | <0.001 | 2.8 (1.6–4.9) | 3.4 (1.6–7.0) |
| Staphylococcus aureus | 23 (19.5) | 19 (19.4) | 0.98 | 1.0 (0.5–2.0) | - |
| Haemophilus influenza | 8 (6.8) | 6 (6.2) | 0.84 | 1.1 (0.4–3.3) | - |
| Mycoplasma spp. | 0 (0) | 1 (1.0) | 0.27 | NE | - |
| Chlamydia spp. | 0 (0) | 0 (0) | - | NE | - |
| Viruses | |||||
| Human metapneumovirus | 12 (10.2) | 1 (1.0) | 0.005 | 11.0 (1.4–86.0) | 17.2 (2.0–151.4) |
| Coronavirus NL63 | 1 (0.8) | 4 (4.1) | 0.12 | 0.2 (0.02–1.8) | - |
| Coronavirus 229E | 2 (1.7) | 0 (0) | 0.19 | NE | - |
| Coronavirus OC43 | 2 (1.7) | 4 (4.1) | 0.29 | 0.4 (0.07–2.3) | - |
| Coronavirus HKU 1 | 6 (5.1) | 2 (2.0) | 0.24 | 2.6 (0.5–13.0) | - |
| Adenovirus | 11 (9.3) | 6 (6.1) | 0.39 | 1.6 (0.6–4.4) | - |
| Enterovirus | 12 (10.2) | 14 (14.3) | 0.35 | 0.7 (0.3–1.5) | - |
| Parechovirus | 0 (0) | 0 (0) | - | NE | - |
| Rhinovirus | 27 (22.9) | 24 (24.5) | 0.78 | 0.9 (0.5–1.7) | - |
| Respiratory syncytial virus | 30 (25.4) | 6 (6.1) | <0.001 | 5.2 (2.1–13.2) | 7.4 (2.3–23.3) |
| Parainfluenzae 1 | 5 (4.2) | 0 (0) | 0.04 | NE | - |
| Parainfluenzae 2 | 1 (0.8) | 0 (0) | 0.36 | NE | - |
| Parainfluenzae 3 | 5 (4.2) | 3 (3.1) | 0.65 | 2.0 (0.4–10.6) | - |
| Parainfluenzae 4 | 2 (1.7) | 4 (4.1) | 0.29 | 0.4 (0.07–2.1) | - |
| Influenza A | 11 (9.3) | 1 (1.0) | 0.008 | 10.0 (1.3–78.7) | 10.7 (1.0–112.2) |
| Infuenza B | 0 (0) | 0 (0) | - | NE | - |
| Influenza A(H1N1) | 3 (2.5) | 0 (0) | 0.11 | NE | - |
| Bocavirus | 13 (11.0) | 12 (12.2) | 0.78 | 0.9 (0.4–2.0) | - |
Abbreviations: NE, non estimable.
a After multivariate logistic regression, adjusted for the presence of other pathogens, gender, age, and period per quarter.
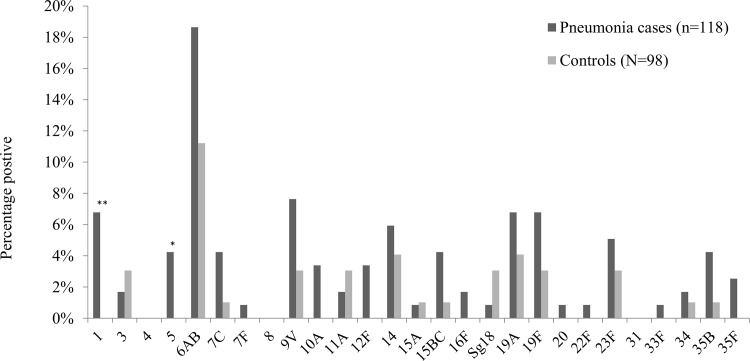
Fig 3 reports the distribution of pneumococcal serotypes detected in nasal swabs from cases and controls. The most prevalent serotype in pneumonia cases and controls was serotype 6A/B (18.6% vs. 11.2%, P = 0.13). Serotypes 1 and 5 were more frequent in pneumonia cases than in the controls: 6.8% vs. 0%, P = 0.009, and 4.2% vs. 0%, P = 0.04, respectively.
Fig 3. Streptococcus pneumoniae serotypes in nasal aspirates from pneumonia cases and controls, Mali, N = 216. *P < .05, **P < .001.
In pneumonia cases, S. pneumoniae was positive in 16 (13.6%) patients, S. aureus in 6 (5.1%) patients and H. influenza in 5 (4.2%) patients by PCR blood sample detection. Most patients with S. pneumoniae detection by PCR had also S. pneumoniae nasal carriage (93.5%, 15/16), while only 17.6% (15/85) of patients with S. pneumoniae nasal carriage had positive detection by PCR (P = 0.04). Concordance of serotype 1 detected in nasal swabs and blood in pneumonia cases was high (κ = 0.70, P<0.001). Coronavirus 63 was identified from pleural effusion in 1 patient. Microbiological findings, including S. pneumoniae serotypes distribution, from PCR nasal swab or blood sample were not different in pneumonia cases according to the result of urine antibiotic testing (data not shown). Blood culture was positive in 36 (30.5%) pneumonia cases; most microorganisms were probably related to contamination of samples. The following bacteria were detected: coagulase-negative staphyloccoci (n = 26), Salmonella spp. (n = 3), Gram-positive bacilli (n = 2), Acinetobacter baumannii (n = 1), Aerococcus viridans (n = 1), Enterococcus faecium (n = 1), Granulicatella elegans (n = 1), and Staphylococcus aureus (n = 1).
Discussion
The primary objective of this prospective case-control study was to assess the etiology and factors associated with community-acquired pneumonia in hospitalized children in Mali. In this non-PCV population, we observed that S. pneumoniae, human metapneumovirus, RSV, and influenza A were the main microbial agents associated with pneumonia among children in Mali, independently of patient age, gender, period, and other pathogens. H. influenzae was not associated with pneumonia, but the vaccination rate against this bacteria, at least for the first dose, was above 85% in both cases and controls. In addition, while most patients with S. pneumoniae by PCR in blood had also nasal carriage, frequently with the same serotype, positive S. pneumoniae test from nasal sample was not highly predictive of S. pneumoniae detection in blood, meaning that the bacteria was frequently not the cause of pneumonia.
Few studies have assessed the etiology of pneumonia in sub-Saharan Africa. Howie et al. recently investigated pneumonia etiology in children in Gambia, observing that S. pneumoniae was the leading cause with a high rate of microbial agent co-detection [15]. Conversely, they found that viral pneumonia was not predominant whereas we observed that 3 viruses, namely human metapneumovirus, RSV, and influenza A virus, were associated with pneumonia in Mali. Major differences between the 2 studies were sample type (lung aspirates vs. nasal swabs in our investigation) and the lack of a comparative group in the study by Howie et al. which did not permit comparison of microbial prevalence in pneumonia patients and healthy subjects [15]. In a Western Kenya case-control study, S. pneumoniae, RSV, and influenza A virus were the predominant causes of pneumonia in children [16]. We noted almost similar results in Mali. Thus, S. pneumoniae and RSV could still be considered as the primary causes of pneumonia in several sub-Sharan African countries.
All patients who died had severe pneumonia, according to WHO criteria, with significant dyspnea, very marked chest indrawing and severe hypoxia. Two patients had confirmed pneumococcal pneumonia. Two patients had acute malnutrition linked with pneumonia. In 2 cases, death occurred within 24 hours of admission. Despite active treatment with antibiotics and oxygen, the management of severe respiratory distress is often difficult in this context because the hospital does not have assisted ventilatory support. Mortality was higher than in a recent birth cohort in South Africa [17], but was similar to what was reported previously in other developing countries [18,19].
Some invasive serotypes were detected selectively in pneumonia cases but not in the controls (i.e., serotypes 1 and 5 were associated with the risk of pneumonia). A previous study of serotypes involved in invasive pneumococcal disease in Mali found that serotype 5 was the most prevalent (54%) [20]. It was also linked with pneumonia in our series. In other sub-Saharan African countries, serotype 1 was described as the most prevalent serotype of invasive pneumococcal disease [21]. It was the second serotype significantly associated with pneumonia in children in Mali. Thus, the introduction of PCV in routine immunization programs in Mali would substantially reduce pneumonia caused by S. pneumoniae because most serotypes eliciting pneumonia would be covered by the vaccine [22]. In addition, pneumococcal pneumonia seasonality was similar to that observed previously in Burkina Faso, a neighbouring country, in 2007–2011 with higher incidence during the dry season between January and May [23].
Interestingly, among the 4 microbial agents associated with pneumonia, 3 were viruses: human metapneumovirus, RSV, and influenza A virus. Self et al. recently observed, in a pneumonia cases-control study implemented in hospitals of Utah, that detection respiratory syncytial virus, human metapneumovirus and influenza from nasopharyngeal or oropharyngeal sample of patients with pneumonia probably indicates an etiologic role [24].
We observed similar results in a different context. Despite the dearth of vaccination against pneumococcus in our population, viral pneumonia is a major segment of the pneumonia burden. Treatment of these infections is often problematic because the empirical use of antibiotics or antivirals is not consensual [25]. Vaccine development, particularly against RSV, is warranted to prevent pneumonia in children. Moreover, the impact of influenza vaccine policies in developing countries should be evaluated because this virus often causes pneumonia [26].
Rhinovirus was detected in almost 25% of pneumonia cases of our series, but the detection rate was not different between cases and controls. However, this observation does formally excludes that rhinovirus could play a role in the etiology of pneumonia. Jain et al. recently observed that rhinovirus was detected in 27% of U.S. children with community-pneumonia requiring hospitalization [27]; this large study had however no control group, it was also not possible to estimate the prevalence of respiratory detection in children without pneumonia. Growing evidence suggests the role of rhinovirus in the etiology of viral pneumonia [28] or related to the risk of secondary bacterial pneumonia [29]. Other viruses, such as bocavirus or adenovirus were also equally prevalent in cases and controls. Again, this finding does not exclude completely their implication in pneumonia etiology in children from Mali. However these viruses were poorly described as causative agents of pneumonia in children from other sub-Saharan African countries.
Several microorganisms have been detected in most pneumonia cases but also in the controls. The clinical significance of microbial detection by PCR is problematic at the individual level because most detected pathogens are not causes of pneumonia. However, number of pathogens is predictive of the risk of pneumonia, suggesting that simple quantification of species detected would permit the evaluation of pneumonia risk. At the population level, nasal samples are interesting because the prevalence of carriage in the controls is considered.
The main strengths of our study were its prospective design with the inclusion of controls that permitted us to assess pneumonia etiologies while taking into account the prevalence of pathogen detection in non-infected patients. Ours is the first case-control investigation of pneumonia etiology in children from Mali (S1 STROBE checklist). The results should serve to better manage pneumonia not only in these children but also in those from neighboring countries. Data quality was enhanced by centralized microbiological analysis in the Emerging Pathogens Laboratory (Lyon, France). In addition, this analysis should serve to better focus multicentric pneumonia studies that will permit us to assess the global etiologies of pneumonia and to compare etiologic agents between countries and continents.
Some limitations should be underlined. First, microbiological diagnosis of pneumonia is difficult due to the lack of single reliable test. Then, at the individual level, it is difficult to establish if a positive nasal swab denotes etiology or nasopharyngeal colonization; particularly for bacterial agents such as S. pneumoniae because of the high asymptomatic carriage. However, at the population level, addition of a control group permits to evaluate and control for the prevalence of carriage in asymptomatic children. In a pneumonia etiology study, it would be interesting to correlate the threshold cycle (Ct) values obtained from RT-PCR with lower airway specimens to establish more precisely the relationship between positive nasal swab and etiology of pneumonia, according to the microorganism [30]. Second, because the study was implemented in one hospital in Mali, external validity might be limited. However, Gabriel Touré University Hospital is a reference center in the country, and then source population is not limited to the city of Bamako. Third, study power was confined to detecting some linkages (i.e. serotypes associated with the risk of pneumonia).
Conclusion
Community-acquired pneumonia was mainly attributable to S. pneumoniae, human metapneumovirus, RSV or influenza A among children in Mali. Increased pneumococcal conjugate vaccine coverage in children would significantly reduce the burden of pneumonia in this country. The addition of a control group to assess etiologies of pneumonia in children is critical to properly interpret the microbiological results of diagnostic testing with high sensitivity.
Supporting Information
(DOC)
(XLS)
(XLS)
Acknowledgments
This protocol was developed on behalf of members of the global approach to biological research, infectious diseases and epidemics in low-income countries (GABRIEL) network: http://gabriel.globe-network.org.
Presented in part: Interscience Conference of Antimicrobial Agents and Chemotherapy / International Congress of Chemotherapy and Infection (ICAAC/ICC 2015), San Diego, California, September 2015. Abstract 1925.
Abbreviations
- CRP
C-reactive protein
- FTD
Fast-track Diagnostics
- IQR
interquartile range
- OR
odds ratio
- PCV
pneumococcal conjugate vaccination
- PCT
procalcitonin
- RSV
respiratory syncytial virus
- RT-PCR
real-time multiplex polymerase chain reaction
- WHO
World Health Organization
Data Availability
All relevant data are within the manuscript and its Supporting Information files.
Funding Statement
This study was funded by Fondation Mérieux. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Walker CLF, Rudan I, Liu L, Nair H, Theodoratou E, Bhutta ZA, et al. Global burden of childhood pneumonia and diarrhoea. Lancet. 2013;381: 1405–1416. 10.1016/S0140-6736(13)60222-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Global Health Observatory Data Repository. In: WHO [Internet]. Available: http://apps.who.int/gho/data/view.main.CM100WORLD-CH9?lang=en. Accessed 16 October 2014.
- 3. Liu L, Johnson HL, Cousens S, Perin J, Scott S, Lawn JE, et al. Global, regional, and national causes of child mortality: an updated systematic analysis for 2010 with time trends since 2000. Lancet. 2012;379: 2151–2161. 10.1016/S0140-6736(12)60560-1 [DOI] [PubMed] [Google Scholar]
- 4. Black RE, Cousens S, Johnson HL, Lawn JE, Rudan I, Bassani DG, et al. Global, regional, and national causes of child mortality in 2008: a systematic analysis. Lancet. 2010;375: 1969–1987. 10.1016/S0140-6736(10)60549-1 [DOI] [PubMed] [Google Scholar]
- 5. McIntosh K. Community-acquired pneumonia in children. N Engl J Med. 2002;346: 429–437. 10.1056/NEJMra011994 [DOI] [PubMed] [Google Scholar]
- 6. Gilani Z, Kwong YD, Levine OS, Deloria-Knoll M, Scott JAG, O’Brien KL, et al. A Literature Review and Survey of Childhood Pneumonia Etiology Studies: 2000–2010. Clin Infect Dis. 2012;54: S102–S108. 10.1093/cid/cir1053 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Rudan I, O’Brien KL, Nair H, Liu L, Theodoratou E, Qazi S, et al. Epidemiology and etiology of childhood pneumonia in 2010: estimates of incidence, severe morbidity, mortality, underlying risk factors and causative pathogens for 192 countries. J Glob Health. 2013;3 10.7189/jogh.03.010401 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Campbell JD, Sow SO, Levine MM, Kotloff KL. The causes of hospital admission and death among children in Bamako, Mali. J Trop Pediatr. 2004;50: 158–163. [DOI] [PubMed] [Google Scholar]
- 9. Deloria-Knoll M, Feikin DR, Scott JAG, O’Brien KL, DeLuca AN, Driscoll AJ, et al. Identification and Selection of Cases and Controls in the Pneumonia Etiology Research for Child Health Project. Clin Infect Dis. 2012;54: S117–S123. 10.1093/cid/cir1066 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Burchett HED, Mounier-Jack S, Torres-Rueda S, Griffiths UK, Ongolo-Zogo P, Rulisa S, et al. The impact of introducing new vaccines on the health system: Case studies from six low- and middle-income countries. Vaccine. 2014;32: 6505–6512. 10.1016/j.vaccine.2014.09.031 [DOI] [PubMed] [Google Scholar]
- 11. Komurian-Pradel F, Grundmann N, Siqueira MM, Chou M, Diallo S, Mbacham W, et al. Enhancing research capacities in infectious diseases: The GABRIEL network, a joint approach to major local health issues in developing countries. Clin Epidemiol Glob Health. 2013;1: 40–43. 10.1016/j.cegh.2012.11.002 [DOI] [Google Scholar]
- 12. Picot VS, Bénet T, Messaoudi M, Telles J- N, Chou M, Eap T, et al. Multicenter case–control study protocol of pneumonia etiology in children: Global Approach to Biological Research, Infectious diseases and Epidemics in Low-income countries (GABRIEL network). BMC Infect Dis. 2014;14: 1–9. 10.1186/s12879-014-0635-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Ayieko P, English M. Case Management of Childhood Pneumonia in Developing Countries. Pediatr Infect Dis J. 2007;26: 432–440. 10.1097/01.inf.0000260107.79355.7d [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Cherian T, Mulholland EK, Carlin JB, Ostensen H, Amin R, de Campo M, et al. Standardized interpretation of paediatric chest radiographs for the diagnosis of pneumonia in epidemiological studies. Bull World Health Organ. 2005;83: 353–359. doi:/S0042-96862005000500011. [PMC free article] [PubMed] [Google Scholar]
- 15. Howie SRC, Morris GAJ, Tokarz R, Ebruke BE, Machuka EM, Ideh RC, et al. Etiology of Severe Childhood Pneumonia in The Gambia, West Africa, Determined by Conventional and Molecular Microbiological Analyses of Lung and Pleural Aspirate Samples. Clin Infect Dis. 2014; ciu384. 10.1093/cid/ciu384 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Feikin DR, Njenga MK, Bigogo G, Aura B, Aol G, Audi A, et al. Viral and Bacterial Causes of Severe Acute Respiratory Illness Among Children Aged Less Than 5 Years in a High Malaria Prevalence Area of Western Kenya, 2007–2010: Pediatr Infect Dis J. 2013;32: e14–e19. 10.1097/INF.0b013e31826fd39b [DOI] [PubMed] [Google Scholar]
- 17. le Roux DM, Myer L, Nicol MP, Zar HJ. Incidence and severity of childhood pneumonia in the first year of life in a South African birth cohort: the Drakenstein Child Health Study. Lancet Glob Health. 2015;3: e95–e103. 10.1016/S2214-109X(14)70360-2 [DOI] [PubMed] [Google Scholar]
- 18. Ayieko P, Okiro EA, Edwards T, Nyamai R, English M. Variations in mortality in children admitted with pneumonia to Kenyan hospitals. PloS One. 2012;7: e47622 10.1371/journal.pone.0047622 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Ramachandran P, Nedunchelian K, Vengatesan A, Suresh S. Risk factors for mortality in community acquired pneumonia among children aged 1–59 months admitted in a referral hospital. Indian Pediatr. 2012;49: 889–895. [DOI] [PubMed] [Google Scholar]
- 20. Campbell JD, Kotloff KL, Sow SO, Tapia M, Keita MM, Keita T, et al. Invasive pneumococcal infections among hospitalized children in Bamako, Mali. Pediatr Infect Dis J. 2004;23: 642–649. [DOI] [PubMed] [Google Scholar]
- 21. Donkor ES. Molecular typing of the pneumococcus and its application in epidemiology in sub-Saharan Africa. Front Cell Infect Microbiol. 2013;3: 12 10.3389/fcimb.2013.00012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Centers for Disease Control and Prevention (CDC). Licensure of a 13-valent pneumococcal conjugate vaccine (PCV13) and recommendations for use among children—Advisory Committee on Immunization Practices (ACIP), 2010. MMWR Morb Mortal Wkly Rep. 2010;59: 258–261. [PubMed] [Google Scholar]
- 23. Chaïbou Y, Sanou I, Congo-Ouedraogo M, Kienou MC, Ouattara K, Somlaré H, et al. Streptococcus pneumoniae invasive infections in Burkina Faso, 2007 to 2011. Médecine Mal Infect. 2014;44: 117–122. 10.1016/j.medmal.2014.01.014 [DOI] [PubMed] [Google Scholar]
- 24. Self WH, Williams DJ, Zhu Y, Ampofo K, Pavia AT, Chappell JD, et al. Respiratory Viral Detection in Children and Adults: Comparing Asymptomatic Controls and Patients With Community-Acquired Pneumonia. J Infect Dis. 2015; jiv323. 10.1093/infdis/jiv323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Anderson LJ, Dormitzer PR, Nokes DJ, Rappuoli R, Roca A, Graham BS. Strategic priorities for respiratory syncytial virus (RSV) vaccine development. Vaccine. 2013;31, Supplement 2: B209–B215. 10.1016/j.vaccine.2012.11.106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Nair H, Brooks WA, Katz M, Roca A, Berkley JA, Madhi SA, et al. Global burden of respiratory infections due to seasonal influenza in young children: a systematic review and meta-analysis. The Lancet. 2011;378: 1917–1930. 10.1016/S0140 [DOI] [PubMed] [Google Scholar]
- 27.6736(11)61051-927. Jain S, Williams DJ, Arnold SR, Ampofo K, Bramley AM, Reed C, et al. Community-Acquired Pneumonia Requiring Hospitalization among U.S. Children. N Engl J Med. 2015;372: 835–845. 10.1056/NEJMoa1405870 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Louie JK, Roy-Burman A, Guardia-Labar L, Boston EJ, Kiang D, Padilla T, et al. Rhinovirus associated with severe lower respiratory tract infections in children. Pediatr Infect Dis J. 2009;28: 337–339. 10.1097/INF.0b013e31818ffc1b [DOI] [PubMed] [Google Scholar]
- 29. Bosch AATM, Biesbroek G, Trzcinski K, Sanders EAM, Bogaert D. Viral and Bacterial Interactions in the Upper Respiratory Tract. PLoS Pathog. 2013;9: e1003057 10.1371/journal.ppat.1003057 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Wurzel DF, Marchant JM, Clark JE, Mackay IM, Wang CYT, Sloots TP, et al. Respiratory virus detection in nasopharyngeal aspirate versus bronchoalveolar lavage is dependent on virus type in children with chronic respiratory symptoms. J Clin Virol Off Publ Pan Am Soc Clin Virol. 2013;58: 683–688. 10.1016/j.jcv.2013.09.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(DOC)
(XLS)
(XLS)
Data Availability Statement
All relevant data are within the manuscript and its Supporting Information files.