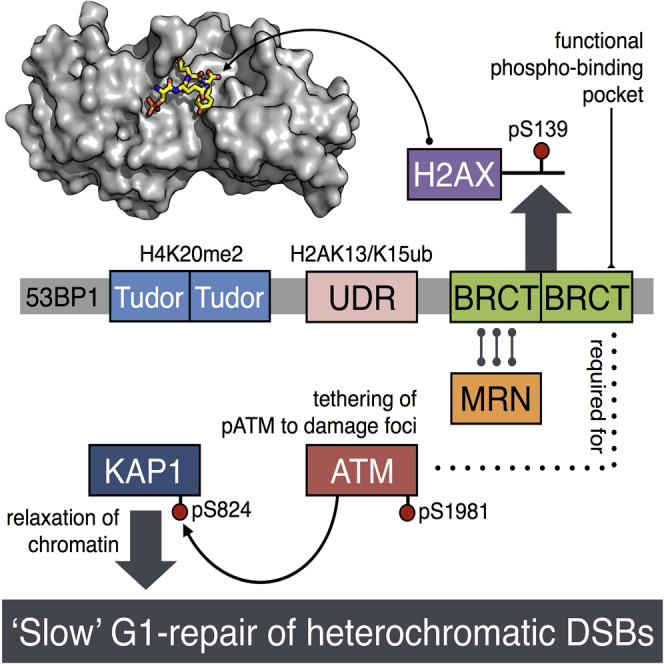
Summary
53BP1 plays multiple roles in mammalian DNA damage repair, mediating pathway choice and facilitating DNA double-strand break repair in heterochromatin. Although it possesses a C-terminal BRCT2 domain, commonly involved in phospho-peptide binding in other proteins, initial recruitment of 53BP1 to sites of DNA damage depends on interaction with histone post-translational modifications—H4K20me2 and H2AK13/K15ub—downstream of the early γH2AX phosphorylation mark of DNA damage. We now show that, contrary to current models, the 53BP1-BRCT2 domain binds γH2AX directly, providing a third post-translational mark regulating 53BP1 function. We find that the interaction of 53BP1 with γH2AX is required for sustaining the 53BP1-dependent focal concentration of activated ATM that facilitates repair of DNA double-strand breaks in heterochromatin in G1.
Graphical Abstract
Highlights
-
•
The BRCT2 domain of 53BP1 binds the DNA damage chromatin mark γH2AX
-
•
Crystal structure of γH2AX bound to 53BP1-BRCT2 reveals the basis of specificity
-
•
53BP1-BRCT2 responds to γH2AX formation by DNA damage in cells
-
•
Disruption of γH2AX binding disrupts pATM foci and DSB repair in heterochromatin
Baldock et al. find that the BRCT2 domain of 53BP1 specifically recognizes γH2AX, the primary chromatin mark at DNA double-strand breaks. Mutational disruption of this recognition in cells affects pATM recruitment into foci in G1 and results in a defect in repair of DNA damage in heterochromatin.
Introduction
TP53 binding protein 1 (53BP1) is a large multi-domain protein with multiple roles in the DNA damage response (Panier and Boulton, 2014, Zimmermann and de Lange, 2014). Following DNA damage and activation of the DNA-damage-responsive protein kinase ATM, 53BP1 is recruited rapidly to nuclear foci (Schultz et al., 2000) containing the primary mark of DNA damage—phosphorylation of Ser139 close to the C terminus of the histone H2A variant—H2AX (Rogakou et al., 1998), generally known as γH2AX. Although 53BP1 has a C-terminal tandem BRCT domain (BRCT2), which in its orthologs, Saccharomyces cerevisiae Rad9p and Schizosaccharomyces pombe Crb2, mediates binding to the equivalents of γH2AX (Hammet et al., 2007, Kilkenny et al., 2008), the role of the 53BP1-BRCT2 domain remains controversial. Although some studies indicated an interaction with γH2AX (Stewart et al., 2003, Ward et al., 2003), others have contradicted this (Stucki et al., 2005, Ward et al., 2006), and a significant role for this domain in the DNA damage response has been largely discounted (Bothmer et al., 2011, Callen et al., 2013).
Current models suggest that 53BP1 recruitment to ionizing radiation induced nuclear foci (IRIF) depends only indirectly on γH2AX and is instead mediated by two other post-translational modifications: (1) H2AK13/15-anchored ubiquitin chains (Fradet-Turcotte et al., 2013) generated by the E3 ubiquitin ligases RNF8 and RNF168, which are themselves recruited by MDC1, whose BRCT2 domain interaction with γH2AX is required for its own recruitment (Bekker-Jensen and Mailand, 2010, Pinder et al., 2013); and (2) direct interaction of the tandem Tudor domains of 53BP1 with dimethylated H4K20 (Botuyan et al., 2006) exposed by release of JMJD2A and L3MBTL1 following their ubiquitylation by RNF8 and RNF168 (Acs et al., 2011, Mallette et al., 2012).
We have re-examined the role of the 53BP1-BRCT2 domain and show unambiguously that it is a competent binding module for phosphorylated peptides with a clear specificity for the DNA-damage marker γH2AX, and in isolation from other parts of 53BP1 is sufficient for localization to sites of DNA damage in cells associated with γH2AX.
Structure-based mutational disruption of γH2AX binding by 53BP1 interferes with the 53BP1-dependent localization of pATM required for repair of DNA damage in regions of heterochromatin and results in a defect in the slow phase of DNA break repair in G1. These data add a third histone post-translational mark to the ligand repertoire of 53BP1, and a clear functional role for phosphopeptide binding by its BRCT2 domain.
Results and Discussion
53BP1-BRCT2 Binds γH2AX In Vitro
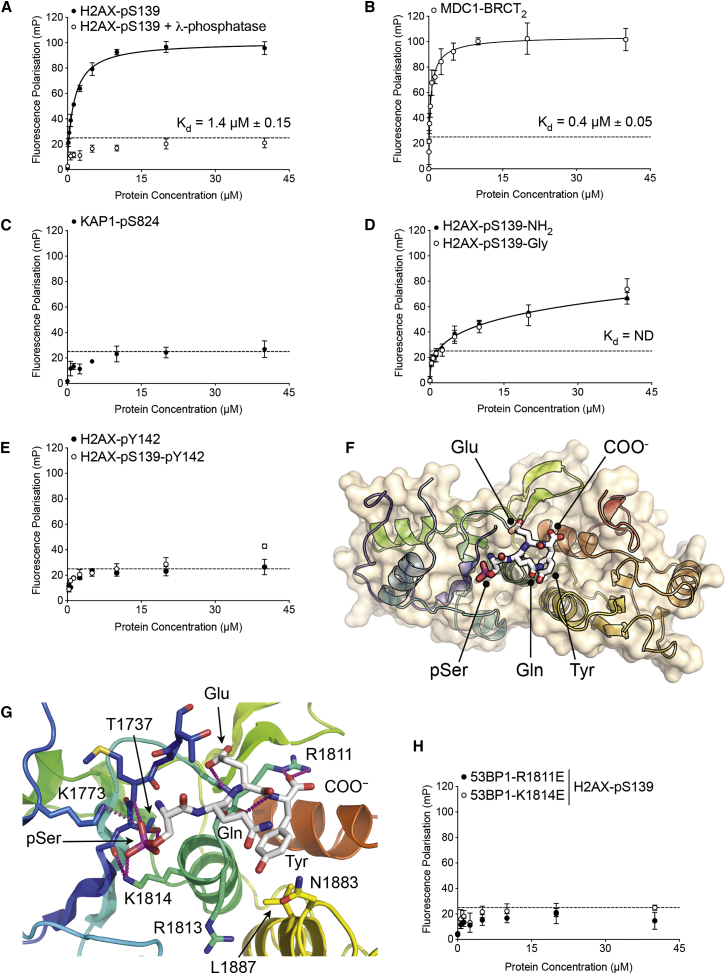
Comparison of the tandem BRCT domains of 53BP1 with those of MDC1 (Rodriguez et al., 2003, Stucki et al., 2005) and Crb2 (Kilkenny et al., 2008) shows strong conservation of residues implicated in specific binding of phosphorylated histone H2A tails. To determine whether 53BP1 shared this property, we measured the binding of the isolated 53BP1-BRCT2 segment to a fluorescently labeled phosphopeptide (fluorescein-SGGKKATQApSQEY) corresponding to the last 13 residues of human γH2AX, by fluorescence polarization (see Experimental Procedures). 53BP1-BRCT2 bound the phosphopeptide with a KD of ∼1.4 μM (Figure 1A), an affinity ∼3.5-fold lower than MDC1-BRCT2 (KD = ∼0.4 μM) when measured under the same experimental conditions (Figure 1B), but well within the range that is highly likely to be physiologically functional. The dephosphorylated H2AX peptide showed no significant binding (Figure 1A), nor did a comparable phosphopeptide from another protein (KAP1-pS824: fluorescein-GYG-SLPGAGLSpSQELSGG), showing the interaction is both phospho-dependent and sequence specific (Figure 1C).
Figure 1.
53BP1 BRCT2 Domain Binds γH2AX
(A) Fluorescence polarization assay (closed circle) showing binding of His6-SUMO-53BP1-BRCT2 to a fluorescein-tagged phosphopeptide (Flu-SGGKKATQApSQEY) corresponding to the last 13 residues of γH2AX. Data are means of four replicates with error bars showing 1 SD, and the KD (1.4 μM ± 0.15) was calculated by least-squares fitting of a one-site binding model. Removal of the phosphate group on Ser139 by addition of λ-phosphatase (open circle) abrogates the interaction. The dashed line indicates a signal level consistent with non-specific interaction.
(B) As (A) but for His6-SUMO-MDC1-BRCT2 binding to the fluorescein-tagged γH2AX phosphopeptide.
(C) As (A) but with a fluorescein-tagged phosphopeptide derived from the major ATM phosphorylation site on the heterochromatin protein KAP-1 (Noon et al., 2010). No binding was observed.
(D) As (A) but for γH2AX peptides in which the C-terminal α-carboxyl group of Tyr142 is modified by either amidation (closed circle) or addition of a further glycine residue (open circle). Both modifications substantially reduce the affinity, and no KD could be determined.
(E) As (A) but for H2AX peptides in which the side chain of Tyr142 is phosphorylated, either alone (closed circle) or in the presence of the γH2AX Ser139 phosphorylation (open circle). In both cases, phosphorylation of Tyr142 substantially diminished binding to His6-SUMO-53BP1-BRCT2.
(F) Overview of crystal structure of a phosphopeptide corresponding to the last four residues of γH2AX bound to the BRCT2 domain of 53BP1. The core DNA-binding domain of P53, which is required for crystallogenesis, makes no interaction with the γH2AX peptide and is omitted for clarity.
(G) Detail of interactions made by the tail of γH2AX with the 53BP1-BRCT2 domain. Carbon atoms in γH2AX are white; those of 53BP1 are rainbow colored to reflect their relative position within the protein sequence. The basic side chains of 53BP1 residues Lys1814 and Arg1811 provide neutralizing hydrogen bonding interactions with the acidic phosphate of γH2AX-pSer139 and the α-carboxyl of Tyr142, which are both required for binding of γH2AX to 53BP1-BRCT2; see (A) and (C).
(H) As (A) but showing binding of the fluorescent γH2AX peptide to His6-SUMO-53BP1-BRCT2 with either an R1811E mutation (closed circle) or a K1814E mutation (open circle).
Given that γH2AX is not the only phosphorylated ligand bound specifically by other BRCT2 domains, we further defined the parameters of phosphopeptide recognition by 53BP1-BRCT2. A γH2AX peptide with an amidated C terminus (H2AX-pS139-NH2 or with an additional glycine (H2AX-pS139-Gly) bound 53BP1-BRCT2 far less tightly than the peptide with the free charged α-carboxyl (Figure 1D). (Although the phosphorylated serine in human H2AX is encoded by codon 140 of the H2AFX gene, the initiator methionine is removed, and therefore not generally counted in the prevalent literature. For consistency, we here refer to this residue as Ser139.) We also explored the effect on binding to 53BP1-BRCT2 of phosphorylation of H2AX-Tyr142, believed to mediate a switch between DNA repair and apoptosis (Cook et al., 2009). An H2AX peptide phosphorylated on Tyr142 bound far less tightly than the γH2AX peptide, as did a peptide bis-phosphorylated on both Ser139 and Tyr142 (Figure 1E). These data reinforce the conclusion that the canonical DNA-damage-responsive mark γH2AX is a specific ligand of 53BP1-BRCT2.
To further characterize the interaction, we determined the crystal structure of 53BP1-BRCT2 in complex with the core DNA-binding domain of P53 and a short “pSQEY” peptide corresponding to the last four residues of γH2AX (Figure 1F; Table S1). The phosphopeptide bound in a similar conformation to its interaction with the BRCT2 domain of MDC1, with the phosphate group on γH2AX-Ser139 bound by the side chain of 53BP1-Thr1737, the peptide backbone of Met1738, and the side chains of Lys1773 and Lys1814 (Figure 1G). The side-chain carboxyl of γH2AX-Glu141 interacts with the peptide backbone of 53BP1-Arg1811, while the α-carboxyl of γH2AX-Tyr142 forms ionic and hydrogen bonding interactions with the side chain of 53BP1-Arg1811, explaining the strong preference for an unblocked C terminus in our binding assays (Figure 1D). The side chain of Tyr142 protrudes into a hydrophobic pocket lined by the side chains of Leu1887, Asn1883, and the main chain of Arg1813, an interaction that would be precluded by its phosphorylation. Consistent with the structure, charge reversal mutation of Lys1814, which interacts with the phosphate on H2AX-pSer139, or Arg1811, which binds the α-carboxyl of H2AX-Tyr142, abrogated interaction of 53BP1-BRCT2 with the γH2AX phosphopeptide (Figure 1H). As the BRCT2 domain of 53BP1 is a dimer in solution, like that of its yeast homolog Crb2 (Kilkenny et al., 2008), we confirmed that dimerization was not affected by the phospho-binding mutations (Figure S1).
53BP1-BRCT2 Binds γH2AX in Cells
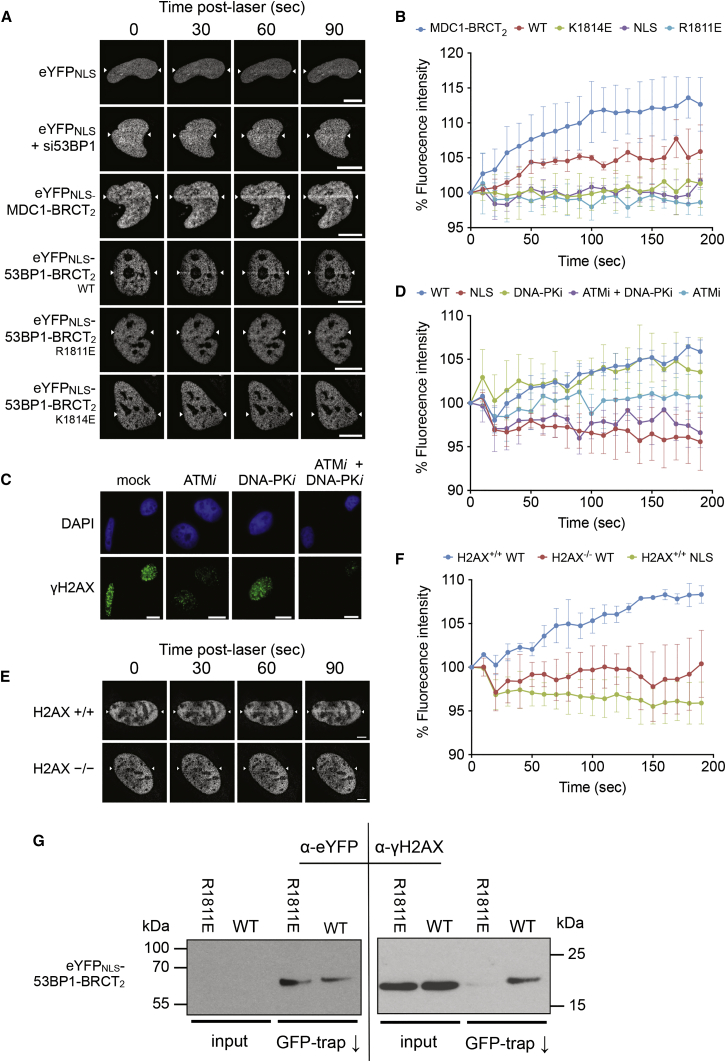
To determine whether the in vitro interaction that we observed for 53BP1-BRCT2 is reflected in its behavior in cells, we transfected HeLa cells with an eYFPNLS-53BP1-BRCT2 construct. Laser micro-irradiation of the nuclei of live transfected cells caused a distinct time-dependent accumulation of fluorescence along the laser track, consistent with recruitment of the tagged 53BP1-BRCT2 construct to sites of DNA damage; a construct lacking the BRCT2 segment or with mutations that abolish γH2AX binding in vitro did not (Figures 2A and 2B; Movies S1 and S2). Although it was not possible to directly image γH2AX formation in live HeLa cells, recruitment of MDC1-BRCT2 to the laser tracks confirms the presence of γH2AX (Movies S3), and clear γH2AX foci could be readily visualized in fixed cells following irradiation. The lower intensity of the recruited 53BP1-BRCT2 compared to MDC1-BRCT2 is consistent with the lower affinity of 53BP1-BRCT2 for γH2AX we measured in vitro. Pre-treatment of the cells with KU55933, a specific inhibitor of ATM, substantially diminished γH2AX focus formation on irradiation, and this was further decreased by a DNA-PK inhibitor NU7441 (Figure 2C). When HeLa cells expressing the eYFPNLS-53BP1-BRCT2 construct were similarly pre-treated, recruitment of fluorescence to the laser stripe was greatly diminished, and with addition of NU7441, effectively abolished (Figure 2D). Consistent with this, the eYFPNLS-53BP1-BRCT2 construct did not localize to sites of DNA damage in mouse embryonic fibroblasts lacking H2AX (Figures 2E, 2F, and S2). Finally, we immunoprecipitated the wild-type eYFPNLS-53BP1-BRCT2 construct from HeLa cell extracts and were able to detect a co-precipitating γH2AX signal in western blots that was greatly diminished in immunoprecipitates of eYFPNLS-53BP1-BRCT2 with the R1811E mutation that abrogates γH2AX binding in vitro (Figure 2G).
Figure 2.
53BP1 BRCT2 Domain Localizes to DNA Damage through Phospho-Specific Interactions
(A) HeLa cells were reverse transfected with 53BP1 siRNA. 48 hr later, cells were transfected with eYFP-SV40NLS or eYFP-SV40NLS-53BP1-BRCT2 constructs containing either wild-type or mutant BRCT domains, or eYFP-SV40NLS-MDC1-BRCT2. 16 hr after this cells were damaged using laser micro-irradiation along a line indicated by the white arrowheads, and images were recorded over 3 min. Scale bar, 10 μm.
(B) eYFP fluorescence was tracked over 3 min to observe localization. Profiles of fluorescence intensity along the laser track in (A) were generated using Slidebook6 software (n = 30 from three experimental repeats). Error bars, 1 SD.
(C) Cells were treated with either KU55933 (ATMi), NU7441 (DNA-PKi), or both for 1 hr prior to irradiation with 3 Gy of IR. Cells were incubated for a further 30 min before fixing and staining for γH2AX. Scale bar, 10 μm.
(D) Cells were processed as before with the addition of either DNA-PKi, ATMi, or both for 1 hr prior to laser micro-irradiation. eYFP-fluorescence was tracked over 3 min to observe localization. Fluorescence intensity profiles were generated using Slidebook6 software (n = 30 from three experimental repeats). Error bars, 1 SD.
(E) H2AX+/+ and H2AX−/− mouse embryonic fibroblasts were reverse transfected with 53BP1 siRNA. 24 hr later, cells were transfected with either the eYFP-SV40NLS-53BP1-BRCT2 construct or the eYFP-SV40NLS control. 16 hr after this, cells were damaged using laser micro-irradiation. eYFP fluorescence was tracked over 3 min to observe localization. Scale bar, 10 μm.
(F) Fluorescence intensity profiles from (E) were generated using Slidebook6 software (n = 30 from three experimental repeats). Error bars, 1 SD.
(G) eYFP-SV40NLS-53BP1-BRCT2 was immunoprecipitated using GFP-trap beads (ChromoTek), from benzonase-treated, γ-irradiated HeLa cell lysates, and eYFP-SV40NLS-53BP1-BRCT2 or γH2AX detected by western blot. Inputs represent 1/1,000 of the total sample used in the immunoprecipitation.
Taken together, these structural, biochemical, and cellular observations provide compelling evidence that the BRCT2 segment of 53BP1 has an inherent ability to localize to sites of DNA damage independently of other regions of the protein, and that this recruitment, which depends on phosphorylation of H2AX by DNA-damage-responsive PIKK kinases, is mediated by the clearly demonstrated ability of 53BP1-BRCT2 to bind γH2AX both specifically and directly.
Phospho-Binding by 53BP1-BRCT2 Regulates pATM Focal Localization
Having demonstrated the ability of 53BP1 to bind γH2AX directly, we sought to determine whether this ability contributed to any of the known roles of 53BP1 in the DNA damage response.
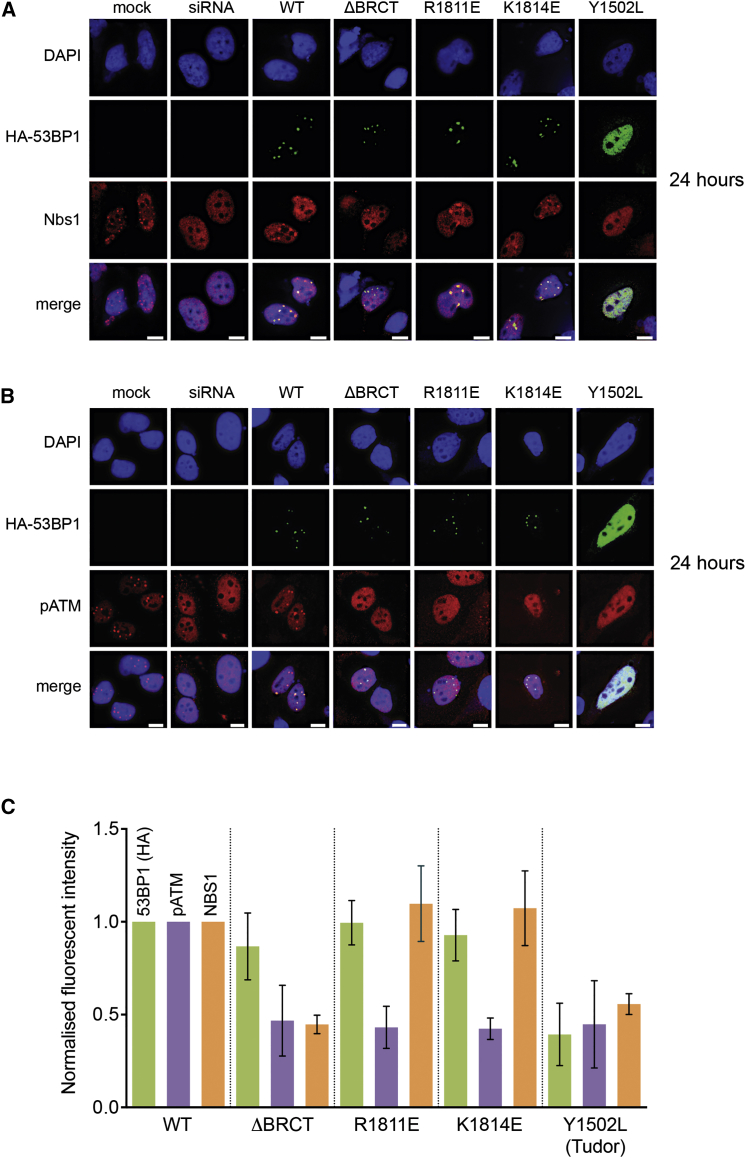
53BP1 is required for the focal localization of activated ATM, autophosphorylated on Ser1981 (Bakkenist and Kastan, 2003), and MRE11-RAD50-NBS1 (MRN) complexes at sites of DNA damage, and in the consequent activation of ATM checkpoint signaling (DiTullio et al., 2002, Mochan et al., 2004, Lee et al., 2010, Panier and Boulton, 2014). Consistent with this, small interfering RNA (siRNA) knockdown of 53BP1 in HeLa cells substantially diminished recruitment of activated pATM and MRN (visualized by NBS1), to nuclear ionizing radiation-induced foci (IRIF), with both of these proteins displaying a diffuse pan-nuclear distribution following irradiation (Figures 3 and S3).
Figure 3.
Co-localization of pATM Is Disrupted in 53BP1-BRCT2 Phospho-Binding Mutants
(A) HeLa cells were reverse transfected with 53BP1 siRNA. 48 hr later, cells were transfected with an siRNA-resistant construct containing wild-type HA-53BP1, one of the BRCT mutants, or a Tudor domain mutant (Y1502L). 16 hr after this, cells were exposed to either 8 Gy for mock and wild-type (WT) or 3 Gy of ionizing radiation for the mutants and allowed to recover for 24 hr before staining for HA and NBS1. Scale bar, 5 μm.
(B) As (A) but stained for pATM.
(C) Cells processed in (A) and (B) were analyzed by measuring foci intensity of HA, pATM, and NBS1 (n = 30). Foci intensities were normalized to the average wild-type foci intensity for their respective channel. Normalized HA foci intensities from cells transfected with either WT or BRCT mutants indicates comparable expression of the plasmid constructs. Graph shows average of three experiments with 1 SD.
Wild-type siRNA-resistant HA-tagged 53BP1 expressed in these knockdown cells, formed distinct foci itself following irradiation, and restored NBS1 and pATM foci that co-localized with those of 53BP1. However, a 53BP1 construct completely lacking the BRCT2 domain still formed foci of its own but failed to restore NBS1 and pATM foci. By contrast 53BP1 constructs in which the BRCT2 domain was present but contained mutations that disrupt binding to γH2AX (R1811E, K1814E) were able to recruit NBS1 into co-incident foci but had no effect on pATM, which retained the diffuse staining evident in the 53BP1 knockdown cells. A defect in pATM focus formation was evident with the K1814E mutant as early as 30 min post-irradiation (Figure S3B). A 53BP1 construct with a Tudor domain mutation (Y1502L) that abolishes recruitment to DNA damage (Huyen et al., 2004, Botuyan et al., 2006) eliminated 53BP1 foci and did not restore focal concentration of either NBS1 or pATM.
These data confirm previous observations that 53BP1 facilitates focal concentration of NBS1 (and thereby MRN) and pATM at DNA breaks in G1 but show that in both cases this is dependent on the BRCT2 domain, and that focal recruitment of pATM is specifically dependent on the ability of that domain to interact with γH2AX. These data also indicate a function for the 53BP1-BRCT2 in mediating an additional link between MRN complexes and chromatin modification, via its phosphorylation-independent interaction with RAD50 (Paull, 2015) and its phosphorylation-dependent interaction with γH2AX.
Phospho-Binding by 53BP1-BRCT2 Contributes to Heterochromatin Repair
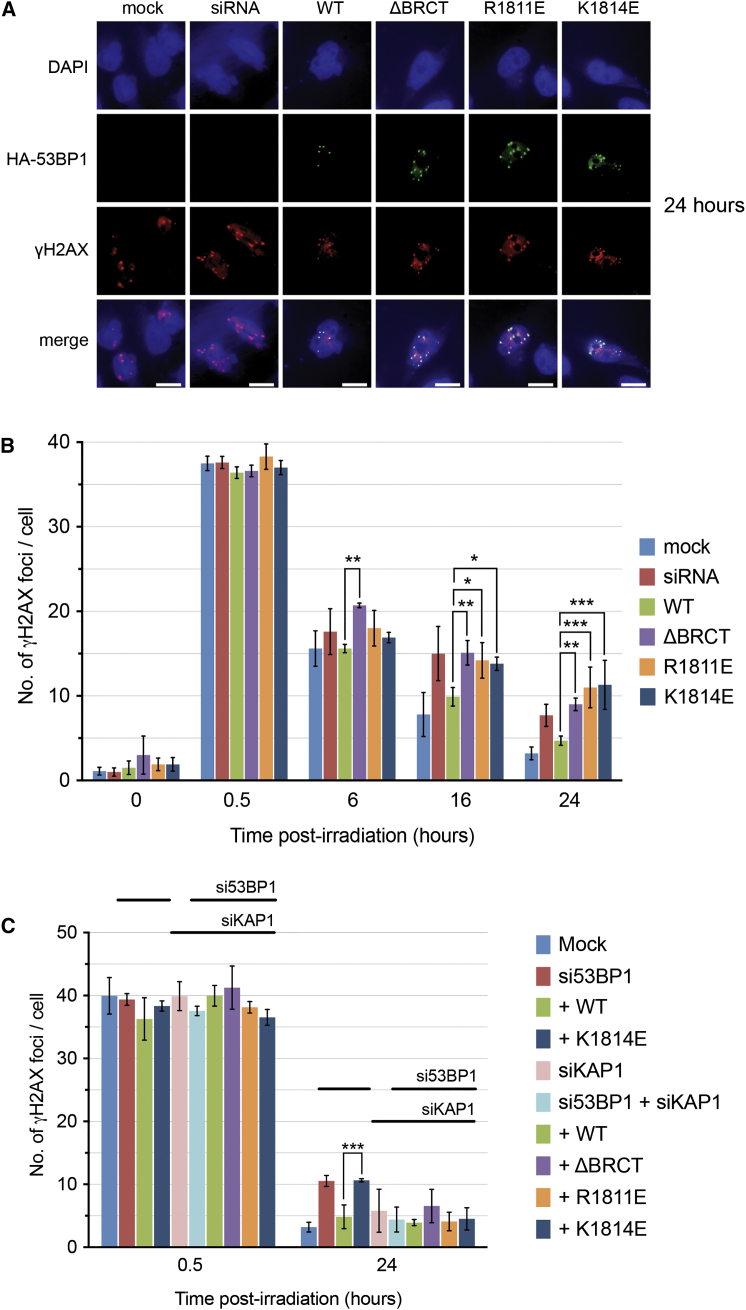
Double-strand break (DSB) repair in heterochromatin occurs more slowly than in euchromatin and requires 53BP1-dependent retention of pATM and consequent phosphorylation of the silencing factor KAP1/TRIM28 (Noon et al., 2010), which promotes its release with consequent relaxation of the heterochromatin. HeLa cells contact-inhibited in G0/G1, and transfected with 53BP1 siRNA, displayed significantly higher numbers of γH2AX foci 16–24 hr after irradiation than mock-transfected cells, or knockdown cells expressing a wild-type 53BP1 construct (Figure 4A). Consistent with their inability to localize pATM (Figure 3B), 53BP1 constructs lacking the BRCT2 domain or with γH2AX-binding defective mutations failed to rescue the knockdown phenotype and resulted in significantly increased levels of γH2AX foci at later time points (Figures 4B and S4). Simultaneous knockdown of KAP1, whose phosphorylation depends on pATM localization, reduced the number of persistent γH2AX foci seen with these mutants back down to wild-type levels (Figure 4C), confirming that these persistent foci were due to damage sites in heterochromatin (Goodarzi et al., 2008).
Figure 4.
Slow Repair of Damage in Heterochromatin Is Defective in 53BP1-BRCT2 Phospho-Binding Mutants
(A) HeLa cells were reverse transfected with 53BP1 siRNA. 48 hr later, cells were transfected with an siRNA-resistant construct containing wild-type HA-tagged 53BP1 or one of the BRCT mutants. 16 hr later, cells were exposed to 3 Gy of ionizing radiation and allowed to recover for the indicated time before staining for HA-53BP1 and γH2AX. Higher numbers of γH2AX foci were evident 24 hr after irradiation in siRNA-treated cells and siRNA-treated cells transfected with 53BP1 mutant constructs.
(B) Quantitation of γH2AX focus persistence. Graphs show the mean of three experiments; error bars, 1 SD. 53BP1 constructs lacking the BRCT domain or with point mutations that abrogate γH2AX binding in vitro show significantly higher levels of persistent γH2AX foci than wild-type (one-way ANOVA).
(C) As (A) but with 53BP1 and/or KAP-1 siRNA. Persistence of significantly higher levels of γH2AX foci in the presence of 53BP1 mutants is suppressed when KAP-1 is also knocked down, indicating that persistence is due to a defect in KAP-1 phosphorylation by ATM as a result of the failure of the 53BP1 mutants to localize pATM to heterochromatinized sites of damage.
Biological Roles of 53BP1 Binding to γH2AX
To date, BRCT2 domains of three mammalian proteins—MCPH1, PTIP, and MDC1—have been found to specifically recognize the primary mark of DNA damage, γH2AX (Yan et al., 2011, Stucki et al., 2005, Singh et al., 2012). Our data show unambiguously that this is a property shared by the BRCT2 domain of 53BP1, and that this interaction plays a role in the biological functions of 53BP1—a very similar conclusion was reached by Kleiner et al. (2015). This observation further strengthens the idea that 53BP1 is the functional ortholog of the fission yeast and budding yeast proteins Crb2 and Rad9p (Hammet et al., 2007, Kilkenny et al., 2008).
Although the ability of the BRCT2 domain of 53BP1 to bind γH2AX contributes to efficient repair of DNA damage, it is not required for initial recruitment of 53BP1 to IRIF in the immediate response to DNA damage in G1. 53BP1 recruitment is downstream of recognition of γH2AX by MDC1, whose BRCT2 domain has a higher affinity for γH2AX. Nonetheless, once recruited to damaged chromatin 53BP1’s interaction with γH2AX would be favored by its interaction with the other post-translational modifications it recognizes, and the γH2AX interaction might only become functionally significant at a later stage of the IRIF-centered repair processes or in specific situations such as the repair of damage in heterochromatin, as we show here.
Recruitment and retention of 53BP1 at sites of DNA damage facilitates co-localization and retention of other factors required for signaling and repair (Panier and Boulton, 2014). In the case of MRN, this is mediated by a phospho-independent interaction between RAD50 and the 53BP1-BRCT2 (Lee et al., 2010). Regions within the MRN complex have, in turn, been implicated in recruiting and activating ATM at sites of DNA damage (Paull, 2015), but it is not clear what role these play in retaining pATM in damage foci downstream of initial activation. Our data indicate that focal concentration and retention of pATM at slowly repaired sites of DNA damage in G1 (Noon et al., 2010) is independent of MRN recruitment but dependent on 53BP1 recruitment and, significantly, the histone modifications it engages with. Thus, while removal of the ability of 53BP1 to bind γH2AX does not affect focal co-localization of MRN and 53BP1, it substantially reduces pATM localization, which, in turn, results in a defect in DSB repair in heterochromatin. These data highlight a role for the 53BP1-BRCT2 domain in reinforcing links between MRN complexes and chromatin modification, via its phosphorylation-independent interaction with RAD50 (Noon et al., 2010) and its phosphorylation-dependent interaction with γH2AX.
The means by which 53BP1 facilitates pATM concentration and retention in IRIFs remains unclear. There is little indication in the literature of a direct interaction of ATM with 53BP1, although both proteins have well-documented interactions with other proteins in common. The dependence we demonstrate here on 53BP1’s ability to bind γH2AX suggests that this is likely to involve complex features of local chromatin conformation and histone modifications and further work will be required to define this. It is highly likely that other of the myriad functions of 53BP1 will also involve this hitherto disregarded property of 53BP1, and work is ongoing to uncover and characterize these.
Experimental Procedures
See the Supplemental Experimental Procedures for full details.
Protein Expression and Purification
Proteins for biochemical and structural analysis were expressed in E. coli and purified by affinity and conventional chromatography.
Fluorescence Polarization Experiments
Binding to BRCT2 domains was determined using fluorescein-labeled peptides and His6-SUMO-BRCT2 fusion proteins. Dissociation constants (KD) were determined by non-linear regression to a one site-specific binding model.
Crystallization, X-Ray Diffraction Data Collection, Phasing, Model Building, and Refinement
Crystals of P53Core/53BP1-BRCT2 were grown by vapor diffusion and soaked with the γH2AX-pS139 peptide (pSQEY) for 60 min prior to plunge-freezing in liquid nitrogen. Diffraction data were collected at the Diamond Synchrotron Lightsource, and the structure was determined by molecular replacement.
Tissue Culture, Cell Lines, and Reagents
HeLa, H2AX+/+, and H2AX−/− mouse embryonic fibroblast (MEF) cells were cultured in DMEM supplemented with 10% (v/v) fetal calf serum (FCS), penicillin, streptomycin, and L-glutamine. Caffeine (Sigma-Aldrich) was used at a final concentration of 10 mM. ATMi (KU55933, Abcam), DNA-PKi (NU7441, Santa Cruz Biotechnology), and ATRi (ATR-kinase inhibitor II, Merck Millipore) were all used at a concentration of 10 μM, except for the CHK1 inhibitor UCN-01 (Sigma-Aldrich) that was used at a concentration of 200 nM. Cells were irradiated using a Caesium137 gamma source.
siRNA Depletion of 53BP1/KAP-1 and siRNA-Resistant Expression of 53BP1
HeLa cells were seeded at high confluence onto 35-mm plates and reverse transfected with either 53BP1-siRNA or negative control oligonucleotides diluted into serum free media Cells were cultured for a further 48 hr for efficient depletion. Subsequently cells were transfected with 53BP1 constructs rendered siRNA-resistant by three silent point mutations in the 53BP1 cDNA clone in pCMH6K (A231G, A234G, and A237C) (Noon et al., 2010) and incubated for a further 16 hr before irradiation.
Antibodies
All antibodies used were from standard commercial sources (see the Supplemental Experimental Procedures for full details).
Immunofluorescence
Cells were fixed on coverslips and washed three times before incubation at room temperature with the primary antibodies. Cells were washed a further three times before incubation with the secondary, fluorophore-coupled, antibodies, and washed a final three times before mounting on glass slides with DAPI mounting media.
Live-Cell Imaging and UV-Laser Microirradiation
Cells were seeded at low confluence before 53BP1 depletion by siRNA, and eYFP-SV40NLS-53BP1-BRCT2 plasmid constructs were transfected into cells. Cells were incubated for 20 min with 100 μg/ml Hoechst 34580 prior to excitation with a 405-nm laser. Protein localization was tracked over 3 min using a 488-nm laser.
GFP-Trap
Stable cell lines were generated for HeLa cells transfected with wild-type or R1811E mutant forms of the eYFP-SV40NLS-53BP1-BRCT2 expression construct. Cellular lysates were generated by re-suspension of frozen pellets, and incubated with GFP-Trap A resin. Retained protein was detected by chemiluminescent western blot.
Author Contributions
Conceptualization: A.W.O., F.Z.W., and L.H.P.; Methodology: P.A.J., A.W.O., F.Z.W., and L.H.P.; Investigation: R.A.B., M.D., O.J.W., R.C., and A.W.O.; Writing–Original Draft: L.H.P.; Writing–Review & Editing: R.A.B., M.D., P.A.J., A.W.O., F.Z.W., and L.H.P.; Visualization R.A.B., A.W.O., and L.H.P.; Supervision: A.W.O., F.Z.W., and L.H.P.; Funding Acquisition: F.Z.W. and L.H.P.
Acknowledgments
We thank Mark Roe for assistance with X-ray data collection, Stuart Rulten for expertise with UVA laser microirradiation experiments, Lihong Zhou for assistance with cell culture, and Tony Carr and Jessica Downs for useful discussion. We thank Diamond Light Source, Didcot, for access to synchrotron radiation and the Wellcome Trust for support for X-ray diffraction facilities at the University of Sussex. This work was supported by Cancer Research UK Project Grant C1206/A11978 (F.Z.W. and L.H.P.) and Cancer Research UK Programme Grant C302/A14532 (L.H.P. and A.W.O.).
Published: November 25, 2015
Footnotes
This is an open access article under the CC BY-NC-ND license (http://creativecommons.org/licenses/by-nc-nd/4.0/).
Supplemental Information includes Supplemental Experimental Procedures, four figures, one table, and three movies and can be found with this article online at http://dx.doi.org/10.1016/j.celrep.2015.10.074.
Contributor Information
Felicity Z. Watts, Email: f.z.watts@sussex.ac.uk.
Laurence H. Pearl, Email: laurence.pearl@sussex.ac.uk.
Accession Numbers
The accession number for the crystallographic data reported here is PDB: 5ECG.
Supplemental Information
Example time-lapse of fluorescence microscope field of HeLa cells showing of eyfp-mdc1-brct2 recruitment to laser stripe damage, positive control for formation of DNA double-strand breaks and γH2AX by laser microirradiation. Crosshairs mark start and end of laser path(s).
As for Movie S1 but for eYFP-NLS-53BP1-BRCT2 showing wild-type BRCT2 mediates recruitment to DNA damage.
As for Movie S1 but for eYFP-NLS-53BP1-BRCT2 with K1814E mutation, which abrogates interaction with γH2AX and prevents recruitment to laser stripe.
References
- Acs K., Luijsterburg M.S., Ackermann L., Salomons F.A., Hoppe T., Dantuma N.P. The AAA-ATPase VCP/p97 promotes 53BP1 recruitment by removing L3MBTL1 from DNA double-strand breaks. Nat. Struct. Mol. Biol. 2011;18:1345–1350. doi: 10.1038/nsmb.2188. [DOI] [PubMed] [Google Scholar]
- Bakkenist C.J., Kastan M.B. DNA damage activates ATM through intermolecular autophosphorylation and dimer dissociation. Nature. 2003;421:499–506. doi: 10.1038/nature01368. [DOI] [PubMed] [Google Scholar]
- Bekker-Jensen S., Mailand N. Assembly and function of DNA double-strand break repair foci in mammalian cells. DNA Repair (Amst.) 2010;9:1219–1228. doi: 10.1016/j.dnarep.2010.09.010. [DOI] [PubMed] [Google Scholar]
- Bothmer A., Robbiani D.F., Di Virgilio M., Bunting S.F., Klein I.A., Feldhahn N., Barlow J., Chen H.T., Bosque D., Callen E. Regulation of DNA end joining, resection, and immunoglobulin class switch recombination by 53BP1. Mol. Cell. 2011;42:319–329. doi: 10.1016/j.molcel.2011.03.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Botuyan M.V., Lee J., Ward I.M., Kim J.E., Thompson J.R., Chen J., Mer G. Structural basis for the methylation state-specific recognition of histone H4-K20 by 53BP1 and Crb2 in DNA repair. Cell. 2006;127:1361–1373. doi: 10.1016/j.cell.2006.10.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Callen E., Di Virgilio M., Kruhlak M.J., Nieto-Soler M., Wong N., Chen H.T., Faryabi R.B., Polato F., Santos M., Starnes L.M. 53BP1 mediates productive and mutagenic DNA repair through distinct phosphoprotein interactions. Cell. 2013;153:1266–1280. doi: 10.1016/j.cell.2013.05.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cook P.J., Ju B.G., Telese F., Wang X., Glass C.K., Rosenfeld M.G. Tyrosine dephosphorylation of H2AX modulates apoptosis and survival decisions. Nature. 2009;458:591–596. doi: 10.1038/nature07849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DiTullio R.A., Jr., Mochan T.A., Venere M., Bartkova J., Sehested M., Bartek J., Halazonetis T.D. 53BP1 functions in an ATM-dependent checkpoint pathway that is constitutively activated in human cancer. Nat. Cell Biol. 2002;4:998–1002. doi: 10.1038/ncb892. [DOI] [PubMed] [Google Scholar]
- Fradet-Turcotte A., Canny M.D., Escribano-Díaz C., Orthwein A., Leung C.C., Huang H., Landry M.C., Kitevski-LeBlanc J., Noordermeer S.M., Sicheri F., Durocher D. 53BP1 is a reader of the DNA-damage-induced H2A Lys 15 ubiquitin mark. Nature. 2013;499:50–54. doi: 10.1038/nature12318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodarzi A.A., Noon A.T., Deckbar D., Ziv Y., Shiloh Y., Löbrich M., Jeggo P.A. ATM signaling facilitates repair of DNA double-strand breaks associated with heterochromatin. Mol. Cell. 2008;31:167–177. doi: 10.1016/j.molcel.2008.05.017. [DOI] [PubMed] [Google Scholar]
- Hammet A., Magill C., Heierhorst J., Jackson S.P. Rad9 BRCT domain interaction with phosphorylated H2AX regulates the G1 checkpoint in budding yeast. EMBO Rep. 2007;8:851–857. doi: 10.1038/sj.embor.7401036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huyen Y., Zgheib O., Ditullio R.A., Jr., Gorgoulis V.G., Zacharatos P., Petty T.J., Sheston E.A., Mellert H.S., Stavridi E.S., Halazonetis T.D. Methylated lysine 79 of histone H3 targets 53BP1 to DNA double-strand breaks. Nature. 2004;432:406–411. doi: 10.1038/nature03114. [DOI] [PubMed] [Google Scholar]
- Kilkenny M.L., Doré A.S., Roe S.M., Nestoras K., Ho J.C., Watts F.Z., Pearl L.H. Structural and functional analysis of the Crb2-BRCT2 domain reveals distinct roles in checkpoint signaling and DNA damage repair. Genes Dev. 2008;22:2034–2047. doi: 10.1101/gad.472808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kleiner R.E., Verma P., Molloy K.R., Chait B.T., Kapoor T.M. Chemical proteomics reveals a γH2AX-53BP1 interaction in the DNA damage response. Nat. Chem. Biol. 2015;11:807–814. doi: 10.1038/nchembio.1908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee J.H., Goodarzi A.A., Jeggo P.A., Paull T.T. 53BP1 promotes ATM activity through direct interactions with the MRN complex. EMBO J. 2010;29:574–585. doi: 10.1038/emboj.2009.372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mallette F.A., Mattiroli F., Cui G., Young L.C., Hendzel M.J., Mer G., Sixma T.K., Richard S. RNF8- and RNF168-dependent degradation of KDM4A/JMJD2A triggers 53BP1 recruitment to DNA damage sites. EMBO J. 2012;31:1865–1878. doi: 10.1038/emboj.2012.47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mochan T.A., Venere M., DiTullio R.A., Jr., Halazonetis T.D. 53BP1, an activator of ATM in response to DNA damage. DNA Repair (Amst.) 2004;3:945–952. doi: 10.1016/j.dnarep.2004.03.017. [DOI] [PubMed] [Google Scholar]
- Noon A.T., Shibata A., Rief N., Löbrich M., Stewart G.S., Jeggo P.A., Goodarzi A.A. 53BP1-dependent robust localized KAP-1 phosphorylation is essential for heterochromatic DNA double-strand break repair. Nat. Cell Biol. 2010;12:177–184. doi: 10.1038/ncb2017. [DOI] [PubMed] [Google Scholar]
- Panier S., Boulton S.J. Double-strand break repair: 53BP1 comes into focus. Nat. Rev. Mol. Cell Biol. 2014;15:7–18. doi: 10.1038/nrm3719. [DOI] [PubMed] [Google Scholar]
- Paull T.T. Mechanisms of ATM Activation. Annu. Rev. Biochem. 2015;84:711–738. doi: 10.1146/annurev-biochem-060614-034335. [DOI] [PubMed] [Google Scholar]
- Pinder J.B., Attwood K.M., Dellaire G. Reading, writing, and repair: the role of ubiquitin and the ubiquitin-like proteins in DNA damage signaling and repair. Front. Genet. 2013;4:45. doi: 10.3389/fgene.2013.00045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez M., Yu X., Chen J., Songyang Z. Phosphopeptide binding specificities of BRCA1 COOH-terminal (BRCT) domains. J. Biol. Chem. 2003;278:52914–52918. doi: 10.1074/jbc.C300407200. [DOI] [PubMed] [Google Scholar]
- Rogakou E.P., Pilch D.R., Orr A.H., Ivanova V.S., Bonner W.M. DNA double-stranded breaks induce histone H2AX phosphorylation on serine 139. J. Biol. Chem. 1998;273:5858–5868. doi: 10.1074/jbc.273.10.5858. [DOI] [PubMed] [Google Scholar]
- Schultz L.B., Chehab N.H., Malikzay A., Halazonetis T.D. p53 binding protein 1 (53BP1) is an early participant in the cellular response to DNA double-strand breaks. J. Cell Biol. 2000;151:1381–1390. doi: 10.1083/jcb.151.7.1381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh N., Basnet H., Wiltshire T.D., Mohammad D.H., Thompson J.R., Héroux A., Botuyan M.V., Yaffe M.B., Couch F.J., Rosenfeld M.G., Mer G. Dual recognition of phosphoserine and phosphotyrosine in histone variant H2A.X by DNA damage response protein MCPH1. Proc. Natl. Acad. Sci. USA. 2012;109:14381–14386. doi: 10.1073/pnas.1212366109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stewart G.S., Wang B., Bignell C.R., Taylor A.M., Elledge S.J. MDC1 is a mediator of the mammalian DNA damage checkpoint. Nature. 2003;421:961–966. doi: 10.1038/nature01446. [DOI] [PubMed] [Google Scholar]
- Stucki M., Clapperton J.A., Mohammad D., Yaffe M.B., Smerdon S.J., Jackson S.P. MDC1 directly binds phosphorylated histone H2AX to regulate cellular responses to DNA double-strand breaks. Cell. 2005;123:1213–1226. doi: 10.1016/j.cell.2005.09.038. [DOI] [PubMed] [Google Scholar]
- Ward I.M., Minn K., Jorda K.G., Chen J. Accumulation of checkpoint protein 53BP1 at DNA breaks involves its binding to phosphorylated histone H2AX. J. Biol. Chem. 2003;278:19579–19582. doi: 10.1074/jbc.C300117200. [DOI] [PubMed] [Google Scholar]
- Ward I., Kim J.E., Minn K., Chini C.C., Mer G., Chen J. The tandem BRCT domain of 53BP1 is not required for its repair function. J. Biol. Chem. 2006;281:38472–38477. doi: 10.1074/jbc.M607577200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan W., Shao Z., Li F., Niu L., Shi Y., Teng M., Li X. Structural basis of γH2AX recognition by human PTIP BRCT5-BRCT6 domains in the DNA damage response pathway. FEBS Lett. 2011;585:3874–3879. doi: 10.1016/j.febslet.2011.10.045. [DOI] [PubMed] [Google Scholar]
- Zimmermann M., de Lange T. 53BP1: pro choice in DNA repair. Trends Cell Biol. 2014;24:108–117. doi: 10.1016/j.tcb.2013.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Example time-lapse of fluorescence microscope field of HeLa cells showing of eyfp-mdc1-brct2 recruitment to laser stripe damage, positive control for formation of DNA double-strand breaks and γH2AX by laser microirradiation. Crosshairs mark start and end of laser path(s).
As for Movie S1 but for eYFP-NLS-53BP1-BRCT2 showing wild-type BRCT2 mediates recruitment to DNA damage.
As for Movie S1 but for eYFP-NLS-53BP1-BRCT2 with K1814E mutation, which abrogates interaction with γH2AX and prevents recruitment to laser stripe.