Abstract
The epithelial-to-mesenchymal transition (EMT) is a cellular process that functions during embryonic development and tissue regeneration, thought to be aberrantly activated in epithelial-derived cancer and play an important role in the process of metastasis. The TGFβ signaling pathway is a key inducer of EMT and we have elucidated a post-transcriptional mechanism by which TGFβ modulates expression of select transcripts via the RNA binding protein hnRNP E1 during EMT. One such transcript inhibin βA is a member of the TGFβ superfamily. Here, we show by polysome profiling that inhibin βA is translationally regulated by TGFβ via hnRNP E1. TGFβ treatment or knockdown of hnRNP E1 relieves silencing of the inhibin βA transcript, resulting in increased protein expression and secreted levels of the inhibin βA homodimer, activin A. Our data indicates that the translational up-regulation of inhibin βA enhances the migration and invasion of cells that have undergone an EMT and promotes cancer progression in vivo.
Keywords: TGFβ-induced EMT, Activin A signaling, breast cancer, metastasis
Introduction
The epithelial-to-mesenchymal transition (EMT) is a cellular process that is thought to play a key role in the metastatic progression of epithelial-derived cancers (1). This transition is associated with reorganization of the actin cytoskeleton and microtubule network, a loss of cell-cell contacts and an increase in the migratory and invasive capabilities of transitioned cells (2). A key inducer of EMT is the TGFβ signaling pathway, which signals through the type II receptor TβRII and the type I receptor Alk5, activating the canonical Smad2/3 pathway and non-canonical pathways including AKT, ERK and p38 MAPK (3). The induction of EMT by TGFβ is associated with increased migration, invasion and acquisition of cancer stem cell properties that are linked to metastatic potential (2, 4). This process is regulated not only at the transcriptional level through factors such as Zeb1/2, Snail and Slug, but also post-transcriptionally (5).
Our laboratory has delineated a mechanism through which EMT is post-transcriptionally regulated by the RNA binding protein hnRNP E1 (6, 7). We have shown that hnRNP E1 binds to select mRNAs and silences their translation in the absence of TGFβ signaling. In a global unbiased approach to identify transcripts silenced by this mechanism, using polysome profiling of normal mouse mammary NMuMG cells, inhibin βA was identified as translationally regulated by TGFβ (8).
Inhibin βA forms the homodimer activin A, which is a member of the TGFβ superfamily. This ligand functions in diverse processes and was originally identified for its role in reproduction through the modulation of follicle stimulating hormone (FSH) levels (9). Activin A also functions during embryogenesis and in several differentiation processes including hematopoiesis and osteogenesis (10–12). Similar to TGFβ, activin A signals through the Smad2/3 pathway albeit through different receptors. Activin A interacts with the type II activin receptors, ActRIIA and ActRIIB, leading to the recruitment and activation of the type I receptor Alk4 and subsequent phosphorylation of Smad2/3. Smad4 associates with activated Smad2/3 forming a complex which translocates to the nucleus and activates transcription (13).
Similar to the role of TGFβ in tumorigenesis, activin A has been shown to exert both tumor suppressing and promoting effects. Activin A is reported to inhibit cancer cell proliferation in certain models and to promote cell migration, invasion and acquisition of cancer stem cell traits in others (14–16). In vivo, increased inhibin βA expression is observed in cancer tissue compared to normal tissue in several cancers, including breast (17–19). Furthermore, inhibin βA is overexpressed in cancers with a propensity to metastasize with increased levels of secreted activin A observed in cancer patients with metastasis of the bone (20–22), indicating a cancer promoting effect of activin A in advanced stages of the disease.
In this study, we show that TGFβ treatment or knockdown of hnRNP E1 relieves silencing of the inhibin βA transcript, resulting in increased protein levels and secreted levels of the inhibin βA homodimer, activin A. Activin A treatment enhanced the invasion of TGFβ-treated NMuMG cells, while inhibin βA silencing inhibited NMuMG cell migration and invasion in vitro. In a xenograft mouse model, inhibin βA knockdown attenuated the metastatic progression of hnRNP E1 silenced cells. Thus, inhibin βA translational up-regulation induced by TGFβ or hnRNP E1 knockdown appears to promote the migratory and invasive phenotype observed in these cells.
Results
Inhibin βA is translationally regulated by TGFβ in a hnRNP E1 dependent manner
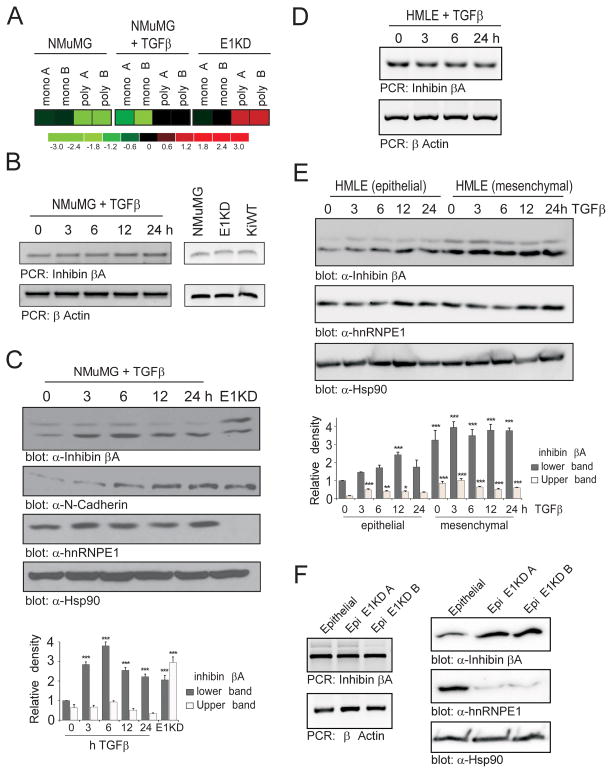
We have previously identified inhibin βA (InhβA) as a candidate target of the RNA binding protein hnRNP E1 through combined polysome profiling and Affymetrix array (Figure 1A). To validate these findings, transcript and protein expression levels were initially assessed in normal murine mammary gland epithelial cells (NMuMG), hnRNP E1 knockdown cells (E1KD) and also in the KiWT cell line (E1KD cells that express the hnRNP E1 open reading frame, not targeted by shRNA-mediated silencing). No change in inhibin βA mRNA was observed following TGFβ treatment in NMuMG cells, indicating that the inhibin βA transcript is not transcriptionally regulated by TGFβ (Figure 1B). In addition, no significant change in transcript was observed between the NMuMG, E1KD and KiWT cell lines (Figure 1B). In contrast, inhibin βA protein levels were significantly increased within 3 h of TGFβ treatment in NMuMG cells and were 2-fold higher in E1KD cells in the absence of TGFβ (Figure 1C). An antibody that detects the precursor form of inhibin βA, which has a molecular weight of 47kDa, was used in this study. The lower band in inhibin βA immunoblots corresponds to 47kDa and the upper band is believed to be the mono-glycosylated form of this precursor, which has been described previously (23).
Figure 1. Inhibin βA protein levels increase following TGFβ treatment or hnRNP E1 knockdown.
(a) Heatmap of Affymetrix array data showing expression levels (log2 fold) of the inhibin βA transcript in monosomal (mono) and polysomal (poly) fractions in duplicate samples from control and 24 h TGFβ-treated NMuMG cells and hnRNP E1 silenced NMuMG cells (E1KD). (b) Expression of inhibin βA and βActin (loading control) transcript following TGFβ treatment in the NMuMG cell line (left panel) and in NMuMG, E1KD and KiWT cells (right panel), as assessed by semi-quantitative PCR. (c) Immunoblot analysis of inhibin βA, N-Cadherin, hnRNP E1 and Hsp90 (loading control) protein levels in NMuMG cells treated with TGFβ compared to untreated E1KD cells. Graph illustrating relative density of bands observed in inhibin βA immunoblot compared to 0 h sample (bottom panel; n=3, ***P<0.001) (d) Expression of inhibin βA and βActin (loading control) transcript following TGFβ treatment in the HMLE cell line, as assessed by semi-quantitative PCR. (e) Immunoblot analysis of inhibin βA, hnRNP E1 and Hsp90 (loading control) protein levels in HMLE epithelial and mesenchymal cells treated with TGFβ. Graph illustrating relative density of bands observed in inhibin βA immunoblot compared to 0 h epithelial sample (bottom panel; n=3, *P<0.05, **P<0.01, ***P<0.001) (f) Inhibin βA and βActin (loading control) transcript expression in HMLE epithelial cells and epithelial E1KD cells (left panel). Immunoblot analysis of inhibin βA, hnRNP E1 and Hsp90 (loading control) protein levels in HMLE epithelial and Epi E1KD cells (right panel).
The regulation of inhibin βA by hnRNP E1 was also assessed in the human mammary epithelial HMLE cell line which consists of a mixed population of epithelial cells (CD44-low/CD24-high sorted cells) and a stably transitioned mesenchymal subpopulation (CD44-high/CD24-low sorted cells) (Supplementary Figure S1A). We hypothesized that inhibin βA would be translationally up-regulated following TGFβ treatment in the epithelial subpopulation. Consistent with our data using NMuMG cells, we observed no change in inhibin βA transcript levels (Figure 1D) and significantly higher protein levels in HMLE epithelial cells (Figure 1E) upon TGFβ treatment. Interestingly, higher levels of inhibin βA protein were observed in the mesenchymal subpopulation (Figure 1E). Next, we assessed the effect of hnRNP E1 knockdown on inhibin βA expression in the HMLE epithelial subpopulation. Similar to hnRNP E1 knockdown in NMuMG cells, silencing of hnRNP E1 led to an increase in inhibin βA protein levels whereas no change in transcript level was observed (Figure 1F).
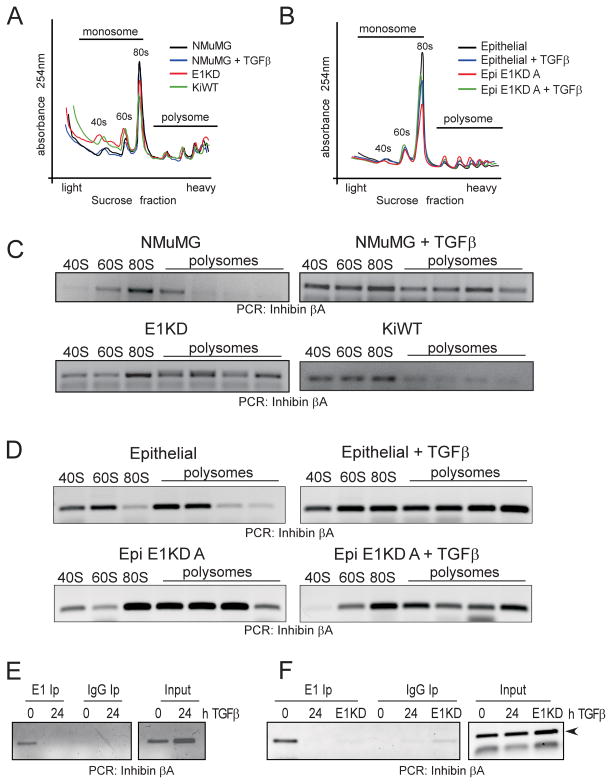
We utilized polysome profiling to confirm that increased inhibin βA protein levels were due to translational activation (Figure 2). Global translation levels were only modestly affected by either TGFβ treatment or hnRNP E1 knockdown in NMuMG cells (Figure 2A) and the HMLE epithelial subpopulation (Figure 2B) as assessed by quantitation of RNA concentration (abs 254nm) across polysome fractions. In contrast, polysomal analysis in the mesenchymal HMLE subpopulation indicated that the increase in inhibin βA protein levels observed in this subpopulation was due to an increase in global translation levels when compared to epithelial HMLE cells (Supplementary Figure S1B).
Figure 2. Inhibin βA is translationally up-regulated following TGFβ treatment in a hnRNP E1 dependent manner.
(a) Polysome profiling of control NMuMG cells, NMuMG cells treated for 24 h with TGFβ, E1KD and KiWT cells. Quantitation of RNA concentration (abs 254nm) across sucrose gradient fractions; transcripts associated with the 40S ribosomal subunit or 80S monosome are detected in lighter fractions, transcripts bound to multiple ribosomes sediment in denser sucrose fractions. (b) Polysome profiling of control and 24 h TGFβ-treated Epithelial HMLE and Epi E1KD A cells. (c) Semi-quantitative PCR analysis of inhibin βA transcript levels across monosomal (40, 60 and 80S) and polysomal fractions from control and 24 h TGFβ–treated NMuMG cells, E1KD and KiWT cells. (d) Semi-quantitative PCR analysis of inhibin βA transcript levels across monosomal (40, 60 and 80S) and polysomal fractions from control and 24 h TGFβ–treated Epithelial HMLE and Epi E1KD A cells. (e) RNA immunoprecipitation of inhibin βA transcript using anti-hnRNP E1 antibody or isotype control IgG in NMuMG cells either untreated or treated with TGFβ for 24 h. (f) RNA immunoprecipitation of inhibin βA transcript using anti-hnRNP E1 antibody or isotype control IgG in control or 24 h TGFβ-treated Epithelial HMLE cells and untreated Epi E1KD A cells.
Upon transcript-selective analysis, we observed a shift of the inhibin βA transcript from the monosomal to polysomal fractions upon TGFβ treatment of both NMuMG (Figure 2C) and HMLE epithelial (Figure 2D) cells, indicative of active translation. Inhibin βA transcript was present in denser polysomal fractions, irrespective of TGFβ treatment in hnRNP E1-silenced NMuMG cells (E1KD; Figure 2C) and hnRNP E1-silenced HMLE epithelial cells (Epi E1KD; Figure 2D). Additionally, rescue of hnRNP E1 silencing in the E1KD cells by expression of non-targeted human hnRNP E1 resulted in a shift of the inhibin βA transcript back to the monosomal fractions (KiWT; Figure 2C). Further, RNA immunoprecipitation analysis using anti-hnRNP E1 antibody demonstrates that in both NMuMG (Figure 2E) and HMLE (Figure 2F) cells, inhibin βA transcript associates with hnRNP E1 and this interaction is lost upon TGFβ treatment, indicating that direct interaction between hnRNP E1 and the inhibin βA transcript is required for translational regulation. Overall, these data indicate that the induction of inhibin βA following TGFβ treatment is mediated by hnRNP E1 which functions as a translational silencer in our model.
Secreted levels of Activin A are elevated following TGFβ treatment and hnRNP E1 knockdown
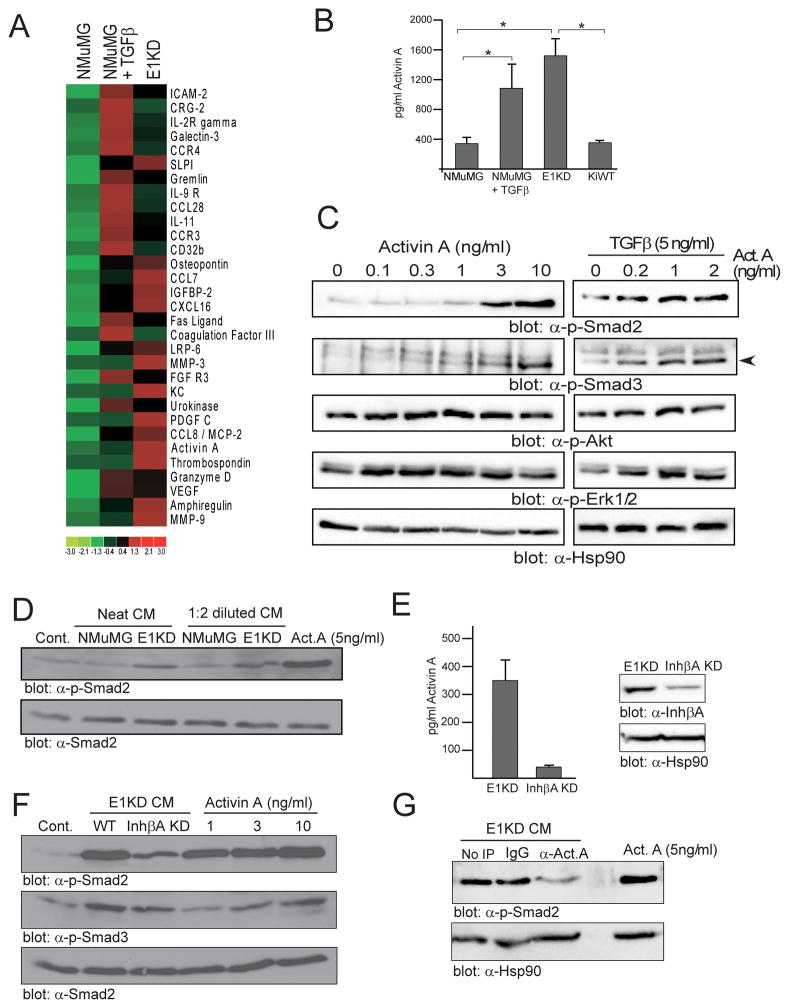
As inhibin βA expression is translationally up-regulated following TGFβ treatment, we next sought to characterize the levels of the secreted inhibin βA homodimer, activin A, as part of an array of growth factors, chemokines and cytokines (Figure 3A, Supplementary Figure S2A). Conditioned medium (CM) from control and TGFβ-treated NMuMG and E1KD cells was assayed in this experiment. Up-regulation of a number of factors, including activin A, VEGF, Osteopontin and the matrix metalloproteinases MMP-3 and MMP-9 was observed upon TGFβ treatment and hnRNP E1 silencing (Figure 3A). The up-regulation of activin A was confirmed by ELISA with increased levels observed following TGFβ treatment of NMuMG cells and in the E1KD cell line. Rescue of hnRNP E1 expression in the E1KD cell line resulted in a reduction in activin A levels (Figure 3B).
Figure 3. Secretion of the active inhibin βA homodimer activin A is increased following TGFβ treatment or hnRNP E1 knockdown.
(a) Heatmap of antibody array data (log2 fold expression) representing factors up-regulated at least 3 fold in the conditioned medium (CM) of TGFβ treated NMuMG and E1KD cells compared to control NMuMG cells. (b) Activin A ELISA of control and TGFβ–treated NMuMG cells, E1KD and KiWT cells (n≥3, *P<0.05). (c) Immunoblot of phospho-Smad2/3, phospho-AKT, phospho-ERK1/2 and Hsp90 (loading control) following treatment of NMuMG cells with 0–10 ng/ml activin A or 5ng/ml TGFβ combined with 0–2 ng/ml activin A for 30 min. (d) Immunoblot of phospho-Smad2 and total Smad 2 (loading control) levels in serum-starved NMuMG cells treated for 30 min with control DMEM, NMuMG or E1KD CM (neat or 1:2 diluted in DMEM) or 5 ng/ml recombinant activin A. (e) Activin A ELISA of CM from control and inhibin βA-silenced E1KD cells (left panel). Immunoblot of inhibin βA and Hsp90 (loading control) levels in control and inhibin βA-silenced E1KD cells (right panel). (f) Immunoblot of phospho-Smad2/3 and total Smad 2 (loading control) levels in serum-starved NMuMG cells treated for 30 min with DMEM (cont.), E1KD and inhibin βA-silenced E1KD CM or 1–10 ng/ml recombinant activin A. (g) Immunoblot of phospho-Smad2 and Hsp90 (loading control) levels in serum-starved NMuMG cells treated for 30 min with E1KD CM immunodepleted using anti-activin A (α-Act. A) antibody or an IgG control or treated with 5 ng/ml recombinant activin A.
To determine whether the secreted activin A was active, we first established in serum-starved NMuMG cells that recombinant activin A is capable of inducing phospho-Smad2/3 (Figure 3C), recombinant activin A treatment was also capable of further increasing phospho-Smad2/3 levels induced in TGFβ-treated cells (Figure 3C). Induction of the non-canonical MAPK pathway, as determined by ERK1/2 phosphorylation, was also observed in single and combination treatments, whereas the Akt pathway was not significantly activated (Figure 3C). Next the activity of E1KD CM, which contains significantly higher levels of activin A compared to NMuMG cells, was characterized. As depicted in Figure 3D, phospho-Smad2 levels were induced in serum-starved NMuMG cells upon addition of E1KD CM compared to NMuMG CM and to DMEM control. Recombinant activin A was used as a positive control in this experiment.
Next, the contribution of activin A to Smad2/3 activation was assessed. We silenced inhibin βA in E1KD cells resulting in a reduction in secreted levels of activin A (Figure 3E) and an attenuation of Smad2/3 activation when NMuMG cells were treated with CM from inhibin βA KD cells compared to E1KD CM (Figure 3F). We also immunodepleted activin A from E1KD CM (Supplementary Figure S2B) and similarly observed a reduction in Smad2 activation compared to IgG control CM (Figure 3G). These data indicate that the activin A ligand significantly contributes to Smad2/3 activation induced by E1KD CM.
Translational up-regulation of inhibin βA promotes cell migration and invasion following TGFβ treatment and hnRNP E1 silencing
We wished to elucidate the functional significance of inhibin βA up-regulation in response to TGFβ treatment. We hypothesized that inhibin βA may either promote EMT or function in promoting some aspect of the mesenchymal phenotype, such as migration or invasion. Previous studies have reported a lack of EMT induction in activin A treated NMuMG cells due to limited receptor levels, therefore, we initially characterized the expression of activin receptors in NMuMG cells, in addition to the effect of activin A and combined activin A/TGFβ treatment on EMT. The type II receptor ActRIIA and the type I receptor Alk4, which form heteromeric complexes in NMuMG cells (24), were detected in cell lysates with higher levels of the type II receptor observed in E1KD cells. An increase in the type I receptor Alk4 was observed within 3 h of TGFβ treatment, which returned to basal levels within 24 h of treatment. In contrast, expression of the type II receptor did not change upon TGFβ treatment (Supplementary Figure S3A&B).
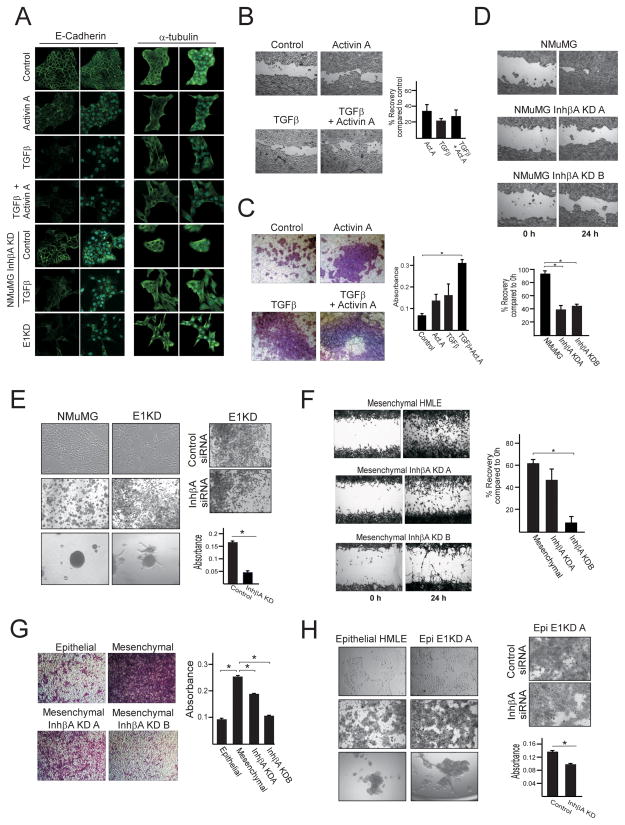
Morphologically, the transition to a more spindle-shaped, fibroblast-like cell that occurs during TGFβ–induced EMT was not observed following activin A exposure (Supplementary Figure S3C). A slight loss of the epithelial marker E-cadherin at the cell membrane was detected in activin A treated cells, however, no microtubule reorganization was observed as determined by α-tubulin immunofluorescence (Figure 4A). In contrast, TGFβ and combined TGFβ/activin A treatment resulted in complete loss of E-cadherin at the cell membrane and reorganization of the microtubule network. shRNA-mediated silencing of inhibin βA had a modest effect on TGFβ-induced EMT, as determine by partial loss of E-cadherin at the cell membrane following TGFβ treatment compared to control TGFβ-treated cells (Figure 4A). Finally, induction of the EMT-associated transcription factors Zeb1/2, the smad target GADD45b and the mesenchymal marker N-cadherin was not observed in activin A-treated cells compared to TGFβ-treated cells (Supplementary Figure S3D&E). These data indicate that exogenous activin A, or TGFβ-induced inhibin βA, is not sufficient to induce an EMT in NMuMG cells.
Figure 4. Inhibin βA enhances migration and invasion of TGFβ-treated and hnRNP E1 silenced mammary epithelial cells.
(a) Immunofluorescence analysis of E-cadherin and α-tubulin expression in NMuMG cells treated with TGFβ (5ng/ml), activin A (10ng/ml) or a combination of both for 48 h, inhibin βA-silenced NMuMG cells treated with TGFβ for 48 h and E1KD cells. All micrographs are 60× magnifications. Dual-colored micrographs (green/blue) represent merged Alexa Fluor 488 and Dapi images. (b) Wound healing assay demonstrating an increase in cell migration when NMuMG cells are pre-treated with TGFβ (5ng/ml), activin A (10ng/ml) or a combination of both for 72 h. Following reseeding of pre-treated cells in wells containing wound healing inserts and the formation of a confluent monolayer, inserts were removed and wound recovery was assayed 12 h later. Quantitation, compared to control, was performed using ImageJ software (right panel; n=7). (c) Invasion of NMuMG cells across matrigel-coated membranes was assayed in control (untreated) cells and cells pre-treated with TGFβ (5ng/ml), Activin A (10ng/ml) or combined TGFβ and activin A for 72 h. Quantitation of invasion was performed by measuring the absorbance of extracted cell stain from cells that invaded across matrigel membranes (right panel; n=3 *P<0.05). (d) Wound healing bright-field images and quantitation (% wound recovery; n=3 *P<0.05) using control and inhibin βA-silenced NMuMG cells. (e) Bright-field images of NMuMG and hnRNP E1-silenced NMuMG (E1KD) cell morphology (top left), stained cells that have invaded across matrigel-coated membranes (middle left) and spheres grown in invasion matrix (bottom left panel). Invasion of E1KD cells across matrigel-coated membranes is attenuated by inhibin βA silencing (right panel). Quantitation of invasion (right panel; n=3 *P<0.05). (f) Wound healing bright-field images and quantitation (% wound recovery; n=3 *P<0.05) using control and inhibin βA-silenced mesenchymal HMLE cells. (g) Bright-field images of epithelial HMLE cells and control or inhibin βA-silenced mesenchymal HMLE cells which have invaded across matrigel membranes (left panel), quantitation of invasion (right panel; n=3 *P<0.05). (h) Bright-field images of epithelial HMLE and hnRNP E1-silenced epithelial (Epi E1KD A) cell morphology (top left), stained cells that have invaded across matrigel membranes (middle left) and spheres grown in invasion matrix (bottom left panel). Invasion of Epi E1KD A cells across matrigel-coated membranes is attenuated by inhibin βA silencing (right panel). Quantitation of invasion (right panel; n=3 *P<0.05).
To investigate whether activin A promotes invasion and migration in control and TGFβ-treated cells we performed wound healing and invasion assays. The results demonstrate that treatment of NMuMG cells with either TGFβ or activin A promoted cell migration (Figure 4B) and invasion (Figure 4C). Combined treatment of activin A and TGFβ did not significantly alter cell migration, but did enhance cell invasion (Figure 4B&C). Furthermore, migration and invasion were significantly attenuated when inhibin βA was silenced using two different shRNAs in NMuMG cells (Figure 4D and Supplementary Figure S4). Additionally, in the spontaneously arising mesenchymal subpopulation of HMLE cells, which exhibits enhanced invasiveness compared to their epithelial counterparts (Figure 4G), silencing of inhibin βA attenuated migration and invasion (Figure 4F and 4G).
Our results demonstrate that knockdown of hnRNP E1 in both NMuMG (E1KD) and HMLE (Epi E1KD) cells results in the induction of inhibin βA (Figure 1) with concomitant altered cell morphology, loss of cell-cell contacts and increased motility and invasiveness (Figure 4E and 4H). To determine the contribution of inhibin βA induction to these cellular phenotypes, silencing of inhibin βA in both EIKD (Figure 4E) and Epi E1KD (Figure 4H) cells was performed resulting in a significant reduction in invasion. These data suggest that enhanced invasiveness observed following TGFβ treatment or hnRNP E1 knockdown are in part due to an up-regulation of inhibin βA.
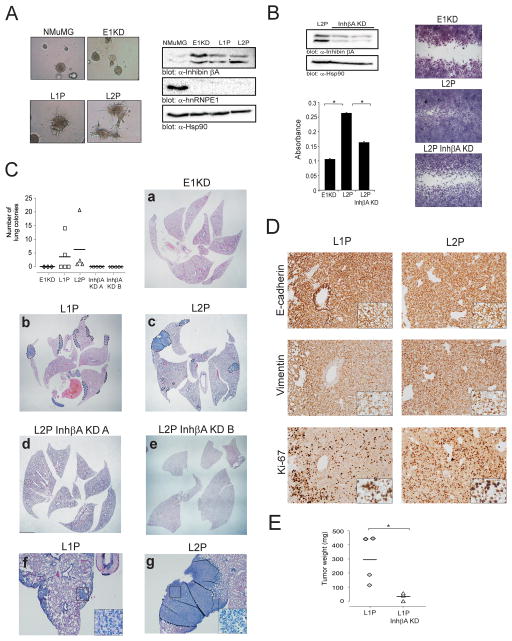
To establish the relative contribution of inhibin βA to the invasive and metastatic phenotype in vivo, we initially assessed tumor formation and lung colonization using control and inhibin βA-silenced E1KD cells (Figure 3E). E1KD cells formed primary tumors when injected into the mammary fat pad of NOD/SCID mice and formed lung colonies when tail vein injections were performed, albeit with low efficiency (data not shown). Due to the low number of metastatic outgrowths observed in E1KD-injected animals, we generated a more aggressive form of the E1KD cells using a process of in vivo selection. Briefly, lung metastases generated from mammary fat pad injection of E1KD cells were isolated and cells from these metastases were cultured in selective media generating the L1P cell line, the L2P cell line was isolated from the lungs metastases of L1P-injected mice. We hypothesized that if inhibin βA is necessary for the invasive phenotype of hnRNP E1 cells, silencing of inhibin βA in these serially selected cells that aggressively metastasize to lung would attenuate this invasive phenotype. As shown in Figure 5A, in vivo selected cells demonstrate enhanced invasive ability, are still silenced for hnRNP E1 and exhibit increased levels of inhibin βA compared to parental NMuMG cells.
Figure 5. Inhibin βA promotes cancer progression in hnRNP E1-silenced mammary epithelial cells.
(a) Bright-field images of NMuMG, E1KD, L1P and L2P spheres grown in invasion matrix (left panel). Immunoblot analysis of inhibin βA, hnRNP E1 and Hsp90 (loading control) protein levels in NMuMG, E1KD and in vivo passaged L1P and L2P cells (right panel). (b) Immunoblot of inhibin βA and Hsp90 (loading control) in control L2P and inhibin βA-silenced L2P cells. Graph showing quantitation of E1KD, control L2P and inhibin βA-silenced L2P invasion assay (n=3 *P<0.05). Bright-field images from a representative wound healing assay using E1KD, control L2P and inhibin βA-silenced L2P cells (right panel). (c) Quantitation of lung colonies observed in NOD/SCID mice tail vein injected with E1KD, L1P, control L2P and inhibin βA-silenced L2P cells. Lung colonies were independently scored by two investigators. Representative images of H&E stained lung tissue sections. Dashed lines delineate lung colonies. Images a-e are stitched from 3–4 micrographs (1.25×) using Microsoft ICE software (Microsoft, Redmond, WA). Images f and g are 4× magnifications, insets are 40× magnifications. (d) Immunohistochemical analyses of E-cadherin, Vimentin and Ki67 levels in L1P and L2P lung colonies. Images are 20× magnifications, insets are 40× magnifications. (e) Graph showing the tumor weight (mg) of mammary fat pad tumors in mice injected with either control L1P or inhibin βA-silenced L1P cells (*P<0.05).
In vitro, the increased migratory and invasive ability observed in L2P cells compared to E1KD cells was attenuated by inhibin βA silencing (L2P InhβA KD cells; Figure 5B). Consistent with these findings, tail vein injection of L1P and L2P cells resulted in an increase in lung colonization in NOD/SCID mice compared to E1KD cells, and silencing of inhibin βA in L2P cells (L2P InhβA KD) using two different shRNAs attenuated lung colonization (Figure 5C). Immunohistochemical analysis of the epithelial marker E-cadherin and the mesenchymal marker Vimentin in metastatic outgrowths revealed expression of both markers indicative of a partial reversal of the EMT phenotype, which has been described previously in metastases (25). The proliferative marker Ki67 was also analyzed with an increase in nuclear ki67 staining observed in L2P lung colonies compared to L1P colonies (Figure 5D). Inhibin βA knockdown also attenuated tumor growth in our model as inhibin βA silenced L1P cells (L1P InhβA KD) formed mammary fat pad tumors that were significantly smaller in size than control L1P tumors (Figure 5E).
Inhibin βA is up-regulated in breast cancer and is associated with poor prognosis
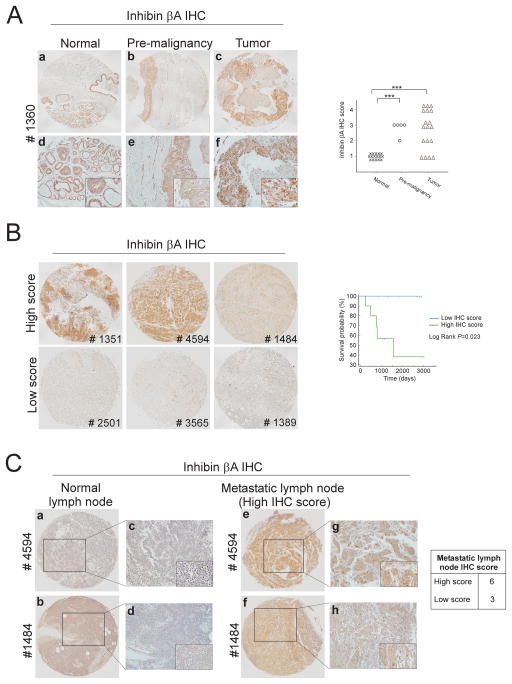
In order to correlate our findings with human breast cancer data, we assessed inhibin βA protein expression by immunohistochemistry in human breast tissue microarrays (TMAs) obtained from the Biorepository and Tissue Analysis core of MUSC (Supplementary Table S2). Inhibin βA expression was localized predominantly to the cytoplasm of cells and was detected in mammary gland epithelial cells of normal tissue biopsies (Figure 6A). A significant increase in inhibin βA expression was observed in pre-malignant and tumor tissue compared to normal breast tissue (Figure 6A). Furthermore, when tumor biopsies were divided into two groups based on inhibin βA expression levels, a significant correlation between overall survival and inhibin βA expression was observed, with high inhibin βA expression associated with poor prognosis (Figure 6B; P=0.023). No correlation between inhibin βA expression and tumor size or lymph node involvement was observed, however, high expression of inhibin βA was observed in 67% of metastatic lymph node biopsies (Figure 6C).
Figure 6.
(a) Representative images of inhibin βA immunohistochemistry (IHC) performed on normal, premalignant and tumor breast tissue microarrays. Images a-c are stitched from 2–4 micrographs (10×) using Microsoft ICE software. Images d-f are 20× magnifications, insets are 40× magnifications. Graph showing quantitation of inhibin βA staining in normal, pre-malignant and tumor breast tissue biopsies. The intensity and proportion of inhibin βA staining was scored on a scale of 1 (low/negative staining) to 4 (intense staining). Independent scoring by a second investigator was blinded to sample type and clinicopathological data. (***P<0.001). (b) Representative images of breast tumor specimens which were divided into high inhibin βA scoring biopsies (score= 3 or 4) or low scoring biopsies (score= 1 or 2). Kaplan–Meier survival analysis of low and high scoring biopsies (right panel; log rank P=0.023). (c) Immunohistochemical analyses of inhibin βA levels in normal and malignant lymph node biopsies. Images a, b, e and f are stitched from 2–4 micrographs (10×) using Microsoft ICE software. Images c, d, g and h are 20× magnifications, insets are 40× magnifications.
Discussion
During TGFβ-induced EMT, the transition to a mesenchymal phenotype requires not only TGFβ-induced transcriptional regulation, through factors such as Zeb1/2, but also coordinated post-transcriptional regulation, via microRNAs and RNA-binding proteins, such as hnRNP E1 (5, 6). We have shown previously that hnRNP E1 is a key regulator of TGFβ-induced EMT. Silencing of this protein induces a transition of epithelial cells to a mesenchymal phenotype, resulting in enhanced migration, invasion and tumorigenesis (6, 7). Here we show that the inhibin βA transcript is translationally regulated by hnRNP E1 during TGFβ-induced EMT and contributes to the enhanced migration and invasion observed in transitioned cells.
Transcriptional regulation of the inhibin βA gene has been reported previously, with IL-1β, NFκB and MAPK signaling being linked to this regulation (26, 27). To our knowledge, this is the first study demonstrating translational regulation of inhibin βA. An up-regulation of inhibin βA protein occurs within 3 h of TGFβ treatment with an increase in secreted levels of the inhibin βA homodimer, activin A, observed within 24 h. Polysome profiling in NMuMG and HMLE cells confirmed translational activation of inhibin βA following TGFβ treatment or hnRNP E1 silencing. Furthermore, the enhancement of inhibin βA translation coincided with the release of the RNA binding protein from the transcript. This finding is consistent with our previous data which shows loss of interaction of hnRNP E1 with ILEI and Dab2 mRNA following TGFβ treatment, two transcripts that are translationally regulated by this RNA binding protein (6). Further work is required to characterize the cis-element in the inhibin βA transcript through which hnRNP E1 interacts.
The functional significance of inhibin βA up-regulation was assessed in NMuMG and HMLE cell lines. Activin A alone was not capable of inducing an EMT nor did this ligand appear to enhance the transition induced by TGFβ. These findings are consistent with previous studies which report no induction of EMT in NMuMG cells treated with activin A (24, 28). This lack of induction is thought to be due to limited receptor levels and weak Smad activation, as expression of constitutively active ALK4 induces an EMT (28). The ability of activin A signaling to alter cell migration and invasion but not EMT in our model may be due to the degree or duration of Smad2/3 activation. TGFβ maintained Smad2 activation throughout a 24 h time course, whereas activin A induced phosphorylated Smad2 to a lesser degree and this activation was lost 12 h following treatment. During embryonic development, Smad2 binds to and activates transcription of target gene subsets in a dose-dependent manner (29), it is possible that a certain threshold of Smad activation is also required in our model to promote transcription of genes necessary for EMT induction.
The ability of activin A signaling to alter cell migration and invasion may also be due to non-canonical pathways, such as the MAPK pathway. Induction of the MAPK pathway has been linked previously to enhanced cell migration and invasion in TGFβ-treated epithelial cells (30), and we observed sustained ERK1/2 phosphorylation in activin A treated cells comparable to levels observed in TGFβ treated cells (Supplementary Figure 3F). Thus, non-canonical Activin signaling may play an important role in the invasive phenotype.
In vivo, inhibin βA appears to promote cancer progression as silencing of inhibin βA in our model resulted in an attenuation of xenograft cancer progression (Figure 5C-E). Consistent with these findings, analysis of secreted activin A levels from the 4T1 progression model revealed significantly elevated levels of activin A in the metastatic 4T1 cell line compared to non-invasive 67NR cells and invasive but non-metastatic 4T07 cells (Supplementary Figure S5). These data are consistent with our immunohistochemical analysis of inhibin βA levels in human breast tissue in which inhibin βA protein levels were up-regulated in tumor compared to normal breast tissue. In addition, inhibin βA expression was significantly higher in premalignant biopsies, indicating that up-regulation of inhibin βA is an early event in cancer progression. We found that high levels of inhibin βA in tumor specimens were associated with poor prognosis. This is consistent with previous studies which report an association between inhibin βA protein levels and overall survival in patients with lung and gastric cancer (19, 31).
In summary, we have shown that inhibin βA is translationally repressed by the RNA binding protein hnRNP E1. During TGFβ-induced EMT, we observe a loss of interaction between hnRNP E1 and the inhibin βA transcript, concomitant with an up-regulation of inhibin βA translation, which contributes to the enhanced invasive phenotype observed in transitioned cells. Future research will be necessary to characterize this regulatory mechanism in processes, such as development, inflammation and immunity, in which inhibin βA functions.
Materials and Methods
Cell lines and treatments
NMuMG cells, obtained from the American Type Culture Collection (ATCC, Manassas, VA, USA), and the 4T1 progression model, acquired from Dr. William Schiemann, were cultured in DMEM high glucose supplemented with 10% FBS and 1% antibiotic/antimycotic solution (penicillin G, streptomycin, amphotericin B). HMLE cells were obtained from Dr. Sendurai Mani and cultured in DMEM:F12 supplemented with 5% calf serum, 0.5 μg/ml hydrocortisone, 10 μg/ml insulin, 20 ng/ml EGF and 1% antibiotic/antimycotic. The HMLE subpopulations were initially isolated as described previously (4, 32) and subsequently FACS sorted using CD44 and CD24 antibodies (see supplementary methods). All cells were cultured in a 37°C, 5% CO2 incubator. Recombinant activin A was purchased from R&D systems (Minneapolis, MN, USA). TGFβ2 was a generous gift from Genzyme Corporation (Cambridge, MA, USA).
Polysome profiling
Cytosolic cell extracts were layered onto sucrose gradients (10–50%) and centrifuged at 35 000 rpm in a SW40Ti rotor for 3 h at 4°C. Fractions were collected and RNA was isolated using Trizol as per manufacturer’s instructions (Life Technologies, Carlsbad, CA, USA). Monosomal and polysomal fractions were determined by analysis of 18s and 28s rRNA levels using denaturing agarose gel electrophoresis (see supplementary methods). The levels of inhibin βA transcript across fractions were assessed by semi-quantitative PCR (see supplementary methods).
Migration assay
Migration was assessed using the CytoSelect™ 24-Well Wound Healing Assay (Cell Biolabs, San Diego, CA, USA). Wound healing inserts were removed from confluent monolayers and cells were fixed and stained at 0, 12 h or 24 h. Wound area was determined using ImageJ software (NIH, Bethesda, MO, USA) in order to calculate % wound recovery compared to control or 0 h samples.
2D invasion assay
Invasion across a basement membrane was performed using BD BioCoat™ Matrigel™ Invasion Chambers (BD Biosciences). Briefly, a volume of 0.5 ml cell suspension at a density of 2 x 105 cells/ml in serum-free DMEM was added to invasion chambers, a volume of 750 μl DMEM supplemented with 10% FBS was added to wells that contained invasion chambers. After 16–22 h, invasive cells located on the underside of chambers were stained (cell stain solution, Cell Biolabs), imaged and quantitated by measuring absorbance at 570nm of extracted cell stain following incubation of inserts in 400 μl extraction solution (Cell Biolabs) for 10 min.
In vivo animal studies
Nonobese diabetic, severe combined immunodeficient (NOD/SCID) mice (NOD.CB17-PrkdcSCID/J) were supplied by the National Cancer Institute (NCI, Frederick, MD, USA). All procedures were approved by the Animal Care and Use Committees of the Medical University of South Carolina. No randomization was used to select animals for each group and studies were not blinded. For lung colonization experiments, a cell suspension of 105 cells in 100 μl PBS was injected into the tail vein of 6–9 week old NOD/SCID females. Mammary fat pad injections into the right inguinal mammary fat pad of 6–9 week old NOD/SCID females were performed using 5 x 104 cells in 100 μl PBS. Mice were sacrificed 9 weeks following tail vein inoculation; at this stage all animals in the study were sacrificed in order to compare lung colonization between cell lines. In the case of mammary fat pad injection of control and InhβA KD L1P cells, mice were sacrificed 4 weeks after injection. Lungs or primary tumors were removed from mice and either formalin fixed and embedded in paraffin or collagenase treated in order to isolate in vivo passaged cells (see supplementary methods). Formalin fixed, paraffin embedded lung sections were cut at 5 μm and stained with hematoxylin and eosin for histopathological evaluation by the laboratory core in the Centre for Oral Health Research at MUSC. Micrographs of stained sections were taken using an Olympus DP72 8-bit RGB camera with Cellsens acquisition software.
Tissue microarray and immunohistochemistry
Assembly and IHC staining of human breast tissue microarrays (TMAs) were performed by the Biorepository and Tissue Analysis core at MUSC. TMAs consisted of matched normal and tumor tissue as well as normal and metastatic lymph node biopsies from patients for which survival data was available (Supplementary Table S2). For IHC staining of formalin-fixed, paraffin-embedded TMAs and mouse lung tissue, sections were deparaffinized in xylene, rehydrated in alcohol, and processed as follows: The sections were incubated with target retrieval solution (S2367; Dako, Carpinteria, CA, USA) in a steamer for 45 min followed by 3% hydrogen peroxide solution for 10 min and protein block (X0909; Dako) for 20 min at room temperature. Sections were incubated overnight in a humid chamber at 4°C with antibodies against Inhibin βA [HPA020031] (1:3200; Sigma), E-cadherin [#3195], Vimentin [#5741] or Ki-67 [#12202] (1:400; Cell Signaling) followed by biotinylated secondary antibody (PK6106; Vector laboratories, Burlingame, CA, USA) for 30 min and ABC reagent for 30 min. Immunocomplexes of horseradish peroxidase were visualized by DAB reaction (K3468; Dako), and sections were counterstained with hematoxylin before mounting. Micrographs of stained sections were taken using an Olympus DP72 8-bit RGB camera with Cellsens acquisition software.
Statistical analyses
Data are presented as the mean ± s.e.m. and experiments have been independently repeated at least twice. Statistical analysis was performed by two-tailed Student’s t-test of pair-wise comparisons or one-way ANOVA followed by Tukey’s post hoc test. Kaplan–Meier survival curves were analyzed using MedCalc Statistical Software (MedCalc Software, Ostend, Belgium) with significance determined by the Logrank test. P-values <0.05 were considered statistically significant.
All other techniques are described in the Supplementary Material and Methods section.
Supplementary Material
Acknowledgments
This work was supported by Grants CA055536 and CA154663 from the National Cancer Institute. BVH was supported by funding from the Abney Foundation. GSH was supported by an American Heart Association Pre-doctoral Fellowship 10PRE3870024. This study used the services of the MUSC Center for Oral Health Research (COHR), which is partially supported by the National Institute of General Medicine grant P30GM103331, the MUSC Flow Cytometry Facility which is supported by P30GM103342, the Cell & Molecular Imaging Shared Resource of MUSC supported by P30CA138313, the Biorepository & Tissue Analysis Shared Resource, Hollings Cancer Center, MUSC and the Gene Targeting and Knockout Shared Resource at MUSC. We would like to thank Maya El Sabban for assisting us with xenograft experiments.
Footnotes
Disclosure of potential conflicts of interest: No potential conflicts of interest were disclosed.
Conflict of Interest: The authors declare no conflict of interest.
Supplementary Information accompanies the paper on the Oncogene website (http://www.nature.com/onc)
References
- 1.Radisky DC. Epithelial-mesenchymal transition. J Cell Sci. 2005;118:4325–4326. doi: 10.1242/jcs.02552. [DOI] [PubMed] [Google Scholar]
- 2.Polyak K, Weinberg RA. Transitions between epithelial and mesenchymal states: acquisition of malignant and stem cell traits. Nat Rev Cancer. 2009;9:265–273. doi: 10.1038/nrc2620. [DOI] [PubMed] [Google Scholar]
- 3.Massagué J, Blain SW, Lo RS. TGFβ signaling in growth control, cancer, and heritable disorders. Cell. 2000;103:295–309. doi: 10.1016/s0092-8674(00)00121-5. [DOI] [PubMed] [Google Scholar]
- 4.Scheel C, Eaton EN, Li SH-J, Chaffer CL, Reinhardt F, Kah K-J, et al. Paracrine and autocrine signals induce and maintain mesenchymal and stem cell states in the breast. Cell. 2011;145:926–940. doi: 10.1016/j.cell.2011.04.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Thiery JP, Acloque H, Huang RYJ, Nieto MA. Epithelial-mesenchymal transitions in development and disease. Cell. 2009;139:871–890. doi: 10.1016/j.cell.2009.11.007. [DOI] [PubMed] [Google Scholar]
- 6.Chaudhury A, Hussey GS, Ray PS, Jin G, Fox PL, Howe PH. TGF-[beta]-mediated phosphorylation of hnRNP E1 induces EMT via transcript-selective translational induction of Dab2 and ILEI. Nat Cell Biol. 2010;12:286–293. doi: 10.1038/ncb2029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hussey GS, Chaudhury A, Dawson AE, Lindner DJ, Knudsen CR, Wilce MCJ, et al. Identification of an mRNP complex regulating tumorigenesis at the translational elongation step. Mol Cell. 2011;41:419–431. doi: 10.1016/j.molcel.2011.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hussey GS, Link LA, Brown AS, Howley BV, Chaudhury A, Howe PH. Establishment of a TGFβ-induced post-transcriptional EMT gene signature. PLoS ONE. 2012;7:e52624. doi: 10.1371/journal.pone.0052624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Li R, Phillips DM, Mather JP. Activin promotes ovarian follicle development in vitro. Endocrinology. 1995;136:849–856. doi: 10.1210/endo.136.3.7867593. [DOI] [PubMed] [Google Scholar]
- 10.Matzuk MM, Kumar TR, Vassalli A, Bickenbach JR, Roop DR, Jaenisch R, et al. Functional analysis of activins during mammalian development. Nature. 1995;374:354–357. doi: 10.1038/374354a0. [DOI] [PubMed] [Google Scholar]
- 11.Ogawa Y, Schmidt DK, Nathan RM, Armstrong RM, Miller KL, Sawamura SJ, et al. Bovine bone activin enhances bone morphogenetic protein-induced ectopic bone formation. J Biol Chem. 1992;267:14233–14237. [PubMed] [Google Scholar]
- 12.Pearson S, Sroczynska P, Lacaud G, Kouskoff V. The stepwise specification of embryonic stem cells to hematopoietic fate is driven by sequential exposure to Bmp4, activin A, bFGF and VEGF. Development. 2008;135:1525–1535. doi: 10.1242/dev.011767. [DOI] [PubMed] [Google Scholar]
- 13.Schmierer B, Hill CS. TGF[beta]-SMAD signal transduction: molecular specificity and functional flexibility. Nat Rev Mol Cell Biol. 2007;8:970–982. doi: 10.1038/nrm2297. [DOI] [PubMed] [Google Scholar]
- 14.Le Bras GF, Loomans HA, Taylor CJ, Revetta FL, Andl CD. Activin A balance regulates epithelial invasiveness and tumorigenesis. Lab Invest. 2014;94:1134–1146. doi: 10.1038/labinvest.2014.97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Burdette JE, Jeruss JS, Kurley SJ, Lee EJ, Woodruff TK. Activin A mediates growth inhibition and cell cycle arrest through smads in human breast cancer cells. Cancer Res. 2005;65:7968–7975. doi: 10.1158/0008-5472.CAN-04-3553. [DOI] [PubMed] [Google Scholar]
- 16.Lonardo E, Hermann PC, Mueller M-T, Huber S, Balic A, Miranda-Lorenzo I, et al. Nodal/activin signaling drives self-renewal and tumorigenicity of pancreatic cancer stem cells and provides a target for combined drug therapy. Cell Stem Cell. 2011;9:433–446. doi: 10.1016/j.stem.2011.10.001. [DOI] [PubMed] [Google Scholar]
- 17.Reis FM, Cobellis L, Tameirão LC, Anania G, Luisi S, Silva ISB, et al. Serum and tissue expression of activin A in postmenopausal women with breast cancer. J Clin Endocrinol Metab. 2002;87:2277–2282. doi: 10.1210/jcem.87.5.8512. [DOI] [PubMed] [Google Scholar]
- 18.Wildi S, Kleeff J, Maruyama H, Maurer CA, Büchler MW, Korc M. Overexpression of activin A in stage IV colorectal cancer. Gut. 2001;49:409–417. doi: 10.1136/gut.49.3.409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wang Q, Wen Y-G, Li D-P, Xia J, Zhou C-Z, Yan D-W, et al. Upregulated INHBA expression is associated with poor survival in gastric cancer. Med Oncol. 2012;29:77–83. doi: 10.1007/s12032-010-9766-y. [DOI] [PubMed] [Google Scholar]
- 20.Leto G, Incorvaia L, Badalamenti G, Tumminello F, Gebbia N, Flandina C, et al. Activin A circulating levels in patients with bone metastasis from breast or prostate cancer. Clin Exp Metastasis. 2006;23:117–122. doi: 10.1007/s10585-006-9010-5. [DOI] [PubMed] [Google Scholar]
- 21.Kang H-Y, Huang H-Y, Hsieh C-Y, Li C-F, Shyr C-R, Tsai M-Y, et al. Activin A enhances prostate cancer cell migration through activation of androgen receptor and is overexpressed in metastatic prostate cancer. J Bone Miner Res. 2009;24:1180–1193. doi: 10.1359/jbmr.090219. [DOI] [PubMed] [Google Scholar]
- 22.Kim H, Watkinson J, Varadan V, Anastassiou D. Multi-cancer computational analysis reveals invasion-associated variant of desmoplastic reaction involving INHBA, THBS2 and COL11A1. BMC Med Genomics. 2010;3:51. doi: 10.1186/1755-8794-3-51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Antenos M, Stemler M, Boime I, Woodruff TK. N-Linked Oligosaccharides Direct the Differential Assembly and Secretion of Inhibin α- and βA-Subunit Dimers. Mol Endocrinol. 2007;21:1670–1684. doi: 10.1210/me.2007-0050. [DOI] [PubMed] [Google Scholar]
- 24.Piek E, Moustakas A, Kurisaki A, Heldin CH, Dijke Pt. TGF-(beta) type I receptor/ALK-5 and Smad proteins mediate epithelial to mesenchymal transdifferentiation in NMuMG breast epithelial cells. J Cell Sci. 1999;112:4557–4568. doi: 10.1242/jcs.112.24.4557. [DOI] [PubMed] [Google Scholar]
- 25.Gunasinghe NPAD, Wells A, Thompson E, Hugo H. Mesenchymal–epithelial transition (MET) as a mechanism for metastatic colonisation in breast cancer. Cancer Metastasis Rev. 2012;31:469–478. doi: 10.1007/s10555-012-9377-5. [DOI] [PubMed] [Google Scholar]
- 26.Arai KY, Ono M, Kudo C, Fujioka A, Okamura R, Nomura Y, et al. IL-1β stimulates activin βA mRNA expression in human skin fibroblasts through the MAPK pathways, the nuclear factor-κB pathway, and prostaglandin E2. Endocrinology. 2011;152:3779–3790. doi: 10.1210/en.2011-0255. [DOI] [PubMed] [Google Scholar]
- 27.Wamsley JJ, Kumar M, Allison DF, Clift SH, Holzknecht CM, Szymura SJ, et al. Activin upregulation by NF-κB is required to maintain mesenchymal features of cancer stem-like cells in non-small cell lung cancer. Cancer Res. 2014 doi: 10.1158/0008-5472.can-2013-2702. e-pub ahead of print 28 November 2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Valcourt U, Kowanetz M, Niimi H, Heldin C-H, Moustakas A. TGF-β and the smad signaling pathway support transcriptomic reprogramming during epithelial-mesenchymal cell transition. Mol Biol Cell. 2005;16:1987–2002. doi: 10.1091/mbc.E04-08-0658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lee KL, Lim SK, Orlov YL, Yit LY, Yang H, Ang LT, et al. Graded nodal/activin signaling titrates conversion of quantitative phospho-smad2 levels into qualitative embryonic stem cell fate decisions. PLoS Genet. 2011;7:e1002130. doi: 10.1371/journal.pgen.1002130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kim E-S, Kim M-S, Moon A. Transforming growth factor (TGF)-β in conjunction with H-ras activation promotes malignant progression of MCF10A breast epithelial cells. Cytokine. 2005;29:84–91. doi: 10.1016/j.cyto.2004.10.001. [DOI] [PubMed] [Google Scholar]
- 31.Seder CW, Hartojo W, Lin L, Silvers AL, Wang Z, Thomas DG, et al. Upregulated INHBA Expression May Promote Cell Proliferation and Is Associated with Poor Survival in Lung Adenocarcinoma. Neoplasia. 2009;11:388–396. doi: 10.1593/neo.81582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Walia V, Elble RC. Enrichment for breast cancer cells with stem/progenitor properties by differential adhesion. Stem Cells Dev. 2010;19:1175–1182. doi: 10.1089/scd.2009.0430. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.