Abstract
In the normal mammary gland, the basal epithelium is known to be bi-potent and can generate either basal or luminal cells, whereas the luminal epithelium has not been demonstrated to contribute to the basal compartment in an intact and normally developed mammary gland. It is not clear whether cellular heterogeneity within a breast tumor results from transformation of bi-potent basal cells or from transformation and subsequent basal conversion of the more differentiated luminal cells. Here, we used a retroviral vector to express an oncogene specifically in a small number of the mammary luminal epithelial cells and tested their potential to produce basal cells during tumorigenesis. This in vivo lineage tracing work demonstrates that luminal cells are capable of producing basal cells upon activation of either Polyoma Middle T antigen (PyMT) or ErbB2 signaling. These findings reveal the plasticity of the luminal compartment during tumorigenesis and provide an explanation for cellular heterogeneity within a cancer.
Introduction
Human breast tumors take many years to form and are usually clonal (1, 2). The majority of human breast cancers are comprised primarily of cells resembling the luminal layer of the normal mammary epithelium, and are thought to have an origin in the luminal epithelium. However, some human breast tumors also harbor both luminal- and basal-like cells, or have a basal-like expression profile (3, 4). Therefore, heterogeneous tumors must be derived from basal cells which become luminal cells, or vice versa.
In early mammary development, both the luminal and basal epithelial compartments are generated from bipotent progenitors expressing markers of both lineages (5, 6). After birth, while it is controversial whether basal cells can give rise to luminal cells in an unperturbed mammary gland, they retain the capability of regenerating a complete ductal tree after isolation and transplantation into an epithelia-cleared fat pad. However, luminal cells have been repeatedly demonstrated to be lineage-restricted and incapable of generating basal cells in either the intact mammary gland or upon transplantation (5–9). Unclear is whether this differentiated state in luminal cells precludes the development of basal cells during tumorigenesis.
There have not been any studies which fully address this question. Previous studies in transgenic rodent models congenitally expressed oncogenes, often impacting mammary gland development, and further lacked highly lineage-defined promoters (10–13). Other studies relied on ex vivo lineage enrichment and alteration followed by transplantation (14–17). Clinical BRCA1 patients have an expanded luminal progenitor population and develop basal-like tumors, and this correlation was interpreted to suggest a luminal cell origin for these tumorss (18). However, later studies indicate that the luminal compartment in these patients show a basal-like profile and may not have properly differentiated in the first place (19).
Therefore, it is unclear whether committed luminal cells in a fully developed and intact mammary gland – upon the gain of oncogenic mutations – can generate basal cells in their evolution to breast cancer. Here, we sought a direct answer to this question using a retroviral vector to selectively infect a small number of mammary luminal cells in vivo and to trace clonal tumor initiation events, according the emerging standard for tumor cell-of-origin studies (20–24). Resolving this issue will help explain the source of cellular heterogeneity in human breast cancer, which contributes to therapy resistance.
Results and Discussion
RCAS viral integration is restricted to the luminal compartment in WAP-tva mice
Here, we tested whether during tumorigenesis committed luminal cells can give rise to basal cells by utilizing an avian leukosis virus vector (RCAS) (25) to introduce two oncogenes (PyMT or ErbB2) into whey acidic protein (WAP)-positive committed luminal mammary epithelial cells in a fully developed mammary gland and evaluated the lineage potential during tumorigenesis. WAP is produced selectively by committed luminal cells within mammary ducts and alveoli (26, 27). We recently generated a transgenic FVB mouse line using the luminal WAP promoter to drive the expression of cDNA encoding the TVA receptor (28), which is both necessary and sufficient for infection by RCAS (25).
To reconfirm that RCAS integration is restricted to the WAP+ luminal cell population, RCAS-βactin-HA (29) was injected intraductally into eight adult WAP-tva mice (14–18 weeks of age; 1x107 IUs per gland; one set of #2–4 glands per mouse). Uninjected glands were retained for each mouse as an internal control in this and all subsequent experiments, as opposed to treatment randomization. Mammary glands were collected at 2.5 days post-injection for co-immunofluorescence staining for the viral HA tag in addition to the luminal marker keratin 8 (K8), or the basal marker K5 or p63. Approximately 99% of HA+ cells co-stained for K8 (Figures 1A,J & S1A). We counted a single K5 or p63 positive basal cell per 800–1100 infected cells over 8 mice quantified (0.1%; Figures 1B–C,J & S1B–C). Our previously study of the infection rate of this virus detected approximately 3,000 infected cells per gland (28). Thus, the total number of HA+ basal cells are estimated to be 3 per mammary gland in this current study. These data demonstrate that in the WAP-tva mouse line viral integration – and therefore oncogene expression – is highly selective for the luminal epithelium.
Figure 1. Viral integration is restricted to the luminal epithelial compartment.
(A–I) Immunofluorescence staining for the HA tag of RCAS-βactin-HA at 2.5 days (A–C), 14 days (D–F), and 6 weeks (G–I) shows that viral integration is restricted to the K8+ luminal population (A,D), with extremely rare K5+ (B,E,H) or p63+ (C,F,I) basal cells marked by HA. Solid arrows represent cells double-positive for the HA tag and a lineage marker. Hollow arrows represent cells positive for only the HA tag. (J–L) Quantification of the immunostaining shown above, n=8 mice per time point to ensure adequate HA+ cell numbers and to account for possible estrus cycle effects. See also Figure S1. The WAP-tva transgenic mouse line has been previously described (28). All mice were bred and maintained in accordance with the animal protocol and guidelines approved by the BCM Institutional Animal Care and Use Committee (IACUC). RCAS viral culture and mammary intraductal injection has been previously described (44). For immunofluorescence staining, antibodies used are as follows: mouse anti-HA (1:500; Covance, MMS-101P), rabbit anti-K5 (1:200; Covance, PRB-160P), rabbit anti-p63 (1:200; Biolegend, 619002), and rat anti-K8 (1:200; University of Iowa Hybridoma Bank, Troma-I). Cells were quantified using the WCIF ImageJ package (www.uhnresearch.ca/wcif). Thresholds for each channel were individually determined by evaluation against control duct. HA+ cells were manually selected and quantified using the Nucleus Counter plugin on the DAPI+ channel. Protein colocalization was ascertained using the Colocalization Highlighter plugin. Colocalized protein was required to encircle a nucleus to be considered a positive cell. For nuclear stains, cellular colocalization was manually quantified using thresholded images in cells containing both the nuclear marker and encircled by the cytoplasmic marker.
Next, we evaluated the lineage plasticity of the HA-tagged cells in an unperturbed mammary gland. The RCAS-βactin-HA-infected mammary glands of 8 WAP-tva mice each were collected 2 and 6 weeks post-infection for co-immunofluorescence for the above markers. Again, 99% of the HA+ cells co-stained for K8 at both time points, but none colocalized with K5 and only one with p63 at 2 weeks while only 5 co-stained with either K5 or p63 at 6 weeks (Figures 1D–I,K–L & S1D–I). These data indicate that the infected luminal cells remained luminal-restricted even after extended periods of time (equivalent to approximately 14 estrus cycles at the 6 week time point), confirming previous reports that in postnatal mammary glands luminal cells contribute only to the luminal compartment (6, 7).
PyMT-initiated tumorigenesis from luminal cells leads to the formation of early lesions and tumors that both harbor basal cells
To test whether oncogenic stress could cause committed luminal cells to generate basal cells, eight adult WAP-tva mice (12–14 weeks of age) were intraductally injected with RCAS virus expressing the gene encoding HA-tagged polyoma middle T antigen (RCAS-PyMT-HA) (30). PyMT is a potent oncoprotein widely utilized as a model for breast cancer development that activates Src and PI3K (31, 32). It can rapidly induce mammary tumors in mice either transgenic for PyMT (33) or infected with RCAS-PyMT-HA (25), and the resultant tumors most closely resemble the clinical LumB subtype (34, 35). In our model, we find that the lesions and tumors generated have a solid-cribiform and cribiform-micropapillary pathology, respectively (Figure S2A–B,F). Early lesions can be detected by seven days following RCAS-PyMT-HA injection (25), so at this point, three mice were euthanized, and their mammary glands were stained by co-immunofluorescence for the HA tag and either K8 or K5. In the 30 lesions evaluated, the majority of HA+ cells produced K8, as expected (median=67.3%; Figure 2A,C; Figure S4A); however, a subset of HA+ cells in the majority of lesions stained for K5 (median=1.64%; Figure 2B,D; Figure S4B). Of note, RCAS-PyMT-HA-driven tumorigenesis engenders a multitude of distinct early lesions (~102) from the approximately 3000 initially infected cells (Figure S3). As only a few basal cells existed among this infected population, the overwhelming majority of lesions – and the basal cells within them – must have arisen from committed luminal cells.
Figure 2. PyMT lesions and tumors are comprised of both luminal and basal lineages.
Immunofluorescence staining of RCAS-PyMT-HA precancerous lesions (A&B) and mature tumors (E–G). While luminal K8+ cells are predominant in both early lesions (A&C) and tumors (E & H), K5+ basal cells develop in many precancerous lesions (B&D), and persist and/or continue to develop in the mature tumors (F&H). Basal conversion is confirmed with p63 staining of the mature tumors (G–J). Solid arrows represent cells double-positive for HA tag and K8, K5, or p63. Open-ended arrows represent cells positive for only K5 or p63. WAP-tva mice (11–22 weeks) were intraductally injected with 10 μL RCAS-PyMT virus (107 IU), previously described (25, 30), into each of the 2nd, 3rd, and 4th left mammary glands. Cellular colocalization was quantified as described in Figure 1. Quantifications represent 10 precancerous lesions from each of 3 mice and 5 mature tumors, to ensure detection of cellular conversion occurred in multiple mice at each stage. See also Figure S4.
Tumors that developed in the remaining five mice were collected for co-immunofluorescence analysis for the HA tag in PyMT and either a luminal or a basal marker. As expected, the majority of tumor cells were positive for K8 (median=79.8%; Figure 2E,H; Figure S4C), and they appeared to be well differentiated with a low nuclear grade (Figure S2B). However, again a small subset of HA+ tumor cells stained for K5 (median=1.17%; Figure 2F,I; Figure S4D). These K5+ cells were predominantly, though not exclusively, localized in close proximity to the infiltrating stroma and were well organized like a basal epithelium. This finding was confirmed by staining for p63 (median=2.49%, Figure 2G,J, Figure S4E). Together, these data suggest that PyMT expression in luminal cells leads to the formation of early lesions and tumors that both harbor a small subset of well-differentiated and organized basal cells.
ErbB2-initiated tumorigenesis from luminal cells also leads to the formation of early lesions and tumors that harbor basal cells
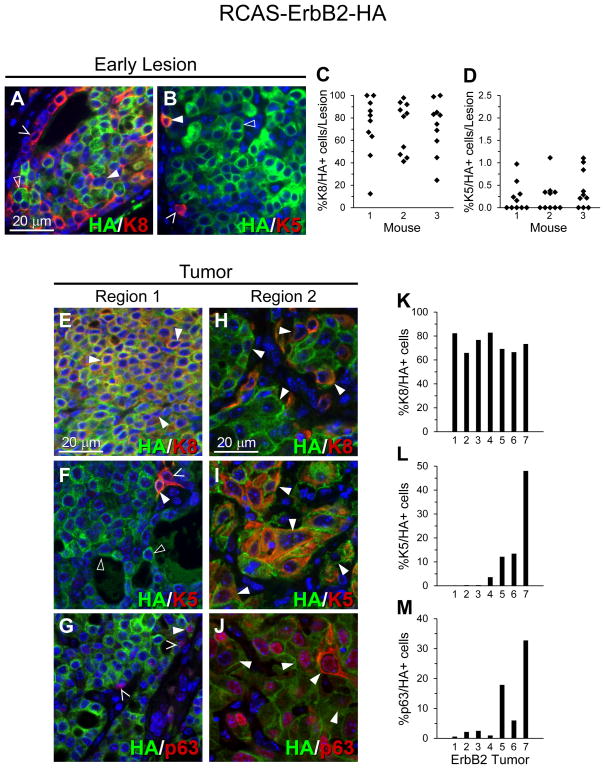
Next, we tested whether a cellular oncogene commonly altered in human breast cancer can also cause committed luminal cells to produce basal cells. ErbB2 encodes a member of the epidermal growth factor receptor family of tyrosine kinases and is amplified and overexpressed in approximately 25% of human breast cancers (36, 37). Although ErbB2 tumors cluster together and are considered their own subtype, the category can be further divided into those tumors which are more luminal-like and those which are more basal-like (3, 38). In our system, we find that the ErbB2 precancerous lesions have a solid or solid-cribirorm pathology, but often develops squamous metaplastic pockets at the tumor stage (Figure S2C–F), making this oncogene a suitable candidate for the study of tumor heterogeneity. Ten adult (13–16 weeks of age) WAP-tva mice were injected intraductally with RCAS-ErbB2-HA, which carries a constitutively activated ErbB2 (25). RCAS-ErbB2-HA leads to early lesions in two weeks (29, 39), and therefore three mice at this time point were euthanized and their infected mammary glands were co-stained for HA and either K8 or K5. Most of the resulting early lesions maintained a high percentage of luminal K8+ cells, as expected, although some down-regulated or lost K8 (median=81.8%; Figure 3A,C; Figure S5A). As in the PyMT-induced early lesions, some of these early lesions also gained a small subset of K5+ cells (median=0.21%; Figure 3B,D; Figure S5B), which existed mainly as isolated single events.
Figure 3. ErbB2 lesions and tumors are mainly cells of the luminal lineage, but some tumors develop large pockets of basal lineage cells.
RCAS-ErbB2-HA mostly generates precancerous lesions with predominantly K8+ cells (A&C) and a minor population of K5+ cells (B&D). Some tumors are predominantly K8+ luminal-like cells (E) that are accompanied by rare and isolated K5+ and/or p63+ cells (F–G). However, others develop also large pockets of K5+ and/or p63+ cells (I&J). Region 1 (E–G) and Region 2 (H–J) represent separate views of the same tumor for each staining. Solid arrows represent cells double-positive for the HA tag and K8, K5, or p63. Hollow arrows represent cells positive for only the HA tag. Open-ended arrows represent cells positive for only K8, K5, or p63. WAP-tva mice (11–22 weeks) were intraductally injected with 10 μL RCAS-ErbB2 virus (107 IU), previously described (29), into each of the 2nd, 3rd, and 4th left mammary glands. Cellular colocalization was quantified as described in Figure 1. Quantifications represent 10 precancerous lesions from each of 3 mice and 7 tumors, to ensure detection of cellular conversion occurred in multiple mice at each stage. See also Figure S5.
Tumors arising in the remaining seven infected mice appeared heterogeneous, as we reported previously (40). The majority of the tumor mass was glandular and comprised of dense luminal K8+ cells that were accompanied by few K5+ basal-like cells (Figure 3E–F; Figure S5C–D). In contrast, large pockets of K5+ squamous metaplastic cells developed in four of the seven tumor (Figure 3J), suggesting a clonal expansion and transdifferentation of basal cells within the ErbB2 tumor mass. These regions precipitously lost K8+ cells (Figure 3H–I; Figure S5F–G). This staining pattern was confirmed by staining for p63 (Figure 3G,J, Figure S5E–H). Squamous metaplasia represents a rare subtype of basal breast cancers with poor prognosis (41, 42). The observations in this study suggest that squamous metaplastic cells within breast cancer may have an origin in the luminal epithelium.
Flow cytometry analysis of cellular lineages
We next wanted to evaluate the lineage profiles by flow cytometry for both PyMT and ErbB2 lesion-bearing mammary glands. Normal uninfected control mammary gland cells clearly showed both luminal (CD24highCD49f+) and basal (CD24lowCD49f+) lineages, as reported (8). For both the PyMT and ErbB2 lesions, while the majority of the HA+ cells remained within the luminal gate, a significant subset was detected within the basal gate, and a substantial proportion of cells massed between the basal and luminal gates (Figure 4A–B, Figure S6). This observation confirmed basal conversion of luminal cells during tumorigenesis and further suggests that even cells which appear in a luminal or basal state by standard immunofluorescence methods may actually be in a transitioning state detectable via flow cytometry. Previous studies have also shown that tumorigenesis can induce the formation of a new cytometric profile which is enriched for tumor stem cells (12). Future studies may elucidate the role for these transitioning cells.
Figure 4. RCAS-PyMT-HA and RCAS-ErbB2-HA driven tumorigenesis exhibit evidence of cellular plasticity.
(A) Representative flow cytometry profile of luminal and basal gating for Lin(−) cells from uninjected mammary glands and Lin(−)HA(+) cells from early lesion-bearing glands. Lesion-bearing mammary glands were collected at appropriate timepoints along with uninjected control mammary glands, and single-cell suspensions were prepared and then serially stained for hematopeoetic lineage markers (CD45, Ter-119, CD31) (BD#553672, BD#553086, Biolegend#102504) and CD24 (BD#562563), and CD49f (BD#555736), and SytoxRed (Life Technologies #S34859). Cells were permeabilized in 2% Tween20 in PBS for 15 minutes at room temperature, followed by HA.11 antibody addition (BioLegend#A488-101L). See also Figure S6. (B) Quantification of cellular population frequencies in PyMT and ErbB2 lesion-bearing mammary glands as analyzed by flow cytometry (Mean±SEM). Plots represent results from 4 independent mice for each oncogene. (C–I) Tri-immunofluorescence staining for K5/K8/HA. The lesion type and the initiating oncogene are as indicated. Solid arrows represent cells triple-positive for K8/K5/HA. Hollow arrows represent cells positive K5/HA but not K8. Cellular colocalization was quantified as described in Figure 1. Quantifications represent percentage (Mean±SEM) of K5+/K8+ double-positive cells and K5−/K8− double-negative cells over total HA+ cells for 10 precancerous lesions from each of 3 mice and 5 (PyMT) and 7 (ErbB2) mature tumors, to ensure detection of cellular conversion occurred in multiple mice at each stage. P-value is calculated using a non-parametric kruskal-wallis test to account for large variability in the data.
Taken together, both PyMT and ErbB2 can stimulate significant luminal-to-basal transition. Therefore, oncogenic stress can cause committed luminal cells to form additional cell lineages, including basal cells as well as squamous metaplastic cells, unveiling oncogene-driven plasticity of committed luminal cells during carcinogenesis. These findings provide further support for the idea that luminal cells can be the cells of origin of heterogeneous breast cancers including those that are basal-like, and suggest that intratumoral heterogeneity does not need to arise from transformation of bipotential progenitor cells.
K8/K5 double-positive cells exist in both early lesions and frank tumors
The detection of both a large population of K5+ cells and a large population of K8+ cells in some caErB2-induced tumors suggests that some of the tumor cells may be double-positive for both basal and luminal markers. Therefore, tri-immunofluorescence staining for K8/K5/HA was used to identify double lineage cells among the provirus+ population (based on HA staining). In PyMT-induced early lesions and tumors, K8+K5+ cells were detected in 2.26±1.0% and 2.17±0.7% of the provirus+ cells, respectively (Figure 4C–E). In ErbB2-induced early lesions and tumors, K8+K5+ cells were found in 0.84±0.3% and 5.11±1.8% of provirus+ cells, respectively (Figure 4F–I; p<0.03). These data demonstrate that K8/K5 double-positive cells exist in both early lesions and frank tumors induced by both PyMT and ErbB2. These double-positive cells may represent a transitional state between the luminal and basal compartments. Alternately, cells positive for both lineage markers may have stem cell properties or be cancer stem cells (CSC) (6, 43).
We further quantified the numbers of double negative cells, as these may represent an alternate and possibly more dedifferentiated transition state. Here we found that PyMT lesions and tumor had 0.54±0.1% and 0.51±0.2% K5/K8 double negative cells among provirus+ cells. ErbB2 lesions and tumors had 0.94±0.4% and 1.68±0.5% double negative cells among provirus+ cells (Figure 4E,I).
Neither the lineage double-positive nor the double-negative cell population size significantly increased with the progression from precancerous lesions to tumors in the PyMT model, but there was a significant increase in the numbers of double positive cells in the ErbB2 model. These cells may represent a transitioning or possibly stem-like cellular state, suggested by previous literature (12). However, this emergent cellular population would necessitate further study to fully elucidate the function.
Conclusion
Using retrovirus-mediated in vivo lineage tracing, we show that oncogenic signaling can cause committed luminal cells to form additional cellular lineages, exposing the plasticity of formerly committed and lineage-restricted mammary luminal epithelial cells. This observation suggests that intratumoral heterogeneity does not need to arise from transformation of a cell with multipotent potential. Rather, heterogeneity may be a natural consequence of oncogene activation regardless of the cell of origin. Consequently, breast cancer prevention should not solely focus on targeting a specific multipotent cell population. Understanding the molecular mechanism of luminal cell plasticity under oncogenic stress may lead to new molecular targets for preventing basal-like and metaplastic breast cancer.
Supplementary Material
Acknowledgments
We thank Dr. Jeffrey Rosen for critical reading of this manuscript. This work was supported in part by funds from NIH CA124820 (to Y.L) and U54CA149196 (to Y. L; PI: Stephan Wong); from DOD CDMRP BC085050 (to Y. L), BC112704 (to Y.L), and BC073703 (Y.L.); and from the Nancy Owens Memorial Foundation (to Y. L); as well as by the resources from the Dan L. Duncan Cancer Center (P30CA125123) and the Sue & Lester Breast Center (P50-CA186784). S. M. H. was supported by the CPRIT Training Program (RP101499) and is supported by NIH training award T32AG000183. A.N.J is supported by NIH training award T32GM088129. This project was supported by the Cytometry and Cell Sorting Core at Baylor College of Medicine with funding from the NIH (P30 AI036211, P30 CA125123, and S10 RR024574) and the expert assistance of Joel M. Sederstrom.
Footnotes
Conflict of Interest
The authors have no competing interests to declare.
References
- 1.Navin N, Kendall J, Troge J, Andrews P, Rodgers L, McIndoo J, et al. Tumour evolution inferred by single-cell sequencing. Nature. 2011;472(7341):90–4. doi: 10.1038/nature09807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Greaves M, Maley CC. Clonal evolution in cancer. Nature. 2012;481(7381):306–13. doi: 10.1038/nature10762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Perou CM, Sorlie T, Eisen MB, van de Rijn M, Jeffrey SS, Rees CA, et al. Molecular portraits of human breast tumours. Nature. 2000;406(6797):747–52. doi: 10.1038/35021093. [DOI] [PubMed] [Google Scholar]
- 4.Malzahn K, Mitze M, Thoenes M, Moll R. Biological and prognostic significance of stratified epithelial cytokeratins in infiltrating ductal breast carcinomas. Virchows Archiv : an international journal of pathology. 1998;433(2):119–29. doi: 10.1007/s004280050226. [DOI] [PubMed] [Google Scholar]
- 5.van Amerongen R, Bowman Angela N, Nusse R. Developmental Stage and Time Dictate the Fate of Wnt/β-Catenin-Responsive Stem Cells in the Mammary Gland. Cell Stem Cell. 2012;11(3):387–400. doi: 10.1016/j.stem.2012.05.023. [DOI] [PubMed] [Google Scholar]
- 6.Van Keymeulen A, Rocha AS, Ousset M, Beck B, Bouvencourt G, Rock J, et al. Distinct stem cells contribute to mammary gland development and maintenance. Nature. 2011;479(7372):189–93. doi: 10.1038/nature10573. [DOI] [PubMed] [Google Scholar]
- 7.Rios AC, Fu NY, Lindeman GJ, Visvader JE. In situ identification of bipotent stem cells in the mammary gland. Nature. 2014;506(7488):322–7. doi: 10.1038/nature12948. [DOI] [PubMed] [Google Scholar]
- 8.Stingl J, Eirew P, Ricketson I, Shackleton M, Vaillant F, Choi D, et al. Purification and unique properties of mammary epithelial stem cells. Nature. 2006;439(7079):993–7. doi: 10.1038/nature04496. [DOI] [PubMed] [Google Scholar]
- 9.Shackleton M, Vaillant F, Simpson KJ, Stingl J, Smyth GK, Asselin-Labat ML, et al. Generation of a functional mammary gland from a single stem cell. Nature. 2006;439(7072):84–8. doi: 10.1038/nature04372. [DOI] [PubMed] [Google Scholar]
- 10.Rosner A, Miyoshi K, Landesman-Bollag E, Xu X, Seldin DC, Moser AR, et al. Pathway pathology: histological differences between ErbB/Ras and Wnt pathway transgenic mammary tumors. Am J Pathol. 2002;161(3):1087–97. doi: 10.1016/S0002-9440(10)64269-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Li Y, Welm B, Podsypanina K, Huang S, Chamorro M, Zhang X, et al. Evidence that transgenes encoding components of the Wnt signaling pathway preferentially induce mammary cancers from progenitor cells. Proceedings of the National Academy of Sciences. 2003;100(26):15853–8. doi: 10.1073/pnas.2136825100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ling H, Jolicoeur P. Notch-1 signaling promotes the cyclinD1-dependent generation of mammary tumor-initiating cells that can revert to bi-potential progenitors from which they arise. Oncogene. 2013;32(29):3410–9. doi: 10.1038/onc.2012.341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Molyneux G, Geyer FC, Magnay F-A, McCarthy A, Kendrick H, Natrajan R, et al. BRCA1 Basal-like Breast Cancers Originate from Luminal Epithelial Progenitors and Not from Basal Stem Cells. Cell Stem Cell. 2010;7(3):403–17. doi: 10.1016/j.stem.2010.07.010. [DOI] [PubMed] [Google Scholar]
- 14.Gastaldi S, Sassi F, Accornero P, Torti D, Galimi F, Migliardi G, et al. Met signaling regulates growth, repopulating potential and basal cell-fate commitment of mammary luminal progenitors: implications for basal-like breast cancer. Oncogene. 2013;32(11):1428–40. doi: 10.1038/onc.2012.154. [DOI] [PubMed] [Google Scholar]
- 15.Guo W, Keckesova Z, Donaher JL, Shibue T, Tischler V, Reinhardt F, et al. Slug and Sox9 cooperatively determine the mammary stem cell state. Cell. 2012;148(5):1015–28. doi: 10.1016/j.cell.2012.02.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Keller PJ, Arendt LM, Skibinski A, Logvinenko T, Klebba I, Dong S, et al. Defining the cellular precursors to human breast cancer. Proceedings of the National Academy of Sciences. 2012;109(8):2772–7. doi: 10.1073/pnas.1017626108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ince TA, Richardson AL, Bell GW, Saitoh M, Godar S, Karnoub AE, et al. Transformation of Different Human Breast Epithelial Cell Types Leads to Distinct Tumor Phenotypes. Cancer Cell. 2007;12(2):160–70. doi: 10.1016/j.ccr.2007.06.013. [DOI] [PubMed] [Google Scholar]
- 18.Lim E, Vaillant F, Wu D, Forrest NC, Pal B, Hart AH, et al. Aberrant luminal progenitors as the candidate target population for basal tumor development in BRCA1 mutation carriers. Nat Med. 2009;15(8):907–13. doi: 10.1038/nm.2000. [DOI] [PubMed] [Google Scholar]
- 19.Proia TA, Keller PJ, Gupta PB, Klebba I, Jones AD, Sedic M, et al. Genetic Predisposition Directs Breast Cancer Phenotype by Dictating Progenitor Cell Fate. Cell Stem Cell. 2011;8(2):149–63. doi: 10.1016/j.stem.2010.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Youssef KK, Van Keymeulen A, Lapouge G, Beck B, Michaux C, Achouri Y, et al. Identification of the cell lineage at the origin of basal cell carcinoma. Nature cell biology. 2010;12(3):299–305. doi: 10.1038/ncb2031. [DOI] [PubMed] [Google Scholar]
- 21.Holland EC, Celestino J, Dai C, Schaefer L, Sawaya RE, Fuller GN. Combined activation of Ras and Akt in neural progenitors induces glioblastoma formation in mice. Nature genetics. 2000;25(1):55–7. doi: 10.1038/75596. [DOI] [PubMed] [Google Scholar]
- 22.Desai TJ, Brownfield DG, Krasnow MA. Alveolar progenitor and stem cells in lung development, renewal and cancer. Nature. 2014;507(7491):190–4. doi: 10.1038/nature12930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Schepers AG, Snippert HJ, Stange DE, van den Born M, van Es JH, van de Wetering M, et al. Lineage Tracing Reveals Lgr5+ Stem Cell Activity in Mouse Intestinal Adenomas. Science. 2012;337(6095):730–5. doi: 10.1126/science.1224676. [DOI] [PubMed] [Google Scholar]
- 24.Driessens G, Beck B, Caauwe A, Simons BD, Blanpain C. Defining the mode of tumour growth by clonal analysis. Nature. 2012;488(7412):527–30. doi: 10.1038/nature11344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Du Z, Podsypanina K, Huang S, McGrath A, Toneff MJ, Bogoslovskaia E, et al. Introduction of oncogenes into mammary glands in vivo with an avian retroviral vector initiates and promotes carcinogenesis in mouse models. Proceedings of the National Academy of Sciences. 2006;103(46):17396–401. doi: 10.1073/pnas.0608607103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Robinson GW, McKnight RA, Smith GH, Hennighausen L. Mammary epithelial cells undergo secretory differentiation in cycling virgins but require pregnancy for the establishment of terminal differentiation. Development. 1995;121(7):2079–90. doi: 10.1242/dev.121.7.2079. [DOI] [PubMed] [Google Scholar]
- 27.Chang TH, Kunasegaran K, Tarulli GA, De Silva D, Voorhoeve PM, Pietersen AM. New insights into lineage restriction of mammary gland epithelium using parity-identified mammary epithelial cells. Breast Cancer Res. 2014;16(1):R1. doi: 10.1186/bcr3593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Haricharan S, Dong J, Hein S, Reddy JP, Du Z, Toneff M, et al. Mechanism and preclinical prevention of increased breast cancer risk caused by pregnancy. eLife. 2013;2:e00996. doi: 10.7554/eLife.00996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Toneff MJ, Du Z, Dong J, Huang J, Sinai P, Forman J, et al. Somatic expression of PyMT or activated ErbB2 induces estrogen-independent mammary tumorigenesis. Neoplasia (New York, NY) 2010;12(9):718–26. doi: 10.1593/neo.10516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Holland EC, Li Y, Celestino J, Dai C, Schaefer L, Sawaya RA, et al. Astrocytes give rise to oligodendrogliomas and astrocytomas after gene transfer of polyoma virus middle T antigen in vivo. Am J Pathol. 2000;157(3):1031–7. doi: 10.1016/S0002-9440(10)64615-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Fluck MM, Schaffhausen BS. Lessons in signaling and tumorigenesis from polyomavirus middle T antigen. Microbiology and molecular biology reviews : MMBR. 2009;73(3):542–63. doi: 10.1128/MMBR.00009-09. Table of Contents. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ichaso N, Dilworth SM. Cell transformation by the middle T-antigen of polyoma virus. Oncogene. 2001;20(54):7908–16. doi: 10.1038/sj.onc.1204859. [DOI] [PubMed] [Google Scholar]
- 33.Guy CT, Cardiff RD, Muller WJ. Induction of mammary tumors by expression of polyomavirus middle T oncogene: a transgenic mouse model for metastatic disease. Molecular and Cellular Biology. 1992;12(3):954–61. doi: 10.1128/mcb.12.3.954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Pfefferle AD, Herschkowitz JI, Usary J, Harrell JC, Spike BT, Adams JR, et al. Transcriptomic classification of genetically engineered mouse models of breast cancer identifies human subtype counterparts. Genome biology. 2013;14(11):R125. doi: 10.1186/gb-2013-14-11-r125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Herschkowitz JI, Simin K, Weigman VJ, Mikaelian I, Usary J, Hu Z, et al. Identification of conserved gene expression features between murine mammary carcinoma models and human breast tumors. Genome biology. 2007;8(5):R76. doi: 10.1186/gb-2007-8-5-r76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Slamon DJ, Clark GM, Wong SG, Levin WJ, Ullrich A, McGuire WL. Human breast cancer: correlation of relapse and survival with amplification of the HER-2/neu oncogene. Science. 1987;235(4785):177–82. doi: 10.1126/science.3798106. [DOI] [PubMed] [Google Scholar]
- 37.Revillion F, Bonneterre J, Peyrat JP. ERBB2 oncogene in human breast cancer and its clinical significance. European journal of cancer (Oxford, England : 1990) 1998;34(6):791–808. doi: 10.1016/s0959-8049(97)10157-5. [DOI] [PubMed] [Google Scholar]
- 38.Cancer Genome Atlas Network. Comprehensive molecular portraits of human breast tumours. Nature. 2012;490(7418):61–70. doi: 10.1038/nature11412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Reddy JP, Peddibhotla S, Bu W, Zhao J, Haricharan S, Du Y-CN, et al. Defining the ATM-mediated barrier to tumorigenesis in somatic mammary cells following ErbB2 activation. Proceedings of the National Academy of Sciences. 2010;107(8):3728–33. doi: 10.1073/pnas.0910665107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Haricharan S, Hein SM, Dong J, Toneff MJ, Aina OH, Rao PH, et al. Contribution of an alveolar cell of origin to the high-grade malignant phenotype of pregnancy-associated breast cancer. Oncogene. 2013 doi: 10.1038/onc.2013.521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Rakha EA, Reis-Filho JS, Ellis IO. Basal-Like Breast Cancer: A Critical Review. Journal of Clinical Oncology. 2008;26(15):2568–81. doi: 10.1200/JCO.2007.13.1748. [DOI] [PubMed] [Google Scholar]
- 42.Reis-Filho JS, Milanezi F, Steele D, Savage K, Simpson PT, Nesland JM, et al. Metaplastic breast carcinomas are basal-like tumours. Histopathology. 2006;49(1):10–21. doi: 10.1111/j.1365-2559.2006.02467.x. [DOI] [PubMed] [Google Scholar]
- 43.Zhang M, Behbod F, Atkinson RL, Landis MD, Kittrell F, Edwards D, et al. Identification of Tumor-Initiating Cells in a p53-Null Mouse Model of Breast Cancer. Cancer Research. 2008;68(12):4674–82. doi: 10.1158/0008-5472.CAN-07-6353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Reddy J, Li Y. The RCAS-TVA System for Introduction of Oncogenes into Selected Somatic Mammary Epithelial Cells in Vivo. Journal of Mammary Gland Biology and Neoplasia. 2009;14(4):405–9. doi: 10.1007/s10911-009-9157-1. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.