Abstract
Multiple genetic variants have been linked to risk of Parkinson disease (PD), but known mutations do not explain a large proportion of the total PD cases. Similarly, multiple loci have been associated with PD risk by Genome-Wide Association Studies (GWAS). The influence that genetic factors confer upon phenotypic diversity remains unclear. Few studies have been performed to determine whether the GWAS loci are also associated with age at onset (AAO) or motor progression. We used two PD case-control datasets (Washington University and the Parkinson’s Progression Markers Initiative) to determine whether polymorphisms located at the GWAS top hits (GBA, ACMSD/TMEM163, STK39, MCCC1/LAMP3, GAK/TMEM175, SNCA, and MAPT) show association with AAO or motor progression. We found associations between SNPs at the GBA and MAPT loci and PD AAO and progression. These findings reinforce the complex genetic basis of PD and suggest that distinct genes and variants explain the genetic architecture of PD risk, onset, and progression.
Keywords: Parkinson Disease, Age at Onset, Motor Progression, GBA, SCNA, MAPT
INTRODUCTION
Parkinson disease (PD) is a neurodegenerative disorder with a complex etiologic basis including genetic and environmental factors. The discovery of mutations in a small number of genes associated with autosomal dominant and autosomal recessive forms of PD (including SNCA, LRRK2, PRKN, PINK1, DJ-1, and others) have shed considerable light on the pathophysiology of PD, but these disease-causing gene mutations only account for a small minority of PD cases [1-6]. Careful analysis of the remaining majority of PD cases that do not have a clear Mendelian inheritance pattern has demonstrated that variants in multiple genes may influence PD risk rather than cause disease. Over the last decade, multiple genome-wide association studies (GWAS) have identified more than twenty loci that each confer relatively small risk or protective effects [7-12]. Interestingly, several of these loci are near genes that cause autosomal forms of PD (e.g. SNCA, LRRK2), though the majority represent novel associations. Several of these loci are in close proximity to genes associated with other neurodegenerative diseases (MAPT), inflammation (STK39), or neurosecretory function (MCCC1/LAMP3, SYT11/RAB25), providing clues to their potential role in PD pathophysiology.
Despite the substantial progress in discovery of PD risk loci over the last decade, the risk attributable to identified genetic variants still does not entirely account for the genetic heritability observed in PD. This gap indicates that multiple genetic variants not yet described likely play a role in PD pathophysiology. Recently, Keller et al. used a statistical model termed genome-wide complex trait analysis (GCTA) to quantify so-called “missing heritability” in PD. They estimated that genetic factors could explain up to 27% of the PD cases, in contrast to the estimated 3-5% attributable to the top single nucleotide polymorphisms (SNPs) identified in GWAS studies to date [13]. This finding suggests that many more genetic variants yet to be discovered may influence PD risk and could represent novel therapeutic targets.
In addition to influencing risk of PD, genetic variants may also play a role in specific disease characteristics. Phenotypic diversity characterizes PD with variability of the pattern of motor manifestations, the range of age at onset, the difference in rate of progression, responsiveness to dopaminergic treatment, and the presence and progression of comorbid neuropsychiatric features including dementia and depression. Several studies have investigated the genetic basis of this diversity and have identified associations between multiple SNPs and disease phenotypes including age at onset, progression, and motor complications [14-24]. A recent analysis using a composite polygenic risk score compiled from multiple GWAS studies reported a significant association between higher polygenic risk scores and earlier age at onset [25]. Taken together these studies support the hypothesis that common genetic variants regulate specific PD phenotypes and underscore the need for further work in this area to identify trait-specific alleles.
In this study, we investigated the effect of common genetic variants on PD risk and other disease-related phenotypes: age at onset, and progression. We used data from the PDGene database to select 23 SNPs associated with PD. We then analyzed data from two studies of PD patients and healthy controls from the Washington University Movement Disorders Center and the Parkinson’s Progression Markers Initiative (PPMI) to determine associations between these SNPs and PD risk, age at onset, and rate of disease progression.
MATERIALS AND METHODS
Subjects
PD patients and healthy controls were recruited from the Washington University in Saint Louis Movement Disorder Center (WU). Appropriate written informed consent was obtained from all subjects and the study protocol was approved by the Institutional Review Board. A clinical diagnosis of PD satisfied United Kingdom Brain Bank criteria [26], modified for genetic studies [27]. Data from subjects in the Parkinson’s Progression Markers Initiative (PPMI) study were obtained from the PPMI database (www.ppmi-info.org), accessed most recently on December 12, 2014. All individuals were of European descent and written consent was obtained from all participants.
Demographic characteristics have been described previously [28] and are listed in Table 1. The Washington University series contained 418 patients with clinical diagnoses of PD and 306 unaffected and unrelated control subjects. The mean age at onset was 60.4 ± 11.1 years for cases and the mean age at inclusion was 72.4.0 ± 15.1 years for controls. The PPMI series contained 368 patients with clinical diagnoses of PD and 150 unaffected and unrelated control subjects. The mean age at onset was 61.4 ± 9.9 years for cases and the mean age at inclusion was 60.9 ± 11.4 years for controls. In the Washington University series, 25% of cases reported a positive family history of PD, compared to 9% of cases in the PPMI series. Family history was defined as any family history and not restricted to first-degree relatives. The two series had a similar percentage of male cases, whereas the Washington University series had a higher percentage of female controls.
Table 1. Subject Characteristics.
WU, Washington University; PPMI, Parkinson’s Progression Markers Initiative
Demographic data for subjects in the Washington University (WU) and Parkinson’s Progression Markers Initiative (PPMI) datasets are shown.
| WU | PPMI | |||
|---|---|---|---|---|
| Sex, n (%) | Controls | Cases | Controls | Cases |
| Male | 105 (34) | 258 (62) | 100 (67) | 240 (65) |
| Female | 201 (66) | 160 (38) | 50 (33) | 128 (35) |
| Total | 306 | 418 | 150 | 368 |
| Age (years), mean (SD), range | Controls (inclusion) | Cases (onset) | Controls (inclusion) | Cases (onset) |
| 72.4 (15.1) | 60.4 (11.1) | 60.9 (11.4) | 61.4 (9.9) | |
| Ethnicity (%) | Controls | Cases | Controls | Cases |
| Caucasian | 81.7 | 92.8 | 92.7 | 95.4 |
| Hispanic | 0.3 | 0.0 | 1.3 | 2.4 |
| American Indian/Alaska Native | 0.0 | 0.2 | 0.0 | 0.0 |
| African American | 0.0 | 0.0 | 6.0 | 1.6 |
| Unknown | 18.0 | 6.9 | 0.0 | 0.5 |
| PD Family History (%) | Controls | Cases | Controls | Cases |
| Positive | 0.0 | 25.1 | 0.0 | 9.2 |
| Negative | 88.6 | 67.9 | 99.3 | 90.2 |
| Unknown | 11.4 | 6.9 | 0.7 | 0.5 |
Genotyping and Selection of SNPs
All WU and PPMI samples were genotyped using the Illumina Immunochip and NeuroX. Prior to association analysis, all samples and genotypes underwent stringent QC. Genotype data was cleaned using PLINK v1.07 (http://pngu.mgh.harvard.edu/purcell/plink/) by applying a minimum call rate for SNPs and individuals (98%) and minimum minor allele frequencies (MAF=0.02) [29]. SNPs not in Hardy-Weinberg equilibrium (P<1×10−6) were excluded. We tested for unanticipated duplicates and cryptic relatedness (Pihat ≥0.5) using pairwise genome-wide estimates of proportion identity-by-descent using PLINK v1.07. When a pair of identical samples or a pair of samples with cryptic relatedness was identified, the sample with a higher number of SNPs that passed QC was prioritized. Eigenstrat was used for each cohort separately to calculate principal component factors for each sample and confirm the ethnicity of the samples. GBA coding variants were extracted from the NeuroX genotyping array. The NeuroX array underwent the same QC steps as the Immunochip, but removing variants based on the MAF.
Selection of SNPs for Analysis: We extracted data for 23 variants that showed significant association with PD risk based in part on recent large-scale meta-analysis of GWAS data [9]. The extracted SNPs tag genome-wide significance signals for the GBA (1q21), ACMSD/TMEM163 (2q21), STK39 (2q24), MCCC1/LAMP3 (3q27), GAK/TMEM175 (4p16), SNCA (4q21), and MAPT (17q21) loci.
Statistical Analyses
We used the PLINK whole genome association analysis toolset to analyze the association of variants with PD risk. Odds Ratios (OR) and p-values with risk for PD were calculated using logistic regression including APOE, sex, study site, and the two principal component factors as covariates. Association with AAO was carried out using the Kaplan-Meier method and tested for significant differences, using a log-rank test, with study site, sex, and family history included as covariates using the PROC LIFETEST model (SAS Institute, Inc., Cary, NC). Association with rate of disease progression was evaluated as described previously [30]. Briefly, progression of disease was measured by the change in Hoehn and Yahr stage or UPDRS score per year. Only individuals with at least three serial clinic visits spanning at least 1 year were included. The change in Hoehn and Yahr stage or UPDRS score per year fitted a linear model in both series and therefore we used a mixed linear model (PROC MIXED; SAS Institute Inc) to determine whether there is a relationship between the slope of the Hoehn and Yahr stage or UPDRS score and time as a function of genotype after controlling for study site, sex, family history, age at onset, and initial score included as covariates. The intra-class correlation coefficient for UPDRS measurements among raters in the WU study group was at least 0.85. The goal of this study is to analyze whether the genetic variants associated with PD risk associated with age at onset or progression. To do this we tested three different phenotypes: age at onset, change in UDPRS and change of H/Y scores. For this reason we performed a multiple test correction based on the number of phenotypes tested: α=0.05/3=0.0167
RESULTS
Replication of Association between SNPs in GBA, SNCA and PD Risk
After combining the two datasets, we found significant association between PD risk and multiple SNPs that have been previously implicated in PD (Table 2, n=456 controls, 786 cases). Consistent with magnitudes reported in previous studies [9, 31], we found the largest risk effect for variants in GBA, with odds ratios of 4.46 and 4.975 for rs76763715 (N370S, MAF 0.00665) and rs75548401 (K26R, MAF 0.01173), respectively (Table 2). Four SNPs on chromosome 4 near the SNCA gene, rs356219, rs356220, rs356165 and rs2736990 were associated with increased PD risk (p=0.00467, 0.003729, 0.003201, and 0.002784, respectively, Table 2). We also report a trend towards increased PD risk for rs2390669, a SNP in an intron region of STK39 that has been associated with PD risk in previous studies [9].
Table 2. Summary of significant loci.
Location, nearest gene, and PD risk odds ratios for 22 variants in the combined WU and PPMI dataset. Variants with significant association (p<0.05) are indicated in bold. SNP, single nucleotide polymorphism; Chr, chromosome; MAF, minor allele frequency; OR, odds ratio; 95% CI, confidence interval.
| Position (bp) |
Minor allele |
Major allele |
PD Risk | ||||||
|---|---|---|---|---|---|---|---|---|---|
| SNP | Chr | Nearest Gene(s) | MAF | OR | 95% CI | p value | |||
| rs76763715 | 1 | GBA (N370S) | 155205634 | C | T | 0.007 | 4.46 | 0.99-19.88 | 0.050 |
| rs75548401 | 1 | GBA (K26R) | 155206037 | A | G | 0.012 | 4.98 | 1.48-16.66 | 9.25E-03 |
| rs10928513 | 2 | ACMSD/TMEM163 | 135173229 | C | T | 0.430 | 1.13 | 0.95-1.32 | 0.143 |
| rs6723108 | 2 | ACMSD/TMEM163 | 135196450 | G | T | 0.432 | 1.13 | 0.96-1.33 | 0.132 |
| rs2390669 | 2 | STK39 | 168800188 | C | A | 0.149 | 1.28 | 0.99-1.64 | 0.058 |
| rs11711441 | 3 | MCCC1/LAMP3 | 184303969 | A | G | 0.132 | 0.84 | 0.66-1.07 | 0.160 |
| rs6599388 | 4 | GAK/TMEM175 | 929087 | T | C | 0.328 | 1.12 | 0.94-1.32 | 0.207 |
| rs356219 | 4 | SNCA | 90856624 | G | A | 0.410 | 1.28 | 1.08-1.52 | 4.67E-03 |
| rs11931074 | 4 | SNCA | 90858538 | T | G | 0.099 | 1.00 | 0.75-1.31 | 0.983 |
| rs356220 | 4 | SNCA | 90860363 | T | C | 0.416 | 1.29 | 1.08-1.52 | 3.73E-03 |
| rs356165 | 4 | SNCA | 90865909 | A | G | 0.464 | 1.30 | 1.09-1.54 | 3.20E-03 |
| rs3822086 | 4 | SNCA | 90883817 | T | C | 0.098 | 0.99 | 0.75-1.30 | 0.944 |
| rs3857059 | 4 | SNCA | 90894261 | G | A | 0.098 | 0.99 | 0.75-1.30 | 0.944 |
| rs2736990 | 4 | SNCA | 90897564 | T | C | 0.497 | 0.78 | 0.65-0.91 | 2.78E-03 |
| rs2942168 | 17 | CRHR1/MAPT | 41070633 | T | C | 0.194 | 0.99 | 0.80-1.22 | 0.961 |
| rs393152 | 17 | CRHR1/MAPT | 41074926 | G | A | 0.201 | 0.95 | 0.77-1.17 | 0.639 |
| rs7215239 | 17 | CRHR1/MAPT | 41123556 | C | T | 0.221 | 0.96 | 0.78-1.16 | 0.672 |
| rs1981997 | 17 | MAPT | 41412603 | A | G | 0.193 | 0.97 | 0.78-1.19 | 0.784 |
| rs1052553 | 17 | MAPT | 41429726 | G | A | 0.194 | 0.98 | 0.79-1.20 | 0.826 |
| rs17652121 | 17 | MAPT | 41429810 | C | T | 0.193 | 0.97 | 0.78-1.19 | 0.784 |
| rs8070723 | 17 | MAPT | 41436901 | G | A | 0.195 | 0.96 | 0.78-1.18 | 0.734 |
| rs199533 | 17 | NSF/MAPT | 42184098 | T | C | 0.183 | 0.94 | 0.754-1.16 | 0.547 |
rs75548401 (GBA) and the H1/H2 MAPT Haplotype Influence Age at Onset
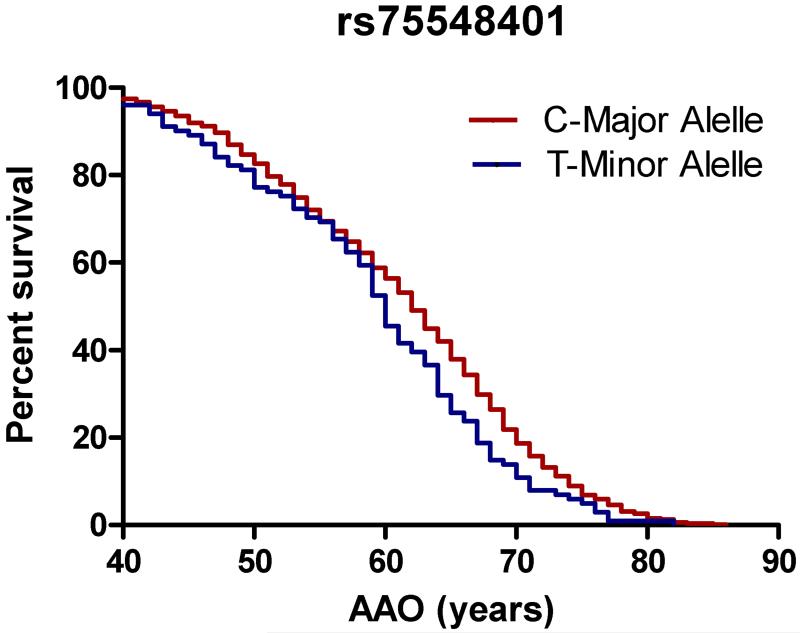
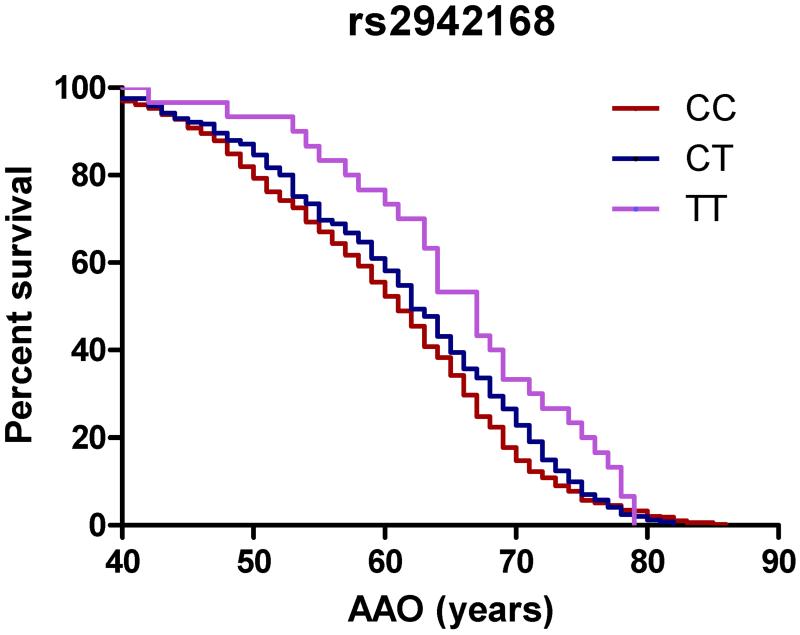
In a subgroup analysis of PD cases only (n=786), we found a significant association between rs75548401 in GBA and younger AAO (p=0.0126; Table 3 and Figure 1). We also found significant associations between older AAO and multiple SNPs on chromosome 17 (Table 3 and Figure 1). This genomic region is near MAPT and has recently also been linked to several other genes including CRHR1 and NSF. In contrast with a previous study reporting association with younger AAO in subjects with rs356165 (SNCA), we did not observe an association with this SNP and AAO in our study population [17].
Table 3. rs75548401 and multiple SNPs at the MAPT locus relate to age at onset.
The age at onset of Parkinson disease motor symptoms was significantly associated with rs75548401 and with multiple SNPs at the MAPT locus. A positive test statistic indicates that the minor allele related to younger age at onset; a negative test statistic indicates that the minor allele related to older age at onset.
| SNP | Chr | Nearest Gene(s) | Test Statistic | Standard Error | p value |
|---|---|---|---|---|---|
| rs76763715 | 1 | GBA (N370S) | 2.646 | 3.284 | 0.420 |
| rs75548401 | 1 | GBA (K26R) | 9.185 | 3.681 | 0.013 |
| rs10928513 | 2 | ACMSD/TMEM163 | −4.540 | 18.923 | 0.810 |
| rs6723108 | 2 | ACMSD/TMEM163 | −1.750 | 18.982 | 0.927 |
| rs2390669 | 2 | STK39 | −3.667 | 13.287 | 0.783 |
| rs11711441 | 3 | MCCC1/LAMP3 | −1.732 | 11.974 | 0.885 |
| rs6599388 | 4 | GAK/TMEM175 | 22.040 | 19.211 | 0.251 |
| rs356219 | 4 | SNCA | −0.537 | 17.916 | 0.976 |
| rs11931074 | 4 | SNCA | 4.383 | 10.994 | 0.690 |
| rs356220 | 4 | SNCA | 4.954 | 18.120 | 0.785 |
| rs356165 | 4 | SNCA | 6.199 | 17.714 | 0.726 |
| rs3822086 | 4 | SNCA | 3.757 | 10.984 | 0.732 |
| rs3857059 | 4 | SNCA | 3.757 | 10.984 | 0.732 |
| rs2736990 | 4 | SNCA | −4.456 | 18.863 | 0.813 |
| rs2942168 | 17 | CRHR1/MAPT | −38.813 | 15.662 | 0.013 |
| rs393152 | 17 | CRHR1/MAPT | −35.708 | 15.692 | 0.023 |
| rs7215239 | 17 | CRHR1/MAPT | −28.984 | 15.795 | 0.067 |
| rs1981997 | 17 | MAPT | −38.052 | 15.650 | 0.015 |
| rs1052553 | 17 | MAPT | −38.849 | 15.655 | 0.013 |
| rs17652121 | 17 | MAPT | −38.052 | 15.650 | 0.015 |
| rs8070723 | 17 | MAPT | −39.225 | 15.659 | 0.012 |
| rs199533 | 17 | NSF/MAPT | −31.585 | 14.611 | 0.031 |
Figure 1. rs75548401 and multiple SNPs at the MAPT locus relate to age at onset.
Kaplan-Meier survival curves for age at onset of Parkinson disease motor symptoms are shown for alleles of the GBA variant rs75548401 (a) and rs2942168 (b), a representative variant at the MAPT locus. Rs75548401: mean survival 62 vs 60 years. Hazard ratio: 0.7497 (0.509-0.9512), p=0.0126. Rs2942168: mean survival 61 vs. 62 vs. 67 years. Hazard ratio: 1.17 (1.007-1.36), p=0.0132.
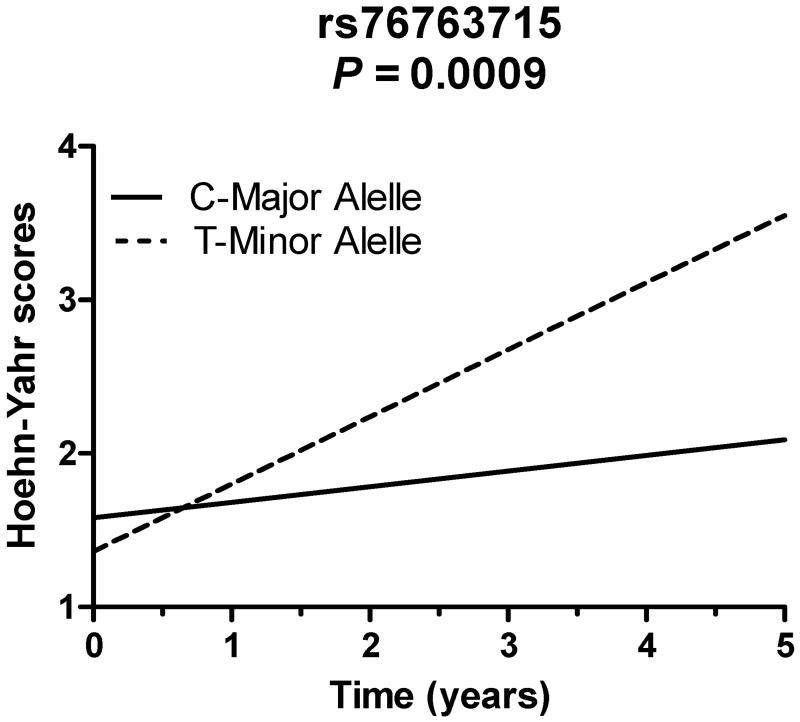
rs76763715 (GBA) is associated with faster Motor Progression
We used serial assessments of Hoehn and Yahr stage to calculate rates of motor progression for PD cases. In the combined WU/PPMI dataset, Hoehn and Yahr scores were analyzed from 425 cases over a mean of 2.7 years. Carriers of the minor allele of rs76763715 (GBA) demonstrated a faster rate of progression of Hoehn and Yahr stage compared to non-carriers (Table 4 and Figure 2, p=9.00 ×10−4). We also analyzed serial measurements of UPDRS-III (motor subset) as another marker of disease progression. To eliminate the potentially confounding effect of dopaminergic medication, we restricted our analysis of UPDRS-III data to scores recorded in the OFF state only. In the WU dataset, UPDRS-III OFF-state scores were analyzed from 110 patients over a mean of 6.8 years. The rate of UPDRS-III progression did not relate to any of the SNPs we examined (Table 5). In similar analyses performed in the WU dataset using UPDRS-III scores measured in both the ON and OFF state, and in the PPMI cohort (in which ON/OFF state was not specified), UPDRS-III score progression also did not relate to any SNPs examined (data not shown). Given the strong contribution of postural stability on the Hoehn and Yahr stage, we performed a subset analysis for progression of UPDRS-III postural stability scores but we did not find an association between rs76763715 (GBA) and postural stability.
Table 4. rs76763715 relates to more rapid progression of Hoehn and Yahr stage.
P values for association of Hoehn and Yahr stage for cases in the combined WU and PPMI dataset are shown. Cases with at least 3 scores measured over a minimum of 1 year were included.
| SNP | Chr | Nearest Gene (s) | p value |
|---|---|---|---|
| rs76763715 | 1 | GBA (N370S) | 9.0E-04 |
| rs75548401 | 1 | GBA (K26R) | 0.682 |
| rs10928513 | 2 | ACMSD/TMEM163 | 0.576 |
| rs6723108 | 2 | ACMSD/TMEM163 | 0.599 |
| rs2390669 | 2 | STK39 | 0.107 |
| rs11711441 | 3 | MCCC1/LAMP3 | 0.495 |
| rs6599388 | 4 | GAK/TMEM175 | 0.206 |
| rs356219 | 4 | SNCA | 0.575 |
| rs11931074 | 4 | SNCA | 0.812 |
| rs356220 | 4 | SNCA | 0.622 |
| rs356165 | 4 | SNCA | 0.591 |
| rs3822086 | 4 | SNCA | 0.820 |
| rs3857059 | 4 | SNCA | 0.821 |
| rs2736990 | 4 | SNCA | 0.287 |
| rs2942168 | 17 | CRHR1/MAPT | 0.456 |
| rs393152 | 17 | CRHR1/MAPT | 0.380 |
| rs7215239 | 17 | CRHR1/MAPT | 0.461 |
| rs1981997 | 17 | MAPT | 0.456 |
| rs1052553 | 17 | MAPT | 0.456 |
| rs17652121 | 17 | MAPT | 0.456 |
| rs8070723 | 17 | MAPT | 0.465 |
| rs199533 | 17 | NSF/MAPT | 0.744 |
Figure 2.
rs76763715 relates to more rapid progression of Hoehn and Yahr stage
Table 5. No association between genetic variants and progression of UPDRS scores.
P values for association of serial measurements of UPDRS-III scores for cases in the WU group are shown for each variant. Cases with at least 3 scores measured over a minimum of 1 year were included. Only UPDRS-III scores measured in the OFF state were included in the final analysis. Inclusion of UPDRS-III scores measured in the ON state did not significantly change the results (not shown).
| SNP | Chr | Nearest Gene(s) | p value |
|---|---|---|---|
| rs76763715 | 1 | GBA (N370S) | N/A |
| rs75548401 | 1 | GBA (K26R) | 0.975 |
| rs10928513 | 2 | ACMSD/TMEM163 | 0.345 |
| rs6723108 | 2 | ACMSD/TMEM163 | 0.918 |
| rs2390669 | 2 | STK39 | 0.620 |
| rs11711441 | 3 | MCCC1/LAMP3 | 0.894 |
| rs6599388 | 4 | GAK/TMEM175 | 0.223 |
| rs356219 | 4 | SNCA | 0.714 |
| rs11931074 | 4 | SNCA | 0.801 |
| rs356220 | 4 | SNCA | 0.768 |
| rs356165 | 4 | SNCA | 0.768 |
| rs3822086 | 4 | SNCA | 0.248 |
| rs3857059 | 4 | SNCA | 0.455 |
| rs2736990 | 4 | SNCA | 0.349 |
| rs2942168 | 17 | CRHR1/MAPT | 0.741 |
| rs393152 | 17 | CRHR1/MAPT | 0.741 |
| rs7215239 | 17 | CRHR1/MAPT | 0.686 |
| rs1981997 | 17 | MAPT | 0.741 |
| rs1052553 | 17 | MAPT | 0.741 |
| rs17652121 | 17 | MAPT | 0.741 |
| rs8070723 | 17 | MAPT | 0.741 |
| rs199533 | 17 | NSF/MAPT | 0.845 |
Discussion
Consistent with previous reports [7, 9, 15, 17-19, 31], we found nominal associations in the WU and PPMI datasets between SNPs in GBA and SNCA and PD risk. While we also observed a trend towards significance with a SNP near STK39, we found no relationship between PD risk in our population and other SNPs across multiple genes previously linked to PD in other populations, suggesting that our study is underpowered to replicate all the known risk loci. On the other hand, these findings indicate that GBA and SNCA present larger effect sizes than the other loci in this predominantly European-American population.
We also investigated the effect that common genetic variants have on specific phenotypic characteristics of PD, including age at onset and motor progression. Previous studies in multiple genetically diverse cohorts of PD patients have found associations between age at onset (AAO) of PD and SNPs in multiple genes including GBA, SNCA, MAPT, and COMT [15, 17-22]. A meta-analysis of GWAS data from three studies of both familial and idiopathic PD cases found several interesting (albeit non-significant at the genome-wide level) associations with AAO and several SNPs near genes implicated in melanin synthesis, protein misfolding pathways, and vesicular transport, all of which are important cell biological processes in PD and other neurodegenerative disorders [32]. A recent study using polygenic score for PD risk found that individuals with earlier onset also have higher polygenic risk, indicating that some of the genetic variants associated with PD risk also affect AAO [25].
Variants in the GBA gene, relatively common among PD patients although rare in the general population, have been shown to relate to earlier age at onset as well as prominent non-motor symptoms in PD patients, including cognitive impairment, depression, sleep disturbance, and autonomic dysfunction [33]. Interestingly, the GBA variant rs76763715 (N370S), common in Ashkenazi Jews and one of the most frequently studied GBA variants, did not relate to AAO in our dataset. However, rs75548401 (K26R), first reported as a novel variant in a study of North African PD patients, was significantly associated in our dataset with both PD risk and younger AAO, but not progression [34].
Several groups have previously reported association between variants at the MAPT locus and AAO in populations of specific ethnic background, including Japanese and Indian subjects, and in families carrying mutations in LRRK2 [20, 21, 35, 36]. To our knowledge, our study is the first to report an association between AAO and variants in MAPT in a non-LRRK2-associated Caucasian population. As in the LRRK2 families, we found that the minor alleles of these MAPT SNPs (H2) were associated with older age at onset of PD. The implications of this finding are not immediately clear, but it should be noted that a recent large meta-analysis found a lower risk of PD with a MAPT variant, consistent with our finding that the H2 haplotype may play a protective role in PD [31]. The fact that we did not find an association between MAPT variants and PD risk in our dataset may suggest that MAPT has a larger effect on AAO than on overall PD risk, or that MAPT influences AAO in patients already at risk of PD due to variants in other genes.
Several studies have also reported that the H2 haplotype is protective for AD risk as well as in progressive supranuclear palsy and cortico-basal degeneration, both of which have pure tau pathology [37-39]. It is not immediately clear why the H2 haplotype is also protective in PD, which has primarily alpha-synuclein pathology with concurrent tau pathology in only a subset of patients [40]. A recent study identified some genetic overlap between AD and PD at the MAPT locus [41], although previous studies failed to identify any genetic overlap at the genome-wide level [42].
It is worth noting that there are several other genes in addition to MAPT located on chromosome 17q21, within the H1/H2 haplotype. While variants in MAPT likely explain the association between the H1 haplotype region and neurodegenerative disorders with prominent tau pathology including progressive supranuclear palsy and cortico-basal degeneration, it is possible that other genes within this region may be important in PD, in which pathological tau aggregates are occasionally present in conjunction with alpha-synuclein aggregates. The CRHR1 gene encodes the corticotropin-releasing hormone receptor and a variant in CRHR1 was recently shown to be associated with decreased PD risk in a large meta-analysis [31]. Brains from PD patients have reduced levels of CRF-like immunoreactivity [43], and urocortin, a CRF-like peptide, reduces nigrostriatal damage in an rat model of parkinsonism [44]. The NSF gene encodes N-ethylmaleimide sensitive fusion protein, an ATPase that plays an integral role in SNARE complex biology and synaptic neurotransmission. A growing literature supports an important link between alpha-synuclein and SNARE proteins [45, 46], and NSF may confer an independent genetic risk factor in PD [47]. Further work is required to clarify the role that genetic variation in CRHR1 and NSF plays in PD.
Genetic variants have also been recently linked to motor progression in PD. A variant in the promoter region of SNCA was linked with faster progression of UPDRS-III scores in a California case-control study [23]. In a longitudinal, community-based study in the United Kingdom, a group of GBA variants (including N370S and L444P) were shown to relate to progression of Hoehn and Yahr stage, although individual analysis was not reported for each variant [24]. A similar but smaller study in a German population also reported more rapid progression of UPDRS-III scores and Hoehn and Yahr stage in patients carrying the same GBA variants, although results were again not reported for each variant individually [16]. In our dataset, we found an association with rs76763715 (N370S) and faster rate of progression of Hoehn and Yahr stage, but no relationship to UPDRS-III scores.
Motor progression is variable in PD and while genetic polymorphisms likely contribute to this variability, modeling these relationships is challenging. Clinical scores of motor symptoms (e.g. UPDRS-III) are routinely collected in movement disorders centers, and many academic centers including ours have good intra-rater and inter-rater reliability of these measurements [48, 49]. However, few studies have reported positive associations between genetic variants and motor progression. There are multiple potential explanations for this, including genetic diversity of the study population, length of follow-up and number of visits, and that disease progression is unlikely to remain linear. In our study, the length of follow up was variable and included subjects with relatively short follow up intervals, which may have compromised our ability to detect significant changes in UPDRS-III scores. Further work is needed to better understand the contribution of genetic variants to disease progression.
This study has several limitations. The number of individuals analyzed in our combined dataset from WU and PPMI is considerably smaller than recent meta-analyses from multicenter collaborations, and as such we were underpowered to detect some effects, particularly those with small effect size. Also, our analysis consisted of a single phase, in contrast to other studies which incorporated separate discovery and replication phases. Given that the variants we analyzed have been reported in previous studies, we feel that the lack of a two-phase approach does not significantly impair our conclusions.
Our data confirm the strong effect of GBA and SNCA on PD risk, and we report novel associations between variants in GBA and MAPT and specific disease phenotypes including AAO and motor progression. Our results also suggest that variants that tag the H1/H2 haplotype may influence AAO. When taken together with other recent reports, our data indicate that specific genes and variants differentially influence aspects of PD risk and phenotype. Certain variants appear to have a strong effect on disease risk, while other variants also modify age at onset or disease progression, suggesting that distinct aspects of PD have a specific genetic architecture. Focusing additional research on these different aspects of PD will likely lead to the discovery of additional genes and/or variants. Further work is necessary to better characterize the genetic architecture of PD, to determine whether disease risk, onset, and progression share common pathophysiologic mechanisms, and to help identify targets for diagnosis and therapeutic intervention.
Parkinson Disease (PD) has a complex genetic etiology.
We identified variants in GBA and SNCA that contribute to PD risk.
We identified variants in GBA, SNCA, and MAPT that influence disease phenotypes including age at onset and motor progression.
Additional studies are likely to identify novel PD genes and facilitate new treatments.
Acknowledgments
The authors thank Susan Loftin and Karen Klumpp for their expert technical assistance. This work was supported by grants from NINDS (NS075321, NS41509, NS058714, and R01AG044546); the Barnes Jewish Hospital Foundation (BJHF); the American Parkinson Disease Association (APDA) Advanced Research Center for Parkinson Disease at Washington University in St. Louis; the Greater St. Louis Chapter of the APDA; the Barnes Jewish Hospital Foundation (Elliot Stein Family Fund and Parkinson Disease Research Fund), The Michael J. Fox Foundation for Parkinson’s Research, Alzheimer’s Association and Weston Brain Institute This research was conducted while CC was a recipient of a New Investigator Award in Alzheimer’s disease from the American Federation for Aging Research. CC is a recipient of a BrightFocus Foundation Alzheimer’s Disease Research Grant (A2013359S).
Data used in the preparation of this article were obtained from the Parkinson’s Progression Markers Initiative (PPMI) database (www.ppmi-info.org/data). For up-to-date information on the study, visit www.ppmi-info.org.
PPMI –a public-private partnership –is funded by the Michael J. Fox Foundation for Parkinson’s Research and funding partners, including Abbvie, Avid Radiopharmaceuticals, Biogen Idec, Bristol-Myers Squibb, Covance, GE Healthcare, Genentech, GlaxoSmithKline, Lilly, Lundbeck, Merck, Meso Scale Discovery, Pfizer, Piramal, Roche, UCB.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Polymeropoulos MH, et al. Mutation in the alpha-synuclein gene identified in families with Parkinson’s disease. Science. 1997;276(5321):2045–7. doi: 10.1126/science.276.5321.2045. [DOI] [PubMed] [Google Scholar]
- 2.Paisan-Ruiz C, et al. Cloning of the gene containing mutations that cause PARK8-linked Parkinson’s disease. Neuron. 2004;44(4):595–600. doi: 10.1016/j.neuron.2004.10.023. [DOI] [PubMed] [Google Scholar]
- 3.Zimprich A, et al. Mutations in LRRK2 cause autosomal-dominant parkinsonism with pleomorphic pathology. Neuron. 2004;44(4):601–7. doi: 10.1016/j.neuron.2004.11.005. [DOI] [PubMed] [Google Scholar]
- 4.Abbas N, et al. French Parkinson’s Disease Genetics Study Group and the European Consortium on Genetic Susceptibility in Parkinson’s Disease A wide variety of mutations in the parkin gene are responsible for autosomal recessive parkinsonism in Europe. Hum Mol Genet. 1999;8(4):567–74. doi: 10.1093/hmg/8.4.567. [DOI] [PubMed] [Google Scholar]
- 5.Bonifati V, et al. Mutations in the DJ-1 gene associated with autosomal recessive early-onset parkinsonism. Science. 2003;299(5604):256–9. doi: 10.1126/science.1077209. [DOI] [PubMed] [Google Scholar]
- 6.Bonifati V, et al. Early-onset parkinsonism associated with PINK1 mutations: frequency, genotypes, and phenotypes. Neurology. 2005;65(1):87–95. doi: 10.1212/01.wnl.0000167546.39375.82. [DOI] [PubMed] [Google Scholar]
- 7.International Parkinson’s Disease Genomics Consortium, W.T.C.C.C. A two-stage meta-analysis identifies several new loci for Parkinson’s disease. PLoS Genet. 2011;7(6):e1002142. doi: 10.1371/journal.pgen.1002142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Maraganore DM, et al. High-resolution whole-genome association study of Parkinson disease. Am J Hum Genet. 2005;77(5):685–93. doi: 10.1086/496902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Nalls MA, et al. Imputation of sequence variants for identification of genetic risks for Parkinson’s disease: a meta-analysis of genome-wide association studies. Lancet. 2011;377(9766):641–9. doi: 10.1016/S0140-6736(10)62345-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Pankratz N, et al. Genomewide association study for susceptibility genes contributing to familial Parkinson disease. Hum Genet. 2009;124(6):593–605. doi: 10.1007/s00439-008-0582-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Satake W, et al. Genome-wide association study identifies common variants at four loci as genetic risk factors for Parkinson’s disease. Nat Genet. 2009;41(12):1303–7. doi: 10.1038/ng.485. [DOI] [PubMed] [Google Scholar]
- 12.Simon-Sanchez J, et al. Genome-wide association study reveals genetic risk underlying Parkinson’s disease. Nat Genet. 2009;41(12):1308–12. doi: 10.1038/ng.487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Keller MF, et al. Using genome-wide complex trait analysis to quantify ‘missing heritability’ in Parkinson’s disease. Hum Mol Genet. 2012;21(22):4996–5009. doi: 10.1093/hmg/dds335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wang J, Liu ZL, Chen B. Association study of dopamine D2, D3 receptor gene polymorphisms with motor fluctuations in PD. Neurology. 2001;56(12):1757–9. doi: 10.1212/wnl.56.12.1757. [DOI] [PubMed] [Google Scholar]
- 15.Aharon-Peretz J, Rosenbaum H, Gershoni-Baruch R. Mutations in the glucocerebrosidase gene and Parkinson’s disease in Ashkenazi Jews. N Engl J Med. 2004;351(19):1972–7. doi: 10.1056/NEJMoa033277. [DOI] [PubMed] [Google Scholar]
- 16.Brockmann K, et al. GBA-associated Parkinson’s disease: Reduced survival and more rapid progression in a prospective longitudinal study. Mov Disord. 2014 doi: 10.1002/mds.26071. [DOI] [PubMed] [Google Scholar]
- 17.Cardo LF, et al. A search for SNCA 3′ UTR variants identified SNP rs356165 as a determinant of disease risk and onset age in Parkinson’s disease. J Mol Neurosci. 2012;47(3):425–30. doi: 10.1007/s12031-011-9669-1. [DOI] [PubMed] [Google Scholar]
- 18.Clark LN, et al. Mutations in the glucocerebrosidase gene are associated with early-onset Parkinson disease. Neurology. 2007;69(12):1270–7. doi: 10.1212/01.wnl.0000276989.17578.02. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Elbaz A, et al. Independent and joint effects of the MAPT and SNCA genes in Parkinson disease. Ann Neurol. 2011;69(5):778–92. doi: 10.1002/ana.22321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gan-Or Z, et al. The age at motor symptoms onset in LRRK2-associated Parkinson’s disease is affected by a variation in the MAPT locus: a possible interaction. J Mol Neurosci. 2012;46(3):541–4. doi: 10.1007/s12031-011-9641-0. [DOI] [PubMed] [Google Scholar]
- 21.Golub Y, et al. Genetic factors influencing age at onset in LRRK2-linked Parkinson disease. Parkinsonism Relat Disord. 2009;15(7):539–41. doi: 10.1016/j.parkreldis.2008.10.008. [DOI] [PubMed] [Google Scholar]
- 22.Klebe S, et al. The Val158Met COMT polymorphism is a modifier of the age at onset in Parkinson’s disease with a sexual dimorphism. J Neurol Neurosurg Psychiatry. 2013;84(6):666–73. doi: 10.1136/jnnp-2012-304475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ritz B, et al. alpha-Synuclein genetic variants predict faster motor symptom progression in idiopathic Parkinson disease. PLoS One. 2012;7(5):e36199. doi: 10.1371/journal.pone.0036199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Winder-Rhodes SE, et al. Glucocerebrosidase mutations influence the natural history of Parkinson’s disease in a community-based incident cohort. Brain. 2013;136(Pt 2):392–9. doi: 10.1093/brain/aws318. [DOI] [PubMed] [Google Scholar]
- 25.Escott-Price V, et al. Polygenic risk of Parkinson disease is correlated with disease age at onset. Ann Neurol. 2015;77(4):582–91. doi: 10.1002/ana.24335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hughes AJ, et al. Accuracy of clinical diagnosis of idiopathic Parkinson’s disease: a clinico-pathological study of 100 cases. J Neurol Neurosurg Psychiatry. 1992;55(3):181–4. doi: 10.1136/jnnp.55.3.181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Racette BA, et al. Evaluation of a screening questionnaire for genetic studies of Parkinson’s disease. Am J Med Genet. 1999;88(5):539–43. [PubMed] [Google Scholar]
- 28.Harms MB, et al. Parkinson disease is not associated with C9ORF72 repeat expansions. Neurobiol Aging. 2013;34(5):1519, e1–2. doi: 10.1016/j.neurobiolaging.2012.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Purcell S, et al. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet. 2007;81(3):559–75. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Cruchaga C, et al. Rare variants in APP, PSEN1 and PSEN2 increase risk for AD in late-onset Alzheimer’s disease families. PLoS One. 2012;7(2):e31039. doi: 10.1371/journal.pone.0031039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Nalls MA, et al. Large-scale meta-analysis of genome-wide association data identifies six new risk loci for Parkinson’s disease. Nat Genet. 2014;46(9):989–93. doi: 10.1038/ng.3043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Latourelle JC, et al. Genomewide association study for onset age in Parkinson disease. BMC Med Genet. 2009;10:98. doi: 10.1186/1471-2350-10-98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Brockmann K, et al. GBA-associated PD presents with nonmotor characteristics. Neurology. 2011;77(3):276–80. doi: 10.1212/WNL.0b013e318225ab77. [DOI] [PubMed] [Google Scholar]
- 34.Nishioka K, et al. Glucocerebrosidase mutations are not a common risk factor for Parkinson disease in North Africa. Neurosci Lett. 2010;477(2):57–60. doi: 10.1016/j.neulet.2009.11.066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Das G, et al. Microtubule-associated protein tau (MAPT) influences the risk of Parkinson’s disease among Indians. Neurosci Lett. 2009;460(1):16–20. doi: 10.1016/j.neulet.2009.05.031. [DOI] [PubMed] [Google Scholar]
- 36.Kobayashi H, et al. Correlation of tau gene polymorphism with age at onset of Parkinson’s disease. Neurosci Lett. 2006;405(3):202–6. doi: 10.1016/j.neulet.2006.06.057. [DOI] [PubMed] [Google Scholar]
- 37.Myers AJ, et al. The H1c haplotype at the MAPT locus is associated with Alzheimer’s disease. Hum Mol Genet. 2005;14(16):2399–404. doi: 10.1093/hmg/ddi241. [DOI] [PubMed] [Google Scholar]
- 38.Pittman AM, et al. Linkage disequilibrium fine mapping and haplotype association analysis of the tau gene in progressive supranuclear palsy and corticobasal degeneration. J Med Genet. 2005;42(11):837–46. doi: 10.1136/jmg.2005.031377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Rademakers R, et al. High-density SNP haplotyping suggests altered regulation of tau gene expression in progressive supranuclear palsy. Hum Mol Genet. 2005;14(21):3281–92. doi: 10.1093/hmg/ddi361. [DOI] [PubMed] [Google Scholar]
- 40.Charlesworth G, et al. Tau acts as an independent genetic risk factor in pathologically proven PD. Neurobiol Aging. 2012;33(4):838, e7–11. doi: 10.1016/j.neurobiolaging.2011.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Desikan RS, et al. Genetic overlap between Alzheimer’s disease and Parkinson’s disease at the MAPT locus. Mol Psychiatry. 2015 doi: 10.1038/mp.2015.6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Moskvina V, et al. Analysis of genome-wide association studies of Alzheimer disease and of Parkinson disease to determine if these 2 diseases share a common genetic risk. JAMA Neurol. 2013;70(10):1268–76. doi: 10.1001/jamaneurol.2013.448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Whitehouse PJ, et al. Reductions in corticotropin releasing factor-like immunoreactivity in cerebral cortex in Alzheimer’s disease, Parkinson’s disease, and progressive supranuclear palsy. Neurology. 1987;37(6):905–9. doi: 10.1212/wnl.37.6.905. [DOI] [PubMed] [Google Scholar]
- 44.Abuirmeileh A, et al. The CRF-like peptide urocortin produces a long-lasting recovery in rats made hemiparkinsonian by 6-hydroxydopamine or lipopolysaccharide. J Neurol Sci. 2008;271(1-2):131–6. doi: 10.1016/j.jns.2008.04.016. [DOI] [PubMed] [Google Scholar]
- 45.Burre J, Sharma M, Sudhof TC. alpha-Synuclein assembles into higher-order multimers upon membrane binding to promote SNARE complex formation. Proc Natl Acad Sci U S A. 2014;111(40):E4274–83. doi: 10.1073/pnas.1416598111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Burre J, et al. Alpha-synuclein promotes SNARE-complex assembly in vivo and in vitro. Science. 2010;329(5999):1663–7. doi: 10.1126/science.1195227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Liu X, et al. Genome-wide association study identifies candidate genes for Parkinson’s disease in an Ashkenazi Jewish population. BMC Med Genet. 2011;12:104. doi: 10.1186/1471-2350-12-104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ramaker C, et al. Systematic evaluation of rating scales for impairment and disability in Parkinson’s disease. Mov Disord. 2002;17(5):867–76. doi: 10.1002/mds.10248. [DOI] [PubMed] [Google Scholar]
- 49.Siderowf A, et al. Test-retest reliability of the unified Parkinson’s disease rating scale in patients with early Parkinson’s disease: results from a multicenter clinical trial. Mov Disord. 2002;17(4):758–63. doi: 10.1002/mds.10011. [DOI] [PubMed] [Google Scholar]