Abstract
The demonstration of DNA damage excision and repair replication by Setlow, Howard-Flanders, Hanawalt and their colleagues in the early 1960s, constituted the discovery of the ubiquitous pathway of nucleotide excision repair (NER). The serial steps in NER are similar in organisms from unicellular bacteria to complex mammals and plants, and involve recognition of lesions, adducts or structures that disrupt the DNA double helix, removal of a short oligonucleotide containing the offending lesion, synthesis of a repair patch copying the opposite undamaged strand, and ligation, to restore the DNA to its original form. The transcription-coupled repair (TCR) subpathway of NER, discovered nearly two decades later, is dedicated to the removal of lesions from the template DNA strands of actively transcribed genes. In this review I will outline the essential factors and complexes involved in NER in humans, and will comment on additional factors and metabolic processes that affect the efficiency of this important process.
Keywords: DNA repair, nucleotide excision repair, transcription-coupled repair, global genome repair
1. Introduction
Living organisms across the evolutionary scale protect their genetic material against the constant threats posed by environmental agents and byproducts of cellular metabolic processes. Even the simplest unicellular beings possess mechanisms for the prevention and repair of damage to their DNA.
Here I will review the nucleotide excision repair (NER) pathway, a versatile mechanism that removes helix-distorting DNA lesions and structures from the genome. Several other dedicated pathways that have evolved to deal with different classes of lesions or non-canonical forms of DNA will be addressed in other sections in this volume.
The NER process begins with the recognition of a DNA lesion. Then, dual incisions of the damaged DNA strand, one on either side of the lesion, are produced. The lesion-bearing oligonucleotide is removed, a patch is synthesized using the undamaged complementary strand as a template, and the patch is ligated to the contiguous strand.
NER removes lesions from the entire genome, albeit with varying efficiencies; in the early 1980s it was discovered in mammalian cells that certain lesions such as UVC-induced cyclobutane pyrimidine dimers (CPD) were excised more rapidly from the transcribed strands of active genes than from the opposite strands. This process was named transcription-coupled repair (TCR), and has been defined as a subpathway of NER; repair in the global genome is known as global genomic repair (GGR). TCR was subsequently shown to operate for other so-called bulky adducts, and in E. coli, in yeast and in other organisms, reviewed in [1, 2].
2. Global and transcription-coupled repair in human cells
NER in eukaryotes has been the subject of extensive reviews, for example [3, 4], and human disorders caused by defective NER have been described [5-7]. The sequence of events in human GGR is the same as that in unicellular prokaryotes, but the process is more complicated and the number of proteins involved is much larger than in E. coli, as shown in Figure 1. Importantly, in E. coli the same six NER factors are required for GGR and for TCR, while in humans TCR can be completed in the absence of the GGR damage recognition factors XPE, XPC or hRAD23b [1].
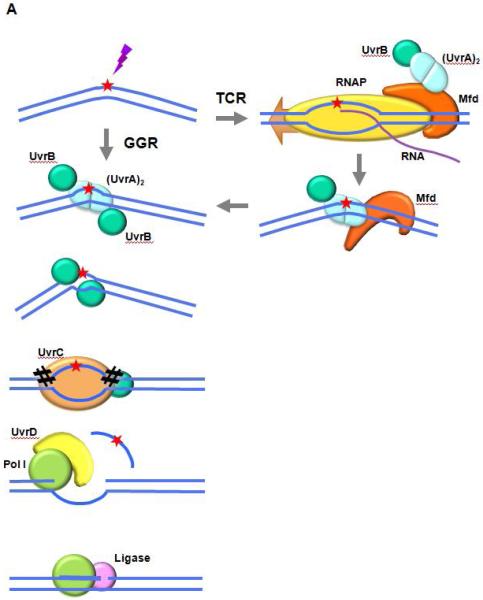
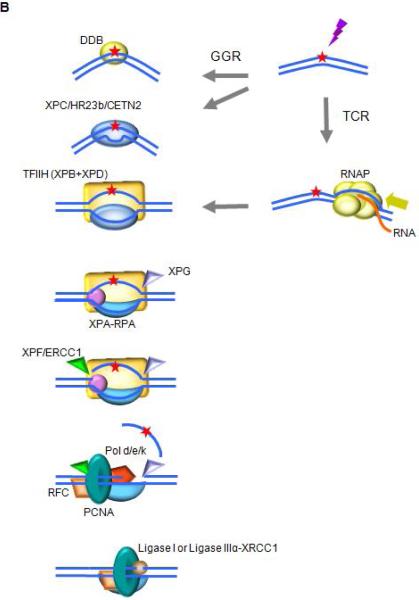
Fig. 1. Schematics of NER in the prototypic bacterium E. coli, and in humans.
A. NER in E. coli
The UvrAB complex binds to the DNA and recognizes and verifies the damage to be repaired. UvrA dissociates from the preincision complex leaving one or two molecules of UvrB bound to the DNA. UvrC interacts with UvrB and catalyzes two nicks in the DNA, one on either side of the lesion. The combined action of UvrD (helicase I) and DNA polymerase I removes the oligonucleotide containing the lesion, as well as UvrB and UvrC, from the DNA and results in the synthesis of a patch using the undamaged complementary strand as a template. DNA ligase I seals the patch to the contiguous DNA strand. In TCR, Mfd is activated by binding to a stalled RNAP. It dissociates the RNAP and nascent transcript from the DNA and recruits UvrA complexed with UvrB. Repair then proceeds through the same reaction sequence as GGR.
B. NER in humans
Higher eukaryotes utilize different mechanisms for detecting DNA alterations in actively transcribed genes and in the genome as a whole. In TCR, RNAPII is arrested at a lesion. TCR factors are recruited; the polymerase is removed or backtracked to allow access to TFIIH and other NER repair enzymes. In GGR, a helix distorting lesion or structure can be directly recognized by XPC complexed with hRAD23B and centrin 2 (CETN2). Certain lesions such as CPD, which do not significantly destabilize DNA duplexes, are first recognized by DDB2 (XPE) in complex with DDB1, creating a kink that is recognized by XPC. The XPC-hRAD23b-CETN2 complex melts the DNA around the lesion and attracts the multiunit complex TFIIH. TCR and GGR converge; XPB and XPD unwind the DNA to create a ~30-nucleotide bubble. Once the pre-incision complex is assembled, XPA, RPA and XPG are recruited and the XPC complex is released. XPA binds the DNA near the 5′ side of the bubble, and RPA binds the ssDNA opposite the lesion, protecting it from degradation and coordinating excision and repair events. XPG and ERCC1-XPF associate with TFIIH. ERCC1-XPF makes the first incision, and repair synthesis proceeds for several nucleotides displacing the damaged strand; XPG incises the 3’ single/double strand junction, and ligase I or ligase IIIα-XRCC1 seal the DNA.
2.1 Damage recognition
2.1.1 Global genomic repair
In GGR, a helix distorting lesion or structure can be directly recognized by XPC, complexed with hRAD23B and centrin 2 (CETN2). Certain lesions such as CPD, which do not significantly destabilize DNA duplexes, are first recognized by DDB2 (XPE) in complex with DDB1, creating a kink that is recognized by XPC. DDB1 and DDB2 are part of the CUL4-ROC1 ubiquitin ligase complex that ubiquitylates DDB2, XPC and histones after DNA damage has occurred. XPC has been shown to bind the strand opposite the lesion, thus explaining its universal capacity for recognition of diverse types of lesions [8]. The XPC-hRAD23b-CETN2 complex melts the DNA around the lesion and recruits the multiprotein complex TFIIH.
2.1.2 Transcription-coupled repair
Humans have four RNA polymerases (RNAP), the nuclear RNAPI, II and III, and a single-polypeptide mitochondrial mtRNAP that is similar to that of bacteriophage T7. RNAPI synthesizes ribosomal RNA precursors, RNAPII synthesizes mRNA, snRNA and micro RNA, and RNAPIII produces ribosomal 5S rRNA, tRNA and other small RNAs. Although TCR in RNAPI-transcribed genes has been shown in yeast [9], no strand bias for repair of UV-induced CPD was found in ribosomal RNA genes in mammalian cells [10], although interestingly the TCR factor CSB has been shown to form a complex with RNAPI, TFIIH and XPG in nucleoli, providing stability to the complex to promote efficient rRNA synthesis [11]. To our knowledge, no TCR has been documented for RNAPIII-transcribed genes (see elsewhere in this volume). Thus we will limit our comments to repair of sequences transcribed by RNAPII.
A translocating RNAP can be blocked by bulky adducts such as the photoadducts CPD and 6-4 pyrimidine-pyrimidones (6-4PP), cis-platin intrastrand crosslinks, BPDE (Benzo(a)pyrene diolepoxide) and other polycyclic aromatic hydrocarbons; by discontinuities in the template strand (nicks, gaps, abasic sites); and by collisions with replication complexes. Some sequences in DNA that are prone to forming non-canonical DNA structures, or those that can form super-stable DNA-RNA hybrids, can also arrest transcription when the RNA polymerase attempts translocation through the suspect sequence; the current hypothesis is that R-loops that occur “behind” the polymerase impede its progress, reviewed in [12]. R-loops can be converted to double strand breaks in a process that requires the NER endonucleases XPF and XPG and the TCR factor CSB, but not XPC [13].
A blocked RNAPII constitutes the first step for damage recognition in TCR. The arrested elongation complex recruits CSB (ERCC6), a transcription elongation factor that translocates along template DNA with RNAPII. CSB strongly binds the polymerase when it is blocked at a lesion, and changes the DNA conformation by wrapping the DNA around itself, altering the interface between RNAPII and DNA [14]. CSB recruits the CSA complex, NER factors (not including the GGR recognition factors XPC and XPE) and p300 to sites of arrested RNAPII, and has been considered the master coordinator of TCR in humans, with roles similar to those carried out by Mfd in E. coli. CSA (ERCC8) is the substrate recognition factor in the DCX E3 ubiquitin ligase complex, which contains CSA, RBX1, and CUL4A. This complex interacts with the COP9 signalosome upon UV irradiation. CSA is required for recruitment of XAB2, the nucleosome remodeling factor HMGN1 and TFIIS to sites of arrested RNAPII. CSA-dependent degradation of CSB is required for recovery of RNA synthesis after UV damage [15].
The recently identified UVSSA protein and its partner USP7 [16-18] are loosely associated with elongating RNAPII. Upon transcription arrest, these factors bind strongly to RNAPII, and USP7 deubiquitylates CSB to stabilize it. Together UVSSA and USP7 prevent DNA damage-induced degradation of CSB, facilitate CSA and CSB-dependent ubiquitylation of arrested RNAPIIo (the phosphorylated form of RNAPII during elongation), its remodeling, and USP7-dependent deubiquitylation, and its recycling to non-phosphorylated RNAPIIa for transcription initiation (reviewed in [1, 19]).
An arrested RNAPII may be targeted for degradation, or it may bypass the lesion with possible misincorporation of ribonucleotides, a phenomenon termed transcriptional mutagenesis [20]. We favor a model in which the RNAPII reverses translocation with cleavage of the nascent transcript, also called backtracking, to reveal the offending lesion and to allow space for the repair complex to operate. TFIIS is a transcription elongation factor that stimulates mRNA cleavage by RNAPII, is required for recovery of RNA synthesis after UV-induced damage, and is recruited to sites of damage by CSA [21]. RNAPII backtracking would result in re-annealing of the 8 to 11-nucleotide bubble that RNAP forms as it elongates; it is not known which factor(s) keep this bubble open, or reopen it, to facilitate the next steps in NER, a job performed by XPE and the XPC complex in the GGR pathway.
2.2 Incision, excision, repair synthesis and ligation
The TCR and GGR subpathways converge when TFIIH is recruited to the repair site (Fig. 1). TFIIH is a transcription initiation complex that comprises 10 proteins; two of them are the ATPases/helicases XPB and XPD. Only the ATPase activity of XPB is required for NER, while XPD must be active as both ATPase and helicase, suggesting that XPD translocates along the DNA and detects lesions when its motion is impeded [22]. XPB and XPD unwind the DNA to create a 20 to 30-nucleotide bubble. Other components of TFIIH participate in NER: p8, the smallest subunit, is an absolute requirement for NER and it is defective in trichothiodystrophy complementation group A (TTD-A); p52 stimulates XPB, and p44 stimulates XPD [23]. Once the pre-incision complex is assembled, XPA, RPA and XPG are recruited. XPA binds the DNA near the 5′ side of the bubble, where it interacts with TFIIH, RPA, PCNA, XPC, DDB2 and ERCC1-XPF. RPA is composed of three subunits, and binds the ssDNA opposite the lesion, protecting it from degradation and helping to coordinate excision and repair events [3]. XPG associates with TFIIH and lends structural support, but its endonuclease activity is not triggered until ERCC1-XPF has been recruited by XPA to the 5′ end of the bubble. The order in which the dual incisions are made had been the subject of discussion; Fagbemi and colleagues demonstrated that ERCC1-XPF makes the first incision, and that repair synthesis can be initiated and will proceed for several nucleotides in the absence of XPG incision [24]. The DNA replication machinery pol δ/ε/κ-PCNA-RFC-RPA synthesizes a patch displacing the damaged strand and TFIIH; pol ε is active for NER in replicating cells and pol δ and κ are the main NER polymerases in non-replicating cells [25, 26]. XPG incises the 3’ single/double strand junction, and ligase I seals the DNA in replicating cells, while ligase IIIα and its cofactor XRCC1 carry out this step in quiescent cells [3].
3. NER in chromatin
The complicated NER process operates within the nucleus, where eukaryotic DNA is bound to histones that need to be modified or rearranged to allow transcription, replication or repair activities, as proposed by Smerdon and Lieberman in their “access-repair-restore” model [27]. The subject of NER in chromatin has been recently reviewed [28] and is addressed in this volume.
3.1 TCR in chromatin
Nucleosome-bound DNA poses significant barriers to RNA polymerases, while avoiding exposure of cryptic promoters within ORFs. Tightly controlled and extremely complex sets of histone modifiers have evolved to regulate each step during transcription. The situation becomes much more complicated when repair must occur in conjunction with transcription. In addition to histone acetylases, histone chaperones, deacetylases (HDACs) and other chromatin modifying enzymes that regulate transcription, some factors are specifically recruited by TCR proteins, or act exclusively during TCR. For example, the HAT p300 and the nucleosomal nonhistone binding protein HMGN1 (see section in this volume), which stimulates HAT activity [21] and unwinds chromatin, are recruited to TCR sites in a CSA- and CSB-dependent manner [29], and UVSSA also interacts with HMGN1 [16; reviewed in [30]. CAF-1 is involved in chromatin assembly associated with DNA replication and repair, but to our knowledge it has not been found in association with TCR complexes. The newly identified FACT (facilitates chromatin transcription) subunit SPT16 specifically binds to TCR sites in chromatin in UV-irradiated cells, while RNAPII stalling per se does not elicit binding of SPT16. Knock-down of SPT16 results in decreased recovery of RNA synthesis after UV [31]. The backtracking model for the TCR mechanism requires removal of the nucleosomes that reassemble behind RNAPII. The p300 and HMGN1 chromatin factors, which have been found to co-precipitate with stalled RNAPII complexes, facilitate nucleosome sliding upstream of RNAPII so that the polymerase can backtrack [1, 32, 33].
As the nascent RNA exits RNAPII, various protein complexes process the RNA readying it for export to the cytoplasm. Of these, the THO, TREX and THSC/TREX-2 complexes have been shown to be required for TCR, suggesting that proper biogenesis of export-competent mRNA is important for transcription processivity [34]. Other factors that modulate transactions between transcription and mRNA biogenesis also play a role in TCR; these include the PAF/Paf1 and the Ccr4-Not complexes, reviewed in [35].
4. The eukaryotic response to DNA damage
As has been shown for the SOS genomic stress response for bacteria, DNA damaged in human cells results in altered expression of hundreds of genes; however, different human cell types respond to damage differently, suggesting that cells in each organ may have evolved unique sensitivities to different types of DNA damage [36].
4.1 DNA damage checkpoints, effects on NER
An important factor in regulation of NER is the abundance of the proteins involved. In E. coli, DNA damage leads to the induction of a set of genes in a process known as the SOS response [37, 38]. It is generally thought that NER proteins are kept at low levels to inhibit unspecific “gratuitous repair” events in undamaged DNA. Although there is no direct equivalent to the SOS response in humans, a series of reactions termed the DNA damage response (DDR) is induced upon DNA damage (see section in this volume): cell cycle checkpoints at the G1/S and G2/M boundaries are activated, pausing the cell cycle and giving the cell time to repair the damage before continuing to divide. The use of the term, DDR, has usually been applied specifically to double-strand breaks and replication fork arrest, and for different types of genomic stress the scenario may be quite different. Checkpoint activation is controlled by two master kinases, ataxia telangiectasia mutated (ATM) and ataxia telangiectasia and Rad3 related (ATR). These kinases phosphorylate downstream targets in a signal transduction cascade, eventually leading to cell cycle arrest [39]. Among the numerous protein-protein interactions of ATM and ATR, illustrated in a SPIKE interactive map (http://www.cs.tau.ac.il/~spike/), an important downstream target is p53, a tumor suppressor that can arrest the cell cycle at the G1/S checkpoint when the DNA is damaged, and is required for inducing apoptosis following DNA damage; p53 can activate a large number of genes (http://www.cs.tau.ac.il/~spike/), including genes coding for the NER lesion recognizing proteins DDB2 (XPE) and XPC [40, 41].
4.2 Other factors that regulate NER
In addition to p53, it has been reported that DDB2 and XPC are targeted for stimulation by poly(ADP-ribose) polymerase-1 (PARP-1) (see section in this volume). PARP-1 is activated by DNA damage in mammalian cells, using NAD+ to form ADP-ribose polymers (PAR; PARylation) that modify numerous enzymes including itself. Among its targets are several proteins involved in BER, homologous recombination and non-homologous end joining, reviewed in [42]. Robu et al. have now shown that PARP-1 PARylates DDB2 and stimulates recruitment of XPC by DDB2, significantly increasing the efficiency of NER [43].
DDB2, XPC and GADD45 (a protein involved in growth arrest in response to DNA damage) can also be simulated by BRCA1 (Breast Cancer 1) independently of p53, as shown by Hartman and Ford [44]. BRCA1 is a tumor suppressor that forms part of the BRCA1-associated genome surveillance complex, or BASC; it plays roles in transcription, in repair of double strand breaks, and repair of DNA interstrand crosslinks through the Fanconi anemia pathway [45]. Interestingly, and in parallel with the bacterial SOS response, activation of NER by p53, PARP-1 and BRCA1 occurs through stimulation of the initial steps of the pathway, namely damage recognition by UvrA and UvrB in E. coli and by DDB2 and XPC in humans.
In addition to phosphorylation and PARylation, ubiquitylation plays a central role in regulation of NER [46, 47]. Ubiquitin is a protein that can be attached to lysine residues within itself or in other proteins; the particular residues that are modified and how many ubiquitins are attached can determine the fate of the targeted protein. The targets for ubiquitylation in GGR are DDB2, which forms an ubiquitin ligase complex with DDB1, and XPC as mentioned above; XPC is also targeted for ubiquitylation by the RING finger protein 111 (RNF111). In TCR, CSB is ubiquitylated by an ubiquitin ligase complex containing CSA, and the UVSSA-USP7 complex stabilizes CSB by deubiquitylating it. RNAPII can be ubiquitylated by UVSSA, by NEDD4, by elongin A, B or C, and by CUL5 and RING-box protein 2, reviewed in [4]. In addition to core NER proteins, PCNA is monoubiquitylated when DNA lesions block progression of the replication fork, facilitating the switch from replicative to translesion synthesis DNA polymerases [48].
Other post-translation protein modifications include sumoylation, in which small ubiquitin-related modifier (SUMO) proteins can be reversibly attached to proteins; and neddylation, the attachment of ubiquitin-like protein NEDD8 [49]. Several and various modifications may affect the same protein in a coordinated fashion, for example XPC is sumoylated in response to UV damage, and then polyubiquitylated by RNF111, and UV-induced PARylation of DDB2 inhibits its ubiquitiylation, thus protecting it from degradation, reviewed in [4].
In contrast to the stimulation of DDB2 and XPC by DNA damage, XPA synthesis is not affected by UV irradiation, but the protein is stabilized so that its half-life is 4-fold longer than in undamaged cells. Curiously, levels of this core NER protein vary by as much as 10-fold in response to circadian rhythms, increasing during daytime and declining at night, in a process regulated at the transcriptional level by cryptochrome and at the posttranslational level by the HERC2 ubiquitin ligase. This rhythmicity may have evolved in the primordial seas to enhance repair of sunlight-induced CPDs and 6-4PP, and to protect undamaged DNA from gratuitous attack by NER in the dark [50].
Among the genes induced by DNA damage it is important to mention those that code for translesion synthesis DNA polymerases, both in bacteria and in eukaryotes. These enzymes, characterized by low fidelity when copying DNA, increase the tolerance to damage and increased rates of survival at the cost of greater accumulation of mutations. Human polymerases Rev1, η, ι and κ are members of the Y family of translesion DNA polymerases present during global response to DNA damage and are responsible for enhanced mutagenesis during a global response to DNA damage [51].
5. Concluding remarks and future directions
The universality of the NER repair pathway among all life forms points to the necessity of a mechanism for the elimination of obstacles for replication and transcription, and of structures that can affect the stability or integrity of DNA. An important feature of NER is that it is able to repair a large variety of lesions with very different structures, without the need for lesion-specific specialized enzymes such as the glycosylases that initiate the base excision repair pathway. The efficiency of NER depends, among other factors, on the degree of distortion caused by the lesion, adduct or unusual structure, which range in size from abasic sites to multi-ring aromatic hydrocarbons and protein-DNA crosslinks. Other examples of heterogeneous levels of repair include slower or deficient repair of chemical adducts in tandemly repeated αDNA of African green monkey cells [52], slower GGR in the inactive loci 754 and coagulation factor IX [53], and attenuation of GGR in terminally differentiated cells except for transcriptionally active regions where the transcribed strand is subject to TCR and both strands are repaired by GGR, a phenomenon termed domain associated repair (DAR) [54]. Moreover, as mentioned above, the efficiency of NER varies in different tissues, reviewed in [55]. Some chemicals such as irofulven, illudin S and aristolochic acid [56-58] induce lesions that are invisible to GGR but are repaired by TCR; it is not yet known why lesions that can obstruct RNAPII and thus must cause significant alterations in DNA are not detected by XPE or XPC. Of course, the mechanism for TCR lesion recognition deals with the ease of a single strand attempting to pass through the RNAP, while for GGR the recognition is based upon the stability of the double-strand DNA structure; this suggests that this class of lesions do not cause significant distortions in double stranded DNA. Although some NER factors reside in the cytoplasm until they are called to action, NER is exclusively nuclear. Mitochondria possess a robust BER system, but evidence for other repair pathways such as MMR is limited, and NER has not been detected [59].
In addition to short ribonucleotide chains made by primases to initiate DNA synthesis, DNA polymerases can incorporate ribonucleotides with frequencies that vary with the particular polymerase and with the relative concentrations of ribo- and deoxyribonucleotides in the pools; a ratio of 1 ribonucleotide per ~7000 bases was found in yeast and mouse cells, reviewed in [60]. Ribonucleotides are mainly removed from DNA by a process termed Ribonucleotide Excision Repair (RER) that involves RNase H2, pol δ, PCNA, RFC, FEN1 and ligase 1. NER has an active role in ribonucleotide removal in E. coli [61]; however NER cannot substitute for the loss or decrease of RNase H2 activity, which can result in Aicardi-Goutières syndrome in humans.
Most of the DNA lesions that are recognized by NER are considered strong mutagens and carcinogens. Ironically, some agents that induce these lesions are regularly used in chemotherapeutic regimens to combat cancer; a problem often found is that the highly mutable tumor cells develop resistance to the drug. Synthetic lethality, originally defined as the combination of mutations in two or more separate genes leading to cell death, has been adopted as a strategy for overcoming drug resistance; it consists of combining two or more drugs, for example cisplatin to damage DNA and the NER inhibitor UCN-01 [62].
Although the basic mechanism of NER has been well defined, there remain a wealth of discoveries to be made, including detailed analyses of protein crystal structures that may allow the design of small molecules that can enhance or inhibit DNA repair activities, identification of small non-coding RNAs that can modulate NER, high-definition microscopy combined with quantum dot single molecule labeling for studies of protein-protein interactions, and others.
Highlights.
NER is a mechanism for removal of a variety of structurally unrelated DNA lesions
NER is prevalent in bacteria, archea, yeast, and in all higher eukaryotes
NER efficiency is modulated by damage type and modification of relevant enzymes
Transcription-coupled repair deals with the unique problems of transcription arrest
Acknowledgements
I would like to thank Phil Hanawalt for his generous and insightful contributions to the field of DNA repair, and to my personal career and life, and to honor his mentor and co-discoverer of NER Dick Setlow, who passed away April 6, 2015.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Conflict of interest statement
None
References
- 1.Spivak G, Ganesan AK. The complex choreography of transcription-coupled repair. DNA Repair (Amst) 2014;19:64–70. doi: 10.1016/j.dnarep.2014.03.025. [DOI] [PubMed] [Google Scholar]
- 2.Ganesan A, Spivak G, Hanawalt PC. Transcription-coupled DNA repair in prokaryotes. Prog Mol Biol Transl Sci. 2012;110:25–40. doi: 10.1016/B978-0-12-387665-2.00002-X. [DOI] [PubMed] [Google Scholar]
- 3.Scharer OD. Nucleotide excision repair in eukaryotes. Cold Spring Harb Perspect Biol. 2013;5(10):a012609. doi: 10.1101/cshperspect.a012609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Marteijn JA, et al. Understanding nucleotide excision repair and its roles in cancer and ageing. Nature Reviews Molecular Cell Biology. 2014;15(7):465–481. doi: 10.1038/nrm3822. [DOI] [PubMed] [Google Scholar]
- 5.Oh DH, Spivak G. Hereditary photodermatoses. Adv Exp Med Biol. 2010;685:95–105. doi: 10.1007/978-1-4419-6448-9_9. [DOI] [PubMed] [Google Scholar]
- 6.Menck CFM, Munford V. DNA repair diseases: What do they tell us about cancer and aging? Genetics and Molecular Biology. 2014;37(1):220–233. doi: 10.1590/s1415-47572014000200008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Spivak G, Hanawalt PC. Photosensitive human syndromes. Mutation Research/Fundamental and Molecular Mechanisms of Mutagenesis. 2015 doi: 10.1016/j.mrfmmm.2014.11.003. In press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lee YC, et al. The relationships between XPC binding to conformationally diverse DNA adducts and their excision by the human NER system: is there a correlation? DNA Repair (Amst) 2014;19:55–63. doi: 10.1016/j.dnarep.2014.03.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Conconi A, Bespalov VA, Smerdon MJ. Transcription-coupled repair in RNA polymerase I-transcribed genes of yeast. Proc Natl Acad Sci U S A. 2002;99(2):649–54. doi: 10.1073/pnas.022373099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Christians FC, Hanawalt PC. Lack of transcription-coupled repair in mammalian ribosomal RNA genes. Biochemistry. 1993;32(39):10512–8. doi: 10.1021/bi00090a030. [DOI] [PubMed] [Google Scholar]
- 11.Bradsher J, et al. CSB is a component of RNA pol I transcription. Mol Cell. 2002;10(4):819–29. doi: 10.1016/s1097-2765(02)00678-0. [DOI] [PubMed] [Google Scholar]
- 12.Belotserkovskii BP, Hanawalt PC. Anchoring nascent RNA to the DNA template could interfere with transcription. Biophys J. 2011;100(3):675–84. doi: 10.1016/j.bpj.2010.12.3709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sollier J, et al. Transcription-coupled nucleotide excision repair factors promote R-loopinduced genome instability. Mol Cell. 2014;56(6):777–85. doi: 10.1016/j.molcel.2014.10.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Beerens N, et al. The CSB protein actively wraps DNA. J Biol Chem. 2005;280(6):4722–9. doi: 10.1074/jbc.M409147200. [DOI] [PubMed] [Google Scholar]
- 15.Brooks PJ. Blinded by the UV light: how the focus on transcription-coupled NER has distracted from understanding the mechanisms of Cockayne syndrome neurologic disease. DNA Repair (Amst) 2013;12(8):656–71. doi: 10.1016/j.dnarep.2013.04.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Schwertman P, et al. UV-sensitive syndrome protein UVSSA recruits USP7 to regulate transcription-coupled repair. Nat Genet. 2012;44(5):598–602. doi: 10.1038/ng.2230. [DOI] [PubMed] [Google Scholar]
- 17.Zhang X, et al. Mutations in UVSSA cause UV-sensitive syndrome and destabilize ERCC6 in transcription-coupled DNA repair. Nat Genet. 2012;44(5):593–7. doi: 10.1038/ng.2228. [DOI] [PubMed] [Google Scholar]
- 18.Nakazawa Y, et al. Mutations in UVSSA cause UV-sensitive syndrome and impair RNA polymerase IIo processing in transcription-coupled nucleotide-excision repair. Nat Genet. 44(5):586–92. doi: 10.1038/ng.2229. [DOI] [PubMed] [Google Scholar]
- 19.Schwertman P, Vermeulen W, Marteijn JA. UVSSA and USP7, a new couple in transcription-coupled DNA repair. Chromosoma. 2013;122(4):275–84. doi: 10.1007/s00412-013-0420-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Saxowsky TT, et al. 8-Oxoguanine-mediated transcriptional mutagenesis causes Ras activation in mammalian cells. Proc Natl Acad Sci U S A. 2008;105(48):18877–82. doi: 10.1073/pnas.0806464105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Fousteri M, et al. Cockayne syndrome A and B proteins differentially regulate recruitment of chromatin remodeling and repair factors to stalled RNA polymerase II in vivo. Mol Cell. 2006;23(4):471–82. doi: 10.1016/j.molcel.2006.06.029. [DOI] [PubMed] [Google Scholar]
- 22.Mathieu N, et al. DNA quality control by a lesion sensor pocket of the xeroderma pigmentosum group D helicase subunit of TFIIH. Curr Biol. 2013;23(3):204–12. doi: 10.1016/j.cub.2012.12.032. [DOI] [PubMed] [Google Scholar]
- 23.Singh A, et al. TFIIH subunit alterations causing xeroderma pigmentosum and trichothiodystrophy specifically disturb several steps during transcription. Am J Hum Genet. 2015;96(2):194–207. doi: 10.1016/j.ajhg.2014.12.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Fagbemi AF, Orelli B, Scharer OD. Regulation of endonuclease activity in human nucleotide excision repair. DNA Repair (Amst) 2011;10(7):722–9. doi: 10.1016/j.dnarep.2011.04.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ogi T, et al. Three DNA polymerases, recruited by different mechanisms, carry out NER repair synthesis in human cells. Mol Cell. 2010;37(5):714–27. doi: 10.1016/j.molcel.2010.02.009. [DOI] [PubMed] [Google Scholar]
- 26.Lehmann AR. DNA polymerases and repair synthesis in NER in human cells. DNA Repair (Amst) 2011;10(7):730–3. doi: 10.1016/j.dnarep.2011.04.023. [DOI] [PubMed] [Google Scholar]
- 27.Smerdon MJ, Lieberman MW. Nucleosome rearrangement in human chromatin during UV-induced DNA-reapir synthesis. Proc Natl Acad Sci U S A. 1978;75(9):4238–41. doi: 10.1073/pnas.75.9.4238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Peterson CL, Almouzni G. Nucleosome dynamics as modular systems that integrate DNA damage and repair. Cold Spring Harb Perspect Biol. 2013;5(9) doi: 10.1101/cshperspect.a012658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Fousteri M, et al. Cockayne syndrome A and B proteins differentially regulate recruitment of chromatin remodeling and repair factors to stalled RNA polymerase II in vivo. Molecular cell. 2006;23(4):471–82. doi: 10.1016/j.molcel.2006.06.029. [DOI] [PubMed] [Google Scholar]
- 30.Vermeulen W, Fousteri M. Mammalian transcription-coupled excision repair. Cold Spring Harb Perspect Biol. 2013;5(8) doi: 10.1101/cshperspect.a012625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Dinant C, et al. Enhanced Chromatin Dynamics by FACT Promotes Transcriptional Restart after UV-Induced DNA Damage. Mol Cell. 2013;51(4):469–79. doi: 10.1016/j.molcel.2013.08.007. [DOI] [PubMed] [Google Scholar]
- 32.Hanawalt PC, Spivak G. Transcription-coupled DNA repair: two decades of progress and surprises. Nat Rev Mol Cell Biol. 2008;9(12):958–70. doi: 10.1038/nrm2549. [DOI] [PubMed] [Google Scholar]
- 33.Lans H, Marteijn JA, Vermeulen W. ATP-dependent chromatin remodeling in the DNAdamage response. Epigenetics Chromatin. 2012;5:4. doi: 10.1186/1756-8935-5-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Gaillard H, Wellinger RE, Aguilera A. A new connection of mRNP biogenesis and export with transcription-coupled repair. Nucleic Acids Res. 2007;35(12):3893–906. doi: 10.1093/nar/gkm373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Gaillard H, Aguilera A. Transcription coupled repair at the interface between transcription elongation and mRNP biogenesis. Biochim Biophys Acta. 2013;1829(1):141–50. doi: 10.1016/j.bbagrm.2012.09.008. [DOI] [PubMed] [Google Scholar]
- 36.Fry RC, Begley TJ, Samson LD. Genome-wide responses to DNA-damaging agents. Annu Rev Microbiol. 2004;59:357–377. doi: 10.1146/annurev.micro.59.031805.133658. [DOI] [PubMed] [Google Scholar]
- 37.Radman M. SOS repair hypothesis: phenomenology of an inducible DNA repair which is accompanied by mutagenesis. Basic Life Sci. 1975;5A:355–67. doi: 10.1007/978-1-4684-2895-7_48. [DOI] [PubMed] [Google Scholar]
- 38.Smith BT, Walker GC. Mutagenesis and more: umuDC and the Escherichia coli SOS response. Genetics. 1998;148(4):1599–610. doi: 10.1093/genetics/148.4.1599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Marechal A, Zou L. DNA damage sensing by the ATM and ATR kinases. Cold Spring Harb Perspect Biol. 2013;5(9) doi: 10.1101/cshperspect.a012716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hwang BJ, et al. Expression of the p48 xeroderma pigmentosum gene is p53-dependent and is involved in global genomic repair. Proc Natl Acad Sci U S A. 1999;96(2):424–8. doi: 10.1073/pnas.96.2.424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Adimoolam S, Ford JM. p53 and DNA damage-inducible expression of the xeroderma pigmentosum group C gene. Proc Natl Acad Sci U S A. 2002;99(20):12985–90. doi: 10.1073/pnas.202485699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.De Vos M, Schreiber V, Dantzer F. The diverse roles and clinical relevance of PARPs in DNA damage repair: current state of the art. Biochem Pharmacol. 2012;84(2):137–46. doi: 10.1016/j.bcp.2012.03.018. [DOI] [PubMed] [Google Scholar]
- 43.Robu M, et al. Role of poly(ADP-ribose) polymerase-1 in the removal of UV-induced DNA lesions by nucleotide excision repair. Proc Natl Acad Sci U S A. 2013;110(5):1658–63. doi: 10.1073/pnas.1209507110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hartman AR, Ford JM. BRCA1 induces DNA damage recognition factors and enhances nucleotide excision repair. Nat Genet. 2002;32(1):180–4. doi: 10.1038/ng953. [DOI] [PubMed] [Google Scholar]
- 45.D'Andrea AD. BRCA1: a missing link in the Fanconi anemia/BRCA pathway. Cancer Discov. 2013;3(4):376–8. doi: 10.1158/2159-8290.CD-13-0044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.van Cuijk L, Vermeulen W, Marteijn JA. Ubiquitin at work: the ubiquitous regulation of the damage recognition step of NER. Exp Cell Res. 2014;329(1):101–9. doi: 10.1016/j.yexcr.2014.07.018. [DOI] [PubMed] [Google Scholar]
- 47.Dijk M, et al. Insight in the multilevel regulation of NER. Exp Cell Res. 2014;329(1):116–23. doi: 10.1016/j.yexcr.2014.08.010. [DOI] [PubMed] [Google Scholar]
- 48.Niimi A, et al. Regulation of proliferating cell nuclear antigen ubiquitination in mammalian cells. Proceedings of the National Academy of Sciences of the United States of America. 2008;105(42):16125–16130. doi: 10.1073/pnas.0802727105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Brown JS, Jackson SP. Ubiquitylation, neddylation and the DNA damage response. Open Biol. 2015;5(4) doi: 10.1098/rsob.150018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kang TH, Reardon JT, Sancar A. Regulation of nucleotide excision repair activity by transcriptional and post-transcriptional control of the XPA protein. Nucleic Acids Research. 2011;39(8):3176–3187. doi: 10.1093/nar/gkq1318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Goodman MF, Woodgate R. Translesion DNA polymerases. Cold Spring Harb Perspect Biol. 2013;5(10):a010363. doi: 10.1101/cshperspect.a010363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Zolan ME, et al. Deficient Repair of Chemical Adducts in Alpha-DNA of Monkey Cells. Cell. 1982;28(3):613–619. doi: 10.1016/0092-8674(82)90216-1. [DOI] [PubMed] [Google Scholar]
- 53.Venema J, et al. Transcription Affects the Rate but Not the Extent of Repair of Cyclobutane Pyrimidine Dimers in the Human Adenosine-Deaminase Gene. Journal of Biological Chemistry. 1992;267(13):8852–8856. [PubMed] [Google Scholar]
- 54.Nouspikel TP, Hyka-Nouspikel N, Hanawalt PC. Transcription domain-associated repair in human cells. Mol Cell Biol. 2006;26(23):8722–30. doi: 10.1128/MCB.01263-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Dion V. Tissue specificity in DNA repair: lessons from trinucleotide repeat instability. Trends in Genetics. 2014;30(6):220–229. doi: 10.1016/j.tig.2014.04.005. [DOI] [PubMed] [Google Scholar]
- 56.Koeppel F, et al. Irofulven cytotoxicity depends on transcription-coupled nucleotide excision repair and is correlated with XPG expression in solid tumor cells. Clin Cancer Res. 2004;10(16):5604–13. doi: 10.1158/1078-0432.CCR-04-0442. [DOI] [PubMed] [Google Scholar]
- 57.Jaspers NG, et al. Anti-tumour compounds illudin S and Irofulven induce DNA lesions ignored by global repair and exclusively processed by transcription- and replication-coupled repair pathways. DNA Repair (Amst) 2002;1(12):1027–38. doi: 10.1016/s1568-7864(02)00166-0. [DOI] [PubMed] [Google Scholar]
- 58.Sidorenko VS, et al. Lack of recognition by global-genome nucleotide excision repair accounts for the high mutagenicity and persistence of aristolactam-DNA adducts. Nucleic Acids Res. 2012;40(6):2494–505. doi: 10.1093/nar/gkr1095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Copeland WC, Longley MJ. Mitochondrial genome maintenance in health and disease. DNA Repair (Amst) 2014;19:190–8. doi: 10.1016/j.dnarep.2014.03.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Williams JS, Kunkel TA. Ribonucleotides in DNA: Origins, repair and consequences. DNA Repair. 2014;19:27–37. doi: 10.1016/j.dnarep.2014.03.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Vaisman A, et al. Removal of Misincorporated Ribonucleotides from Prokaryotic Genomes: An Unexpected Role for Nucleotide Excision Repair. Plos Genetics. 2013;9(11) doi: 10.1371/journal.pgen.1003878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.van Midwoud PM, Sturla SJ. Improved efficacy of acylfulvene in colon cancer cells when combined with a nuclear excision repair inhibitor. Chem Res Toxicol. 2013;26(11):1674–82. doi: 10.1021/tx400255f. [DOI] [PMC free article] [PubMed] [Google Scholar]