Abstract
Fanconi anemia (FA) is a rare human genetic disease characterized by bone marrow failure, cancer predisposition, and genomic instability. It has been known for many years that FA patient-derived cells are exquisitely sensitive to DNA interstrand cross-linking agents such as cisplatin and mitomycin C. On this basis, it was widely assumed that failure to repair endogenous interstrand cross-links (ICLs) causes FA, although the endogenous mutagen that generates these lesions remained elusive. Recent genetic evidence now suggests that endogenous aldehydes are the driving force behind FA. Importantly, aldehydes cause a variety of DNA lesions, including ICLs and DNA protein cross-links (DPCs), re-kindling the debate about which DNA lesions cause FA. In this review, we discuss new developments in our understanding of DPC and ICL repair, and how these findings bear on the question of which DNA lesion underlies FA.
Introduction
In 1927, the Swiss pediatrician Guido Fanconi described three brothers who presented with developmental birth defects and died of a condition resembling pernicious anemia [1,2]. He soon realized that the disease affected all blood lineages, and that it also involves cancer predisposition. Fanconi’s anemia (now referred to as Fanconi anemia; FA) was subsequently recognized as a rare genetic disorder inherited as a Mendelian recessive trait that affects 1 in every ~100,000 births. So far, 19 FANC genes have been identified, mutations in which cause FA. While mutations in most complementation groups cause the full spectrum of FA-associated phenotypes (congenital abnormalities, early onset bone marrow failure (BMF), predisposition to acute myeloid leukemia and solid tumors), some complementation groups (e.g. FANCD1, N, O, S and R) exhibit a subset of these features.
In the 1970s, researchers discovered that FA cells undergo chromosome breakage upon treatment with crosslinking agents such as mitomycin C (MMC) or diepoxybutane [3,4], suggesting that an inability to repair DNA interstrand cross-links (ICLs) underlies FA. ICLs are cytotoxic lesions that covalently link the two strands of the double helix, thereby inhibiting any process that requires DNA unwinding, including DNA replication and transcription. Two distinct mechanisms of ICL repair have been described. One mechanism is tightly coupled to DNA replication and requires the FANC proteins [5-9]. The other operates outside of S phase, involves nucleotide excision repair but not the FANC proteins, and may sometimes be coupled to transcription [10,11].
The FA pathway
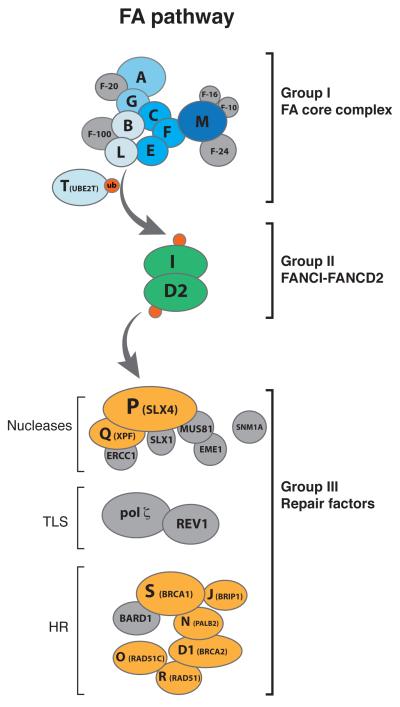
The proteins encoded by the 19 FANC genes coordinate different steps of ICL-repair and can be placed into 3 groups based on their functions [12]. The group I proteins FANCA, B, C, E, F, G, L, and M, together with three Fanconi associated proteins (FAAP20, FAAP24 and FAAP100), assemble into a large FA core complex, which functions as an E3 ubiquitin ligase (Figure 1, group I). The core complex associates with chromatin upon DNA damage or replication stress [13,14], and it monoubiquitylates the group II proteins FANCI and FANCD2, which form a heterodimer called the ID2 complex [15,16] (Figure 1, group II). Mono-ubiquitylated ID2 binds to chromatin and is required to suppress ICL sensitivity. Extensive evidence suggests the existence of distinct functional modules within the core complex. FANCM interacts with FAAP24 and a dimer of histone-fold containing proteins, MHF1 and MHF2 (also known as FAAP16 and FAAP10) [17-19]. This heterotetrameric FANCM subcomplex recognizes model DNA structures that resemble replication forks [17], and this binding is thought to recruit the core complex to chromatin [14,20]. The FANCM subcomplex also regulates downstream repair and checkpoint signaling [21,22], presumably by remodeling stalled replication forks through FANCM’s ATPase activity [23,24]. FANCB, FANCL, and FAAP100 form a minimal catalytic module in which the RING domain of FANCL ubiquitylates ID2 [25-29]. UBE2T (recently identified as FANCT [30-32]) functions as the E2 ubiquitin-conjugating enzyme, and its interaction with FANCL is required for ID2 monoubiquitination [33,34]. FANCA, FANCG, FAAP20 and FANCC, FANCE, FANCF form two other subcomplexes that are proposed to assist the catalytic subcomplex in binding to chromatin [26].
Figure 1. The Fanconi anemia pathway.
The FA pathway comprises 19 proteins that have been classified into three groups [12]. Upon detection of the crosslink, the FA core complex (group I, blue spheres) ubiquitylates the heterodimer FANCI-FANCD2 (ID2) (group II, green spheres). Ubiquitylated ID2 then coordinates processing by downstream repair factors (group III, orange spheres). Proteins shaded in grey are important for ICL repair and can be classified as group I-III, but they have not been found to be mutated in patients with FA. Although BRCA1 and RAD51 are considered to fall into group III, they also have functions upstream of ID2 ubiquitylation [40,61].
Mono-ubiquitylated ID2 promotes repair of the ICL by group III proteins, which include the nuclease XPF (FANCQ) [35], the scaffolding protein SLX4 (FANCP) [36,37], and the homologous recombination (HR) factors PALB2 (FANCN) [38], BRCA2 (FANCD1) [39], RAD51 (FANCR) [40], RAD51C (FANCO) [41], BRCA1 (FANCS) [42] and FANCJ (BRIP1) [43-45] (Figure 1, group III). Finally, the modified ID2 complex is deubiquitinated by the ubiquitin specific peptidase 1 (USP1) [46] and its activating partner UAF1 [47]. Importantly many other factors participate in ICL repair including the nucleases SNM1A, SNM1B, FAN1, MUS81-EME1, SLX1, MRN and CTIP, and translesion (TLS) polymerases REV1 and polymerase ζ (pol ζ) [11]. In most cases, these factors were identified because their deficiency causes cellular sensitivity to ICLs. Whether all the above proteins actually operate in the FA pathway of ICL repair is presently unclear. For example, current evidence suggests that FAN1, originally identified as a nuclease that is recruited to sites of damage by ubiquitylated ID2 [48-50], probably does not operate in the FA pathway of ICL repair [51]. Consistent with this view, mutations in FAN1 are associated with karyomegalic interstitial nephritis, a form of chronic kidney disease, instead of FA [52].
Some Fanconi proteins stabilize stressed replication forks. A new report identified a dominant negative RAD51 (FANCR) mutation that causes a Fanconi-like phenotype [40]. While FANCR patient cells are HR proficient, they are sensitive to crosslinking agents, apparently due to over-resection of nascent strands by DNA2 [40,53-55]. Interestingly, when forks are arrested with hydroxyurea, nascent strands are protected from Mre11-dependent degradation by BRCA1, BRCA2, and RAD51 [56,57], as well as FANCA and ubiquitylated FANCD2 [58]. However, it seems unlikely that all FA genes, particularly those required for endonucleoytic cleavage of DNA (SLX4 and XPF), will be required to protect stalled forks in the absence of damage. As such, it seems unlikely that fork protection by FA proteins in the absence of damage is central to suppression of the FA phenotype.
Replication-coupled ICL repair: how does it work?
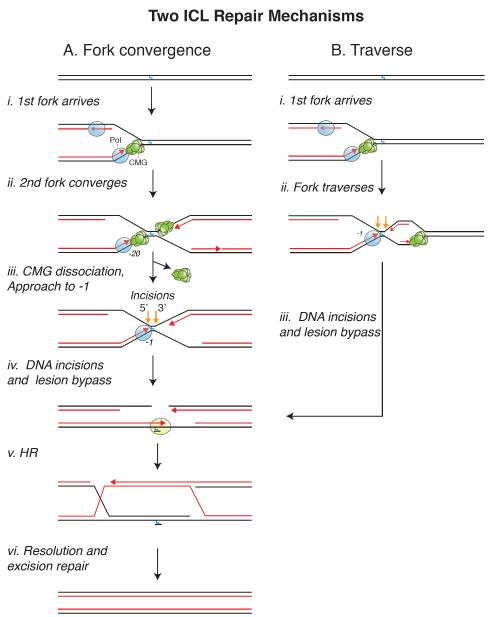
The participation of structure-specific endonucleases, TLS polymerases, and the HR machinery led to a model of ICL repair in which collision of a replication fork triggers incisions on either side of the ICL, followed by TLS, and HR-mediated fork restart [12,59]. However, a detailed mechanism, as well as the function of the FA pathway, remained elusive until replication-coupled ICL repair was recapitulated in Xenopus egg extracts [6]. When a plasmid containing a site-specific ICL is incubated in egg extract, replication initiates at a random location on the plasmid, and two replisomes quickly converge on the ICL (Figure 2A, i-ii). Upon collision with the ICL, leading strands of each replisome initially stall ~20 nucleotides from the lesion due to steric hindrance by the CMG helicase (a complex of Cdc45, MCM2-7, and GINS), which travels along the leading strand template and thus stalls at the lesion [60]. The first detectable event in repair is the active unloading of the stalled CMGs, which requires ubiquitin signaling and the BRCA1 (FANCS)-BARD1 tumor suppressor complex [60,61]. The mechanism of CMG unloading is still unknown but may involve ubiquitylation of the MCM2-7 complex [62,63] by BRCA1-BARD1. Once CMG is unloaded, leading strands are extended to within 1 nucleotide of the crosslink (-1 position), probably by the leading strand DNA polymerase ε [64] (Figure 2A, iii). Concurrent with this “approach” of the leading strand, ubiquitylated ID2 localizes to the ICL and promotes dual incisions on either side of the ICL (“unhooking”) [65] by recruiting a complex of the scaffolding protein SLX4 and the 3′ flap endonuclease XPF (FANCQ)-ERCC1 [66,67] (Figure 2A, iv). Interestingly, in the absence of XPF, neither the 5′ nor the 3′ incision takes place [67], suggesting that XPF might perform both incisions, as recently proposed [68]. Alternatively, one or more 5′ flap endonucleases might incise the 5′ side but depend on prior action of XPF on the 3′ side. Indeed, the 5′ flap endonuclease SLX1 could be recruited through an interaction with SLX4 [69-72] while CTIP, a newly recognized 5′ flap endonuclease [73,74], might be recruited directly via ID2 [75,76]. Analogous coupling between 3′ and 5′ flap endonucleases has been observed between XPF and XPG during nucleotide excision repair (NER) [77]. Dual incisions generate a two-ended double strand break (DSB) in one of the sister chromatids while leaving a DNA adduct on the other sister. The adducted chromatid is restored by TLS in a two-step reaction. First a nucleotide is inserted across from the damage base by an unknown polymerase. The abnormal primer template is then extended by a complex of REV1 and pol ζ, whose recruitment to chromatin requires the FA core complex [6,64,78] (Figure 2A, iv). Finally, the DSB is repaired by Rad51-mediated HR utilizing the intact sister chromatid as a homology donor [79] (Figure 2A, v). In egg extracts, the remaining mono-adduct persists, but in cells, the adduct is likely removed by excision repair (Figure 2A, vi).
Figure 2. Mechanisms of ICL repair.
Two mechanisms of ICL repair are depicted. (A) The fork convergence pathway, in which ICL repair is triggered when two forks converge on the lesion. (B) The fork traverse pathway, in which a single fork bypasses an ICL without unhooking the parental strands. Incisions are represented by orange arrows.
An important question is whether ICL repair requires the convergence of two forks on the lesion or whether one fork suffices, as proposed in most classical models of ICL repair [59]. Indeed, during chromosomal DNA replication, where the inter-origin distance is roughly 100 kb [80], one fork will often encounter an ICL long before a second fork arrives. Importantly, when only one fork was allowed to strike the ICL in egg extracts (due to a barrier that prevented arrival of the second fork), the lone fork did not initiate ICL repair and its CMG was never unloaded [81]. However, CMG unloading and repair were restored when the second fork arrived as much as one hour after the first fork (due to timed dissolution of the barrier). Although the requirement for two forks needs to be confirmed in cells, the data in egg extracts show that a single fork, while inactive for ICL repair, remains stable and competent for repair until a second fork arrives. Consistent with this view, stalled replication forks are generally very stable in vivo, and collapse only after prolonged treatment with hydroxyurea [82] or following global exhaustion of the single strand DNA binding protein, RPA [83]. The advantage of coupling CMG unloading to fork convergence is that it avoids inadvertent replisome disassembly at single forks that have stalled transiently. This is especially important given the absence of de novo CMG assembly pathways in the S phase of the cell cycle [84]. In summary, it appears that waiting for two forks to converge on an ICL is a viable strategy to initiate ICL repair.
Recently, Seidman and colleagues investigated the collision of replication forks with fluorescently marked psoralen ICLs in cells using DNA combing [85]. Although they observed many instances of fork convergence, more often single forks bypassed ICLs without unhooking them (Figure 2B, i-ii, “traverse”). In this scenario, ICL repair is thought to occur after the traversed fork has moved beyond the lesion. The mechanism of traverse, including the identity of the helicase that unwinds DNA distal to the ICL, is currently unclear. However, one attractive possibility is that the CMG ring transiently opens and is pushed past the ICL by FANCM, whose translocase activity is required for traverse [85]. In a mechanism that may be analogous to ICL traverse, the large T antigen DNA helicase was shown to bypass a covalent DNA-protein cross-link (DPC) on the translocation strand, probably via ring opening [86]. Importantly, whether two forks converge on an ICL (Figure 2A, iii) or a single fork undergoes traverse (Figure 2B, ii), a similar X-shaped DNA structure is generated around the lesion, which may be the critical trigger for ICL unhooking [87]. At present, it is unclear how the balance between traverse and fork convergence in cells is governed. Nevertheless, the work in extracts and cells suggests that a single fork stalled at an ICL is unable to promote repair and that an X-shaped structure, generated by fork convergence or traverse, is the key substrate of the endonucleases that unhook the ICL.
FA and reactive aldehydes
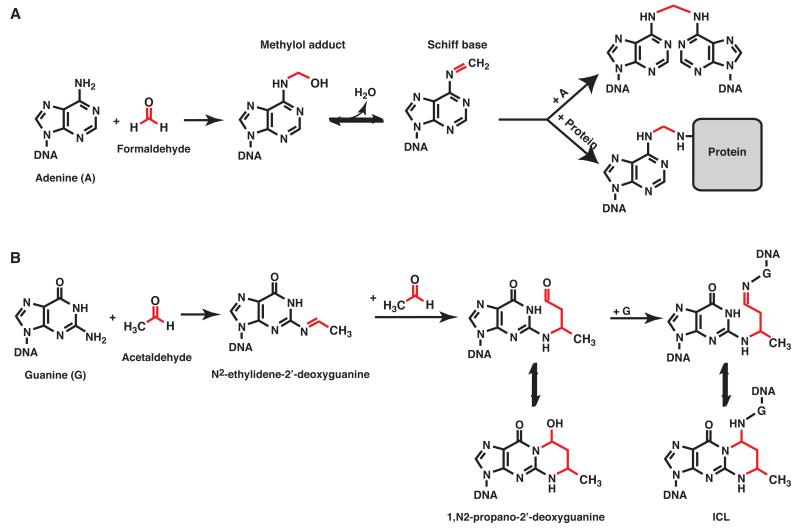
Based on the sensitivity of FA cells to agents such as cisplatin and mitomycin C, it has been widely assumed that FA is caused by defective repair of endogenously produced ICLs. However, which metabolites produce such ICLs has been a matter of conjecture. The best candidates are reactive aldehydes, since they are known to form a wide variety of DNA adducts including ICLs [88]. For example, formaldehyde is generated during histone demethylation and dealkylation of methylated DNA [89,90]. It is also an intermediate required for the biosynthesis of purines and certain amino acids and is found at high concentration in human plasma (~ 30-90 μM) [91,92]. Formaldehyde generates ICLs via a methylene bridge formed between the exocyclic amino groups of adjacent DNA bases [93,94] (Figure 3A, top pathway). Acetaldehyde is a byproduct of ethanol oxidation and an intermediate of carbohydrate metabolism [95], and it forms ICLs, mainly by reacting with guanines (Figure 3B) [96-98]. Other reactive aldehydes including 4-hydroxynonenal (4-HNE), acrolein, malondialdehyde and crotonaldehyde are produced through lipid peroxidation [88], and most of these agents are able to form ICLs [98-102]. Despite evidence that reactive aldehydes induce ICLs in the test tube, for many years there was no evidence to suggest they underlie the etiology of FA.
Figure 3. Aldehyde mediated DNA cross-links.
Reactions of DNA with formaldehyde (A) and acetaldehyde (B) are depicted. (A) Formaldehyde reacts with primary amines of DNA bases to form a methylol adduct. Dehydration results in the formation of a Schiff base intermediate that can react with another base to form an interstrand cross-link (top) or a lysine to form a DNA-protein cross-link (bottom). (B) Acetaldehyde mainly reacts with the exocyclic amine of deoxyguanine to form N2-ethylidene-2′-deoxyguanine adducts [97]. Through the reaction of a second acetaldehyde molecule N2-ethylidene-2′-deoxyguanine is converted to 1,N2-propano-2′-deoxyguanine [97,132]. 1,N2-propano-2′-deoxyguanine adducts exist in equilibrium between the open and closed form. The ring open form which is favored in double strand DNA can induce ICLs by reacting with another dG on the complementary strand [96-98]. It can also form DPCs by reacting with primary amines of proteins (not depicted) [111].
This picture has changed dramatically, as recent genetic experiments provide powerful evidence for a connection between endogenous aldehydes and FA. Initially, it was reported that chicken cells deficient in FANC proteins are highly sensitive to low doses of formaldehyde [103]. Patel and colleagues then explored the interplay of aldehyde metabolism and the Fanconi pathway in mice, by deleting the acetaldehyde detoxifying enzyme, aldehyde dehydrogenase 2 (ALDH2). While Aldh2−/− and Fancd2−/− single mutant mice were viable and exhibited no severe phenotypes, Aldh2−/− Fancd2−/− mice were born only from mothers carrying at least one wild-type allele of Aldh2, demonstrating that aldehyde catabolism in utero is essential for embryonic development [104]. Viable Aldh2−/− Fancd2−/− mice displayed developmental abnormalities and died of acute lymphoblastic leukemia within the first 6 months of life. The few mice that did not get leukemia eventually developed spontaneous BMF [105]. When challenged with ethanol soon after birth, mice rapidly developed severe BMF [104]. Underlying these phenotypes was a profound reduction in the hematopoietic and progenitor stem cell pool as observed in FA patients [105]. More recently, Patel and colleagues also examined the connection of the FA pathway with alcohol dehydrogenase 5 (ADH5), thought to be the major formaldehyde catabolizing enzyme [106]. In contrast to the situation for ALDH2, Adh5−/− Fancd2−/− mice were born irrespectively of maternal Adh5 status. However, these mice developed BMF and hematopoietic stem cell depletion much earlier than Aldh2−/− Fancd2−/− mice, suggesting that endogenous formaldehyde is more cytotoxic than acetaldehyde. Accordingly, in chicken cells mutations in FANC genes are synthetically lethal with Adh5 but not Aldh2 mutations [104,107]. The greater toxicity of formaldehyde could be due to a higher reactivity of formaldehyde and/or a greater abundance of endogenous formaldehyde versus acetaldehyde. The fact that Aldh2−/− Fancd2−/− and Adh5−/− Fancd2−/− double mutant mice recapitulate the key phenotypes of FA suggests that BMF in FA patients is caused by aldehyde toxicity. Consistent with this view, in Japanese FA patients, the severity of the disease correlates with the Asian flushing mutation, a dominant negative allele of Aldh2 present in 36% of the population in East Asia [108]. In the future, it will be important to track the maternal Aldh2 status for a wider cohort of FA patients to determine whether maternal aldehyde detoxification protects human FA patients from DNA damage [104,109]. Together, these experiments indicate that eukaryotic cells use a two-pronged approach to avoid the genotoxic effects of reactive aldehydes [110]. On the one hand, their levels are kept in check by aldehyde catabolizing enzymes. On the other hand, the damage created by these agents is neutralized by the FA pathway. In mice, only the absence of both pathways causes severe toxicitiy, whereas in humans, neutralizing the FA pathway alone is sufficient to cause disease.
What do these studies teach us about the endogenous lesions that cause FA? The first question is which specific aldehyde(s) are relevant to FA. Since mutations in Aldh2 and Adh5 both cause synthetic sickness with mutations in FANC genes, both acetaldehyde and formaldehyde might be able to cause the offending lesions. Alternatively, mutations in Aldh2 and Adh5 may lead to an increase in a common, unique metabolite that drives FA. Given its greater toxicity in chicken cells and mice, formaldehyde might be the primary culprit [106,107]. Even if this is the case, there is a further ambiguity, since formaldehyde causes not only ICLs, but also DPCs [111,112]. In fact, formaldehyde is known to favor DPC lesions [92] by forming a methylene bridge between nucleophilic amino acid side chains and exocyclic amines of DNA bases [113]. (Figure 3A, bottom pathway). To complicate matters further, most chemicals traditionally considered to be ICL-inducing agents such as nitrogen mustards or platinum compounds can also cause DPCs [114-117]. Thus, when FA cells are treated with aldehydes, cisplatin, or other bifunctional compounds, both ICLs and DPCs are likely to form, making it impossible to determine which lesion drives toxicity. In conclusion, although the identification of aldehydes as the endogenous metabolites underlying FA represents an important breakthrough, the nature of the offending DNA lesions remains unknown.
The FA pathway and DPC repair
One approach to resolve the above ambiguity is to examine the requirement for the FA pathway in the repair of chemically-defined DNA lesions. Importantly, extract and cell-based studies showed that the FA pathway is essential for the repair of synthetic ICLs [9,65]. What about DPCs?
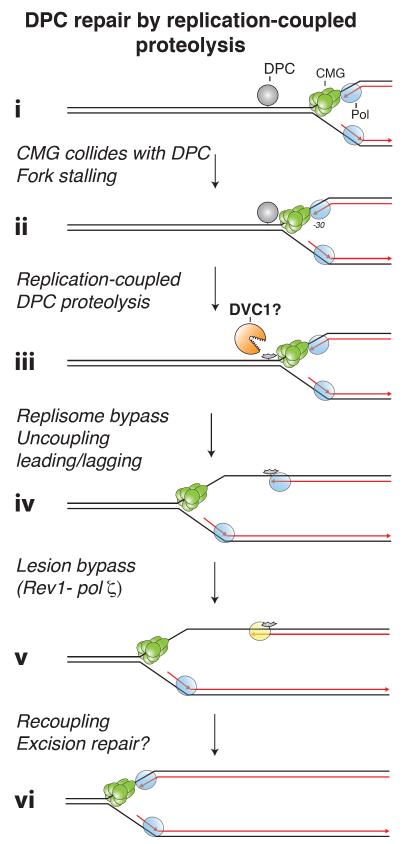
The repair of chemically-defined DPC lesions was recently recapitulated in egg extracts [118]. A bacterial DNA methyltransferase (M.HpaII) was covalently linked to a specific location in a plasmid (via the C6 position on cytosine), and the resulting DPC was incubated in egg extracts, whereupon it was repaired in a replication-dependent manner. When the DPC is encountered on the leading strand template, replication stalls due to collision of CMG with the lesion, as seen during ICL repair (Figure 4, i-ii). However, in contrast to ICL repair, DPC repair does not require fork convergence, and CMG is not unloaded. Instead, the DPC is degraded on DNA via a replication-coupled protease, yielding a peptide-DNA adduct that is bypassed by CMG (Figure 4, iii-iv). The leading strand is subsequently extended and stalls again at the peptide adduct. Finally, a complex of REV1 and Pol ζ allows the leading strand to bypass the peptide adduct (Figure 4, v). A DPC on the lagging strand template only transiently stalls the replisome, but it too is degraded to a peptide, allowing Okazaki fragment bypass. Importantly, DPC repair proceeds without incision of parental strands and therefore does not involve a DSB intermediate. Consistent with this observation, M. HpaII DPC-repair does not require the ID2 complex. If DPCs are formed by reactive aldehydes in vivo, the chemistry of the protein-DNA linkage will be different from that of the M. HpaII DPC. However, the attachment chemistry is unlikely to affect the initial processing of the DPC, but rather dictate which translesion DNA polymerase is employed to bypass the remaining peptide adduct. To test this assumption, it will be critical to examine the role of FANC proteins in the repair of aldehyde-induced DPCs. In conclusion, when the repair of chemically defined DPCs and ICLs is compared in the same cell-free system, only the latter requires the FA pathway. This observation supports the notion that failure to repair ICLs created by endogenous aldehydes is the underlying cause of FA.
Figure 4. DNA-protein cross-link repair by replication-coupled proteolysis.
A replication fork stalls when it encounters a DNA-protein crosslink (DPC). This stalling can be relieved by the degradation of the DPC to a peptide by a replication-dependent protease. A potential candidate for this protease is DVC1, which has homology to the yeast DPC protease Wss1.
DPC repair and human disease
To determine how defective DPC repair impacts human health, it will be critical to identify factors that are specifically dedicated to DPC repair. In an elegant study, Jentsch and colleagues identified budding yeast Wss1 as a DPC protease [119]. Wss1 contains an N-terminal metalloprotease domain, SHP and VIM domains that mediate binding to the Cdc48 ATPase (known as p97 or VCP in higher eukaryotes), and tandem SUMO interaction motifs (SIMs). Wss1 removes covalently trapped topoisomerase I complexes and it confers formaldehyde resistance. Both of these functions require the metalloprotease and Cdc48 binding domains, and to a lesser extent, the SIMs. Interestingly, purified Wss1 contains DNA binding activity and it degrades proteins only when these are bound to DNA. Collectively, these and other data indicate that Wss1 functions as a DPC protease that removes DPCs during replication [119]. Further analysis will be required to elucidate the role of Cdc48 in this process, and to determine how Wss1’s activity is regulated to avoid degrading DNA binding proteins that are not covalently linked to DNA. One possibility is that Wss1 activity is dependent on replication fork stalling, as suggested by the work in egg extracts [118].
Before the discovery of Wss1 and a replication-coupled DPC repair mechanism [118,119], the main pathways implicated in DPC repair were NER and HR. NER provides resistance to DPC-inducing agents such as formaldehyde, and is proposed to remove small (<11 kDa) DPCs outside of S phase [120-123]. In contrast, larger DPCs, which evade NER, were thought to depend on HR during replication [120,121]. In yeast, Wss1 and the recombinase Rad52 are not epistatic with regard to formaldehyde sensitivity, and in the absence of Wss1, Rad52 repair foci and gross chromosomal rearrangements increase, arguing that DPC proteolysis and HR represent alternative mechanisms of DPC repair during S phase [119]. Given that it does not involve a DSB intermediate [118], proteolysis-dependent DPC repair probably represents the preferred means of eliminating DPCs, while HR might act on a subset of DPCs that cannot be degraded. Alternatively, the requirement for HR in formaldehyde resistance might involve the repair of formaldehyde-induced ICLs [93,94], or result from such a large load of DPCs that Wss1 becomes limiting.
The closest vertebrate homolog of Wss1 is DVC1 (also known as Spartan), which also contains an N-terminal metalloprotease domain and an SHP p97 binding motif [124,125]. Instead of SIM domains, DVC1 contains a ubiquitin binding motif and a PCNA-interaction protein (PIP) motif, which it uses to bind ubiquitylated PCNA [126]. DVC1 participates in the response to UV irradiation [124,126,127]. Although this may reflect a role for DVC1 in regulating TLS at UV-induced lesions, it is also consistent with a role for DVC1 in the repair of DPCs, which can be caused by UV light [128,129]. Consistent with the latter view, DVC1 knockdown causes sensitivity to camptothecin, a drug that traps topoisomerase I on DNA [127]. Strikingly, DVC1 was recently identified as the causative mutation in an atypical Werner-like progeroid syndrome with clinical features distinct from those of FA [130]. Patients harboring biallelic germline mutations in DVC1, including one that resides in the protease domain, exhibited premature aging features such as graying hair, muscular atrophy and cataracts. The patients also developed early onset hepatocellular carcinomas. Cells from these patients contained DNA damage and signs of replication stress, consistent with DVC1 acting in a replication-coupled repair pathway. In mice, DVC1 null mutations cause embryonic lethality, implying that DVC1 is essential to repair a highly toxic endogenous lesion. Consistent with the human phenotypes, hypomorphic DVC1 alleles cause premature aging in mice, although no cancers were detected [131]. DVC1 conditional knockout MEFs do not proliferate. Before they die, they display replication stress, which is rescued by introduction of wild-type DVC1 but not DVC1 harboring a mutation in its conserved protease domain. Given the parallels between DVC1 and Wss1, and the requirement for DVC1s protease domain to relieve replication stress and suppress aging and cancer, the simplest interpretation of these results is that DVC1 functions as a DPC protease. If this is the case, it would show that failure to repair DPCs causes a disease that is phenotypically distinct from FA, further disfavoring the idea that defective DPC repair underlies FA. The question then arises whether endogenous aldehydes also underlie the DVC1-deficiency syndrome, which can be addressed by crossing DVC1 hypomorphic mice with Aldh2 deficient mice.
Conclusions
Nearly a century after Guido Fanconi’s description of FA, the field has made great progress in identifying and understanding the properties of 19 FANC gene products and their roles in repairing ICLs. The recent identification of reactive aldehydes as the likely mutagen underlying FA is a major advance, but in itself, does not identify the offending DNA lesion. Importantly, in DNA repair assays using a limited number of chemically defined DNA lesions, the FA pathway is required for ICL repair but not DPC repair, supporting the original idea that failure to repair ICLs causes FA. This conclusion is further strengthened by emerging evidence that defective DPC repair causes a genetic disease that is phenotypically distinct from FA. One plausible scenario is that reactive aldehydes cause ICLs and DPCs, and failure to repair each class of lesion causes a different clinical manifestation. However, the real situation may be more complex. For example, aldehydes might link a protein to both strands of the DNA, generating a DPC that effectively mimics an ICL. If the protein is attached to the two DNA strands at closely spaced amino acids, the lesion may not be amenable to DVC1 processing and thus may have to be dealt with via ID2-dependent incisions. To further clarify these issues, it will be important to better define the aldehydes whose upregulation in Aldh2−/− and Adh5−/− cells cause toxicitiy. In addition, more sensitive analytical tools will also have to be developed to identify endogenous lesions that accumulate on the chromosomes of FA cells. Ultimately, understanding the molecular etiology of FA will not only enhance our view of the cell’s varied DNA repair pathways but also lay the foundation for targeted therapies. Thus, researchers may one day be able to neutralize the offending mutagen or shunt the lesion it causes into an alternative DNA repair pathway that is still intact in FA patients.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Fanconi G. Familiäre infantile perniziosaartige Anämie (perniziöses Blutbild und Konstitution) 1927. [no volume]
- 2.Lobitz S, Velleuer E. Guido Fanconi (1892-1979): a jack of all trades. 2006. [DOI] [PubMed]
- 3.Sasaki MS, Tonomura A. A high susceptibility of Fanconi's anemia to chromosome breakage by DNA cross-linking agents. Cancer Research. 1973;33:1829–1836. [PubMed] [Google Scholar]
- 4.Auerbach AD, Wolman SR. Susceptibility of Fanconi's anaemia fibroblasts to chromosome damage by carcinogens. Nature. 1976;261:494–496. doi: 10.1038/261494a0. [DOI] [PubMed] [Google Scholar]
- 5.Akkari YM, Bateman RL, Reifsteck CA, Olson SB, Grompe M. DNA replication is required To elicit cellular responses to psoralen-induced DNA interstrand cross-links. Mol. Cell. Biol. 2000;20:8283–8289. doi: 10.1128/mcb.20.21.8283-8289.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Räschle M, Knipscheer P, Enoiu M, Angelov T, Sun J, Griffith JD, Ellenberger TE, Schärer OD, Walter JC. Mechanism of Replication-Coupled DNA Interstrand Crosslink Repair. Cell. 2008;134:969–980. doi: 10.1016/j.cell.2008.08.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Rothfuss A, Grompe M. Repair kinetics of genomic interstrand DNA cross-links: evidence for DNA double-strand break-dependent activation of the Fanconi anemia/BRCA pathway. Mol. Cell. Biol. 2004;24:123–134. doi: 10.1128/MCB.24.1.123-134.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Taniguchi T, Garcia-Higuera I, Andreassen PR, Gregory RC, Grompe M, D'Andrea AD. S-phase-specific interaction of the Fanconi anemia protein, FANCD2, with BRCA1 and RAD51. Blood. 2002;100:2414–2420. doi: 10.1182/blood-2002-01-0278. [DOI] [PubMed] [Google Scholar]
- 9.Nakanishi K, Cavallo F, Perrouault L, Giovannangeli C, Moynahan ME, Barchi M, Brunet E, Jasin M. Homology-directed Fanconi anemia pathway cross-link repair is dependent on DNA replication. Nature Structural & Molecular Biology. 2011;18:500–503. doi: 10.1038/nsmb.2029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Williams HL, Gottesman ME, Gautier J. The differences between ICL repair during and outside of S phase. Trends Biochem. Sci. 2013;38:386–393. doi: 10.1016/j.tibs.2013.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Clauson C, Schärer OD, Niedernhofer L. Advances in understanding the complex mechanisms of DNA interstrand cross-link repair. Cold Spring Harb Perspect Biol. 2013;5 doi: 10.1101/cshperspect.a012732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wang W. Emergence of a DNA-damage response network consisting of Fanconi anaemia and BRCA proteins. Nature Publishing Group. 2007;8:735–748. doi: 10.1038/nrg2159. [DOI] [PubMed] [Google Scholar]
- 13.Qiao F, Moss A, Kupfer GM. Fanconi anemia proteins localize to chromatin and the nuclear matrix in a DNA damage- and cell cycle-regulated manner. J. Biol. Chem. 2001;276:23391–23396. doi: 10.1074/jbc.M101855200. [DOI] [PubMed] [Google Scholar]
- 14.Kim JM, Kee Y, Gurtan A, D'Andrea AD. Cell cycle-dependent chromatin loading of the Fanconi anemia core complex by FANCM/FAAP24. Blood. 2008;111:5215–5222. doi: 10.1182/blood-2007-09-113092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Garcia-Higuera I, Taniguchi T, Ganesan S, Meyn MS, Timmers C, Hejna J, Grompe M, D'Andrea AD. Interaction of the Fanconi anemia proteins and BRCA1 in a common pathway. Mol. Cell. 2001;7:249–262. doi: 10.1016/s1097-2765(01)00173-3. [DOI] [PubMed] [Google Scholar]
- 16.Smogorzewska A, Matsuoka S, Vinciguerra P, McDonald ER, Hurov KE, Luo J, Ballif BA, Gygi SP, Hofmann K, D'Andrea AD, et al. Identification of the FANCI protein, a monoubiquitinated FANCD2 paralog required for DNA repair. Cell. 2007;129:289–301. doi: 10.1016/j.cell.2007.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ciccia A, Ling C, Coulthard R, Yan Z, Xue Y, Meetei AR, Laghmani EH, Joenje H, McDonald N, de Winter JP, et al. Identification of FAAP24, a Fanconi anemia core complex protein that interacts with FANCM. Mol. Cell. 2007;25:331–343. doi: 10.1016/j.molcel.2007.01.003. [DOI] [PubMed] [Google Scholar]
- 18.Singh TR, Saro D, Ali AM, Zheng X-F, Du C-H, Killen MW, Sachpatzidis A, Wahengbam K, Pierce AJ, Xiong Y, et al. MHF1-MHF2, a histone-fold-containing protein complex, participates in the Fanconi anemia pathway via FANCM. Mol. Cell. 2010;37:879–886. doi: 10.1016/j.molcel.2010.01.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yan Z, Delannoy M, Ling C, Daee D, Osman F, Muniandy PA, Shen X, Oostra AB, Du H, Steltenpool J, et al. A histone-fold complex and FANCM form a conserved DNA-remodeling complex to maintain genome stability. Mol. Cell. 2010;37:865–878. doi: 10.1016/j.molcel.2010.01.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Deans AJ, West SC. FANCM Connects the Genome Instability Disorders Bloom's Syndrome and Fanconi Anemia. Mol. Cell. 2009;36:943–953. doi: 10.1016/j.molcel.2009.12.006. [DOI] [PubMed] [Google Scholar]
- 21.Collis SJ, Ciccia A, Deans AJ, Horejsi Z, Martin JS, Maslen SL, Skehel JM, Elledge SJ, West SC, Boulton SJ. FANCM and FAAP24 function in ATR-mediated checkpoint signaling independently of the Fanconi anemia core complex. Mol. Cell. 2008;32:313–324. doi: 10.1016/j.molcel.2008.10.014. [DOI] [PubMed] [Google Scholar]
- 22.Huang M, Kim JM, Shiotani B, Yang K, Zou L, D'Andrea AD. The FANCM/FAAP24 complex is required for the DNA interstrand crosslink-induced checkpoint response. Mol. Cell. 2010;39:259–268. doi: 10.1016/j.molcel.2010.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gari K, Décaillet C, Stasiak AZ, Stasiak A, Constantinou A. The Fanconi anemia protein FANCM can promote branch migration of Holliday junctions and replication forks. Mol. Cell. 2008;29:141–148. doi: 10.1016/j.molcel.2007.11.032. [DOI] [PubMed] [Google Scholar]
- 24.Gari K, Décaillet C, Delannoy M, Wu L, Constantinou A. Remodeling of DNA replication structures by the branch point translocase FANCM. Proc Natl Acad Sci U S A. 2008;105:16107–16112. doi: 10.1073/pnas.0804777105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Meetei AR, de Winter JP, Medhurst AL, Wallisch M, Waisfisz Q, van de Vrugt HJ, Oostra AB, Yan Z, Ling C, Bishop CE, et al. A novel ubiquitin ligase is deficient in Fanconi anemia. Nat Genet. 2003;35:165–170. doi: 10.1038/ng1241. [DOI] [PubMed] [Google Scholar]
- 26.Huang Y, Leung JWC, Lowery M, Matsushita N, Wang Y, Shen X, Do Huong, Takata M, Chen J, Li L. Modularized Functions of the Fanconi Anemia Core Complex. Cell Rep. 2014;7:1849–1857. doi: 10.1016/j.celrep.2014.04.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ling C, Ishiai M, Ali AM, Medhurst AL, Neveling K, Kalb R, Yan Z, Xue Y, Oostra AB, Auerbach AD, et al. FAAP100 is essential for activation of the Fanconi anemia-associated DNA damage response pathway. The EMBO Journal. 2007;26:2104–2114. doi: 10.1038/sj.emboj.7601666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Medhurst AL, Laghmani EH, Steltenpool J, Ferrer M, Fontaine C, de Groot J, Rooimans MA, Scheper RJ, Meetei AR, Wang W, et al. Evidence for subcomplexes in the Fanconi anemia pathway. Blood. 2006;108:2072–2080. doi: 10.1182/blood-2005-11-008151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Rajendra E, Oestergaard VH, Langevin F, Wang M, Dornan GL, Patel KJ, Passmore LA. The Genetic and Biochemical Basis of FANCD2 Monoubiquitination. Mol. Cell. 2014;54:858–869. doi: 10.1016/j.molcel.2014.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hira A, Yoshida K, Sato K, Okuno Y, Shiraishi Y, Chiba K, Tanaka H, Miyano S, Shimamoto A, Tahara H, et al. Mutations in the gene encoding the E2 conjugating enzyme UBE2T cause Fanconi anemia. Am. J. Hum. Genet. 2015;96:1001–1007. doi: 10.1016/j.ajhg.2015.04.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rickman KA, Lach FP, Abhyankar A, Donovan FX, Sanborn EM, Kennedy JA, Sougnez C, Gabriel SB, Elemento O, Chandrasekharappa SC, et al. Deficiency of UBE2T, the E2 Ubiquitin Ligase Necessary for FANCD2 and FANCI Ubiquitination, Causes FA-T Subtype of Fanconi Anemia. Cell Rep. 2015;12:35–41. doi: 10.1016/j.celrep.2015.06.014. 2015, Cell Rep. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Virts EL, Jankowska A, MacKay C, Glaas MF, Wiek C, Kelich SL, Lottmann N, Kennedy FM, Marchal C, Lehnert E, et al. AluY-mediated germline deletion, duplication and somatic stem cell reversion in UBE2T defines a new subtype of Fanconi anemia. Hum. Mol. Genet. 2015;24:5093–108. doi: 10.1093/hmg/ddv227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Machida YJ, Machida Y, Chen Y, Gurtan AM, Kupfer GM, D'Andrea AD, Dutta A. UBE2T is the E2 in the Fanconi anemia pathway and undergoes negative autoregulation. Mol. Cell. 2006;23:589–596. doi: 10.1016/j.molcel.2006.06.024. [DOI] [PubMed] [Google Scholar]
- 34.Alpi AF, Pace PE, Babu MM, Patel KJ. Mechanistic insight into site-restricted monoubiquitination of FANCD2 by Ube2t, FANCL, and FANCI. Mol. Cell. 2008;32:767–777. doi: 10.1016/j.molcel.2008.12.003. [DOI] [PubMed] [Google Scholar]
- 35.Bogliolo M, Schuster B, Stoepker C, Derkunt B, Su Y, Raams A, Trujillo JP, Minguillón J, Ramírez MJ, Pujol R, et al. Mutations in ERCC4, Encoding the DNA-Repair Endonuclease XPF, Cause Fanconi Anemia. The American Journal of Human Genetics. 2013;92:800–806. doi: 10.1016/j.ajhg.2013.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kim Y, Lach FP, Desetty R, Hanenberg H, Auerbach AD, Smogorzewska A. Mutations of the SLX4 gene in Fanconi anemia. Nat Genet. 2011;43:142–146. doi: 10.1038/ng.750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Stoepker C, Hain K, Schuster B, Hilhorst-Hofstee Y, Rooimans MA, Steltenpool J, Oostra AB, Eirich K, Korthof ET, Nieuwint AWM, et al. SLX4, a coordinator of structure-specific endonucleases, is mutated in a new Fanconi anemia subtype. Nat Genet. 2011;43:138–141. doi: 10.1038/ng.751. [DOI] [PubMed] [Google Scholar]
- 38.Reid S, Schindler D, Hanenberg H, Barker K, Hanks S, Kalb R, Neveling K, Kelly P, Seal S, Freund M, et al. Biallelic mutations in PALB2 cause Fanconi anemia subtype FA-N and predispose to childhood cancer. Nat Genet. 2007;39:162–164. doi: 10.1038/ng1947. [DOI] [PubMed] [Google Scholar]
- 39.Howlett NG, Taniguchi T, Olson S, Cox B, Waisfisz Q, De Die-Smulders C, Persky N, Grompe M, Joenje H, Pals G, et al. Biallelic inactivation of BRCA2 in Fanconi anemia. Science. 2002;297:606–609. doi: 10.1126/science.1073834. [DOI] [PubMed] [Google Scholar]
- 40.Wang AT, Kim T, Wagner JE, Conti BA, Lach FP, Huang AL, Molina H, Sanborn EM, Zierhut H, Cornes BK, et al. A Dominant Mutation in Human RAD51 Reveals Its Function in DNA Interstrand Crosslink Repair Independent of Homologous Recombination. Mol. Cell. 2015;59:478–490. doi: 10.1016/j.molcel.2015.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Vaz F, Hanenberg H, Schuster B, Barker K, Wiek C, Erven V, Neveling K, Endt D, Kesterton I, Autore F, et al. Mutation of the RAD51C gene in a Fanconi anemia-like disorder. Nat Genet. 2010;42:406–409. doi: 10.1038/ng.570. [DOI] [PubMed] [Google Scholar]
- 42.Sawyer SL, Tian L, Kähkönen M, Schwartzentruber J, Kircher M, University of Washington Centre for Mendelian Genomics. FORGE Canada Consortium. Majewski J, Dyment DA, Innes AM, et al. Biallelic mutations in BRCA1 cause a new Fanconi anemia subtype. Cancer Discov. 2015;5:135–142. doi: 10.1158/2159-8290.CD-14-1156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Levitus M, Waisfisz Q, Godthelp BC, de Vries Y, Hussain S, Wiegant WW, Elghalbzouri-Maghrani E, Steltenpool J, Rooimans MA, Pals G, et al. The DNA helicase BRIP1 is defective in Fanconi anemia complementation group J. Nat Genet. 2005;37:934–935. doi: 10.1038/ng1625. [DOI] [PubMed] [Google Scholar]
- 44.Levran O, Attwooll C, Henry RT, Milton KL, Neveling K, Rio P, Batish SD, Kalb R, Velleuer E, Barral S, et al. The BRCA1-interacting helicase BRIP1 is deficient in Fanconi anemia. Nat Genet. 2005;37:931–933. doi: 10.1038/ng1624. [DOI] [PubMed] [Google Scholar]
- 45.Litman R, Peng M, Jin Z, Zhang F, Zhang J, Powell S, Andreassen PR, Cantor SB. BACH1 is critical for homologous recombination and appears to be the Fanconi anemia gene product FANCJ. Cancer Cell. 2005;8:255–265. doi: 10.1016/j.ccr.2005.08.004. [DOI] [PubMed] [Google Scholar]
- 46.Nijman SMB, Huang TT, Dirac AMG, Brummelkamp TR, Kerkhoven RM, D'Andrea AD, Bernards R. The deubiquitinating enzyme USP1 regulates the Fanconi anemia pathway. Mol. Cell. 2005;17:331–339. doi: 10.1016/j.molcel.2005.01.008. [DOI] [PubMed] [Google Scholar]
- 47.Cohn MA, Kowal P, Yang K, Haas W, Huang TT, Gygi SP, D'Andrea AD. A UAF1-containing multisubunit protein complex regulates the Fanconi anemia pathway. Mol. Cell. 2007;28:786–797. doi: 10.1016/j.molcel.2007.09.031. [DOI] [PubMed] [Google Scholar]
- 48.Kratz K, Schöpf B, Kaden S, Sendoel A, Eberhard R, Lademann C, Cannavo E, Sartori AA, Hengartner MO, Jiricny J. Deficiency of FANCD2-associated nuclease KIAA1018/FAN1 sensitizes cells to interstrand crosslinking agents. Cell. 2010;142:77–88. doi: 10.1016/j.cell.2010.06.022. [DOI] [PubMed] [Google Scholar]
- 49.Liu T, Ghosal G, Yuan J, Chen J, Huang J. FAN1 acts with FANCI-FANCD2 to promote DNA interstrand cross-link repair. Science. 2010;329:693–696. doi: 10.1126/science.1192656. [DOI] [PubMed] [Google Scholar]
- 50.MacKay C, Déclais A-C, Lundin C, Agostinho A, Deans AJ, MacArtney TJ, Hofmann K, Gartner A, West SC, Helleday T, et al. Identification of KIAA1018/FAN1, a DNA Repair Nuclease Recruited to DNA Damage by Monoubiquitinated FANCD2. Cell. 2010;142:65–76. doi: 10.1016/j.cell.2010.06.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Yoshikiyo K, Kratz K, Hirota K, Nishihara K, Takata M, Kurumizaka H, Horimoto S, Takeda S, Jiricny J. KIAA1018/FAN1 nuclease protects cells against genomic instability induced by interstrand cross-linking agents. Proc Natl Acad Sci U S A. 2010;107:21553–21557. doi: 10.1073/pnas.1011081107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Zhou W, Otto EA, Cluckey A, Airik R, Hurd TW, Chaki M, Diaz K, Lach FP, Bennett GR, Gee HY, et al. FAN1 mutations cause karyomegalic interstitial nephritis, linking chronic kidney failure to defective DNA damage repair. Nat Genet. 2012;44:910–915. doi: 10.1038/ng.2347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Karanja KK, Cox SW, Duxin JP, Stewart SA, Campbell JL. DNA2 and EXO1 in replication-coupled, homology-directed repair and in the interplay between HDR and the FA/BRCA network. Cell Cycle. 2012;11:3983–3996. doi: 10.4161/cc.22215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Karanja KK, Lee EH, Hendrickson EA, Campbell JL. Preventing over-resection by DNA2 helicase/nuclease suppresses repair defects in Fanconi anemia cells. Cell Cycle. 2014;13:1540–1550. doi: 10.4161/cc.28476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Thangavel S, Berti M, Levikova M, Pinto C, Gomathinayagam S, Vujanovic M, Zellweger R, Moore H, Lee EH, Hendrickson EA, et al. DNA2 drives processing and restart of reversed replication forks in human cells. The Journal of Cell Biology. 2015;208:545–562. doi: 10.1083/jcb.201406100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Hashimoto Y, Chaudhuri AR, Lopes M, Costanzo V. Rad51 protects nascent DNA from Mre11-dependent degradation and promotes continuous DNA synthesis. Nature Structural & Molecular Biology. 2010;17:1305–1311. doi: 10.1038/nsmb.1927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Schlacher K, Christ N, Siaud N, Egashira A, Wu H, Jasin M. Double-strand break repair-independent role for BRCA2 in blocking stalled replication fork degradation by MRE11. Cell. 2011;145:529–542. doi: 10.1016/j.cell.2011.03.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Schlacher K, Wu H, Jasin M. A distinct replication fork protection pathway connects Fanconi anemia tumor suppressors to RAD51-BRCA1/2. Cancer Cell. 2012;22:106–116. doi: 10.1016/j.ccr.2012.05.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Niedernhofer LJ, Lalai AS, Hoeijmakers JHJ. Fanconi anemia (cross)linked to DNA repair. Cell. 2005;123:1191–1198. doi: 10.1016/j.cell.2005.12.009. [DOI] [PubMed] [Google Scholar]
- 60.Fu YV, Yardimci H, Long DT, Guainazzi A, Bermudez VP, Hurwitz J, van Oijen A, Schärer OD, Walter JC, Ho247 TV. Selective Bypass of a Lagging Strand Roadblock by the Eukaryotic Replicative DNA Helicase. Cell. 2011;146:931–941. doi: 10.1016/j.cell.2011.07.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Long DT, Joukov V, Budzowska M, Walter JC. BRCA1 Promotes Unloading of the CMG Helicase from a Stalled DNA Replication Fork. Mol. Cell. 2014;56:174–185. doi: 10.1016/j.molcel.2014.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Maric M, Maculins T, de Piccoli G, Labib K. Cdc48 and a ubiquitin ligase drive disassembly of the CMG helicase at the end of DNA replication. Science. 2014;346:1253596. doi: 10.1126/science.1253596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Moreno SP, Bailey R, Campion N, Herron S, Gambus A. Polyubiquitylation drives replisome disassembly at the termination of DNA replication. Science. 2014;346:477–481. doi: 10.1126/science.1253585. [DOI] [PubMed] [Google Scholar]
- 64.Budzowska M, Graham TG, Sobeck A, Waga S, Walter JC. Regulation of the Rev1-pol complex during bypass of a DNA interstrand cross-link. The EMBO Journal. 2015;34:1971–85. doi: 10.15252/embj.201490878. 2015, The EMBO Journal. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Knipscheer P, Raschle M, Smogorzewska A, Enoiu M, Ho TV, Scharer OD, Elledge SJ, Walter JC. The Fanconi Anemia Pathway Promotes Replication-Dependent DNA Interstrand Cross-Link Repair. Science. 2009;326:1698–1701. doi: 10.1126/science.1182372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Yamamoto KN, Kobayashi S, Tsuda M, Kurumizaka H, Takata M, Kono K, Jiricny J, Takeda S, Hirota K. Involvement of SLX4 in interstrand cross-link repair is regulated by the Fanconi anemia pathway. Proc Natl Acad Sci U S A. 2011;108:6492–6496. doi: 10.1073/pnas.1018487108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Klein Douwel D, Boonen RACM, Long DT, Szypowska AA, Räschle M, Walter JC, Knipscheer P. XPF-ERCC1 Acts in Unhooking DNA Interstrand Crosslinks in Cooperation with FANCD2 and FANCP/SLX4. Mol. Cell. 2014;54:460–471. doi: 10.1016/j.molcel.2014.03.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Hodskinson MRG, Silhan J, Crossan GP, Garaycoechea JI, Mukherjee S, Johnson CM, Schärer OD, Patel KJ. Mouse SLX4 Is a Tumor Suppressor that Stimulates the Activity of the Nuclease XPF-ERCC1 in DNA Crosslink Repair. Mol. Cell. 2014;54:472–484. doi: 10.1016/j.molcel.2014.03.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Andersen SL, Bergstralh DT, Kohl KP, LaRocque JR, Moore CB, Sekelsky J. Drosophila MUS312 and the vertebrate ortholog BTBD12 interact with DNA structure-specific endonucleases in DNA repair and recombination. Mol. Cell. 2009;35:128–135. doi: 10.1016/j.molcel.2009.06.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Fekairi S, Scaglione S, Chahwan C, Taylor ER, Tissier A, Coulon S, Dong M-Q, Ruse C, Yates JR, Russell P, et al. Human SLX4 is a Holliday junction resolvase subunit that binds multiple DNA repair/recombination endonucleases. Cell. 2009;138:78–89. doi: 10.1016/j.cell.2009.06.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Muñoz IM, Hain K, Déclais A-C, Gardiner M, Toh GW, Sanchez-Pulido L, Heuckmann JM, Toth R, Macartney T, Eppink B, et al. Coordination of structure-specific nucleases by human SLX4/BTBD12 is required for DNA repair. Mol. Cell. 2009;35:116–127. doi: 10.1016/j.molcel.2009.06.020. [DOI] [PubMed] [Google Scholar]
- 72.Svendsen JM, Smogorzewska A, Sowa ME, O'Connell BC, Gygi SP, Elledge SJ, Harper JW. Mammalian BTBD12/SLX4 assembles a Holliday junction resolvase and is required for DNA repair. Cell. 2009;138:63–77. doi: 10.1016/j.cell.2009.06.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Wang H, Li Y, Truong LN, Shi LZ, Hwang PY-H, He J, Do J, Cho MJ, Li H, Negrete A, et al. CtIP Maintains Stability at Common Fragile Sites and Inverted Repeats by End Resection-Independent Endonuclease Activity. Mol. Cell. 2014;56:1012–21. doi: 10.1016/j.molcel.2014.04.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Makharashvili N, Tubbs AT, Yang S-H, Wang H, Barton O, Zhou Y, Deshpande RA, Lee J-H, Lobrich M, Sleckman BP, et al. Catalytic and Noncatalytic Roles of the CtIP Endonuclease in Double-Strand Break End Resection. Mol. Cell. 2014;54:1022–3. doi: 10.1016/j.molcel.2014.04.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Unno J, Itaya A, Taoka M, Sato K, Tomida J, Sakai W, Sugasawa K, Ishiai M, Ikura T, Isobe T, et al. FANCD2 binds CtIP and regulates DNA-end resection during DNA interstrand crosslink repair. Cell Rep. 2014;7:1039–1047. doi: 10.1016/j.celrep.2014.04.005. [DOI] [PubMed] [Google Scholar]
- 76.Murina O, Aesch von C, Karakus U, Ferretti LP, Bolck HA, Hänggi K, Sartori AA. FANCD2 and CtIP Cooperate to Repair DNA Interstrand Crosslinks. Cell Rep. 2014;7:1030–8. doi: 10.1016/j.celrep.2014.03.069. [DOI] [PubMed] [Google Scholar]
- 77.Staresincic L, Fagbemi AF, Enzlin JH, Gourdin AM, Wijgers N, Dunand-Sauthier I, Giglia-Mari G, Clarkson SG, Vermeulen W, Schärer OD. Coordination of dual incision and repair synthesis in human nucleotide excision repair. The EMBO Journal. 2009;28:1111–1120. doi: 10.1038/emboj.2009.49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Kim H, Yang K, Dejsuphong D, D'Andrea AD. Regulation of Rev1 by the Fanconi anemia core complex. Nature Structural & Molecular Biology. 2012;19:164–170. doi: 10.1038/nsmb.2222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Long DT, Raschle M, Joukov V, Walter JC. Mechanism of RAD51-Dependent DNA Interstrand Cross-Link Repair. Science. 2011;333:84–87. doi: 10.1126/science.1204258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Méchali M. Eukaryotic DNA replication origins: many choices for appropriate answers. Nature Reviews Molecular Cell Biology. 2010;11:728–738. doi: 10.1038/nrm2976. [DOI] [PubMed] [Google Scholar]
- 81.Zhang J, Dewar JM, Budzowska M, Motnenko A, Cohn MA, Walter JC. DNA interstrand cross-link repair requires replication-fork convergence. Nature Structural & Molecular Biology. 2015;22:242–247. doi: 10.1038/nsmb.2956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Petermann E, Orta ML, Issaeva N, Schultz N, Helleday T. Hydroxyurea-Stalled Replication Forks Become Progressively Inactivated and Require Two Different RAD51-Mediated Pathways for Restart and Repair. Mol. Cell. 2010;37:492–502. doi: 10.1016/j.molcel.2010.01.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Toledo LI, Altmeyer M, Rask M-B, Lukas C, Larsen DH, Povlsen LK, Bekker-Jensen S, Mailand N, Bartek J, Lukas J. ATR Prohibits Replication Catastrophe by Preventing Global Exhaustion of RPA. Cell. 2013;155:1088–1103. doi: 10.1016/j.cell.2013.10.043. [DOI] [PubMed] [Google Scholar]
- 84.Siddiqui K, On KF, Diffley JFX. Regulating DNA Replication in Eukarya. Cold Spring Harb Perspect Biol. 2013;5 doi: 10.1101/cshperspect.a012930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Huang J, Liu S, Bellani MA, Thazhathveetil AK, Ling C, de Winter JP, Wang Y, Wang W, Seidman MM. The DNA Translocase FANCM/MHF Promotes Replication Traverse of DNA Interstrand Crosslinks. Mol. Cell. 2013;52:1–13. doi: 10.1016/j.molcel.2013.09.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Yardimci H, Wang X, Loveland AB, Tappin I, Rudner DZ, Hurwitz J, van Oijen AM, Walter JC. Bypass of a protein barrier by a replicative DNA helicase. Nature. 2012;492:205–209. doi: 10.1038/nature11730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Zhang J, Walter JC. Mechanism and regulation of incisions during DNA interstrand cross-link repair. DNA Repair. 2014;19:135–142. doi: 10.1016/j.dnarep.2014.03.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Voulgaridou G-P, Anestopoulos I, Franco R, Panayiotidis MI, Pappa A. DNA damage induced by endogenous aldehydes: current state of knowledge. Mutation Research/Fundamental and Molecular Mechanisms of Mutagenesis. 2011;711:13–27. doi: 10.1016/j.mrfmmm.2011.03.006. [DOI] [PubMed] [Google Scholar]
- 89.Mosammaparast N, Shi Y. Reversal of histone methylation: biochemical and molecular mechanisms of histone demethylases. Annu. Rev. Biochem. 2010;79:155–179. doi: 10.1146/annurev.biochem.78.070907.103946. [DOI] [PubMed] [Google Scholar]
- 90.Shen L, Song C-X, He C, Zhang Y. Mechanism and function of oxidative reversal of DNA and RNA methylation. Annu. Rev. Biochem. 2014;83:585–614. doi: 10.1146/annurev-biochem-060713-035513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Luo W, Li H, Zhang Y, Ang CY. Determination of formaldehyde in blood plasma by high-performance liquid chromatography with fluorescence detection. J. Chromatogr. B Biomed. Sci. Appl. 2001;753:253–257. doi: 10.1016/s0378-4347(00)00552-1. [DOI] [PubMed] [Google Scholar]
- 92.Heck HD, Casanova-Schmitz M, Dodd PB, Schachter EN, Witek TJ, Tosun T. Formaldehyde (CH2O) concentrations in the blood of humans and Fischer-344 rats exposed to CH2O under controlled conditions. Am Ind Hyg Assoc J. 1985;46:1–3. doi: 10.1080/15298668591394275. [DOI] [PubMed] [Google Scholar]
- 93.Chaw YF, Crane LE, Lange P, Shapiro R. Isolation and identification of cross-links from formaldehyde-treated nucleic acids. Biochemistry. 1980;19:5525–5531. doi: 10.1021/bi00565a010. [DOI] [PubMed] [Google Scholar]
- 94.Huang H, Hopkins PB. DNA interstrand cross-linking by formaldehyde: nucleotide sequence preference and covalent structure of the predominant cross-link formed in synthetic oligonucleotides. J. Am. Chem. Soc. 1993;115:9402–9408. [Google Scholar]
- 95.Brooks PJ, Zakhari S. Acetaldehyde and the genome: Beyond nuclear DNA adducts and carcinogenesis. J. Am. Chem. Soc. 2003;125:50–61. doi: 10.1002/em.21824. [DOI] [PubMed] [Google Scholar]
- 96.Matsuda T, Kawanishi M, Yagi T, Matsui S, Takebe H. Specific tandem GG to TT base substitutions induced by acetaldehyde are due to intra-strand crosslinks between adjacent guanine bases. Nucleic Acids Res. 1998;26:1769–1774. doi: 10.1093/nar/26.7.1769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Wang M, McIntee EJ, Cheng G, Shi Y, Villalta PW, Hecht SS. Identification of DNA adducts of acetaldehyde. Chem. Res. Toxicol. 2000;13:1149–1157. doi: 10.1021/tx000118t. [DOI] [PubMed] [Google Scholar]
- 98.Kozekov ID, Nechev LV, Moseley MS, Harris CM, Rizzo CJ, Stone MP, Harris TM. DNA interchain cross-links formed by acrolein and crotonaldehyde. J. Am. Chem. Soc. 2003;125:50–61. doi: 10.1021/ja020778f. [DOI] [PubMed] [Google Scholar]
- 99.Wang H, Kozekov ID, Harris TM, Rizzo CJ. Site-specific synthesis and reactivity of oligonucleotides containing stereochemically defined 1,N2-deoxyguanosine adducts of the lipid peroxidation product trans-4-hydroxynonenal. J. Am. Chem. Soc. 2003;125:5687–5700. doi: 10.1021/ja0288800. [DOI] [PubMed] [Google Scholar]
- 100.Summerfield FW, Tappel AL. Detection and measurement by high-performance liquid chromatography of malondialdehyde crosslinks in DNA. Anal. Biochem. 1984;143:265–271. doi: 10.1016/0003-2697(84)90662-6. [DOI] [PubMed] [Google Scholar]
- 101.Kozekov ID, Nechev LV, Sanchez A, Harris CM, Lloyd RS, Harris TM. Interchain cross-linking of DNA mediated by the principal adduct of acrolein. Chem. Res. Toxicol. 2001;14:1482–1485. doi: 10.1021/tx010127h. [DOI] [PubMed] [Google Scholar]
- 102.Cho Y-J, Wang H, Kozekov ID, Kurtz AJ, Jacob J, Voehler M, Smith J, Harris TM, Lloyd RS, Rizzo CJ, et al. Stereospecific formation of interstrand carbinolamine DNA cross-links by crotonaldehyde- and acetaldehyde-derived alpha-CH3-gamma-OH-1,N2-propano-2′-deoxyguanosine adducts in the 5′-CpG-3′ sequence. Chem. Res. Toxicol. 2006;19:195–208. doi: 10.1021/tx050239z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Ridpath JR, Nakamura A, Tano K, Luke AM, Sonoda E, Arakawa H, Buerstedde JM, Gillespie DAF, Sale JE, Yamazoe M, et al. Cells Deficient in the FANC/BRCA Pathway Are Hypersensitive to Plasma Levels of Formaldehyde. Cancer Research. 2007;67:11117–11122. doi: 10.1158/0008-5472.CAN-07-3028. [DOI] [PubMed] [Google Scholar]
- 104.Langevin F, Crossan GP, Rosado IV, Arends MJ, Patel KJ. Fancd2 counteracts the toxic effects of naturally produced aldehydes in mice. Nature. 2011;475:53–58. doi: 10.1038/nature10192. [DOI] [PubMed] [Google Scholar]
- 105.Garaycoechea JI, Crossan GP, Langevin F, Daly M, Arends MJ, Patel KJ. Genotoxic consequences of endogenous aldehydes on mouse haematopoietic stem cell function. Nature. 2012 doi: 10.1038/nature11368. doi:10.1038/nature11368. [DOI] [PubMed] [Google Scholar]
- 106.Pontel L, Rosado IV, Burgos-Barragan G, Garaycoechea JI, Yu R, et al. Endogenous formaldehyde is a hematopoietic stem cell genotoxin and metabolic carcinogen. Mol. Cell. 2015 doi: 10.1016/j.molcel.2015.08.020. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Rosado IV, Langevin F, Crossan GP, Takata M, Patel KJ. Formaldehyde catabolism is essential in cells deficient for the Fanconi anemia DNA-repair pathway. Nature Structural & Molecular Biology. 2011;18:1432–1434. doi: 10.1038/nsmb.2173. [DOI] [PubMed] [Google Scholar]
- 108.Hira A, Yabe H, Yoshida K, Okuno Y, Shiraishi Y, Chiba K, Tanaka H, Miyano S, Nakamura J, Kojima S, et al. Variant ALDH2 is associated with accelerated progression of bone marrow failure in Japanese Fanconi anemia patients. Blood. 2013;122:3206–2309. doi: 10.1182/blood-2013-06-507962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Oberbeck N, Langevin F, King G, de Wind N, Crossan GP, Patel KJ. Maternal Aldehyde Elimination during Pregnancy Preserves the Fetal Genome. Mol. Cell. 2014;55:807–817. doi: 10.1016/j.molcel.2014.07.010. 2014, Mol. Cell. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Garaycoechea JI, Patel KJ. Why does the bone marrow fail in Fanconi anemia? Blood. 2014;123:26–34. doi: 10.1182/blood-2013-09-427740. [DOI] [PubMed] [Google Scholar]
- 111.Kurtz AJ. 1,N2-Deoxyguanosine Adducts of Acrolein, Crotonaldehyde, and trans-4-Hydroxynonenal Cross-link to Peptides via Schiff Base Linkage. Journal of Biological Chemistry. 2002;278:5970–5976. doi: 10.1074/jbc.M212012200. [DOI] [PubMed] [Google Scholar]
- 112.Tretyakova NY, Groehler A, Ji S. DNA-Protein Cross-Links: Formation, Structural Identities, and Biological Outcomes. Acc. Chem. Res. 2015;48:1631–44. doi: 10.1021/acs.accounts.5b00056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Lu K, Ye W, Zhou L, Collins LB, Chen X, Gold A, Ball LM, Swenberg JA. Structural Characterization of Formaldehyde-Induced Cross-Links Between Amino Acids and Deoxynucleosides and Their Oligomers. J. Am. Chem. Soc. 2010;132:3388–3399. doi: 10.1021/ja908282f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Loeber R, Michaelson E, Fang Q, Campbell C, Pegg AE, Tretyakova N. Cross-linking of the DNA repair protein Omicron6-alkylguanine DNA alkyltransferase to DNA in the presence of antitumor nitrogen mustards. Chem. Res. Toxicol. 2008;21:787–795. doi: 10.1021/tx7004508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Loeber RL, Michaelson-Richie ED, Codreanu SG, Liebler DC, Campbell CR, Tretyakova NY. Proteomic analysis of DNA-protein cross-linking by antitumor nitrogen mustards. Chem. Res. Toxicol. 2009;22:1151–1162. doi: 10.1021/tx900078y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Michaelson-Richie ED, Loeber RL, Codreanu SG, Ming X, Liebler DC, Campbell C, Tretyakova NY. DNA-protein cross-linking by 1,2,3,4-diepoxybutane. J. Proteome Res. 2010;9:4356–4367. doi: 10.1021/pr1000835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Chvalova K, Brabec V, Kasparkova J. Mechanism of the formation of DNA-protein cross-links by antitumor cisplatin. Nucleic Acids Res. 2007;35:1812–1821. doi: 10.1093/nar/gkm032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Duxin JP, Dewar JM, Yardimci H, Walter JC. Repair of a DNA-Protein Crosslink by Replication-Coupled Proteolysis. Cell. 2014;159:346–357. doi: 10.1016/j.cell.2014.09.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Stingele J, Schwarz MS, Bloemeke N, Wolf PG, Jentsch S. A DNA-Dependent Protease Involved in DNA-Protein Crosslink Repair. Cell. 2014;158:327–338. doi: 10.1016/j.cell.2014.04.053. [DOI] [PubMed] [Google Scholar]
- 120.Nakano T, Morishita S, Katafuchi A, Matsubara M, Horikawa Y, Terato H, Salem AMH, Izumi S, Pack SP, Makino K, et al. Nucleotide Excision Repair and Homologous Recombination Systems Commit Differentially to the Repair of DNA-Protein Crosslinks. Mol. Cell. 2007;28:147–158. doi: 10.1016/j.molcel.2007.07.029. [DOI] [PubMed] [Google Scholar]
- 121.Nakano T, Katafuchi A, Matsubara M, Terato H, Tsuboi T, Masuda T, Tatsumoto T, Pack SP, Makino K, Croteau DL, et al. Homologous Recombination but Not Nucleotide Excision Repair Plays a Pivotal Role in Tolerance of DNA-Protein Cross-links in Mammalian Cells. J. Biol. Chem. 2009;284:27065–27076. doi: 10.1074/jbc.M109.019174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Reardon JT, Sancar A. Repair of DNA-polypeptide crosslinks by human excision nuclease. Proc Natl Acad Sci U S A. 2006;103:4056–4061. doi: 10.1073/pnas.0600538103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Baker DJ, Wuenschell G, Xia L, Termini J, Bates SE, Riggs AD, O'Connor TR. Nucleotide Excision Repair Eliminates Unique DNA-Protein Cross-links from Mammalian Cells. Journal of Biological Chemistry. 2007;282:22592–22604. doi: 10.1074/jbc.M702856200. [DOI] [PubMed] [Google Scholar]
- 124.Mosbech A, Gibbs-Seymour I, Kagias K, Thorslund T, Beli P, Lou Povlsen, Nielsen SV, Smedegaard S, Sedgwick G, Lukas C, et al. DVC1 (C1orf124) is a DNA damage-targeting p97 adaptor that promotes ubiquitin-dependent responses to replication blocks. Nature Structural & Molecular Biology. 2012;19:1084–1092. doi: 10.1038/nsmb.2395. [DOI] [PubMed] [Google Scholar]
- 125.Stingele J, Habermann B, Jentsch S. DNA-protein crosslink repair: proteases as DNA repair enzymes. Trends Biochem. Sci. 2014;40:67–71. doi: 10.1016/j.tibs.2014.10.012. [DOI] [PubMed] [Google Scholar]
- 126.Centore RC, Yazinski SA, Tse A, Zou L. Spartan/C1orf124, a reader of PCNA ubiquitylation and a regulator of UV-induced DNA damage response. Mol. Cell. 2012;46:625–635. doi: 10.1016/j.molcel.2012.05.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Davis EJ, Lachaud C, Appleton P, MacArtney TJ, thke INA, Rouse J. DVC1 (C1orf124) recruits the p97 protein segregase to sites of DNA damage. Nature Structural & Molecular Biology. 2012;19:1093–100. doi: 10.1038/nsmb.2394. [DOI] [PubMed] [Google Scholar]
- 128.Peak JG, Peak MJ, Sikorski RS, Jones CA. Induction of DNA-protein crosslinks in human cells by ultraviolet and visible radiations: action spectrum. Photochem. Photobiol. 1985;41:295–302. doi: 10.1111/j.1751-1097.1985.tb03488.x. [DOI] [PubMed] [Google Scholar]
- 129.Banjar ZM, Hnilica LS, Briggs RC, Stein J, Stein G. Crosslinking of chromosomal proteins to DNA in HeLa cells by UV gamma radiation and some antitumor drugs. Biochem. Biophys. Res. Commun. 1983;114:767–773. doi: 10.1016/0006-291x(83)90847-1. [DOI] [PubMed] [Google Scholar]
- 130.Lessel D, Vaz B, Halder S, Lockhart PJ, Marinovic-Terzic I, Lopez-Mosqueda J, Philipp M, Sim JCH, Smith KR, Oehler J, et al. Mutations in SPRTN cause early onset hepatocellular carcinoma, genomic instability and progeroid features. Nat Genet. 2014;46:1239–1244. doi: 10.1038/ng.3103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Maskey RS, Kim MS, Baker DJ, Childs B, Malureanu LA, Jeganathan KB, Machida Y, van Deursen JM, Machida YJ. Spartan deficiency causes genomic instability and progeroid phenotypes. Nat Commun. 2014;5:5744. doi: 10.1038/ncomms6744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Garcia CCM, Angeli JPF, Freitas FP, Gomes OF, de Oliveira TF, Loureiro APM, Di Mascio P, Medeiros MHG. [13C2]-Acetaldehyde promotes unequivocal formation of 1,N2-propano-2′-deoxyguanosine in human cells. J. Am. Chem. Soc. 2011;133:9140–9143. doi: 10.1021/ja2004686. [DOI] [PubMed] [Google Scholar]
Annotated References
- Sasaki MS, Tonomura A. A high susceptibility of Fanconi's anemia to chromosome breakage by DNA cross-linking agents. Cancer Research. 1973;33:1829–1836. [PubMed] [Google Scholar]; •• This study described for the first time that FA patient derived cells are susceptible to chromosome breakage when treated with interstrand crosslinking agents such as nitrogen mustard or mitomycin C.
- Akkari YM, Bateman RL, Reifsteck CA, Olson SB, Grompe M. DNA replication is required To elicit cellular responses to psoralen-induced DNA interstrand cross-links. Mol. Cell. Biol. 2000;20:8283–8289. doi: 10.1128/mcb.20.21.8283-8289.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]; • This study demonstrates that DNA replication is required to trigger a cellular response to ICLs. The authors induced ICLs in human cells at different stages of the cell cycle. ICLs induced in G1 or S phase lead to a late S phase arrest. In contrast ICLs introduced in G2 did not cause arrest until cells entered the next cell cycle, where they again accumulated in late S phase. This was the first indication for a replication-coupled mechanism of ICL repair.
- Räschle M, Knipscheer P, Enoiu M, Angelov T, Sun J, Griffith JD, Ellenberger TE, Schärer OD, Walter JC. Mechanism of Replication-Coupled DNA Interstrand Crosslink Repair. Cell. 2008;134:969–980. doi: 10.1016/j.cell.2008.08.030. [DOI] [PMC free article] [PubMed] [Google Scholar]; •• This paper shows that Xenopus egg extracts recapitulate replication-coupled ICL repair. The paper describes in detail how the convergence of two replication forks on an ICL triggers its repair.
- Nakanishi K, Cavallo F, Perrouault L, Giovannangeli C, Moynahan ME, Barchi M, Brunet E, Jasin M. Homology-directed Fanconi anemia pathway cross-link repair is dependent on DNA replication. Nature Structural & Molecular Biology. 2011;18:500–503. doi: 10.1038/nsmb.2029. [DOI] [PMC free article] [PubMed] [Google Scholar]; • This report describes the generation of a GFP based reporter assay that measures homology directed ICL repair in mammalian cells. The authors demonstrate that ICL repair is dependent on the FA pathway and DNA replication.
- Garcia-Higuera I, Taniguchi T, Ganesan S, Meyn MS, Timmers C, Hejna J, Grompe M, D'Andrea AD. Interaction of the Fanconi anemia proteins and BRCA1 in a common pathway. Mol. Cell. 2001;7:249–262. doi: 10.1016/s1097-2765(01)00173-3. [DOI] [PubMed] [Google Scholar]; •• This study demonstrates that FANCA, C, F, and G form a complex that is required for FANCD2 monoubiquitylation on K561 after DNA damage. Monoubiquitylated FANCD2 is essential for ICL repair and is targeted into nuclear foci that colocalize with BRCA1, linking the FA pathway to HR.
- Smogorzewska A, Matsuoka S, Vinciguerra P, McDonald ER, Hurov KE, Luo J, Ballif BA, Gygi SP, Hofmann K, D'Andrea AD, et al. Identification of the FANCI protein, a monoubiquitinated FANCD2 paralog required for DNA repair. Cell. 2007;129:289–301. doi: 10.1016/j.cell.2007.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]; •• This report identifies the FANCD2 paralog and binding partner, FANCI. Like FANCD2, FANCI is monoubiquitylated upon DNA damage. Monoubiquitylated ID2 localizes to chromatin and is required for downstream repair.
- Meetei AR, de Winter JP, Medhurst AL, Wallisch M, Waisfisz Q, van de Vrugt HJ, Oostra AB, Yan Z, Ling C, Bishop CE, et al. A novel ubiquitin ligase is deficient in Fanconi anemia. Nat Genet. 2003;35:165–170. doi: 10.1038/ng1241. [DOI] [PubMed] [Google Scholar]; • While FANCD2 ubiquitylation was known to be required for ICL repair, it was unknown which protein provided the E3 ligase activity. Meetei et al. discovered FANCL as the missing piece of the puzzle.
- Rajendra E, Oestergaard VH, Langevin F, Wang M, Dornan GL, Patel KJ, Passmore LA. The Genetic and Biochemical Basis of FANCD2 Monoubiquitination. Mol. Cell. 2014;54:858–869. doi: 10.1016/j.molcel.2014.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]; • Rajendra et al. demonstrate that a complex comprising FANCB, FANCL and FAAP100 functions as the monoubiquitination module of the FA core complex.
- Howlett NG, Taniguchi T, Olson S, Cox B, Waisfisz Q, De Die-Smulders C, Persky N, Grompe M, Joenje H, Pals G, et al. Biallelic inactivation of BRCA2 in Fanconi anemia. Science. 2002;297:606–609. doi: 10.1126/science.1073834. [DOI] [PubMed] [Google Scholar]; •• This study identifies a bona fide DNA repair protein, BRCA2, as an FA gene, firmly establishing that FA is caused by a deficiency in DNA repair.
- Yoshikiyo K, Kratz K, Hirota K, Nishihara K, Takata M, Kurumizaka H, Horimoto S, Takeda S, Jiricny J. KIAA1018/FAN1 nuclease protects cells against genomic instability induced by interstrand cross-linking agents. Proc Natl Acad Sci U S A. 2010;107:21553–21557. doi: 10.1073/pnas.1011081107. [DOI] [PMC free article] [PubMed] [Google Scholar]; • This study shows that FAN1 and FANC genes are not epistatic with respect to ICL sensitivity, strongly suggesting that FAN1 does not operate in the classical FA ICL repair pathway.
- Zhou W, Otto EA, Cluckey A, Airik R, Hurd TW, Chaki M, Diaz K, Lach FP, Bennett GR, Gee HY, et al. FAN1 mutations cause karyomegalic interstitial nephritis, linking chronic kidney failure to defective DNA damage repair. Nat Genet. 2012;44:910–915. doi: 10.1038/ng.2347. [DOI] [PMC free article] [PubMed] [Google Scholar]; • This study identifies mutations in FAN1 as a cause of karyomegalic interstitial nephritis (KIN) and shows that FAN1-deficient cells are sensitive to ICL inducing agents but do not exhibit broken chromosomes, unlike FA cells. Together, these observations support the notion that FAN1 participates in ICL repair independently of the FA pathway.
- Wang AT, Kim T, Wagner JE, Conti BA, Lach FP, Huang AL, Molina H, Sanborn EM, Zierhut H, Cornes BK, et al. A Dominant Mutation in Human RAD51 Reveals Its Function in DNA Interstrand Crosslink Repair Independent of Homologous Recombination. Mol. Cell. 2015;59:478–490. doi: 10.1016/j.molcel.2015.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]; • This study identifies a dominant negative mutation in RAD51 that causes FA-like phenotypes. While cells from this patient are proficient in HR, they are deficient in ICL repair due to over-resection of nascent strands by DNA2. Thus, RAD51 appears to have a fork protection function during ICL repair that may be distinct from its role in HR.
- Schlacher K, Wu H, Jasin M. A distinct replication fork protection pathway connects Fanconi anemia tumor suppressors to RAD51-BRCA1/2. Cancer Cell. 2012;22:106–116. doi: 10.1016/j.ccr.2012.05.015. [DOI] [PMC free article] [PubMed] [Google Scholar]; • This report demonstrates a role for FANCA and FANCD2 in protecting forks stalled by hydroxyurea. Thus, FANCA and FANCD2 appear to have genome maintenance functions that are independent of ICL repair.
- Knipscheer P, Raschle M, Smogorzewska A, Enoiu M, Ho TV, Scharer OD, Elledge SJ, Walter JC. The Fanconi Anemia Pathway Promotes Replication-Dependent DNA Interstrand Cross-Link Repair. Science. 2009;326:1698–1701. doi: 10.1126/science.1182372. [DOI] [PMC free article] [PubMed] [Google Scholar]; •• This study shows that monoubiquitylated ID2 promotes replication-coupled ICL repair by facilitating dual incisions on either side of the ICL.
- Klein Douwel D, Boonen RACM, Long DT, Szypowska AA, Räschle M, Walter JC, Knipscheer P. XPF-ERCC1 Acts in Unhooking DNA Interstrand Crosslinks in Cooperation with FANCD2 and FANCP/SLX4. Mol. Cell. 2014;54:460–471. doi: 10.1016/j.molcel.2014.03.015. [DOI] [PMC free article] [PubMed] [Google Scholar]; • Using Xenopus egg extracts, the authors demonstrate that monoubiquitylated ID2 promotes ICL repair by recruiting to ICLs a complex of SLX4 and the 3′ flap endonuclease XPF-ERCC1, which carries out at least one of the two incisions required for unhooking.
- Hodskinson MRG, Silhan J, Crossan GP, Garaycoechea JI, Mukherjee S, Johnson CM, Schärer OD, Patel KJ. Mouse SLX4 Is a Tumor Suppressor that Stimulates the Activity of the Nuclease XPF-ERCC1 in DNA Crosslink Repair. Mol. Cell. 2014;54:472–484. doi: 10.1016/j.molcel.2014.03.014. [DOI] [PMC free article] [PubMed] [Google Scholar]; • Using a combination of mouse genetics and biochemical assays, the authors demonstrate that the N-terminus of SLX4 that only binds to XPF-ERCC1 is sufficient to confer resistance to ICL inducing agents, and promotes dual incisions by XPF-ERCC1 around an ICL. These observations suggest that XPFERCC1 can perform both incisions during ICL repair.
- Zhang J, Dewar JM, Budzowska M, Motnenko A, Cohn MA, Walter JC. DNA interstrand cross-link repair requires replication-fork convergence. Nature Structural & Molecular Biology. 2015;22:242–247. doi: 10.1038/nsmb.2956. [DOI] [PMC free article] [PubMed] [Google Scholar]; • This report demonstrates that a single replication fork encountering an ICL in Xenopus egg extracts is extremely stable but incompetent to trigger repair. When a second fork arrives, even after an extended delay, repair takes place. These data imply that fork convergence is a viable strategy to promote repair in mammalian cells, where there will usually be a significant delay between arrival of the first and second forks.
- Petermann E, Orta ML, Issaeva N, Schultz N, Helleday T. Hydroxyurea-Stalled Replication Forks Become Progressively Inactivated and Require Two Different RAD51-Mediated Pathways for Restart and Repair. Mol. Cell. 2010;37:492–502. doi: 10.1016/j.molcel.2010.01.021. [DOI] [PMC free article] [PubMed] [Google Scholar]; • This report shows that replication forks collapse only after prolonged stalling in hydroxyurea, demonstrating the intrinsic stability of forks in mammalian cells.
- Toledo LI, Altmeyer M, Rask M-B, Lukas C, Larsen DH, Povlsen LK, Bekker-Jensen S, Mailand N, Bartek J, Lukas J. ATR Prohibits Replication Catastrophe by Preventing Global Exhaustion of RPA. Cell. 2013;155:1088–1103. doi: 10.1016/j.cell.2013.10.043. [DOI] [PubMed] [Google Scholar]; • This study demonstrates that ATR signaling following DNA damage prevents RPA exhaustion by restraining origin firing. When ATR signaling is inhibited during hydroxyurea treatment, the nuclear pool of RPA is used up, leading to fork collapse. The work implies that under normal conditions when RPA is in excess, replication forks are very stable even after prolonged arrest.
- Huang J, Liu S, Bellani MA, Thazhathveetil AK, Ling C, de Winter JP, Wang Y, Wang W, Seidman MM. The DNA Translocase FANCM/MHF Promotes Replication Traverse of DNA Interstrand Crosslinks. Mol. Cell. 2013;52:1–13. doi: 10.1016/j.molcel.2013.09.021. [DOI] [PMC free article] [PubMed] [Google Scholar]; •• This paradigm-shifting study demonstrates that in mammalian tissue culture cells, a single DNA replication fork can bypass an intact ICL without unhooking it, and that this “traverse” reaction requires FANCM. The work suggests that a single fork is sufficient to trigger ICL repair, probably by generating a similar X-shaped structure to the one created when two forks converge on an ICL.
- Rosado IV, Langevin F, Crossan GP, Takata M, Patel KJ. Formaldehyde catabolism is essential in cells deficient for the Fanconi anemia DNA-repair pathway. Nature Structural & Molecular Biology. 2011;18:1432–1434. doi: 10.1038/nsmb.2173. [DOI] [PubMed] [Google Scholar]; • This paper demonstrates formaldehyde detoxifying enzyme ADH5 are synthetically lethal. In contrast, ALDH2 (catalyzes mainly acetaldehyde) is not synthetically lethal with FANCC, suggesting that formaldehyde is more genotoxic than acetaldehyde.
- Langevin F, Crossan GP, Rosado IV, Arends MJ, Patel KJ. Fancd2 counteracts the toxic effects of naturally produced aldehydes in mice. Nature. 2011;475:53–58. doi: 10.1038/nature10192. [DOI] [PubMed] [Google Scholar]; •• This study shows that Aldh2−/− Fancd2−/− mice develop acute leukemia and, when exposed to ethanol, bone marrow failure. These results suggest that endogenous aldehydes might trigger bone marrow failure and leukemia in FA patients.
- Garaycoechea JI, Crossan GP, Langevin F, Daly M, Arends MJ, Patel KJ. Genotoxic consequences of endogenous aldehydes on mouse haematopoietic stem cell function. Nature. 2012;489:571–575. doi: 10.1038/nature11368. [DOI] [PubMed] [Google Scholar]; •• This study shows that Aldh2−/− Fancd2−/− mice that do not get leukemia develop aplastic anemia with concomitant depletion of hematopoietic stem and progenitor cells. These observations suggest that aldehyde toxicity causes bone marrow failure by depleting hematopoietic stem and progenitor cell pools.
- Hira A, Yabe H, Yoshida K, Okuno Y, Shiraishi Y, Chiba K, Tanaka H, Miyano S, Nakamura J, Kojima S, et al. Variant ALDH2 is associated with accelerated progression of bone marrow failure in Japanese Fanconi anemia patients. Blood. 2013;122:3206–2309. doi: 10.1182/blood-2013-06-507962. [DOI] [PMC free article] [PubMed] [Google Scholar]; • Hira et al. report a strong correlation between a dominant negative allele of ALDH2 and the age of bone marrow onset in Japanese FA patients. The work provides powerful support for the hypothesis from the Patel laboratory that FA is caused by a failure to counteract the genotoxic effects of endogenous acetaldehyde.
- Duxin JP, Dewar JM, Yardimci H, Walter JC. Repair of a DNA-Protein Crosslink by Replication-Coupled Proteolysis. Cell. 2014;159:346–357. doi: 10.1016/j.cell.2014.09.024. [DOI] [PMC free article] [PubMed] [Google Scholar]; •• Recapitulating replication-coupled DPC repair in Xenopus egg extracts shows that DPCs are repaired by a replication-coupled protease in the absence of double-strand break formation. Accodingly, DPC repair in this system is independent of the ID2 complex.
- Stingele J, Schwarz MS, Bloemeke N, Wolf PG, Jentsch S. A DNA-Dependent Protease Involved in DNA-Protein Crosslink Repair. Cell. 2014;158:327–338. doi: 10.1016/j.cell.2014.04.053. [DOI] [PubMed] [Google Scholar]; •• This paper identifies Wss1 as a DPC-protease in yeast. Cells lacking Wss1 are hypersensitive to formaldehyde, and Wss1 mutations are synthetically lethal with mutations in TDP1, an enzyme that processes TOPO1-DNA adducts. In vitro, Wss1 specifically degrades proteins that bind DNA.
- Lessel D, Vaz B, Halder S, Lockhart PJ, Marinovic-Terzic I, Lopez-Mosqueda J, Philipp M, Sim JCH, Smith KR, Oehler J, et al. Mutations in SPRTN cause early onset hepatocellular carcinoma, genomic instability and progeroid features. Nat Genet. 2014;46:1239–1244. doi: 10.1038/ng.3103. [DOI] [PMC free article] [PubMed] [Google Scholar]; •• This report identifies three patients from two unrelated families suffering from a progeria syndrome caused by mutations in DVC1 (Spartan), including one in the protease domain. These patients also developed early-onset hepatocellular carcinoma. Patient-derived cells contained DNA damage and signs of replication stress, consistent with DVC1 acting in a replication-coupled repair pathway.
- Maskey RS, Kim MS, Baker DJ, Childs B, Malureanu LA, Jeganathan KB, Machida Y, van Deursen JM, Machida YJ. Spartan deficiency causes genomic instability and progeroid phenotypes. Nat Commun. 2014;5:5744. doi: 10.1038/ncomms6744. [DOI] [PMC free article] [PubMed] [Google Scholar]; • Maskey et al. demonstrate that DVC1 (Spartan) knockout mice are embryonic lethal. In contrast, hypomorphic mice with low amounts of DVC1 are viable, but display accelerated aging features.