Abstract
Within many fungal syncytia, nuclei behave independently despite sharing a common cytoplasm. Creation of independent nuclear zones of control in one cell is paradoxical considering random protein synthesis sites, predicted rapid diffusion rates, and well-mixed cytosol. In studying the surprising fungal nuclear autonomy, new principles of cellular organization are emerging. We discuss the current understanding of nuclear autonomy, focusing on asynchronous cell cycle progression where most work has been directed. Mechanisms underlying nuclear autonomy are diverse including mRNA localization, ploidy variability, and nuclear spacing control. With the challenges fungal syncytia face due to cytoplasmic size and shape, they serve as powerful models for uncovering new subcellular organization modes, variability sources among isogenic uninucleate cells, and the evolution of multicellularity.
Introduction
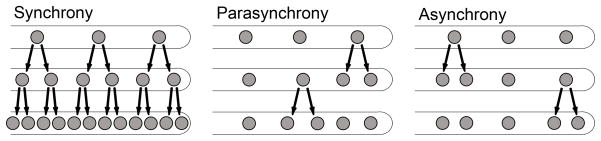
Years of study using the fungi Saccharomyces cerevisiae, Schizosaccharomyces pombe and members of other phyla have revealed cell cycle progression is regulated by periodic changes in the activities of cyclin-cyclin dependent kinase (CDK) complexes. Typically, different cyclins are synthesized and degraded at each transition, regulating progression through protein abundance. One could therefore predict that nuclei sharing cytoplasm divide synchronously. Experiments from the 1970’s support this hypothesis, as mammalian nuclei forced to share cytoplasm synchronize [1]. Some of the first studies done on natural syncytia examined nuclear division in the slime mold Physarum polycephalum where, as would be predicted, nuclei cycle highly synchronously in a common cytoplasm [2–4]. However, subsequent work in fungi revealed that syncytia exhibit a variety of cell cycle strategies. Nuclei within Ceratocystis fagacearum apical compartments divide synchronously [5]. However, Aspergillus nidulans nuclei divide in parasynchronous waves and in other species, including Ashbya gossypii and Neurospora crassa, nuclei divide asynchronously (Fig. 1) [6–11]. The coexistence of multiple, independent oscillators in these latter species poses a perplexing problem, which is especially intriguing given the cytoplasm continuously flows and mixes within a complex network [12,13*]. In this environment nuclei can bypass each other, and sister nuclei move far from each other [13*,14**,15]. In this cellular context, how do nuclei only microns apart behave independently?
Figure 1.
Mitotic strategies utilized by different filamentous fungi.
Synchronous: All nuclei divide at the same time. Parasynchronous: Nuclei divide in waves, with one neighbor dividing after another. Asynchronous: Nuclei divide independently of their neighbors.
Molecular basis of nuclear division autonomy
Nuclear autonomy in division has likely two components in most systems: sources of variability in the division cycle and ways for the cytoplasm to be compartmentalized to limit mixing. Nuclear division timing in A. gossypii is highly variable; cycle times of 40 minutes to over four hours have been observed [9]. Therefore, not only do multiple, independently cycling clocks coexist within this common cytoplasm, but the clocks are also of different durations. Lineage analyses demonstrated positively correlated division times between A. gossypii sister nuclei independent of distance traveled from each other within the cell [14**]. Additionally, nuclei are asynchronous during the first cell cycle after release from arrest [9]. These data support a nuclear intrinsic, heritable division-timing component (Fig 2, top). Similar division time correlations were recently detected between related HeLa cells, suggesting the possibility of conserved mechanisms generating division timing variability [16]. One candidate for a nuclear intrinsic regulator in A. gossypii is ploidy. Nuclei with ploidy between 1N and 4N have been observed, and average ploidy typically increases as cells age [17*]. A nucleus containing more DNA may take more time to replicate DNA, however evidence supports that the bulk of division timing variability originates in G1 [18]. Another source of autonomy may originate with nuclear import differences. Many fungal species exhibit closed mitosis, without nuclear envelope (NE) breakdown. If the NEs of different nuclei differ in import capacity, this could enable nuclear autonomy by restricting regulator access to DNA, preventing it until the appropriate time for each nucleus. Evidence from S. cerevisiae indicates that nuclear import can be differentially regulated, the transcription factor Ace2 is imported by daughter nuclei, but not mother nuclei, during the M/G1 transition [19]. However, differences in import capacity among nuclei in a fungal syncytium have not been assessed.
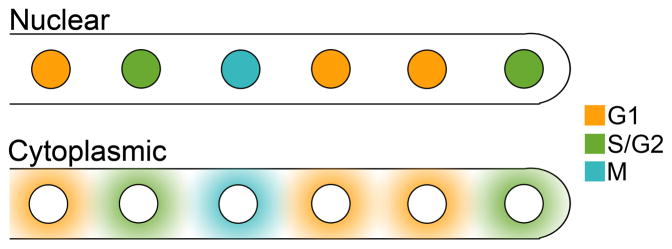
Figure 2.
Potential models for regulation of nuclear autonomy.
Top: Nuclear regulation of division autonomy. Nuclei carry with them the signals controlling their cell cycle stage. Nuclear movement and proximity to neighbors does not influence cell cycle stage.
Bottom: Cytoplasmic regulation of division autonomy. Regulators are spatially organized in the cytoplasm so that each nucleus is only exposed to the appropriate cell cycle signals. Bypassing and/or closely positioned nuclei may influence each others’ states.
Although it is possible that import variability contributes to autonomy, some evidence indicates not all cell cycle regulators are controlled in this manner. The nuclear import variability hypothesis predicts that such regulators, e.g. cyclins, are only imported at specific times in asynchronous syncytia. This does not appear to be the case in A. gossypii, in which all cyclins except the S phase Clb5/6 are detectable in nuclei at every stage [20]. The anaphase promoting complex (APC) is also nuclear localized at all stages, however it appears to be active only at certain times [21]. The mechanism controlling this is currently unclear, however, nuclear-autonomous APC activation is also observed in A. nidulans and, in this species, may be regulated by spindle pole body association [22*]. A similar SPB-based regulatory mechanism may exist in A. gossypii, but this has not been examined. In contrast, in A. gossypii the CDK inhibitor Sic1 cycles between nucleus and cytoplasm and changes localization on the spindle within the nucleus [9]. For this and other regulators that change localization, cytoplasmic protein diffusion may be restricted so that individual nuclei are only exposed to the appropriate cytoplasmic signals (Fig 2, bottom). For example, G1 cyclin CLN3 transcripts are heterogeneously localized via the intrinsically-disordered protein Whi3. When Whi3 function is compromised, CLN3 transcripts are more homogeneously distributed and neighboring nuclei are more synchronous [23**]. The large assemblies of RNA and Whi3 restrict the diffusion of cyclin transcripts and thus somehow limit entrainment between neighboring nuclei. Whether these RNA-protein assemblies locally repress or activate Cln3 activity is not yet clear. In fact, it may be possible that Whi3 can alternately repress or activate translation of the mRNA under different conditions, as has been shown for the RNA-binding protein Puf3 and its target mRNAs in S. cerevisiae [24]. Thus, nuclear autonomy arises from restricted localization of cell cycle regulators that rely both on nuclear structures and the ability of proteins to self assemble into complexes with limited cytosolic diffusivity. However, the respective contributions of nucleus and cytoplasmic regulatory mechanisms to nuclear behavior require substantial further investigation.
Intrinsically-disordered proteins may play an especially vital role in cytoplasmic organization in large cells, like filamentous fungi, by keeping regulators in close proximity to specific nuclei. This can, in turn, enable nuclear autonomy (Fig 2, bottom). Evidence supporting the importance of intrinsically-disordered proteins in cells with large volumes of cytoplasm comes from a study comparing the prevalence of polyN/Q tracts in the proteomes of various species. The social amoeba Dictyostelium discoideum, which exhibits both a uninucleate and a complex multinucleate life stage, was found to have a highly aggregation-prone proteome [25]. In multinucleate fungi, intrinsically-disordered proteins may prevent sharing of regulators among nuclei. Consistent with this, A. gossypii nuclei that bypass each other have positively correlated division times [14**]. During bypassing, cytoplasmic components restricted to the periphery of separate nuclei may intermix and exchange, resulting in more similar concentrations of regulators around these nuclei than before bypassing. Previous work had demonstrated that nuclei can import protein products translated from transcripts synthesized by another nucleus, these may be obtained during bypass events [9]. Combined, these data suggest that the cytoplasm is heterogeneous, and nucleus-cytoplasm and nucleus-nucleus interactions are tightly regulated to control division. Further support for this comes from the parasynchronous species A. nidulans, in which partial nuclear pore complex breakdown occurs during mitosis [26]. Parasynchrony may be achieved by combining the resulting increased diffusion between nucleoplasm and cytoplasm with propagation of a wave of mitotic activators through the cell. How cells regulate the assembly of complexes formed through aggregation-prone proteins, how intrinsically-disordered protein sequences evolve to compartmentalize cytosol of different volumes, and how compartments function to regulate specific biochemistry are critical questions for many aggregation-based compartments that can be studied using nuclear autonomy in syncytia.
Nuclear response to cellular requirements and environmental signals
It is unclear what adaptive advantage nuclear autonomy could confer, though it has been proposed that cells composed of multiple, autonomous genomes were a step during the evolution of eukaryotes and multicellularity [27**]. One benefit of a multinucleate strategy is the buffer provided by multiple genome copies against recessive mutations and environmental insults. Asynchrony of this nuclear population may further increase tolerance for stresses that are more damaging during certain cell cycle stages. Within uninucleate N. crassa conidia, nuclei are in different stages just as within the multinucleate mycelium and therefore, as a population, also utilize this strategy [11]. However, not all multinucleate cells exhibit nuclear autonomous cell cycle progression; it is only one successful strategy. Another potential advantage is maintenance of a constant nucleus/cytoplasm ratio. Synchronous divisions can cause rapid nuclear number changes in a similar cytoplasmic volume. The nuclear/cytoplasmic volume ratio has been shown to be tightly controlled in some uninucleate species, nuclear number could also be regulated in multinucleate species [28]. Consistent with this, A. gossypii nuclei dynamically respond to the amount of available cytoplasm. Nocodazole treatment decreases the number of nuclei per volume cytoplasm; after washout, nuclei more rapidly divide to re-establish a wild-type ratio, though the mechanistic specifics of how the cell senses the altered ratio of cytoplasm remain mysterious [9,14**]. Tight regulation of nuclear number may also be required for nuclear autonomous processes besides mitosis. One example is nuclear-autonomous APC activation in A. nidulans, which has been hypothesized remove damaged nuclei from the replicative pool [22*]. Additionally, A. gossypii nuclei were found to be transcriptionally autonomous using single molecule RNA FISH [23**]. Undoubtedly a variety of nuclear-autonomous processes in these and other species await investigation.
Nuclear autonomy may contribute to phenotypic differences across a colony. Gene expression in N. crassa mycelium differs between areas centimeters apart, though the importance of autonomous behavior of individual nuclei separated by micron-scale distances has not been investigated in this process [29]. Older mycelium upregulates genes required for creating fruiting bodies, while new areas synthesize components for new growth and nutrient absorption [29]. Intriguingly, colony-level gene expression in a heterokaryon can be independent of the proportion of nuclei carrying the respective gene. Strains with different ratios of HIS+:HIS− nuclei produce the same amount of protein product, suggesting nuclear-autonomous gene expression can be modulated according to cellular requirements, and indicating it is possible for a subset of nuclei to contribute sufficient resources for the entire population [30*]. It is possible that the nucleus/cytoplasm ratio discussed previously is important for effective nutrient sharing. In Schizophyllum commune average internuclear distance changes based on growth substrate along with secondary metabolite production, suggesting a link between nuclei per unit cytoplasm and gene expression and indicating nuclei dynamically respond to environmental cues [31]. Another example of nuclei responding to the environment comes from N. crassa, which is typically considered an asynchronous species. However, circadian studies revealed a greater degree of division synchrony is achieved by light/dark cycle entrainment [32**]. Similar circadian rhythm and cell cycle coupling has been observed in mammalian cells [33]. If cells with multiple, autonomous nuclei were an intermediate on the path of eukaryotic evolution as has been suggested, synchronous behavior could then have arisen from coordinate responses to exogenous signals such as the day/night cycle [27**].
Cohabitation of multiple nuclei within one cell allows for resource sharing, but this benefit must be balanced against the threat of intracellular competition between genotypes. If one genotype propagates more rapidly or is enriched within reproductive structures, the fitness of the whole organism could be negatively impacted. For example, N. crassa nuclei with the ‘spore killer’ phenotype render ‘sensitive’ conidia inviable if they share an ascospore. Heterokaryon incompatibility responses may have adapted to prevent vegetative fusion of cells with such incompatible genotypes [34,35]. While it is not possible to definitively state what advantages are conferred by nuclear division autonomy, this growth strategy is clearly a common response in fungi to a complex array of pressures.
Nuclear autonomy and dynamics in other species
Future work in species exhibiting multiple morphologies will provide insight into generation and maintenance of nuclear autonomy. For example, S. cerevisiae diploids grown in limiting nitrogen conditions form pseudohyphae that undergo synchronous cellular divisions [36]. However, this species also exhibits nuclear autonomy; in a binucleate cell, if one nucleus is damaged, the undamaged nucleus divides while the damaged nucleus arrests [37]. Nuclei within true hyphae of the dimorphic Candida albicans divide asynchronously, and nuclear-autonomous G1 arrest can occur depending on nutrient availability [38]. Young Fusarium oxysporum cells are comprised of uninucleate hyphal compartment chains, but as cells age multinucleate compartments arise exhibiting parasynchronous mitosis [39*]. Interestingly, when a binucleate, heterokaryotic nuclear compartment is formed by vegetative fusion, one nucleus is degraded [40*]. Similar phenomena were long ago observed in several basidiomycete species [41–44]. However, in basidiomycetes stable dikaryon formation is necessary for sexual reproduction. Whether there are conditions under which F. oxysporum nuclei harmoniously cohabitate after fusion remains a mystery. Further questions arise from studies demonstrating suppression of heterokaryon incompatibility responses in pathogenic fungal species in the presence of a host [45*]. These data suggest that intracellular and intranuclear interactions are variable even within one species. It is intriguing to speculate on the nature of the influence of environment and growth strategy on nuclear autonomy. Study of organisms exhibiting multiple geometries, variable nuclear number, and pathogenesis strategies will undoubtedly shed light on these interactions.
Nuclear autonomy in a syncytium is not unique to fungi. Nuclei of skeletal muscle syncytia maintain transcriptional autonomy and cell cycle re-entry timing differences [46,47]. In contrast with the fungi discussed, which primarily form by nucleokinesis without cytokinesis, fusion of uninucleate myoblasts produces multinucleate muscle cells. Some phenotypic variability exhibited by the myoblasts is maintained by the nuclei after syncytium formation, suggesting that cytoplasmic fusion is insufficient to coordinate nuclear behavior in this system [48]. However, nuclei can respond to the local cytoplasmic environment and/or other nuclei. During neuromuscular junction formation, nuclei cluster together and coordinately alter their transcriptional activity [49]. The similarities between the behavior of these nuclei and the principles examined in fungi support the hypothesis that nuclear autonomy confers adaptive advantages and that the mechanisms for generating autonomy in a common cytoplasm may be conserved.
Conclusions
Nuclei have been shown to alter cell cycle dynamics, synchrony, and gene expression in response to cellular requirements and environmental cues [31,32**]. Nuclear behavior can also change along with growth strategy and cellular geometry alterations [36,38,39*,45*]. The remarkable advances in light microscopy of the last few decades enable researchers to examine nuclear autonomy in ways never before possible, and these techniques will only continue to improve and advance our capabilities. Combined with the adaptation of CRISPR to increase the genetic tractability of many fungi, this will allow us to further our understanding of generation and regulation of nuclear autonomy in the species discussed here and species that have not been previously examined [50,51]. Investigating mechanisms of cytoplasmic organization and variations on nuclear autonomy in these syncytia will help reveal the spatial regulation of other cellular processes. This research will not only further fungal biology, but will also be invaluable for our understanding of how physical organization regulates functions within all eukaryotic cells.
Highlights.
Nuclei sharing a common cytoplasm can behave autonomously with respect to a variety of cellular processes, including mitosis.
Cytoplasmic organization and nuclear intrinsic factors combine to promote nuclear autonomous behavior.
Mechanisms for insulating nuclei in a common cytoplasm are likely used in many contexts for organizing cytosol, even in uninucleate cells.
Our current understanding of nuclear autonomy is limited and requires further study in a variety of species, both within and outside the fungal kingdom.
Acknowledgments
We apologize to the authors whose work was not cited because of space restrictions. Special thanks to all members of the Gladfelter lab for helpful discussions and especially Molly McQuilken, Anum Khan, and Therese Gerbich for critical reading of the manuscript. ASG and SER were supported by the National Institutes of Health R01-GM081506 and SER was also supported by the Neukom Institute at Dartmouth College.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Johnson RT, Rao PN. Nucleo-cytoplasmic interactions in the achievement of nuclear synchrony in DNA synthesis and mitosis in multinucleate cells. Biol Rev Camb Philos Soc. 1971;46:97–155. doi: 10.1111/j.1469-185x.1971.tb01180.x. [DOI] [PubMed] [Google Scholar]
- 2.Howard FL. Nuclear Division in Plasmodia of Physarum. Ann Bot. 1932;46:461–477. [Google Scholar]
- 3.Nygaard OF, Güttes S, Rusch HP. Nucleic acid metabolism in a slime mold with synchronous mitosis. Biochim Biophys Acta. 1960;38:298–306. doi: 10.1016/0006-3002(60)91245-2. [DOI] [PubMed] [Google Scholar]
- 4.Sudbery PE, Grant WD. The control of mitosis in Physarum polycephalum: the effect of delaying mitosis and evidence for the operation of the control mechanism in the absence of growth. J Cell Sci. 1976;22:59–65. doi: 10.1242/jcs.22.1.59. [DOI] [PubMed] [Google Scholar]
- 5.Aist JR. The mitotic apparatus in fungi, Ceratocystis fagacearum and Fusarium oxysporum. J Cell Biol. 1969;40:120–135. doi: 10.1083/jcb.40.1.120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Clutterbuck AJ. Synchronous nuclear division and septation in Aspergillus nidulans. J Gen Microbiol. 1970;60:133–135. doi: 10.1099/00221287-60-1-133. [DOI] [PubMed] [Google Scholar]
- 7.Rosenberger RF, Kessel M. Synchrony of nuclear replication in individual hyphae of Aspergillus nidulans. J Bacteriol. 1967;94:1464–1469. doi: 10.1128/jb.94.5.1464-1469.1967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Maheshwari R. Nuclear behavior in fungal hyphae. FEMS Microbiol Lett. 2005;249:7–14. doi: 10.1016/j.femsle.2005.06.031. [DOI] [PubMed] [Google Scholar]
- 9.Gladfelter AS, Hungerbuehler AK, Philippsen P. Asynchronous nuclear division cycles in multinucleated cells. J Cell Biol. 2006;172:347–362. doi: 10.1083/jcb.200507003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Minke PF, Lee IH, Plamann M. Microscopic analysis of Neurospora ropy mutants defective in nuclear distribution. Fungal Genet Biol. 1999;28:55–67. doi: 10.1006/fgbi.1999.1160. [DOI] [PubMed] [Google Scholar]
- 11.Serna L, Stadler D. Nuclear division cycle in germinating conidia of Neurospora crassa. J Bacteriol. 1978;136:341–351. doi: 10.1128/jb.136.1.341-351.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lew RR. Mass flow and pressure-driven hyphal extension in Neurospora crassa [Internet] Microbiology. 2005;151:2685–2692. doi: 10.1099/mic.0.27947-0. [DOI] [PubMed] [Google Scholar]
- *13.Roper M, Simonin A, Hickey PC, Leeder A, Glass NL. Nuclear dynamics in a fungal chimera. P Natl Acad Sci USA. 2013;110:12875–12880. doi: 10.1073/pnas.1220842110. This paper demonstrates that nuclei of a N. crassa heterokaryon formed by vegetative fusion rapidly redistribute to mix the two genotypes within the hyphal network via complex fluid flows. [DOI] [PMC free article] [PubMed] [Google Scholar]
- **14.Anderson CA, Eser U, Korndorf T, Borsuk ME, Skotheim JM, Gladfelter AS. Nuclear repulsion enables division autonomy in a single cytoplasm. Curr Biol. 2013;23:1999–2010. doi: 10.1016/j.cub.2013.07.076. This paper demonstrates that nuclei actively maintain a specific internuclear spacing in A. gossypii mycelium via the microtubule cytoskeleton, and this spacing is important for maintaining division autonomy. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Grava S, Philippsen P. Dynamics of multiple nuclei in Ashbya gossypii hyphae depend on control of cytoplasmic microtubules length by Bik1, Kip2, Kip3, and not on a capture/shrinkage mechanism. Mol Biol Cell. 2010;21:3680–3692. doi: 10.1091/mbc.E10-06-0527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sandler O, Mizrahi SP, Weiss N, Agam O, Simon I, Balaban NQ. Lineage correlations of single cell division time as a probe of cell-cycle dynamics. Nature. 2015;519:468–471. doi: 10.1038/nature14318. [DOI] [PubMed] [Google Scholar]
- *17.Anderson CA, Roberts S, Zhang H, Kelly CM, Kendall A, Lee C, Gerstenberger J, Koenig AB, Kabeche R, Gladfelter AS. Ploidy variation in multinucleate cells changes under stress. Mol Biol Cell. 2015;26:1129–1140. doi: 10.1091/mbc.E14-09-1375. This is the first demonstration that A. gossypii nuclei are of varying ploidies within a single cell and the proportion of ploidies is dependent on cell age and growth conditions. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Nair DR, D’Ausilio CA, Occhipinti P, Borsuk ME, Gladfelter AS. A conserved G1 regulatory circuit promotes asynchronous behavior of nuclei sharing a common cytoplasm. Cell Cycle. 2010;9:3771–3779. doi: 10.4161/cc.9.18.12999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mazanka E, Weiss EL. Sequential counteracting kinases restrict an asymmetric gene expression program to early G1. Mol Biol Cell. 2010;21:2809–2820. doi: 10.1091/mbc.E10-02-0174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hungerbuehler AK, Philippsen P, Gladfelter AS. Limited functional redundancy and oscillation of cyclins in multinucleated Ashbya gossypii fungal cells. Eukaryot Cell. 2007;6:473–486. doi: 10.1128/EC.00273-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gladfelter AS, Sustreanu N, Hungerbuehler AK, Voegeli S, Galati V, Philippsen P. The anaphase-promoting complex/cyclosome is required for anaphase progression in multinucleated Ashbya gossypii cells. Eukaryot Cell. 2007;6:182–197. doi: 10.1128/EC.00364-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- *22.Edgerton-Morgan H, Oakley BR. γ-Tubulin plays a key role in inactivating APC/CCdh1 at the G1-S boundary. J Cell Biol. 2012;198:785–791. doi: 10.1083/jcb.201203115. This paper demonstrates that the APC can be autonomously activated in the parasynchronous species A. nidulans. [DOI] [PMC free article] [PubMed] [Google Scholar]
- **23.Lee C, Zhang H, Baker AE, Occhipinti P, Borsuk ME, Gladfelter AS. Protein aggregation behavior regulates cyclin transcript localization and cell-cycle control. Dev Cell. 2013;25:572–584. doi: 10.1016/j.devcel.2013.05.007. The authors show that aggregation behavior of an RNA-binding intrinsically-disordered protein is important for maintaining nuclear division autonomy. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lee C-D, Tu BP. Glucose-regulated phosphorylation of the PUF protein Puf3 regulates the translational fate of its bound mRNAs and association with RNA granules. Cell Rep. 2015;11:1638–1650. doi: 10.1016/j.celrep.2015.05.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Malinovska L, Palm S, Gibson K, Verbavatz J-M, Alberti S. Dictyostelium discoideum has a highly Q/N-rich proteome and shows an unusual resilience to protein aggregation. P Natl Acad Sci USA. 2015;112:E2620–9. doi: 10.1073/pnas.1504459112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Osmani AH, Davies J, Liu H-L, Nile A, Osmani SA. Systematic deletion and mitotic localization of the nuclear pore complex proteins of Aspergillus nidulans. Mol Biol Cell. 2006;17:4946–4961. doi: 10.1091/mbc.E06-07-0657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- **27.Baum DA, Baum B. An inside-out origin for the eukaryotic cell. BMC Biol. 2014;12:76. doi: 10.1186/s12915-014-0076-2. This paper proposes that eukaryotic cells evolved from the nuclear membrane out, and that syncytium with autonomous nuclei represent a step in the evolution of multicellularity. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Neumann FR, Nurse P. Nuclear size control in fission yeast. J Cell Biol. 2007;179:593–600. doi: 10.1083/jcb.200708054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kasuga T, Glass NL. Dissecting colony development of Neurospora crassa using mRNA profiling and comparative genomics approaches. Eukaryot Cell. 2008;7:1549–1564. doi: 10.1128/EC.00195-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- *30.Pitchaimani K, Maheshwari R. Extreme nuclear disproportion and constancy of enzyme activity in a heterokaryon of Neurospora crassa. J Genet. 2003;82:1–6. doi: 10.1007/BF02715873. The authors demonstrate that total cell output of a specific product can be constant despite varying proportions of nuclei with the appropriate genes to produce the product, suggesting nuclear autonomous responses to whole-cell requirements. [DOI] [PubMed] [Google Scholar]
- 31.Schuurs TA, Dalstra HJ, Scheer JM, Wessels JG. Positioning of nuclei in the secondary mycelium of Schizophyllum commune in relation to differential gene expression. Fungal Genet Biol. 1998;23:150–161. doi: 10.1006/fgbi.1997.1028. [DOI] [PubMed] [Google Scholar]
- **32.Hong CI, Zamborszky J, Baek M, Labiscsak L, Ju K, Lee H, Larrondo LF, Goity A, Chong HS, Belden WJ, et al. Circadian rhythms synchronize mitosis in Neurospora crassa. P Natl Acad Sci USA. 2014;111:1397–1402. doi: 10.1073/pnas.1319399111. This is the first demonstration that the cell cycle in the canonically asynchronous species N. crassa is coupled to circadian rhythms. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Feillet C, Krusche P, Tamanini F, Janssens RC, Downey MJ, Martin P, Teboul M, Saito S, Levi FA, Bretschneider T, et al. Phase locking and multiple oscillating attractors for the coupled mammalian clock and cell cycle. P Natl Acad Sci USA. 2014;111:9828–9833. doi: 10.1073/pnas.1320474111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Muirhead CA, Glass NL, Slatkin M. Multilocus self-recognition systems in fungi as a cause of trans-species polymorphism. Genetics. 2002;161:633–641. doi: 10.1093/genetics/161.2.633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Czaran T, Hoekstra RF, Aanen DK. Selection against somatic parasitism can maintain allorecognition in fungi. Fungal Genet and Biol. 2014;73:128–137. doi: 10.1016/j.fgb.2014.09.010. [DOI] [PubMed] [Google Scholar]
- 36.Kron SJ, Styles CA, Fink GR. Symmetric cell division in pseudohyphae of the yeast Saccharomyces cerevisiae. Mol Biol Cell. 1994;5:1003–1022. doi: 10.1091/mbc.5.9.1003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Demeter J, Lee SE, Haber JE, Stearns T. The DNA damage checkpoint signal in budding yeast is nuclear limited. Mol Cell. 2000;6:487–492. doi: 10.1016/s1097-2765(00)00047-2. [DOI] [PubMed] [Google Scholar]
- 38.Barelle CJ, Bohula EA, Kron SJ, Wessels D, Soll DR, Schafer A, Brown AJP, Gow NAR. Asynchronous cell cycle and asymmetric vacuolar inheritance in true hyphae of Candida albicans. Eukaryot Cell. 2003;2:398–410. doi: 10.1128/EC.2.3.398-410.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- *39.Shahi S, Beerens B, Manders EMM, Rep M. Dynamics of the establishment of multinucleate compartments in Fusarium oxysporum. Eukaryot Cell. 2015;14:78–85. doi: 10.1128/EC.00200-14. This is the first demonstration of multinucleate, parasynchronous compartments in F. oxysporum and that sub-apical compartments can re-enter the cell cycle. [DOI] [PMC free article] [PubMed] [Google Scholar]
- *40.Ruiz-Roldan MC, Kohli M, Roncero MIG, Philippsen P, Di Pietro A, Espeso EA. Nuclear dynamics during germination, conidiation, and hyphal fusion of Fusarium oxysporum. Eukaryot Cell. 2010;9:1216–1224. doi: 10.1128/EC.00040-10. This is the first observation of selective nuclear degradation upon hyphal fusion in F. oxysporum. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Todd NK, Caylmore RC. Cytology of hyphal interactions and reactions in Schizophyllum commune. In: Moore D, Casselton LA, Wood DA, Frankland JC, editors. Developmental Biology of Higher Fungi. Cambridge University Press; 1985. [Google Scholar]
- 42.Aylmore RC, Todd NK. Hyphal fusion in Coriolus versicolor. In: Jennings DH, Rayner A, editors. The ecology and physiology of the fungal mycelium. Cambridge University Press; 1984. [Google Scholar]
- 43.Noble M. The Morphology and Cytology of Typhula Trifolii Rostr. Ann Bot. 1937;1:67–98. [Google Scholar]
- 44.Bensaude M. Recherches sur le cycle evolutive et la sexualite chez les Basidiomycetes. 1918. [Google Scholar]
- *45.Ishikawa FH, Souza EA, Shoji J-Y, Connolly L, Freitag M, Read ND, Roca MG. Heterokaryon incompatibility is suppressed following conidial anastomosis tube fusion in a fungal plant pathogen. PLoS ONE. 2012;7:e31175. doi: 10.1371/journal.pone.0031175. This paper demonstrates that the heterokaryon incompatibility response is tunable by environmental conditions, e.g. presence of a host in the case of pathogenic species. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Cardoso MC, Leonhardt H, Nadal-Ginard B. Reversal of terminal differentiation and control of DNA replication: cyclin A and Cdk2 specifically localize at subnuclear sites of DNA replication. Cell. 1993;74:979–992. doi: 10.1016/0092-8674(93)90721-2. [DOI] [PubMed] [Google Scholar]
- 47.Bursztajn S, Berman SA, Gilbert W. Differential expression of acetylcholine receptor mRNA in nuclei of cultured muscle cells. P Natl Acad Sci USA. 1989;86:2928–2932. doi: 10.1073/pnas.86.8.2928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Tsim KW, Greenberg I, Rimer M, Randall WR, Salpeter MM. Transcripts for the acetylcholine receptor and acetylcholine esterase show distribution differences in cultured chick muscle cells. J Cell Biol. 1992;118:1201–1212. doi: 10.1083/jcb.118.5.1201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Duca KA, Chiu KP, Sullivan T, Berman SA, Bursztajn S. Nuclear clustering in myotubes: a proposed role in acetylcholine receptor mRNA expression. Biochim Biophys Acta. 1998;1401:1–20. doi: 10.1016/s0167-4889(97)00118-3. [DOI] [PubMed] [Google Scholar]
- 50.Arazoe T, Miyoshi K, Yamato T, Ogawa T, Ohsato S, Arie T, Kuwata S. Tailor-made CRISPR/Cas system for highly efficient targeted gene replacement in the rice blast fungus. Biotechnol Bioeng. 2015 doi: 10.1002/bit.25662. [DOI] [PubMed] [Google Scholar]
- 51.Nødvig CS, Nielsen JB, Kogle ME, Mortensen UH. A CRISPR-Cas9 system for genetic engineering of filamentous fungi. PLoS ONE. 2015;10:e0133085. doi: 10.1371/journal.pone.0133085. [DOI] [PMC free article] [PubMed] [Google Scholar]