Abstract
Candida albicans is both a prevalent human commensal and the most commonly encountered human fungal pathogen. This lifestyle is dependent on the ability of the fungus to undergo rapid genetic and epigenetic changes, often in response to specific environmental cues. A parasexual cycle in C. albicans has been defined that includes several unique properties when compared to the related model yeast, Saccharomyces cerevisiae. Novel features include strict regulation of mating via a phenotypic switch, enhanced conjugation within a sexual biofilm, and a program of concerted chromosome loss in place of a conventional meiosis. It is expected that several of these adaptations co-evolved with the ability of C. albicans to colonize the mammalian host.
Graphical Abstract
Introduction
Sexual reproduction is a ubiquitous property in eukaryotic cells, having arisen in the common ancestor to this lineage. It involves an alternation between ploidy states, often between that of haploid and diploid; mating (or gamete fusion) leads to a doubling of the genetic material in the cell, whereas meiosis leads to a halving of the DNA content in the cell. Fungal species exhibit highly diverse sexual cycles, from the relatively simple modes of sex seen in the model hemiascomycete S. cerevisiae, to the more complex reproductive cycles seen in the mushroom-forming basidiomycetes (for recent reviews on fungal sex see [1–5]). In the case of C. albicans, this important opportunistic pathogen is capable of causing both debilitating mucosal and life-threatening invasive infections, with the potential to infect almost every organ in the human body. Candida species were originally defined as species that can form pseudophyphae or true hyphae but lacked any form of sexual reproduction [6]. However, experiments have now deciphered an unusual parasexual mating cycle in C. albicans that shows several adaptations to that found in model yeasts.
Regulation of cell type
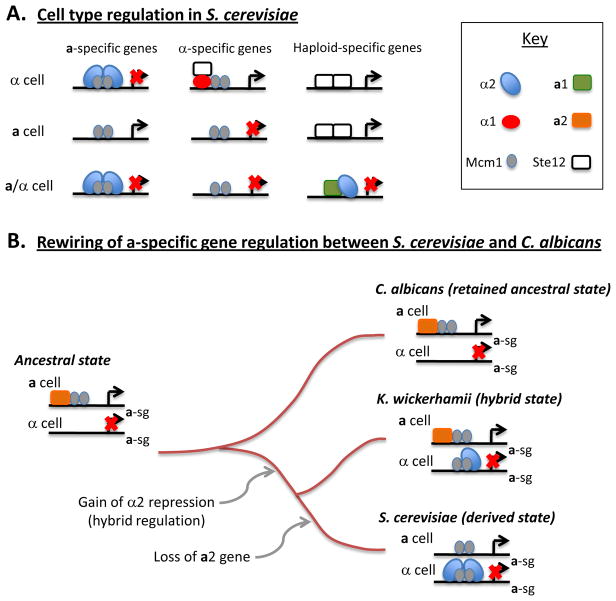
The sexual cycle in S. cerevisiae has served as the primary model for studies into mating and meiosis in yeast. In this species, mating occurs between a and α cells and is regulated by genes encoded at a single genetic locus called the mating-type or MAT locus. Three transcription factors encoded at the MAT locus, a1, α1, and α2, determine whether cells behave as a, α, or a/α cells (Figure 1A). MATa-containing cells express the a1 gene, MATα cells express α1 and α2 genes, and MATa/α cells express all three genes. In α cells, α1 is responsible for the induction of α-specific genes, whereas in a cells, a-specific genes are constitutively expressed. However, a-specific genes are not expressed in α cells due to repression by α2 together with the Mcm1 transcription factor [7]. MATa and MATα cells both express “haploid-specific” genes that include pheromone-signaling factors that promote mating [8,9]. Meiosis occurs only in MATa/α cells, as a complex between a1 and α2 turns off haploid-specific genes including RME1, which is a repressor of meiosis [7]. Thus, the combinatorial control of cell-type expression by MAT transcription factors regulates both mating and meiosis in S. cerevisiae.
Figure 1. Cell type regulation in S. cerevisiae and C. albicans.
(A) Schematics show transcription factors regulating a-specific genes, α-specific genes, and haploid-specific genes in S. cerevisiae. The transcription factors a1, α1, α2, Mcm1, and Ste12 act coordinately to regulate cell type specificity in S. cerevisiae.
(B) Rewiring of the regulation of a-specific genes (a-sg) between hemiascomycetes. The ancestral mode of regulation involves the a2 transcription factor acting in concert with Mcm1 to regulate a-specific genes. This mode of regulation has been retained in C. albicans. In some yeast lineages, expression of the a-specific genes also came under negative control of the α2 transcription factor. In S. cerevisiae, the ancestral form of regulation by a2 was lost, whereas in K. wickerhamii the hybrid form of regulation (involving both a2 and α2) has been retained.
Mating in Candida albicans and rewiring of cell type
C. albicans is a hemiascomycete yeast related to S. cerevisiae, although these species diverged more than 900 million years ago, and are approximately as divergent as humans and fish [10]. Natural isolates of C. albicans are diploid, and it was originally thought that this species could not undergo any form of sexual reproduction. However, diploid a and α strains of C. albicans were engineered from a clinical a/α isolate and shown to be capable of mating to form tetraploids, both in vitro and in a murine model of systemic infection [11,12]. Subsequent experiments established that C. albicans mating shows parallels with that in S. cerevisiae; both species secrete sex-specific pheromones that induce mating responses in cells of the opposite sex, leading to cell-cell conjugation and karyogamy [13–17]. Despite these similarities, the transcriptional circuits regulating mating have diverged since these two species last shared a common ancestor. In particular, where there is negative regulation of the a-specific genes in S. cerevisiae α cells via the α2 protein, there is positive regulation of the a-specific genes in C. albicans via an additional transcription factor, a2, that is absent from S. cerevisiae (Figure 1A and B)[18]. Analysis of multiple yeast species revealed that the transcriptional circuit in C. albicans is the ancestral circuit, whereas that in S. cerevisiae is a derived circuit [19]. Strikingly, some hemiascomycete species (e.g. K. wickerhamii) have retained a hybrid form of the circuit in which a-specific genes are both positively regulated by a2 and negatively regulated by α2 (Figure 1B) [20]. The hybrid state has been resolved by different routes in the lineages giving rise to modern yeast species; in some species regulation reverted to the ancestral state, while in others, such as S. cerevisiae, the ancestral mode of regulation was lost and only the derived mode of regulation was retained [20].
Mating in C. albicans is regulated by the white-opaque switch
Initial studies of C. albicans mating revealed that mating efficiency was low both in vitro and in vivo. However, a major breakthrough came with the discovery that mating is regulated not only by the mating locus, but also by an epigenetic switch between phenotypic states. Certain isolates of C. albicans had long been recognized to undergo phenotypic switching between “white” and “opaque” states. Cells in the white state are round and give rise to bright, dome-shaped colonies whereas cells in the opaque state are elongated and give rise to darker, flatter colonies (see images in Figure 2)[21]. Miller and Johnson revealed two key attributes of the switch. First, that switching to the opaque state occurred in a or α cells, but not a/α cells, due to repression by the a1/α2 complex. Second, that mating between opaque cells occurs a million times more efficiently than that between white cells [22]. These findings revolutionized the field, enabling mechanistic studies of C. albicans mating for the first time.
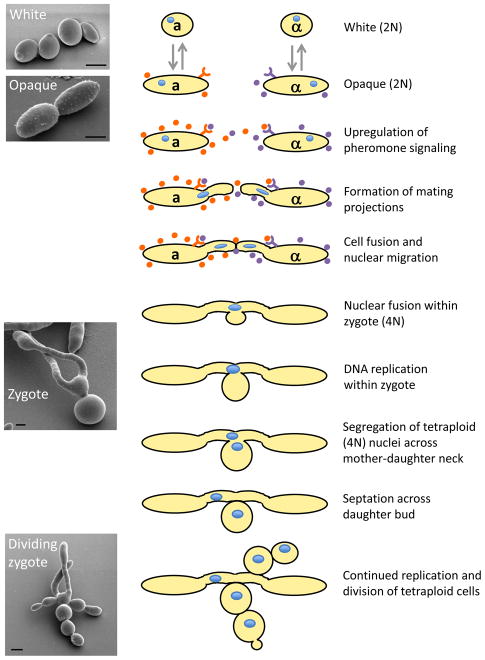
Figure 2. Mating in C. albicans.
Outline of the steps during heterothallic a-α mating in C. albicans. White cells switch to the mating-competent opaque form, and then undergo pheromone signaling between cells of opposite sexes. Mating projections fuse to form a mating zygote within which karyogamy (nuclear fusion) occurs. The nucleus in the tetraploid mating product undergoes replication, with one tetraploid nucleus entering the budding daughter cell. Continued replication and budding of tetraploid cells occurs. Scanning electron micrograph images of white, opaque and zygote cells are shown (scale bar, 2.5 μm, images courtesy of Matthew Hirakawa).
Regulation of the white-opaque switch requires the transcription factor Wor1, whose expression is necessary and sufficient for formation of opaque cells [23–25]. Wor1 acts as part of a complex network of interacting transcription factors to regulate bistable switching in C. albicans [26,27]. A similar white-opaque switch was subsequently uncovered in the related Candida species C. dubliniensis and C. tropicalis [28–30]. It therefore appears that the white-opaque switch evolved in the common ancestor to these three species, all of which are opportunistic human pathogens.
One curious aspect of the white-opaque switch is that C. albicans opaque cells are generally unstable at 37°C and switch back to the white state en masse [31]. It was therefore envisaged that mating takes place outside of the mammalian host, and conjugation events were observed during opaque cell colonization of the skin [32]. However, mating on the skin rarely results in productive daughter cell formation [33], and low mating frequencies were recently reported using this infection model [34]. Other niches may therefore hold greater promise for mating in the host, particularly given that host cues can stabilize opaque cells at body temperature. For example, N-acetyl glucosamine, CO2, anaerobiosis, and cellular stress all promote formation of the opaque state [35–38], and some of these cues can even induce white-to-opaque switching in a/α isolates [39]. Furthermore, at least in one clinical isolate white-to-opaque switching was observed in a murine model of gastrointestinal colonization [40], and occasional mating was also reported from this niche [35]. It therefore appears that mating, at least at low frequency, can occur in multiple sites in animal models of infection.
Concerted chromosome loss
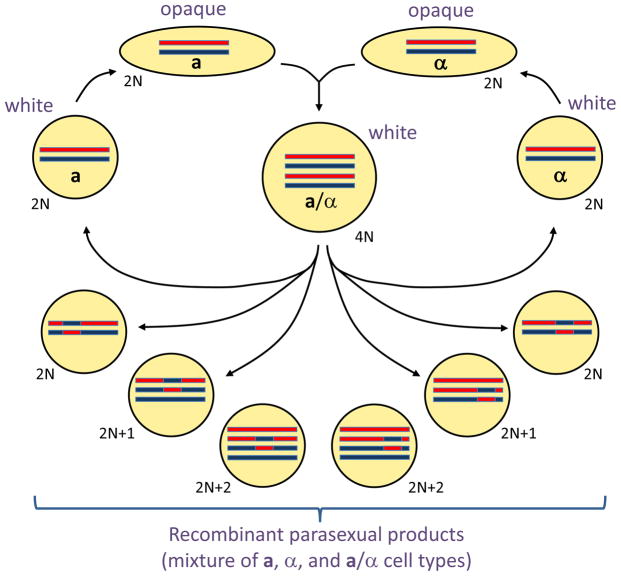
In most eukaryotes, sexual reproduction involves a meiotic program in which DNA replication is followed by two successive DNA divisions, thereby halving the genetic material in the cell. The C. albicans genome contains many conserved genes that function in meiosis in other species [41,42], including some that are functional when heterologously expressed in S. cerevisiae [43]. However, a conventional meiosis has not been described in C. albicans. In its place, tetraploid mating products return to a diploid, or near-diploid state, by a parasexual mechanism of concerted chromosome loss (Figure 3) [44,45]. During parasex, cells undergo limited recombination, which is dependent on the ‘meiotic’ recombinase, Spo11 [46]. This indicates an intriguing mechanistic link between the programs of meiosis and parasexual chromosome loss. Several steps in the parasexual cycle are stimulated by environmental stress, suggesting that parasex might be particularly advantageous under conditions where genetic variation is beneficial [47]. Parasexual chromosome loss is thought to involve chromosome non-disjunction during mitotic divisions, leading to unstable cells of intermediate ploidy, which then further reduce their ploidy back to diploid or near diploid [45,48]. A similar parasexual cycle was observed in C. tropicalis, implying that meiosis is also absent (or at least cryptic) in this species [49].
Figure 3. Genetic rearrangements during the parasexual cycle in C. albicans.
Diploid (2N) a and α cells switch from the white state to the mating-competent opaque state, and subsequently undergo mating to form tetraploid (4N) a/α cells. Tetraploid cells can continue to divide as stable tetraploids or can be induced to undergo concerted chromosome loss, producing diploid a, α, and a/α cells. Chromosome loss is accompanied by genetic recombination and results in large numbers of aneuploid cells (e.g., 2N+1 and 2N+2 cells) as well as diploid cells with different MTL configurations. For clarity, only one of the eight C. albicans chromosomes is shown in each cell.
Despite the absence of a conventional meiosis in C. albicans, the products of the parasexual cycle exhibit diverse genotypes and phenotypes [46]. Diversity is due to a combination of mechanisms, including shuffling of whole chromosome homologs, genetic recombination, and the presence or absence of supernumerary chromosomes (Figure 3). Given that aneuploidy plays a widespread role in generating phenotypic variants and antifungal drug resistance [50–52], it is clear that parasex could generate potentially important traits relevant to fungal pathogenesis.
Recently it was shown that C. albicans not only undergoes a diploid-tetraploid-diploid cycle but is capable of transitioning between diploid and haploid cell types. Haploid forms of the species were thought to be inviable due to the presence of multiple recessive lethal alleles, but rare haploid cells were obtained both in vitro and following infection of an animal host [53]. Haploid cells were formed by a parasexual mechanism of concerted chromosome loss similar to that observed in tetraploid cells. Furthermore, haploid a and α cells could switch to the opaque state and undergo mating to regenerate a/α diploid cells, or could form MTL homozygous diploids by spontaneous auto-diploidization [53]. While viable, all haploid cells showed marked fitness defects that currently limit their use in studying pathogenesis.
Why did C. albicans, as well as closely related species, apparently lose the ability to undergo a conventional meiosis? One possibility is that the formation of sexual spores accompanying a traditional meiosis would be a hindrance to commensal species, as spores are likely to be highly immunogenic and targeted by host defenses [54]. In addition, parasex is capable of generating progeny that are extremely diverse due to cells existing in multiple ploidy states (Figure 3). The plethora of aneuploidies produced by parasex could be important for generating a pool of genetically diverse isolates upon which selection could then act.
It is worth noting that, in contrast to C. albicans, several Candida clade species have retained a full sexual cycle that includes meiosis and sexual sporulation [41]. For example, Candida lusitaniae undergoes meiosis despite containing a similar repertoire of ‘meiosis-specific’ genes to that of C. albicans [55]. However, whereas Ime1 is the master transcriptional regulator of meiosis in S. cerevisiae, an ortholog of this gene is missing in C. lusitaniae and other Candida clade species [41]. Instead, it appears that the transcription factor Ste12 regulates both mating and meiosis in C. lusitaniae, indicating fusion of the regulatory programs controlling these two pathways [56]. These studies reveal that genomic analysis alone cannot determine whether a cryptic meiosis still remains to be discovered in C. albicans.
Unisexual reproduction
Several yeast species, including S. cerevisiae and S. pombe, undergo mating-type switching in which cells alternate between a and α cell types. Mating-type switching evolved independently in these species, and in both cases involves copying of one of two silent cassettes encoding mating type information to the active expression site [57,58]. Recent experiments suggest that this mode of mating-type switching evolved from an ancestral two-locus system, in which switching occurred using a simpler flip-flop inversion mechanism [59]. These studies also suggest that mating-type switching was lost in C. albicans and other Candida clade species; these species do not contain silent mating-type cassettes or undergo mating-type switching, potentially limiting self-fertility [41]. Surprisingly, however, C. albicans was found to be capable of same-sex mating under the appropriate conditions. This result was first observed using strains that lack the barrier protease, Bar1, that inactivates α pheromone [60]. C. albicans opaque a strains express both a and α pheromone; Bar1 normally degrades secreted α pheromone, but in the absence of Bar1 an autocrine feedback loop involving this pheromone promotes fusion between opaque a cells. Same-sex a-a mating also occurs in mixed cultures of wildtype opaque a and α cells, in which α pheromone produced by α cells overwhelms Bar1 activity from a cells [60]. Subsequent experiments revealed that activation of pheromone signaling is sufficient for driving unisexual mating, and even that pheromones from other Candida species can induce same-sex mating in C. albicans [61]. More recently it was shown that white cells can also be induced to secrete pheromone which can promote both opposite-sex and same-sex mating of C. albicans opaque cells [34].
Same-sex mating has been observed in another prevalent human pathogen, Cryptococcus neoformans [62,63], suggesting that unisexual reproduction may be beneficial for diverse fungal pathogens. Unisexual mating could have advantages for a species, including the fact that sex can occur in the absence of an opposite sex partner, and that the costs associated with unisexual reproduction are significantly less than those accompanying bisexual mating [4].
Pheromone signaling drives biofilm formation
C. albicans infections are often a consequence of biofilm formation, in which communities of fungal cells adhere both to one another and to a biological or inert substrate. Biofilm growth is particularly troublesome on implanted medical devices such as catheters, pacemakers, heart valves, or prosthetic joints, as biofilms are resistant to antifungal therapy or immune clearance [64,65]. C. albicans forms biofilms under a variety of culture conditions, and the ability to undergo filamentation is generally associated with biofilm maturation [66,67].
The Soll group revealed that pheromone signaling could act as a novel environmental cue driving biofilm formation in C. albicans. White a or α cells, although unable to mate, responded to pheromones produced by opposite-sex opaque cells by adhering to an inert surface [68]. Furthermore, formation of these ‘sexual’ biofilms could support mating between opaque a and α cells by stabilizing pheromone gradients between opposite-sex opaque partners [68,69]. Studies of sexual biofilms reveals parallels with ‘conventional’ biofilms, with both structures consisting of a basal layer of yeast cells upon which filamentous cells and extracellular matrix develop [68–70].
The transcriptional regulation of sexual biofilm remains controversial. Experiments in our group showed that pheromone signaling in white cells is via the canonical MAPK pathway and its transcription factor, Cph1 (ortholog of ScSte12) [71]. In fact, deletion of Cph1 abolished the complete transcriptional response of white cells to a pheromone challenge [71]. In contrast, the Soll group found that Cph1 was not involved in the response to pheromone in white cells, and instead Tec1 was the key transcription factor mediating sexual biofilm formation [72]. A third independent study showed that Cph1 was critical for MAPK signaling in white cells, but that Tec1 was also required for MAPK-signaling induced adhesion by white cells [73]. It therefore remains to be seen how exactly Cph1/Tec1 promote adherence and biofilm formation in response to pheromone signaling. The role of sexual biofilms in nature has also yet to be addressed; most clinical isolates are a/α strains [74,75] and are able to form conventional biofilms but are not capable of forming pheromone-induced biofilms. In these strains, it is the presence of the a1/α2 complex that (directly or indirectly) inhibits the expression of pheromone, pheromone receptor, pheromone processing and MAPK signaling genes, thereby blocking both the secretion of, and response to, sexual pheromones [18,34,76].
Future perspectives
Identification of a parasexual cycle in C. albicans has raised as many questions as it has answered. Discovery of this unusual cycle reveals an elegant mechanism for mating and genetic rearrangement, and yet population structure analyses suggests that recombination between isolates is rare in nature [75,77]. Sex could be restricted in C. albicans to limit the disruption of genotypes that are already well adapted for commensal growth [54]. However, sex or parasex has now been documented in most Candida clade species [41], indicating that these species, originally labeled as Fungi Imperfecti, have maintained (para)sexual reproduction over millions of years. The retention of facultative sexuality is compelling, and suggests that sex continues to play important roles in these species. Determining the role(s) of sex in C. albicans will therefore continue to shed light on cryptic modes of sex in fungal species, as well as adaptations to this ancestral program that can enhance the commensal or pathogenic lifestyle.
Highlight Points.
C. albicans was long considered to be an obligate asexual species
An unusual parasexual cycle has now been uncovered in C. albicans
Mating competency is dependent on cells undergoing a white-to-opaque switch
Both heterothallic and homothallic modes of mating exist
In place of meiosis cells undergo concerted chromosome loss
Acknowledgments
The author would like to acknowledge Iuliana Ene, Matthew Hirakawa, and Christine Scaduto for comments on the manuscript. Funding in the author’s laboratory is supported by the NIH (RO1AI081704, R21AI081560) and a PATH award from the Burroughs Wellcome Fund. Scanning electron micrograph images of C. albicans cells were provided by Matthew Hirakawa.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Ene IV, Bennett RJ. The cryptic sexual strategies of human fungal pathogens. Nat Rev Microbiol. 2014;12:239–251. doi: 10.1038/nrmicro3236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ni M, Feretzaki M, Sun S, Wang X, Heitman J. Sex in fungi. Annu Rev Genet. 2011;45:405–430. doi: 10.1146/annurev-genet-110410-132536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lee SC, Ni M, Li W, Shertz C, Heitman J. The evolution of sex: a perspective from the fungal kingdom. Microbiol Mol Biol Rev. 2010;74:298–340. doi: 10.1128/MMBR.00005-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Heitman J, Sun S, James TY. Evolution of fungal sexual reproduction. Mycologia. 2013;105:1–27. doi: 10.3852/12-253. [DOI] [PubMed] [Google Scholar]
- 5.Heitman J, Carter DA, Dyer PS, Soll DR. Sexual reproduction of human fungal pathogens. Cold Spring Harb Perspect Med. 2014:4. doi: 10.1101/cshperspect.a019281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Reedy JL, Heitman J. Evolution of MAT in the Candida species complex: sex, ploidy, and complete sexual cycles in C. lusitaniae, C. guilliermondii, and C. krusei. In: Heitman J, editor. Sex in Fungi. ASM Press; 2007. pp. 235–245. [Google Scholar]
- 7.Johnson AD. Molecular mechanisms of cell-type determination in budding yeast. Curr Opin Genet Dev. 1995;5:552–558. doi: 10.1016/0959-437x(95)80022-0. [DOI] [PubMed] [Google Scholar]
- 8.Mitchell AP, Herskowitz I. Activation of meiosis and sporulation by repression of the RME1 product in yeast. Nature. 1986;319:738–742. doi: 10.1038/319738a0. [DOI] [PubMed] [Google Scholar]
- 9.van Werven FJ, Neuert G, Hendrick N, Lardenois A, Buratowski S, van Oudenaarden A, Primig M, Amon A. Transcription of two long noncoding RNAs mediates mating-type control of gametogenesis in budding yeast. Cell. 2012;150:1170–1181. doi: 10.1016/j.cell.2012.06.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dujon B, Sherman D, Fischer G, Durrens P, Casaregola S, Lafontaine I, De Montigny J, Marck C, Neuveglise C, Talla E, et al. Genome evolution in yeasts. Nature. 2004;430:35–44. doi: 10.1038/nature02579. [DOI] [PubMed] [Google Scholar]
- 11.Hull CM, Raisner RM, Johnson AD. Evidence for mating of the “asexual” yeast Candida albicans in a mammalian host. Science. 2000;289:307–310. doi: 10.1126/science.289.5477.307. [DOI] [PubMed] [Google Scholar]
- 12.Magee BB, Magee PT. Induction of mating in Candida albicans by construction of MTLa and MTLalpha strains. Science. 2000;289:310–313. doi: 10.1126/science.289.5477.310. [DOI] [PubMed] [Google Scholar]
- 13.Bennett RJ, Miller MG, Chua PR, Maxon ME, Johnson AD. Nuclear fusion occurs during mating in Candida albicans and is dependent on the KAR3 gene. Mol Microbiol. 2005;55:1046–1059. doi: 10.1111/j.1365-2958.2005.04466.x. [DOI] [PubMed] [Google Scholar]
- 14.Bennett RJ, Uhl MA, Miller MG, Johnson AD. Identification and characterization of a Candida albicans mating pheromone. Mol Cell Biol. 2003;23:8189–8201. doi: 10.1128/MCB.23.22.8189-8201.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Magee BB, Legrand M, Alarco AM, Raymond M, Magee PT. Many of the genes required for mating in Saccharomyces cerevisiae are also required for mating in Candida albicans. Mol Microbiol. 2002;46:1345–1351. doi: 10.1046/j.1365-2958.2002.03263.x. [DOI] [PubMed] [Google Scholar]
- 16.Lockhart SR, Zhao R, Daniels KJ, Soll DR. Alpha-pheromone-induced “shmooing” and gene regulation require white-opaque switching during Candida albicans mating. Eukaryot Cell. 2003;2:847–855. doi: 10.1128/EC.2.5.847-855.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Panwar SL, Legrand M, Dignard D, Whiteway M, Magee PT. MFalpha1, the gene encoding the alpha mating pheromone of Candida albicans. Eukaryot Cell. 2003;2:1350–1360. doi: 10.1128/EC.2.6.1350-1360.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tsong AE, Miller MG, Raisner RM, Johnson AD. Evolution of a combinatorial transcriptional circuit: a case study in yeasts. Cell. 2003;115:389–399. doi: 10.1016/s0092-8674(03)00885-7. [DOI] [PubMed] [Google Scholar]
- 19.Tsong AE, Tuch BB, Li H, Johnson AD. Evolution of alternative transcriptional circuits with identical logic. Nature. 2006;443:415–420. doi: 10.1038/nature05099. [DOI] [PubMed] [Google Scholar]
- **20.Baker CR, Booth LN, Sorrells TR, Johnson AD. Protein modularity, cooperative binding, and hybrid regulatory states underlie transcriptional network diversification. Cell. 2012;151:80–95. doi: 10.1016/j.cell.2012.08.018. This paper demonstrated how regulatory transcription circuits could contain hybrid states that can be resolved by different routes. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Slutsky B, Staebell M, Anderson J, Risen L, Pfaller M, Soll DR. “White-opaque transition”: a second high-frequency switching system in Candida albicans. J Bacteriol. 1987;169:189–197. doi: 10.1128/jb.169.1.189-197.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- **22.Miller MG, Johnson AD. White-opaque switching in Candida albicans is controlled by mating-type locus homeodomain proteins and allows efficient mating. Cell. 2002;110:293–302. doi: 10.1016/s0092-8674(02)00837-1. This work demonstrated that the white-opaque switch was a key mechanism for regulation of mating in C. albicans. [DOI] [PubMed] [Google Scholar]
- 23.Huang G, Wang H, Chou S, Nie X, Chen J, Liu H. Bistable expression of WOR1, a master regulator of white-opaque switching in Candida albicans. Proc Natl Acad Sci U S A. 2006;103:12813–12818. doi: 10.1073/pnas.0605270103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Srikantha T, Borneman AR, Daniels KJ, Pujol C, Wu W, Seringhaus MR, Gerstein M, Yi S, Snyder M, Soll DR. TOS9 regulates white-opaque switching in Candida albicans. Eukaryot Cell. 2006;5:1674–1687. doi: 10.1128/EC.00252-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zordan RE, Galgoczy DJ, Johnson AD. Epigenetic properties of white-opaque switching in Candida albicans are based on a self-sustaining transcriptional feedback loop. Proc Natl Acad Sci U S A. 2006;103:12807–12812. doi: 10.1073/pnas.0605138103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- *26.Hernday AD, Lohse MB, Fordyce PM, Nobile CJ, DeRisi JL, Johnson AD. Structure of the transcriptional network controlling white-opaque switching in Candida albicans. Mol Microbiol. 2013;90:22–35. doi: 10.1111/mmi.12329. A detailed analysis of the transcriptional networks regulating white-opaque switching. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zordan RE, Miller MG, Galgoczy DJ, Tuch BB, Johnson AD. Interlocking transcriptional feedback loops control white-opaque switching in Candida albicans. PLoS Biol. 2007;5:e256. doi: 10.1371/journal.pbio.0050256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- *28.Porman AM, Alby K, Hirakawa MP, Bennett RJ. Discovery of a phenotypic switch regulating sexual mating in the opportunistic fungal pathogen Candida tropicalis. Proc Natl Acad Sci U S A. 2011;108:21158–21163. doi: 10.1073/pnas.1112076109. Demonstration that mating in C. tropicalis is regulated by a white-opaque switch similar to that in C. albicans. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Pujol C, Daniels KJ, Lockhart SR, Srikantha T, Radke JB, Geiger J, Soll DR. The closely related species Candida albicans and Candida dubliniensis can mate. Eukaryot Cell. 2004;3:1015–1027. doi: 10.1128/EC.3.4.1015-1027.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Xie J, Du H, Guan G, Tong Y, Kourkoumpetis TK, Zhang L, Bai FY, Huang G. N-acetylglucosamine induces white-to-opaque switching and mating in Candida tropicalis, providing new insights into adaptation and fungal sexual evolution. Eukaryot Cell. 2012;11:773–782. doi: 10.1128/EC.00047-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rikkerink EH, Magee BB, Magee PT. Opaque-white phenotype transition: a programmed morphological transition in Candida albicans. J Bacteriol. 1988;170:895–899. doi: 10.1128/jb.170.2.895-899.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lachke SA, Lockhart SR, Daniels KJ, Soll DR. Skin facilitates Candida albicans mating. Infect Immun. 2003;71:4970–4976. doi: 10.1128/IAI.71.9.4970-4976.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Soll DR. Why does Candida albicans switch? FEMS Yeast Res. 2009;9:973–989. doi: 10.1111/j.1567-1364.2009.00562.x. [DOI] [PubMed] [Google Scholar]
- *34.Tao L, Cao C, Liang W, Guan G, Zhang Q, Nobile CJ, Huang G. White cells facilitate opposite- and same-sex mating of opaque cells in Candida albicans. PLoS Genet. 2014;10:e1004737. doi: 10.1371/journal.pgen.1004737. This work showed how ‘sterile’ white cells can promote mating between competent opaque cells. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Dumitru R, Navarathna DH, Semighini CP, Elowsky CG, Dumitru RV, Dignard D, Whiteway M, Atkin AL, Nickerson KW. In vivo and in vitro anaerobic mating in Candida albicans. Eukaryot Cell. 2007;6:465–472. doi: 10.1128/EC.00316-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Huang G, Srikantha T, Sahni N, Yi S, Soll DR. CO2 regulates white-to-opaque switching in Candida albicans. Curr Biol. 2009;19:330–334. doi: 10.1016/j.cub.2009.01.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Huang G, Yi S, Sahni N, Daniels KJ, Srikantha T, Soll DR. N-acetylglucosamine induces white to opaque switching, a mating prerequisite in Candida albicans. PLoS Pathog. 2010;6:e1000806. doi: 10.1371/journal.ppat.1000806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Alby K, Bennett RJ. Stress-induced phenotypic switching in Candida albicans. Mol Biol Cell. 2009;20:3178–3191. doi: 10.1091/mbc.E09-01-0040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Xie J, Tao L, Nobile CJ, Tong Y, Guan G, Sun Y, Cao C, Hernday AD, Johnson AD, Zhang L, et al. White-opaque switching in natural MTLa/alpha isolates of Candida albicans: evolutionary implications for roles in host adaptation, pathogenesis, and sex. PLoS Biol. 2013;11:e1001525. doi: 10.1371/journal.pbio.1001525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ramirez-Zavala B, Reuss O, Park YN, Ohlsen K, Morschhauser J. Environmental induction of white-opaque switching in Candida albicans. PLoS Pathog. 2008;4:e1000089. doi: 10.1371/journal.ppat.1000089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Butler G, Rasmussen MD, Lin MF, Santos MA, Sakthikumar S, Munro CA, Rheinbay E, Grabherr M, Forche A, Reedy JL, et al. Evolution of pathogenicity and sexual reproduction in eight Candida genomes. Nature. 2009;459:657–662. doi: 10.1038/nature08064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Tzung KW, Williams RM, Scherer S, Federspiel N, Jones T, Hansen N, Bivolarevic V, Huizar L, Komp C, Surzycki R, et al. Genomic evidence for a complete sexual cycle in Candida albicans. Proc Natl Acad Sci U S A. 2001;98:3249–3253. doi: 10.1073/pnas.061628798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Diener AC, Fink GR. DLH1 is a functional Candida albicans homologue of the meiosis-specific gene DMC1. Genetics. 1996;143:769–776. doi: 10.1093/genetics/143.2.769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- *44.Bennett RJ, Johnson AD. Completion of a parasexual cycle in Candida albicans by induced chromosome loss in tetraploid strains. Embo J. 2003;22:2505–2515. doi: 10.1093/emboj/cdg235. First demonstration that C. albicans completes its sexual cycle by concerted chromosome loss. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Hickman MA, Paulson C, Dudley AM, Berman J. Parasexual ploidy reduction drives population heterogeneity through random and transient aneuploidy in Candida albicans. Genetics. 2015 doi: 10.1534/genetics.115.178020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Forche A, Alby K, Schaefer D, Johnson AD, Berman J, Bennett RJ. The parasexual cycle in Candida albicans provides an alternative pathway to meiosis for the formation of recombinant strains. PLoS Biol. 2008;6:e110. doi: 10.1371/journal.pbio.0060110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Berman J, Hadany L. Does stress induce (para)sex? Implications for Candida albicans evolution. Trends Genet. 2012;28:197–203. doi: 10.1016/j.tig.2012.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Bennett RJ, Forche A, Berman J. Rapid mechanisms for generating genome diversity: whole ploidy shifts, aneuploidy, and loss of heterozygosity. Cold Spring Harbor Perspect Med. 2014;4(10) doi: 10.1101/cshperspect.a019604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Seervai RN, Jones SK, Jr, Hirakawa MP, Porman AM, Bennett RJ. Parasexuality and ploidy change in Candida tropicalis. Eukaryot Cell. 2013;12:1629–1640. doi: 10.1128/EC.00128-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Selmecki A, Bergmann S, Berman J. Comparative genome hybridization reveals widespread aneuploidy in Candida albicans laboratory strains. Mol Microbiol. 2005;55:1553–1565. doi: 10.1111/j.1365-2958.2005.04492.x. [DOI] [PubMed] [Google Scholar]
- 51.Kwon-Chung KJ, Chang YC. Aneuploidy and drug resistance in pathogenic fungi. PLoS Pathog. 2012;8:e1003022. doi: 10.1371/journal.ppat.1003022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Ni M, Feretzaki M, Li W, Floyd-Averette A, Mieczkowski P, Dietrich FS, Heitman J. Unisexual and heterosexual meiotic reproduction generate aneuploidy and phenotypic diversity de novo in the yeast Cryptococcus neoformans. PLoS Biol. 2013;11:e1001653. doi: 10.1371/journal.pbio.1001653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Hickman MA, Zeng G, Forche A, Hirakawa MP, Abbey D, Harrison BD, Wang YM, Su CH, Bennett RJ, Wang Y, et al. The ‘obligate diploid’ Candida albicans forms mating-competent haploids. Nature. 2013;494:55–59. doi: 10.1038/nature11865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Nielsen K, Heitman J. Sex and virulence of human pathogenic fungi. Adv Genet. 2007;57:143–173. doi: 10.1016/S0065-2660(06)57004-X. [DOI] [PubMed] [Google Scholar]
- 55.Reedy JL, Floyd AM, Heitman J. Mechanistic plasticity of sexual reproduction and meiosis in the Candida pathogenic species complex. Curr Biol. 2009;19:891–899. doi: 10.1016/j.cub.2009.04.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Sherwood RK, Scaduto CM, Torres SE, Bennett RJ. Convergent evolution of a fused sexual cycle promotes the haploid lifestyle. Nature. 2014;506:387–390. doi: 10.1038/nature12891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Haber JE. Mating-type genes and MAT switching in Saccharomyces cerevisiae. Genetics. 2012;191:33–64. doi: 10.1534/genetics.111.134577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Klar AJ. Lessons learned from studies of fission yeast mating-type switching and silencing. Annu Rev Genet. 2007;41:213–236. doi: 10.1146/annurev.genet.39.073103.094316. [DOI] [PubMed] [Google Scholar]
- **59.Hanson SJ, Byrne KP, Wolfe KH. Mating-type switching by chromosomal inversion in methylotrophic yeasts suggests an origin for the three-locus Saccharomyces cerevisiae system. Proc Natl Acad Sci U S A. 2014;111:E4851–4858. doi: 10.1073/pnas.1416014111. Studies show that a two-locus system regulates mating-type switching in methylotrophic yeasts, and that Candida clade yeast probably lost this ability after diverging from other Saccharomycotina species. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Alby K, Schaefer D, Bennett RJ. Homothallic and heterothallic mating in the opportunistic pathogen Candida albicans. Nature. 2009;460:890–893. doi: 10.1038/nature08252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Alby K, Bennett RJ. Interspecies pheromone signaling promotes biofilm formation and same-sex mating in Candida albicans. Proc Natl Acad Sci U S A. 2011;108:2510–2515. doi: 10.1073/pnas.1017234108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Lin X, Hull CM, Heitman J. Sexual reproduction between partners of the same mating type in Cryptococcus neoformans. Nature. 2005;434:1017–1021. doi: 10.1038/nature03448. [DOI] [PubMed] [Google Scholar]
- 63.Lin X, Litvintseva AP, Nielsen K, Patel S, Floyd A, Mitchell TG, Heitman J. alpha AD alpha hybrids of Cryptococcus neoformans: evidence of same-sex mating in nature and hybrid fitness. PLoS Genet. 2007;3:1975–1990. doi: 10.1371/journal.pgen.0030186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Finkel JS, Mitchell AP. Genetic control of Candida albicans biofilm development. Nat Rev Microbiol. 2011;9:109–118. doi: 10.1038/nrmicro2475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Fox EP, Nobile CJ. A sticky situation: untangling the transcriptional network controlling biofilm development in Candida albicans. Transcription. 2012;3:315–322. doi: 10.4161/trns.22281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Baillie GS, Douglas LJ. Role of dimorphism in the development of Candida albicans biofilms. J Med Microbiol. 1999;48:671–679. doi: 10.1099/00222615-48-7-671. [DOI] [PubMed] [Google Scholar]
- 67.Ramage G, VandeWalle K, Lopez-Ribot JL, Wickes BL. The filamentation pathway controlled by the Efg1 regulator protein is required for normal biofilm formation and development in Candida albicans. FEMS Microbiol Lett. 2002;214:95–100. doi: 10.1111/j.1574-6968.2002.tb11330.x. [DOI] [PubMed] [Google Scholar]
- *68.Daniels KJ, Srikantha T, Lockhart SR, Pujol C, Soll DR. Opaque cells signal white cells to form biofilms in Candida albicans. Embo J. 2006;25:2240–2252. doi: 10.1038/sj.emboj.7601099. First demonstration that white cells of C. albicans respond to pheromone resulting in adherence and biofilm formation. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Park YN, Daniels KJ, Pujol C, Srikantha T, Soll DR. Candida albicans forms a specialized “sexual” as well as “pathogenic” biofilm. Eukaryot Cell. 2013;12:1120–1131. doi: 10.1128/EC.00112-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Daniels KJ, Park YN, Srikantha T, Pujol C, Soll DR. Impact of environmental conditions on the form and function of Candida albicans biofilms. Eukaryot Cell. 2013;12:1389–1402. doi: 10.1128/EC.00127-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Lin CH, Kabrawala S, Fox EP, Nobile CJ, Johnson AD, Bennett RJ. Genetic control of conventional and pheromone-stimulated biofilm formation in Candida albicans. PLoS Pathog. 2013;9:e1003305. doi: 10.1371/journal.ppat.1003305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Sahni N, Yi S, Daniels KJ, Huang G, Srikantha T, Soll DR. Tec1 mediates the pheromone response of the white phenotype of Candida albicans: insights into the evolution of new signal transduction pathways. PLoS Biol. 2010;8:e1000363. doi: 10.1371/journal.pbio.1000363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Ramirez-Zavala B, Weyler M, Gildor T, Schmauch C, Kornitzer D, Arkowitz R, Morschhauser J. Activation of the Cph1-dependent MAP kinase signaling pathway induces white-opaque switching in Candida albicans. PLoS Pathog. 2013;9:e1003696. doi: 10.1371/journal.ppat.1003696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Lockhart SR, Pujol C, Daniels KJ, Miller MG, Johnson AD, Pfaller MA, Soll DR. In Candida albicans, white-opaque switchers are homozygous for mating type. Genetics. 2002;162:737–745. doi: 10.1093/genetics/162.2.737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Odds FC, Bougnoux ME, Shaw DJ, Bain JM, Davidson AD, Diogo D, Jacobsen MD, Lecomte M, Li SY, Tavanti A, et al. Molecular phylogenetics of Candida albicans. Eukaryot Cell. 2007;6:1041–1052. doi: 10.1128/EC.00041-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Booth LN, Tuch BB, Johnson AD. Intercalation of a new tier of transcription regulation into an ancient circuit. Nature. 2010;468:959–963. doi: 10.1038/nature09560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Bougnoux ME, Pujol C, Diogo D, Bouchier C, Soll DR, d’Enfert C. Mating is rare within as well as between clades of the human pathogen Candida albicans. Fungal Genet Biol. 2008;45:221–231. doi: 10.1016/j.fgb.2007.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]