SUMMARY
We demonstrate using conditional mutagenesis that Pbx1, with and without Pbx2+/− sensitization, regulates regional identity and laminar patterning of the developing mouse neocortex in cortical progenitors (Emx1-Cre) and in newly generated neurons (Nex1-Cre). Pbx1/2 mutants have three salient molecular phenotypes of cortical regional and laminar organization: hypoplasia of the frontal cortex; ventral expansion of the dorsomedial cortex; ventral expansion of Reelin expression in the cortical plate of the frontal cortex, concomitant with an inversion of cortical layering in the rostral cortex. Molecular analyses, including PBX ChIP-Seq, provide evidence that PBX promotes frontal cortex identity by repressing genes that promote dorsocaudal fate.
INTRODUCTION
Understanding the genetic underpinnings that control development of the frontal cortex is particularly important for understanding the evolution of complex computational modules found in higher mammals, and for understanding mechanisms underlying neuropsychiatric disorders such as autism and schizophrenia. In these disorders, there is evidence for alterations in the size and function of the frontal cortex (Amaral et al., 2008; Crespo-Facorro et al., 2000; Gourion et al., 2004; Piven et al., 1995; Yamasue et al., 2004).
Regional patterning of the cerebral cortex is coordinately controlled by secreted factors such as FGF8, 15 and 17, and cell autonomously controlled by transcription factors (TF), among other mechanisms. Loss of Fgf8 and Fgf17 expression leads to preferential deletion/hypoplasia of the frontal cortex (Cholfin and Rubenstein, 2007; Fukuchi-Shimogori and Grove, 2001; Garel et al., 2003). FGF-signaling controls the gradiential expression of multiple TFs that contribute to cortical regional identity. For instance the graded expression of TFs, such as CoupTF1, Emx2, Lef1, Lhx2, Pax6 and Sp8, along the rostral/caudal (R/C) and ventral/dorsal (V/D) axes, imparts regional identities to neuroepithelial cells in the ventricular zone (VZ) (Armentano et al., 2007; Bishop et al., 2000; Borello et al., 2014; Chou et al., 2009; Faedo et al., 2008; Galceran et al., 2000; Mallamaci and Stoykova, 2006; Mangale et al., 2008; Sahara et al., 2007; Yun et al., 2001).
Regional identity is then translated to the subventricular zone (SVZ) and cortical plate (CP). Initially, the CP also exhibits gradients of TFs (i.e. CoupTF1 (PD1), Bhlhb5, Lhx2, Tbr1 and Tbr2) that are gradually converted to patterns with regional boundaries that are correlated with anatomical and functional subdivisions such as the frontal, motor, somatosensory and visual cortex; there is evidence that these TFs also regulate regional fate (Alfano et al., 2014; Bedogni et al., 2010; Elsen et al., 2013; Greig et al., 2013; Joshi et al., 2008; Zembrzycki et al., 2015). At early developmental stages thalamic afferents have little role in regional patterning (Miyashita-Lin et al., 1999; Nakagawa et al., 1999), whereas later in development thalamic afferents contribute to refining cortical areal properties (Chou et al., 2013).
Herein we demonstrate that the Pbx1 TF has a potent role in orchestrating the developmental elaboration of the mouse frontal cortex. We use a Pbx1 conditional allele (Ficara et al., 2008) that was selectively deleted in the cortical ventricular zone (VZ) using Emx1-Cre (Gorski et al., 2002) or in newly generated cortical neurons using Nex1-Cre (Goebbels et al., 2006).
Pbx1 is one of four vertebrate Pbx genes; these are members of the TALE (Three Amino acid Loop Extension) homeodomain transcription factor superfamily of atypical homeodomain-containing transcription factors, which include the invertebrate orthologues exd (Drosophila melanogaster) and ceh-20 (C. elegans) (Burglin, 1997; Capellini et al., 2011b). These proteins have a PBC domain that promotes protein-protein interactions with two other TALE subclasses: MEIS and PREP (PKNOX). PBX/EXD proteins form complexes with HOX proteins, and function upstream of Hox genes, and control patterning of the A-P body axis and the limb bud (Capellini et al., 2011b; Vitobello et al., 2011). In addition, mouse Pbx genes have critical functions in regulating spleen, craniofacial, and skeletal development (Capellini et al., 2011a; Ferretti et al., 2011; Koss et al., 2012).
Pbx1, 2 and 3 are expressed in the developing forebrain (Long et al., 2009; Toresson et al., 2000), the function of these TFs have not been elucidated in these structures. Here, we found that loss of cortical Pbx1 function alone, or in a Pbx1;Pbx2+/− sensitive background, led to hypoplasia and dyslamination of the frontal cortex through three mechanisms. In progenitors Pbx1 regulated rostrocaudal and dorsoventral patterning. Surprisingly, abnormal D/V patterning resulted in ectopic Reelin expression in the rostral cortical plate leading to abnormal laminar patterning. In immature neurons, loss of Pbx1 resulted in loss of molecular features of the rostral cortex. Gene expression analyses identified dysregulated TFs (e.g. Emx2 and Lhx2) that we propose contribute to abnormal cortical patterning through their functions in progenitors. We used PBX-ChIP-Seq to identify genomic loci where PBX proteins bind in the E12.5 and E15.5 cortex. These results yielded evidence that PBX binds near Emx2 and Lhx2 promoters. Furthermore, we identified enhancer elements that are active in the E11.5 cortex that have PBX-binding sites. Informatics approaches defined in vivo PBX binding sites, and provided evidence that these genomic elements also have signatures of combinatorial binding with other TFs.
RESULTS
Expression of Pbx RNA and protein in developing mouse cortex
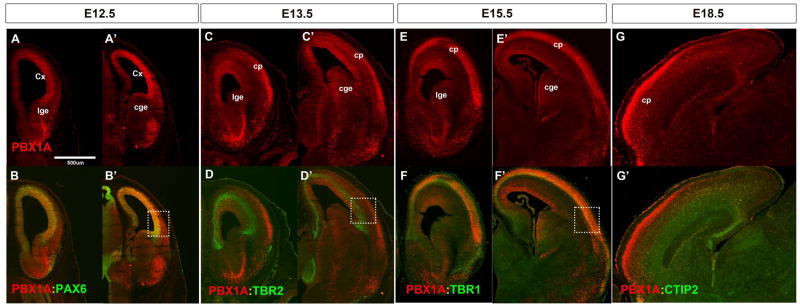
We examined Pbx1 RNA and protein expression in the developing cortex using in situ hybridization (ISH) and immunohistochemistry (IHC) with an antibody specific to PBX1A splice variant of Pbx1 (expression is lost in the Pbx1 mutant; Figure S1Q and R) (Phelan et al., 1995; Shen et al., 1996). PBX1A protein expression in the E12.5 cortical ventricular zone (VZ) showed a caudorostral gradient with low expression in the medial pallium (MP) and cortical plate. Similar results were seen using in situ RNA hybridization (Figure S1A–F; Allen Brain Developmental Atlas at E11.5).
PBX1A and PAX6 proteins were co-expressed in the cortical VZ (Figure1A–B and Figure S1J–K), whereas PBX1a was not detected in secondary progenitors and was not co-expressed with Tbr2 at E13.5 (Figure 1C–D and Figure S1L–M). By E15.5 PBX1a expression in the VZ was reduced, but was extensive in the cortical plate (CP) where it was coincident with TBR1 in deep layers (Figure 1E–F and Figure S1N–P). At E18.5 PBX1A IHC labeled superficial layers of the CP, particularly in the rostral cortex (Figure 1G). Pbx1 RNA expression closely matched protein expression (Figure S1A–F). Two other Pbx family members were expressed in the developing telencephalon. Pbx2 was broadly expressed in progenitors at E12.5, E13.5 and E15.5 except for the MP (Figure S1G–I). Pbx3 expression appeared largely restricted to the basal ganglia [(Toresson et al., 2000) and Allen Brain Atlas]
Figure 1. PBX1 protein is expressed in progenitors and neurons of the embryonic cortex.
Immunofluorescence co-staining at four prenatal ages with indicated antibodies. (A–B′) PBX1A (in red) and PAX6 (in green) co-localize in ventricular zone of the cortex at E12.5; (coronal view). (C–D′) PBX1A (in red) is expressed in ventricular zone and cortical plate at E13.5 PBX1A and TBR2 (in green) do not co-localize in subventricular zone of the cortex (coronal view). (E–F′) PBX1A (in red) and TBR1 (in green) co-localize in the cortical plate at E15.5 in (coronal view). (G, G′) PBX1A (in red) and CTIP2 (in green) at E18.5 (sagittal view). Higher magnification and quantification of the images within the white rectangles are in Supplemental Figure1. Abbreviations: cp: cortical plate; Cx: cortex; cge: caudal ganglionic eminence; lge: lateral ganglionic eminence.
Cre-mediated elimination of Pbx expression in cortical progenitors (Emx1-Cre) and young neurons (Nex-Cre)
Pbx1 null mutants die due to hematopoietic defects in mid-gestation (DiMartino et al., 2001); therefore we used Pbx1 conditional mutants (Pbx1flox allele) to analyze its function during cortical development (Ficara et al., 2008). To distinguish Pbx1’s role in progenitors versus neurons we used two different Cre lines: Emx1-Cre to delete Pbx1 (Pbx;Emx1-Cre) in the VZ beginning at E10.5-E11 (Gorski et al., 2002) and Nex-Cre to remove Pbx1 (Pbx;Nex-Cre) in postmitotic neurons (Goebbels et al., 2006). Deletion was confirmed using Pbx1 ISH (Figure S1Q–R). While the Pbx1 conditional mutants had cortical phenotypes (Figure S2H–K), we augmented the phenotype by reducing Pbx-dosage by including one Pbx2 null allele (Figure S2I–O). Previous studies showed that Pbx2+/− state exacerbated Pbx1 non-brain phenotypes (Capellini et al., 2006), even though Pbx2−/− null mice were viable and had no obvious phenotype (Selleri et al., 2004). We observed an exacerbation of Pbx1−/− cortical molecular phenotypes in a Pbx2+/− background, and therefore performed most of our analyses on the sensitized Pbx2+/− background. We did not observe a patterning phenotype in the Pbx2+/− mice (Figure S2F–G), and thus used Pbx2+/− as the control genotype. Our preliminary analysis suggests that the Pbx2−/− cortex is hypoplastic (not shown).
Pbx;Emx1-Cre mutants were viable and survived into adulthood. The P7 Pbx; Emx1-Cre brain appeared grossly normal; histological analysis showed hypoplasia of telencephalic commissures, mild thinning of the caudal cortex and dyslamination in hippocampal CA fields (Figure S1T–Z). Herein, we focused on the phenotype of the prenatal cortex.
Pbx1 regulates rostrocaudal patterning in both progenitors and in postmitotic neurons, but regulates DV patterning primarily in progenitors
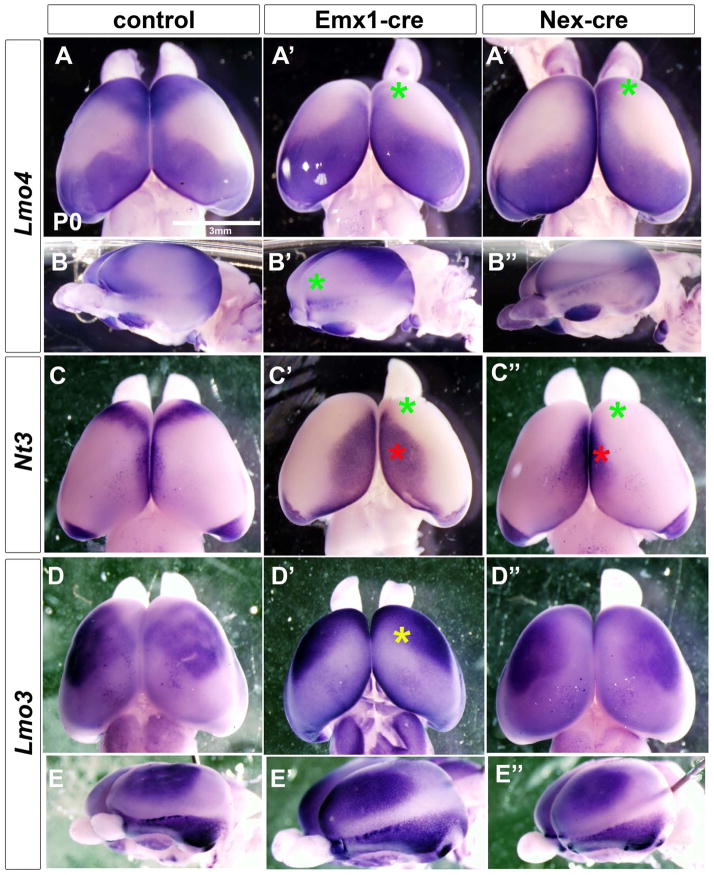
Given PBX1 expression in the cortical VZ, and its function in patterning of other tissues (Capellini et al., 2006; Selleri et al., 2001; Vitobello et al., 2011) we hypothesized that Pbx1 may regulate cortical regionalization. Thus, we performed whole mount ISH (WM-ISH) on P0 brains using Lmo4, Nt3 and Lmo3 probes (Cholfin and Rubenstein, 2007, 2008). In control animals, Lmo4 labels rostral (frontal) and caudal (visual) areas. Pbx;Emx1-Cre mutants lacked the Lmo4+ frontal domain, and there was a rostral shift of the caudal Lmo4 domain (Figure 2A–A′ and B-B′), providing evidence Pbx1 regulates rostrocaudal (RC) cortical patterning.
Figure 2.
Pbx mutants have major alterations in cortical regional patterning including loss of molecular features of the frontal cortex. Analysis of the cortical patterning by whole mount in situ hybridization on P0 brains of control (left column) Pbx;Emx1-cre mutants (middle column) and Pbx;Nex-cre mutants (right column). Lmo4 probe: superior view (A–A″); lateral view (B–B″). Nt3 probe: superior view (C–C″). Lmo3 probe: superior view (D–D″); lateral view (E–E″). Green asterisks represent reduction of frontal cortex expression compared to control. Red asterisks represent expansion of dorsomedial cortical expression as compared to control. Yellow asterisk represents a change in frontal cortex expression pattern as compared to control.
To further assess the rostral phenotype, and to examine dosoventral (DV) patterning in the mutant, we studied Nt3 (Ntf3) expression. At P0, in addition to labeling part of the frontal cortex, Nt3 was expressed dorsally in the cingulate-retrosplenial cortex. Similar to the Lmo4 phenotype, the Pbx;Emx1-cre mutant lacked the frontal Nt3 domain and importantly, the dorsal domain expanded ventrally and rostrally (Figure 2C–C′). Finally, we examined Lmo3 expression (a marker of the somatosensory cortex). The mutants showed a rostroventral expansion of this domain (Figure 2D–D′ and E-E′). Thus, Pbx1 was required to promote rostral and repress dorsal gene expression properties in the developing cortex.
Because Pbx1 is expressed in both the cortical progenitors and neurons, we tested whether loss of Pbx1 expression in postmitotic neurons regulated cortical patterning by studying Pbx;Nex-cre mutants at P0 using WM-ISH. These mutants lost frontal cortex expression of Lmo4 and Nt3 (Figure 2A″ and C″). However, the DV patterning changes of Lmo4 and Nt3 expression in Pbx;Nex-cre mutants were much milder than in the Pbx;Emx1-cre mutants (Figure 2).
We confirmed that the P0 WM-ISH expression changes led to the expected deletion of frontal cortex and expansion of dorsal and caudal cortex by performing ISH on P8 coronal sections of control, Pbx;Emx1-cre and Pbx;Nex-cre brains (Figure S2P–GG″). For instance in the frontal cortex, both mutants lost Nt3 expression, had reduced Cux2 and Er81 expression, and the Pbx;Emx1-cre had greatly reduced Lmo4 expression. The Pbx;Emx1-cre mutant also had ventral expansion of Er81, Nt3 and Nurr1. In all, the data provide evidence that Pbx1 regulates rostrocaudal patterning in both progenitors and in postmitotic neurons, but regulates DV patterning primarily in progenitors.
Abnormal D/V patterning in the Pbx;Emx1-cre mutant leads to ectopic Reelin expression in the rostrodorsal cortex leading to dyslamination
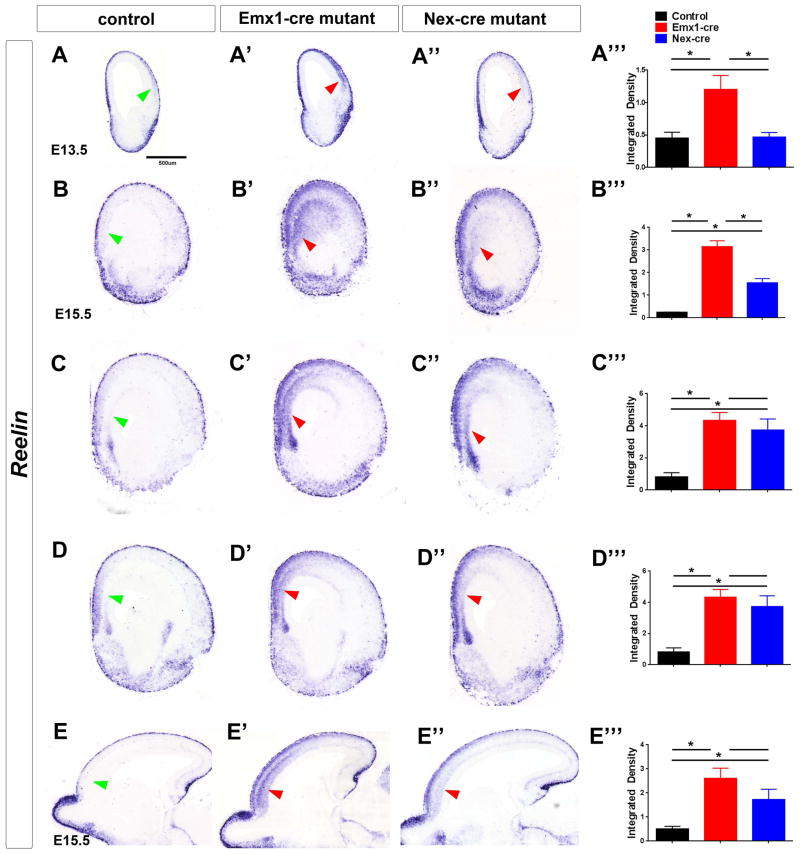
As noted above, loss of Pbx1 function led to ventral expansion of dorsal cortical properties (Nt3 and Lmo3). In the most rostral regions at E13.5 and E15.5, Reelin is expressed in a small domain adjacent to the septum, which is probably the indusium griseum (Figure 3).
Figure 3.
Pbx mutants have ectopic Reelin expression in the rostral cortical plate. Reelin RNA expression analysis by in situ hybridization in control (left column), Pbx;Emx1-cre mutants (middle column) and Pbx;Nex-cre mutants (right column). (A–D) Reelin expression at E13.5 and E15.5 in coronal sections. (E) Reelin expression at E15.5 on sagittal sections. Red arrows point to the increased Reelin expression in the mutant’s rostral cortical plate as compared to the control (green arrows). (A‴, B‴, C‴, D‴, E‴) Reelin in situ signal intensity (Integrated Density) around the regions indicated by arrows, was quantified and analyzed using ImageJ as described (McCloy et al., 2014). At least three brain sections were used for each measurement. * Indicates p<0.05(mean+/−SD)
In E13.5 Pbx;Emx1-cre mutants this Reelin+ domain broadly expanded ventrally in the cortical plate (Figure 3A). By E15.5, a Reelin+ deep layer in the cortical plate extended ventrally from the most dorsal position through roughly half of the cortex, but only in the rostral cortex, as seen in both coronal and sagittal views (Figure 3BE). These Reelin+ cells did not co-express Calretinin, and thus are probably not Cajal Retzius neurons (Figure S3A–C).
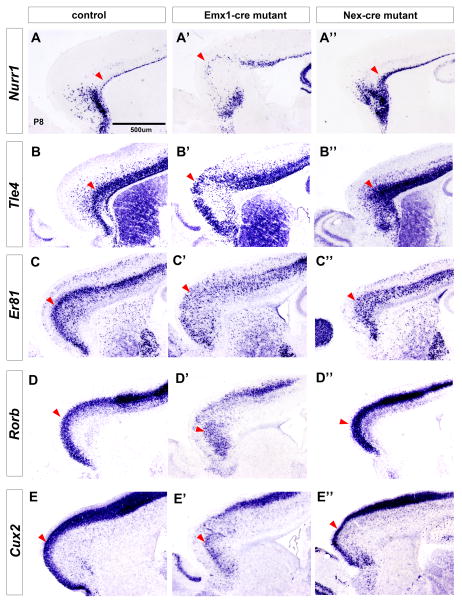
As Reelin regulates laminar positioning of cortical projection neurons (Ogawa et al., 1995), we assessed the expression of molecular markers of the subplate (Nurr1), layer 6 (Tle4), 5 (ER81, Etv1), 4 (RORb) and 2/3 (Cux2) at P8 (Hoerder-Suabedissen et al., 2009; Molyneaux et al., 2007; Nieto et al., 2004; Schaeren-Wiemers et al., 1997). Consistent with the ectopic Reelin expression in the deep cortical plate, we observed an inversion of the cortical layers in the rostral cortex (i.e. the region with the ectopic Reelin) (Figure 4). Particularly note the inverted expression of Nurr1 and Tle4 in the superficial layers (compare Figure 4 A and A′, B and B′), as well as Cux2 and RORb inverted expression in the deep layers (compare Figure 4 D and D′, E-E′). BrdU birthdating analyses support the evidence for inverted lamination in the rostral cortex (Figure S4).
Figure 4.
Pbx mutants have inversion of cortical layers in the rostral cortex. Laminar marker expression in Pbx;Emx1-cre and Pbx;Nex-cre mutants at P8 by in situ hybridization on control (left column), Pbx;Emx1-cre mutants (middle column) and Pbx;Nex-cre mutants (right column). (A–A″) Nurr1, marker of subplate; (B–B″) Tle4, marker of layer VI; (C–C″) Er81, marker of layer V; (D–D″) Rorb, marker of layer IV; (E–E″) Cux2, marker of layers II–III. Red arrowheads point to the superficial boundary of the corresponding layers, showing the laminar inversions in the mutant.
Pbx;Nex-cre mutants did not have the abnormal lamination phenotype (compare Figure 4 A–E and A″-E″). Thus, loss of Pbx function in progenitors, which lead to abnormal D/V patterning in the Pbx;Emx1-cre mutants, caused the ventral spread of Reelin expression into rostral deep cortical layers, with a subsequent inversion of cortical layers of the rostral cortex.
Molecular mechanisms underlying the D/V patterning defects in Pbx mutant cortical progenitors: altered TF expression (Dbx1, Dmrta1, Emx2, Lhx2 and Prep1) and increased SMAD1/5 phosphorylation
We next searched for the mechanisms through which Pbx1 regulates patterning in cortical progenitors. FGF signaling regulates arealization and size of the frontal cortex as exemplified by frontal cortex hypoplasia in Fgf8 hypomorphs and Fgf17 null mice (Cholfin and Rubenstein, 2007, 2008). Therefore we examined the genetic interactions between FGF signaling and Pbx1 function. First, we found that Pbx1 expression appeared normal in Fgf8neo/neo hypomorphs implying that Pbx1 was not strongly regulated by FGF-signaling (Figure S5J). Then we examined expression of FGF-responsive genes (Erm, Pea3 and Sp8) in the VZ of the rostral cortex in E13.5 Pbx;Emx1-cre mutants. We detected no change in their expression suggesting the Pbx1 does not promote rostral identity through promoting FGF-signaling (Figure S5A–C).
Like the Pbx1 mutant, loss of Pax6 function causes RC and DV patterning defects (Stoykova et al., 2000; Yun et al., 2001). However, Pbx1 expression was not altered in Pax6sey/sey mutants at E11.5 and E12.5 (Figure S5K). Furthermore, Pax6 expression was not altered in E13.5 Pbx;Emx1-cre mutants (Figure S2E). Together, these data suggest that Pbx1 exerts its rostral patterning function independently of FGF-signaling or Pax6.
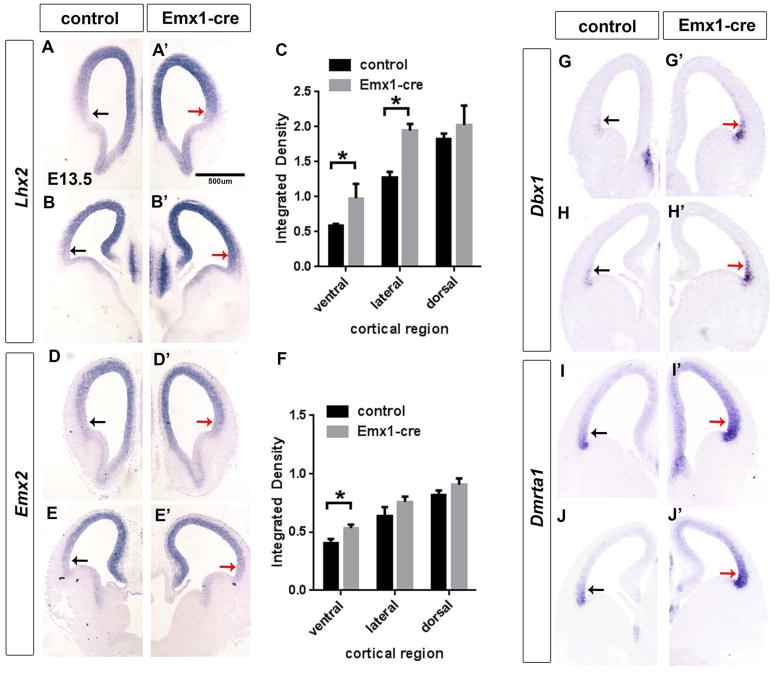
We next turned our attention to Pbx1’s repression of dorsal properties, as up-regulation of this system may alter frontal cortex development. We studied the expression of TFs that control cortical DV patterning: CoupTF1, Emx2, Lhx2, and Lmx1a by ISH at E13.5 in Pbx;Emx1-cre mice. CoupTF1 and Lmx1a had no clear expression changes (Figure S5D and H).
On the other hand, Emx2 and Lhx2 expression were increased, particularly in the ventral cortical VZ (red arrows) (Figure 5 A–D). Lhx2 expression increased about 2-fold in mutant’s ventral cortex and about 1.6 fold in the lateral cortex (Figure 5C). Emx2 expression increased about 1.5-fold in the mutant’s ventral cortex (Figure 5F). Both Emx2 and Lhx2 are critical in specifying cortical identities (Cholfin and Rubenstein, 2008; Chou et al., 2009; Mallamaci et al., 2000; Monuki et al., 2001; Muzio and Mallamaci, 2003). Thus, up-regulation of Emx2 and Lhx2 could contribute to DV and RC patterning shifts in Pbx1 mutants. Lhx2 expression did not change in Nex-cre mutants (Figure S5S–T).
Figure 5.
Pbx;Emx1-cre mutants at E13.5 have changes in the VZ expression of TFs that control cortical patterning shown on coronal sections using in situ hybridization. (A–B′) Lhx2 expression; (D–E′) Emx2 expression; (G–H) Dbx1; (I–J′) Dmrta1. Black arrows indicate normal expression in the ventro-lateral cortex; red arrows point of the increase in expression of patterning genes in the mutant’s ventro-lateral cortex. (C) and (F) Quantification of Lhx2 ( C) and Emx2 ( F) in situ signal in Emx1-cre mutant vs control cortex. Measurements in 4 different brain sections were made in the ventral, lateral and dorsal regions of the cortex. Integrated Density was calculated as described previously (McCloy et al., 2014). * Indicates p<0.05 (mean+/−SD)
Next, to obtain unbiased information on Pbx1-regulated genes, we compared RNA expression (using gene expression array analysis) in the cortex from E12.5 and E15.5 control and Pbx;Emx1-cre brains. RNA expression changes were not strong at E12.5 (data not shown), whereas at E15.5 the Pbx1 mutant had robust changes in RNA levels for several genes (Table 1).
Table 1.
Results of gene expression array analysis of the E15.5 Pbx;Emx1-cre cortex, showing down-regulated and up-regulated genes. A red star indicates which genes have PBX ChIP-Seq peaks in their promoter and/or intragenic regions.
| Genes changed in Pbx1 mutants | |||
|---|---|---|---|
| Symbol | Name | Fold change | FDR |
| Downregulated | |||
| Smoc1* | SPARC related modular calcium binding 1 | 3.8 | 9.9E-05 |
| Rai14* | retinoic acid induced 14 | 2.5 | 1.2E-03 |
| Rbp1 | retinol binding protein 1, cellular | 2.4 | 4.5E-02 |
| Flrt3* | fibronectin leucine rich transmembrane protein 3 | 2.3 | 1.8E-02 |
| Ccbe1* | collagen and calcium binding EGF domains 1 | 2.3 | 3.6E-02 |
| Pdzrn3* | PDZ domain containing RING finger 3 | 2.2 | 1.1E-03 |
| Bmpr1b* | bone morphogenetic protein receptor, type 1B | 2.0 | 1.7E-02 |
| Cxcl12 | chemokine (C-X-C motif) ligand 12 | 2.0 | 5.5E-03 |
| Cux2* | cut-like homeobox 2 | 2.0 | 3.8E-02 |
| Fzd8 | frizzled homolog 8 | 1.8 | 3.5E-03 |
| Figf | c-fos induced growth factor | 1.8 | 2.7E-02 |
| Plxna4* | plexin A4 | 1.6 | 2.4E-02 |
| Ngfr | nerve growth factor receptor | 1.6 | 4.5E-02 |
| Rnd2 | Rho family GTPase 2 | 1.4 | 3.8E-02 |
| Sema6d | semaphorin 6D | 1.4 | 2.3E-02 |
| Upregulated | |||
| Dbx1 | developing brain homeobox 1 | 4.6 | 9.0E-03 |
| Pknox1* | Pbx/knotted 1 homeobox (Prep1) | 3.0 | 9.9E-05 |
| Npr3 | natriuretic peptide receptor 3 | 2.8 | 1.4E-02 |
| Pde1a* | phosphodiesterase 1A, calmodulin-dependent doublesex and mab-3 related transcription factor like | 2.3 | 2.7E-02 |
| Dmrta1* | family A1 | 2.3 | 3.3E-02 |
| Fzd7 | frizzled homolog 7 | 2.0 | 3.6E-02 |
| Lmo3* | LIM domain only 3 | 1.9 | 3.1E-03 |
| Lmo4* | LIM domain only 4 | 1.7 | 3.8E-02 |
| Nr4a2 (Nurr1) | nuclear receptor subfamily 4, group A, member 2 | 1.7 | 2.3E-02 |
| Ngef | neuronal guanine nucleotide exchange factor | 1.6 | 2.7E-02 |
indicates genes that contain promoter or intragenic Pbx ChIP-seq peaks
We focused on TFs with altered expression levels (Dbx1, Dmrta1 and Pknox1) by performing ISH analysis. All three TFs were over-expressed in the cortical VZ (Figures 5G–J and 6F′). The Pknox1 (also known as PREP1) is a co-factor of PBX1 (Berthelsen et al., 1998a; Berthelsen et al., 1998b; Berthelsen et al., 1998c). We performed over-expression experiments, in which Pknox1 was electroporated in utero at E12.5. However, this did not change Lmo4 and NT3 P0 WM-ISH expression (data not shown), suggesting that the increased Pknox1 did not contribute substantively to the Pbx1 mutant phenotype, but rather may reflect compensatory up-regulation.
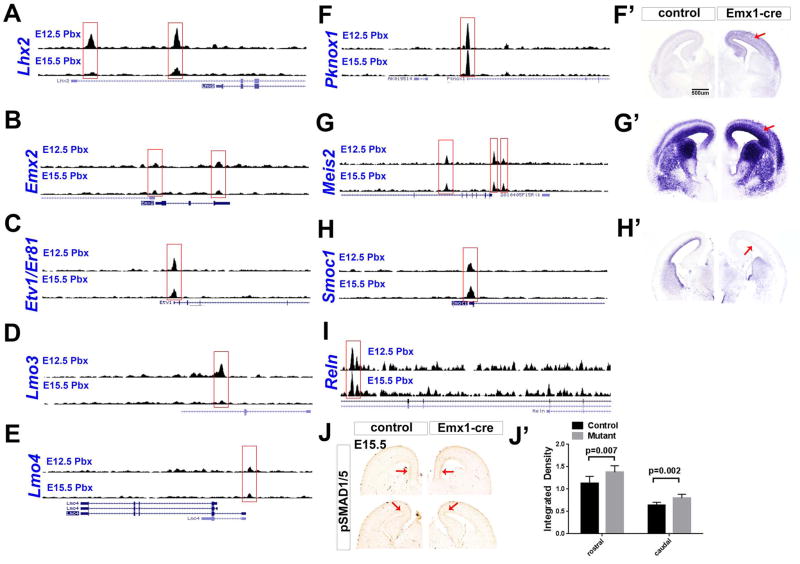
Figure 6.
ChIP-Seq showing PBX binding to promoters, and other gene regions, that are dysregulated in Pbx mutants. Genome browser views showing PBX ChIP-Seq peaks at E12.5 and E15.5. Gene expression changes are shown in Figures 2, 4 and 5, except for Pknox1, Meis2 and Smoc1, which are shown here in panels F–H′. (J,J′) Pbx;Emx1-cre mutants have expanded domain of pSMAD1/5 expression in the dorsal cortex (red arrows) at E15.5. Signal intensity was quantified and expressed as Integrated Density in J′ (mean+/−SD).
Dbx1 and Dmrta1 expression were increased in the Pbx;Emx1-cre cortex; their expression domains expanded dorsally (Figure 5G–J). Dbx1 regulates DV patterning of the spinal cord (Pierani et al., 2001). Dmrta1 loss of function analysis in the cortex has not been reported; however its expression is increased by Pax6, implying that Dmrta1 may promote ventral fate (Kikkawa et al., 2013); Dmrta1’s closely-related family member Dmrta2 regulates cortical dorsoventral pattering (Konno et al., 2012). Thus, we propose that Pbx1 regulates DV patterning, at least in part, by repressing TFs (Dbx1, Dmrta1, Emx2 and Lhx2) that are expressed in VZ cortical progenitors (in either DV or VD gradients).
In addition to molecular defects in the cortical VZ, Pbx;Emx1-cre mutants had dysregulation in the cortical SVZ. There was reduced expression of Svet1 (~2 fold) and Cxcl12 (~2 fold) (Figure S5M–N). The SVZ (but, not the VZ) had a ~40% reduction of M-phase (PH3+) cells at E12.5 in Pbx;Emx1-Cre mutants (and not Nex-cre mutants) (Figure S5L and P-Q). This could account for the reduction in the thickness of the superficial cortical layers. Furthermore, consistent with reduced CXCl12 expression, a known attractant for interneurons (Li et al., 2008; Wang et al., 2011), there were fewer Dlx1+ and Lhx6+ cells in the E15.5 Pbx;Emx1-cre cortex (not shown).
Pbx repressed expression of Cav1 (Figure S5O). Cav1 is normally expressed at low levels in the dorsal-most cortex at E13.5; in the mutant it is dramatically up-regulated throughout the VZ. Cav1 encodes a structural component of caveolae that plays an important role in integrating multiple signaling pathways.
Pbx was essential for the expression of Smoc1 in the ventricular zone of the cortex (Figure 6H′); ChIP-Seq supports it as a PBX-target (see below, Figure 6H). Smoc1 is an extracellular matrix protein that acts as BMP antagonist in early embryogenesis (Thomas et al., 2009). As such, we tested whether BMP signaling may be abnormal in the Pbx;Emx1-cre mutants by measuring SMAD phosphorylation using immunohistochemistry.
Phosphorylation of SMAD increases its ability to signal (Goumans and Mummery, 2000; Kitisin et al., 2007), and regulates dorsoventral patterning of the neural tube (Fernandes et al., 2007). pSMAD1/5 is normally detected in the ventricular zone of the medial pallium with a dorsal to ventral gradient at E15.5. Pbx;Emx1-cre mutants had increased pSMAD1/5, and a ventral spread along ventricular zone (Figure 6J–J′). This increase was detected throughout rostrocaudal extent of the cortex. pSMAD2 levels did not change (data not shown). pSMAD staining did not change in Nex-cre mutants (Figure S5V). Nor did we observe a change in Bmp4 expression at E13.5 (Figure S5I).
WNT-signaling is required for the most-dorsal cortical regions (Lee et al., 2000; Zhou et al., 2006), and participates in neocortical patterning (Caronia-Brown et al., 2014) We assessed expression of Axin2 and Wnt3, transcriptional readouts of WNT-signaling. At E13.5 their expression appeared normal in Pbx;Emx1-cre mutants (Figure S5F–G), providing evidence that Pbx does not mediate cortical patterning through modulating WNT-signaling. Thus, abnormal regional patterning of the Pbx;Emx1-cre cortex appears to be due to the alterations in the gradients of TF expression (Dbx1, Dmrta1, Emx2, Lhx2) and increased SMAD1/5 signaling. We next assessed which of these phenotypes was directly due to Pbx1 chromosomal binding.
PBX ChIP-Seq from embryonic cortex identifies target genes
To determine which of the gene expression changes in the VZ may be directly PBX regulated, we performed ChIP-Seq from E12.5 wild type cortex using pan-PBX antibody. Towards identifying direct PBX targets in progenitors and postmitotic cells, we performed ChIP-Seq from E15.5 cortex. As a specificity control, we added a PBX1 peptide to the chromatin immunoprecipitations to block antibody binding. About ~4100 peaks were identified at E12.5 and ~7600 peaks at E15.5. About ~2500 peaks were the same between e12.5 and E15.5. GREAT analysis [(http://bejerano.stanford.edu/great/public/html/);(McLean et al., 2010)] showed the distribution of PBX binding sites as a function of their distance from the transcription start site; ~35% were near the promoter (+/− 5 kb), whereas the majority (65%) mapped at more distant locations at both E12.5 and E15.5 (Figure S6A–B).
We compared our E12.5 ChIP-Seq peaks with 900 enhancers (Visel et al., 2013) that have reproducible tissue-specific enhancer activity in transgenic assays (Vista enhancer browser, http://enhancer.lbl.gov/). We found that about 30% of these 900 enhancers contained PBX peaks. Of enhancers that contained PBX peaks, about 40% had forebrain expression in the transgenic assay. Examples of four such enhancers are depicted in Supplemental Figure 6C. These data support that PBX is frequently associated with bona fide distant-acting in vivo enhancers in general and with enhancers active in the developing forebrain in particular.
Additionally, we compared p300 ChIP-Seq dataset from E11.5 forebrain (Visel et al., 2009) with our E12.5 ChIP-Seq to evaluate how many of PBX-enriched regions also map to p300-bound enhancers. Out of 2,453 p300 bound forebrain enhancers, 651 (26%) also contained Pbx ChIP-Seq peaks.
Lhx2 and Emx2 were up-regulated in the ventral cortex of Pbx mutants (Figure 5). The promoter regions of these genes had PBX ChIP-Seq peaks (Figure 6A and B). At E12.5 there were two prominent PBX ChIP-Seq peaks at Lhx2 promoters. PBX peaks were present in the same Lhx2 genomic locations at E15.5 cortex, although they were not as pronounced. The Emx2 locus contained two PBX peaks, one at the 5′ end, and the other at the 3′ end of the gene (Figure 6B).
PBX had ChIP-Seq peaks over the proximal promoters of Pknox1 and Meis2 genes (at E12.5 and E15.5) (Figure 6F–G). Pknox1 and Meis2 are members of the TALE homeodomain protein family that cooperatively bind with PBX proteins to promoters of target genes (Bjerke et al., 2011). In Pbx mutants, Pknox1 expression is strongly increased throughout the cortex (Figure 6F′). Expression of Meis2 is also strongly increased in ventricular zone as well as the cortical plate in Pbx mutants (Figure 6G′).
PBX binding sites were also found in proximity of TFs that are preferentially expressed in specific cortical layers and regions. PBX ChIP-Seq mapped to the start sites of Etv1 (ER81) (E12 and E15), Lmo3 (E12) and Lmo4, expression of these three genes was dysregulated in Pbx mutant (Table 1; Figure 4, and not shown).
While our analysis found many other interesting genes that are probable PBX targets, we wish to highlight Reelin. As shown in Figure 3, Reelin RNA expression was upregulated in the Pbx;Emx1-cre mutants (Figure 3). The Reelin locus contains 2 intragenic PBX peaks at E15 and 1 peak at E12 (which is in the same location as one of the E15 peaks) (Figure 6I).
Thus, there is good evidence that PBX binds to regulatory regions of genes whose abnormal expression is implicated in the regional and laminar phenotypes of Pbx mutants.
Nucleotide motifs in genomic loci bound by PBX
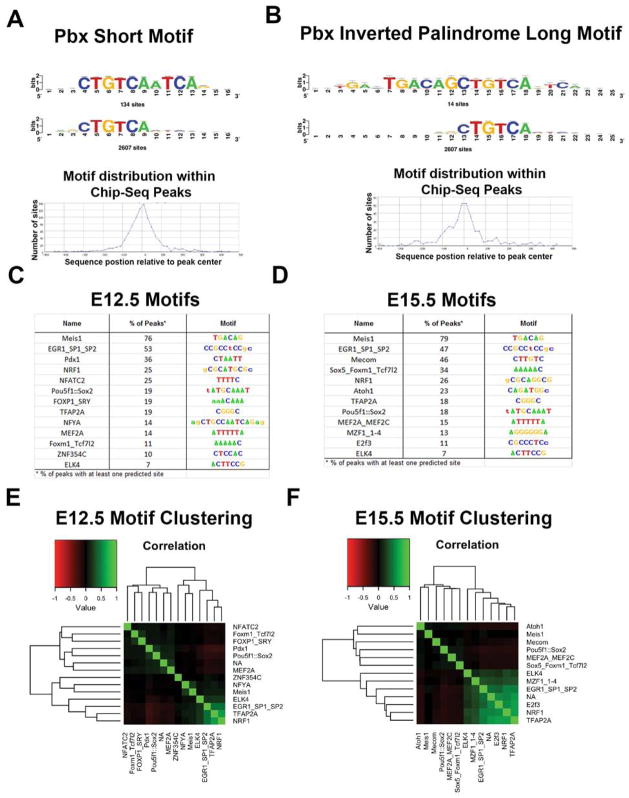
Next we performed computational analyses to identify PBX in vivo binding sequences, and to provide evidence for TFs that interact with the PBX-regulated genomic elements. We identified nucleotide sequences that were over-represented in the PBX-ChIP-Seq peaks using the RSAT peak-motifs tool (Thomas-Chollier et al., 2010) and clustered motifs by motif similarity and co-occurrence within ChIP-seq peaks using pvclust (Suzuki and Shimodaira, 2006). Representative motif logos and frequencies for major identified motifs are shown in Figure 7 and Figure S7A–C. The most common motifs mapped to the MEIS1 motif in the JASPAR database, as well as PBX1 in JASPAR and in the cisPB database. The MEIS1/PBX1-annotated motif family included a long inverted palindromic motif and a short motif that represents half the palindrome (Figure 7A–B). 77% of the identified PBX peaks at E12.5 and 79% of peaks at E15.5 contained at least one predicted motif and these motifs showed strong enrichment at the center of PBX peaks, indicating that this is likely the primary binding motif recognized by PBX. The inverted palindrome motif and half-site have been identified previously using site-selection experiments with MEIS1 (Shen et al., 1997).
Figure 7.
(A,B) PBX short (A) and long (B) motifs identified from the ChIP–seq data. The long motif is an inverted palidrome. The sequence of both the short and long motifs map to the center of the PBX ChIP-Seq peaks. (C, D) Motifs identified in PBX ChIP-Seq peaks at E12.5 (C) and E15.5 (D); their frequency is noted. The MEIS motif is the same as the PBX motif. (E, F): Identification of TF motifs, other than PBX/MEIS at E12.5 (E) and E15.5 (F).
The most common secondary motif at both time-points was a degenerate motif mapping to the SP/EGR families (53% of E12.5 peaks and 47% of E15.5 peaks) (Figure7C–D). At E12.5, the second most frequent secondary motif was a strong PDX1 motif (36% of peaks). At E15.5, the second most frequent secondary motif was MECOM (46%), also correlated with the NFATC motif identified at E12.5. The co-occurrence of PBX1 and PDX1 has been previously reported (Swift et al., 1998).
We tested for differences in the distance to the nearest gene for identified motifs, revealing sets of motifs that are preferentially located proximal or distal to TSSs. There was no difference for MEIS1 along with NFYA (highly correlated with MEIS1/PBX1 binding motifs and also annotated to PBX1 in cisBP), FOXM1/TCF7L2, and ZNF354C. There was significant bias (t-test, p-value < 0.05) towards proximal PBX peaks for SP/EGR, NRF1, ELK4, MZF1_1-4, and TFAP2A motifs and bias towards distal PBX peaks for PDX1, POU5f1::SOX2, NFATC2/MECOM, FOXP1/SRY, MEF2A, ATOH1, and SOX5. Clustering of motif co-occurrence captures the strong enrichment for TSS-proximal motifs, with little minimal structure observed for motifs preferentially found distal to the TSS at both time points (Figure 7E–F, Figure S7D–E).
Finally we tested for motifs that were enriched at one time point but not the other using peak-motifs with the background as the second time point. At E12.5, PDX1/NOBOX motifs are strongly enriched compared to peaks identified at E15.5, with 64% of E12.5 peaks annotated with this site. SP/EGR family motifs are also enriched relative to E15.5. At E15.5, we identified ATOH1, NFIC/HAND1::TCFE2a, and FOXD3, all three of which were preferentially located distal to TSSs. ATOH motifs were found in nearly 70% of E15.5 PBX peaks when analyzed with E12.5 as the background. The increased frequency of occurrence for the NOBOX/PBX1 at E12.5 and ATOH1 at E15.5 is at least in part driven by these motifs clustering with MEIS1 sites in the original analysis using peak-motifs default background.
DISCUSSION
Herein, we demonstrate using conditional mutagenesis that Pbx1 regulates regional identity and laminar patterning of the developing mouse neocortex in cortical progenitors (using Emx1-Cre) and in newly generated neurons (using Nex1-Cre). Because Pbx1 and Pbx2 have similar RNA expression patterns at E11.5 (Figure S1), and because they are known to share functions (Capellini et al., 2006), we amplified the cortical phenotype by eliminating one Pbx2 allele. Note that analyses of Pbx1 mutants with normal Pbx2 dosage were qualitatively the same as the compound mutant (Figure S1). Furthermore, cortical patterning appeared normal in Pbx2+/− (Figure S2).
We found three salient molecular phenotypes of cortical regional and laminar organization: 1) hypoplasia of the frontal cortex in both Pbx;Emx1-Cre and Pbx;Nex1-Cre (Figures 2, S2); 2) ventral expansion of the dorsomedial cortex in Pbx;Emx1-Cre (Figure 2); 3) robust ventral expansion of Reelin expression in the cortical plate of the frontal cortex, concomitant with a partial inversion of cortical layering in Pbx;Emx1-Cre (Figures 3, 4). The latter is a novel phenotype, in which abnormal cortical patterning is coupled with region-specific abnormal laminar patterning. Below we address mechanistic underpinnings of these phenotypes.
Pbx function in cortical progenitors regulates DV patterning by repressing Lhx2 and Emx2 expression
We propose the Pbx regulates cortical regional fate in the cortical VZ in part by repressing expression of TFs that control cortical dorsoventral patterning. Pbx;Emx1-cre mutants have ventral expansion of high Emx2 and Lhx2 expression. The mutants also exhibit dorsal expansion of Dbx1 and Dmrta1 (from the ventral-most cortex) (Figure 5). Concomitant with these changes in the VZ are a dorsal-to-ventral expansion of molecular properties in the cortical plate that is particularly striking for NT3 (Figures 2, S2). There is no clear ventral-to-dorsal expansion of cortical plate properties. We propose that the loss of frontal cortex properties (loss of Lmo4, NT3; gain of Lmo3) is in part due to the rostroventral shift of caudodorsal properties (e.g. Lmo3) (Figure 2).
The changes in expression of Lhx2 and Emx2 could contribute to the Pbx mutant’s patterning phenotype, as each of these TFs has demonstrated functions in cortical patterning. Lhx2 promotes neocortical fate by repressing properties of flanking structures. Lhx2 null mutants illustrate that Lhx2 dorsally represses choroid plexus identity, and that ventrally Lhx2 represses properties of the ventral pallium (also known as antihem) (Bulchand et al., 2001; Mangale et al., 2008; Monuki et al., 2001). In Lhx2;Emx1-cre mutants, lateroventral cortex acquires neocortical fate (Chou et al., 2009). Thus, like Lhx2, Pbx controls the balance of cortical fates along the dorsoventral axis. In Pbx mutants the Lhx2 gradient is changed, where there is up-regulation in the lateral/ventral regions of the cortical VZ, and in the cortical plate. Dorsal properties are expanded (e.g. NT3+ cingulate/retrosplenial) at the expense of more ventral properties (Lmo3+ somatosensory). Thus, we propose that Pbx maintains the correct level of Lhx2 expression, which is crucial in regulating the balance between different cortical regions.
Emx2 over-expression is also likely to contribute to the Pbx mutant phenotype. A ~2 fold increase in Emx2 expression in the VZ repressed rostroventral fate, and led to expansion of caudodorsal cortical areas (Hamasaki et al., 2004). As noted above, this phenotype is similar to that of the Pbx mutant. ChIP-Seq analysis identified two PBX peaks just 5′ of the transcribed region of Lhx2, and two PBX peaks within Emx2’s transcribed domain (Figure 6). Thus, PBX may directly control Lhx2 and Emx2 transcription.
Molecular mechanisms underlying the D/V patterning defects in Pbx mutant cortical progenitors: altered TF expression (Dbx1, Dmrta1, Emx2, Lhx2 and Pknox1) and increased SMAD1/5 phosphorylation
It is likely that additional mechanisms contribute to Pbx1’s control of cortical region fate, in addition to altered Emx2 and Lhx2 expression. Prominent dorsal expansion of Dbx1 and Dmrta1 expression (Figure 5) merits further consideration, but will require better understanding of the functions of these TFs during cortical development.
PBX proteins function in part through forming complexes with other TALE homeodomains, such as the PKNOX (PREP) and MEIS proteins (Bjerke et al., 2011). Pbx;Emx1-Cre mutants have striking over-expression of Pknox1 (Prep1) and Meis2; both genes have PBX ChIP-Seq peaks (Figure 6). It is possible that the increased Pknox1 (Prep1) and Meis2 expression was a compensatory mechanism, or that the increase intensifies the phenotype. Future analyses of Pknox1 (Prep1) and Meis2 mutants, alone or in combination with Pbx1, is needed to elucidate their respective functions.
As noted above, Pbx;Emx1 mutants had a prominent ventral expansion of dorsal properties in the VZ (Emx2 and Lhx2; Figure 5), and in the cortical plate (NT3, Figure 2). These mutants also showed increased levels of activated (phosphorylated) pSMAD1/5 TFs (Figure 6J; J′). BMP signaling activates SMAD1/5 (Itoh et al., 2000; Kitisin et al., 2007). In the forebrain, BMP signaling is known to specify the choroid plexus, most dorsal fate in the telencephalon (Fernandes et al., 2007). Thus it is possible that increased pSMAD1/5 participates in the expansion of dorsal cortical properties in the Pbx;Emx1 mutant that in conjunction with the increased Emx2 and Lhx2 expression, contributes to the loss of the frontal cortex. We are uncertain about the mechanisms for increased pSMAD1/5, but speculate that loss of Smoc1 expression (Figure 6H, H′) may contribute to this. In Xenopus early embryogenesis, there is evidence that Smoc1 acts as a BMP antagonist (Thomas et al., 2009).
Pbx function in newly born cortical neurons regulates cortical patterning
While regional specification of cortical domains is initiated in neuroepithelial stem cells (VZ) by processes that control gene expression, there is evidence, based on loss of CoupTFI, Bhlhb5 and Lhx2 functions (Alfano et al., 2014; Joshi et al., 2008; Zembrzycki et al., 2015), that immature cortical plate neurons maintain plasticity regarding cortical regional/areal identity. Here we found that eliminating Pbx function in newly generated cortical neurons, using Nex-Cre, degraded regional cortical molecular properties, particularly in the frontal cortex, which showed greatly reduced Lmo4 and NT3 expression (Figure 2).
ChIP-Seq analysis identified PBX peaks in the Lmo3 and Lmo4 loci, providing evidence that Pbx expression in postmotitc neurons (Figure 1) directly regulates these markers of cortical areas. Furthermore, Lmo4 function is required in rostral cortical neurons to control the identity of its projection neurons (Cederquist et al., 2013). Thus, our data are consistent with the model that Pbx expression in cortical progenitors controls region fate through repression of Emx2 and Lhx2, and Pbx expression in cortical neurons controls their identity through promoting Lmo4 expression.
Pbx represses Reelin expression in rostral cortical plate neurons – Evidence that ectopic Reelin expression in Pbx mutants leads to dyslamination in the rostral cortex
Reelin regulates radial migration of immature cortical projection neurons, and orchestrates cortical “inside-out” laminar organization (Ogawa et al., 1995). Here, we found that loss of Pbx in cortical progenitors leads to ectopic Reelin expression, that expands ventrally from indusium griseum (where it is normally present), into the rostral neocortex, particularly in early born layers (Figure 3). We suggest that abnormal dorsoventral patterning in the cortical VZ accounts for much of this ectopic expression. On the other hand, loss of Pbx in newly generated neurons (Nex-Cre) leads to a subtle increase in Reelin over-expression (Figure 3), suggesting that Pbx regulates Reelin in both the VZ and in cortical neurons. Consistent with this, there is a prominent PBX ChIP-Seq intragenic peak in Reelin in both the E12.5 and E15.5 analyses (Figure 6I).
The Pbx;Emx1-Cre mutant shows a robust lamination phenotype (only in the rostral cortex, where the ectopic Reelin is evident) (Figure 4), consistent with the model that ectopic Reelin expression disrupts the normal lamination pattern. In the rostral cortex, neurons expressing subplate, layer 6 and layer 5 markers are in a superficial position, whereas neurons expressing Cux2, the layer 2/3 marker, are in a deep position. BrdU pulse-chase analyses support the inversion of these layers in the rostral cortex (Figure S4). Thus, Pbx controls programs that mediate regional and laminar development, particularly in the rostral cortex.
PBX ChIP-Seq from embryonic cortex identifies target genes
The anti-PBX antibody used in the ChIP experiments recognizes PBX1, PBX2 and PBX3 (Ferretti et al., 2011). Pbx3 expression is not detectable in the developing cortex (Allen Brain Atlas). As noted previously, Pbx1 and Pbx2 share similar expression during cortical development (Figure 1, S1), and share functions (Capellini et al., 2006; Capellini et al., 2008). Thus, the PBX ChIP-Seq results most likely reflect both PBX1 and PBX2 genomic binding sites.
We performed PBX ChIP-Seq from E12.5 and E15.5 cortex. At E12.5, the majority of cells in the cortex are progenitors – thus the ~4,100 PBX ChIP-Seq peaks from this age should largely reflect PBX-bound regulatory elements in dividing cells of the VZ and SVZ, some of which are generating neurons destined to deep cortical layers. In the E15.5 cortex there are both progenitors and neurons; thus the ~7,600 PBX ChIP-Seq peaks from this age should reflect a mixture of PBX-bound regulatory elements in progenitors and immature neurons. We predict that PBX binding captured by the ChIP-Seq experiments includes both activating and repressive activity. In the future, region-specific chromatin datasets across the cortex could be used to examine this at a chromatin level. Nonetheless, here we show that PBX binds near genes that are both up-regulated and down-regulated after conditional deletion of Pbx1, evidence for direct PBX regulation of critical patterning genes.
The intersection of the E12.5 and E15.5 ChIP-Seq data identified ~1,600 PBX peaks unique to E12.5 data. These may be enriched for regulatory elements that function in cortical progenitors, and/or are important in the generation of deep layer cortical neurons. Conversely, the intersection of the E12.5 and E15.5 ChIP-Seq data showed ~5,100 PBX peaks unique to E15.5. These may be enriched for regulatory elements that function in immature cortical neurons, and/or are important in the generation of superficial layer cortical neurons. Finally, PBX bound to ~2,500 peaks at both E12.5 and E15.5; these regulatory elements may execute functions common to these stages of corticogenesis.
Roughly 20% of PBX ChIP-Seq peaks were found close (0–5 kb) to genes, particularly 5′ of the exons, and thus represent, in part, binding to promoters (Figure S6). In addition, >65% of the peaks mapped > 5kb away from transcribed genic regions (Figure S6), suggesting that PBX also binds to enhancers. Indeed, we identified PBX peaks on 270 regions that have enhancer activity at E11.5 (Table S1); 120 of these regions have enhancer activity in the E11.5 forebrain (Figure S6D; Table S2) (Visel et al., 2013).
Nucleotide motifs in genomic loci bound by PBX
De novo analysis of motifs from PBX peak sequences identified the likely primary PBX binding motif, which corresponds to a previously described motif for MEIS1 (Knoepfler et al., 1997; Shen et al., 1997). This binding motif occurs as both a full inverted palindromic motif and a set of motifs that are half-sites of the full inverted motif. PBX peaks proximal to TSS were strongly enriched for binding motifs mapped to the SP/EGR family, NRF1, ELK4, MZF1_1-4, and TFAP2A. Thus, PBX proteins may cooperate at promoters with these proteins. Note that the SP family member SP8 has a prominent role in cortical patterning (Borello et al., 2014; Sahara et al., 2007).
Distal PBX peaks were strongly enriched for motifs that are bound by proteins related to PDX1, MECOM/NFATC2, POU5f1::SOX2, FOXP1/SRY, NFATC2, ATOH1, SOX5, and MEF2A. These TFs are likely enhancer regulators, many of which are related to TFs which known functions in cortical development. For instance, the POU5f1::SOX2 complex, which control ES cell pluripotency, is related to the SOX2 and BRN (POU) proteins that promote neural fate (Tanaka et al., 2004). Sox5 function is crucial for development of deep layer neurons (Kwan et al., 2008; Lai et al., 2008; Leone et al., 2008). ATOH1 is a bHLH family member, many of which have fundamental roles in cortical development, including Ngn1, Ngn2, and the NeuroD family (Fode et al., 2000; Mattar et al., 2008; Olson et al., 2001; Sun et al., 2001). The observation that the ATOH1 motif was enriched at E15.5 but not E12.5 (Figure 7C–F), suggests that PBX and bHLH proteins may coordinately bind to enhancers with activity during neurogenesis, neuronal migration and maturation, rather than in neuroepithelial progenitors. Finally, the FOXP1 motif is consistent with known functions of FoxP1 and FoxP2 in neural differentiation (Bacon et al., 2014; Tsui et al., 2013). In sum, these results are an entrée for elucidating the mechanisms whereby combinations of transcription factors interact with PBX proteins on cis-regulatory elements to modulate gene expression during cortical regionalization, laminar patterning and neuronal differentiation.
EXPERIMENTAL PROCEDURES
In situ hybridization on brain sections
20μm frozen sections were dried, wash three times with PBS (5 min each) and fixed with 4% paraformaldehyde in PBS for 10min. Sectioned were then rinsed with PBS three times (3 min each) and treated with 1ug/ml Proteinase K for 17 min. After two quick rinsed in PBS sections were postfixed with 4% Paraformaldehyde for 5min and rinsed again in PBS three times (3 min each). Acetylation was performed for 10min in Acetylation Buffer containing 1.3% triethanolamine, 0.17% HCl and 0.4% acetic anhydride in water. Sections were then rinsed with PBS three times (10 min each) and prehybridized by incubating with hybridization buffer (50% formamide, 5XSSC pH 4.5, 50ug/ml yeast tRNA, 1% SDS, 50ug/ml Heparin) for 2 hours in 67C oven. After prehybridization in situ probes diluted in hybridization buffer at 500ng/ml were added for overnight incubation at 67C. Next day slides were rinsed with prewarmed 5X SSC pH 4.5, washed twice (30min each) with 0.2X SSC pH4.5 at 70C and then wash once (5min) with 0.2X SSC pH4.5 at room temperature followed by a wash with NTT buffer (0.15 M NaCl, 0.1 M Tris pH 8.0 0.1% Tween-20). Sections were blocked with NTT blocking buffer containing 5% heat inactivated horse serum and 2% Blocking buffer (cat# 11096176001, Roche) for 1hour at room temperature followed by an overnight incubation at 4C with Anti-Digoxigenin-AP antibody (1:5000 dilution in NTT blocking buffer). Next day sections were washed three times with NTT buffer (30min each), followed by three 5-min washes with NTTML buffer (0.15 M NaCl, 0.1 M Tris pH 9.5, 0.1% Tween-20, 50 mM MgCl2, 2 mM Levamisole) and incubated with developing reagent BM Purple (cat# 11442074001, Roche) until desired intensity of the signal was reached. Development reaction was stopped with PBS. Sections were allowed to dry, were dehydrated with Xylenes and mounted with Permount.
Whole mount in situ hybridization
The meninges were removed from dissected P0 brains and brains were fixed overnight in 4% paraformaldehyde. After two rinsed with PBS containing 0.1% Tween-20 (10min each) brains were rehydrated through a series of methanol washes in PBS-Tween-20, (25%, 50%, 75%, 100%) and stored at −20C until further processing. On the day of the experiment brains were rehydrated through a series of methanol washes (75%, 50%, 25%), rinsed with PBS-Tween twice and treated with 20um/ml Proteinase K for 30 min. After digestion tissue was rinsed with 100mM Glycine and PBS-Tween and postfixed with 4% paraformaldehyde/0.1% glutaraldehyde for 20min. After postfixation brains were washed once with PBS-Tween and then washed once with 1:1 mixture of PBS-Tween and hybridization buffer (50% formamide, 1.3X SSC pH4.5, 5mM EDTA, 50ug/ml yeast tRNA, 100ug./ml heparin, 0.5% Tween). Solution was then replaced with hybridization buffer and tissue was allowed to prehybridize for 1h at 70C. In situ probes were diluted in hybridization buffer at 500ng/ml and hybridization was performed overnight at 70C. Next day brains were washed three times (30min each, at 70C) with hybridization buffer, once with 1:1 mixture of hybridization buffer and TBST (30min at 70C) and three times (30 min each) with room temperature TBST. Brains were blocked with TBST containing 10% heat inactivated horse serum and 0.1% blocking buffer (Roche) for 2h at room temperature followed by an overnight incubation at 4C with Anti-Digoxigenin-AP antibody (1:4000 dilution). Next day brains were washed with TBST 8 times 30 min each and left in wash buffer overnight. BM Purple (Roche) was used as a developing reagent.
Chromatin Immunoprecipitation
Wt cortices (one litter of E12.5 or E15.5) were dissected, triturated in 1% formaldehyde in PBS and fixed for total of 10min at room temperature. Fixed cell were pelleted and washed with cold PBS. Pellets were lysed in 500ul of lysis buffer (1% SDS, 10mM EDTA and 50mM Tris, pH 8.1) on ice for 10min and lysates were sonicated using Bioruptor (Diagenode) on High Settings for 15 cycles (7.5 min of total sonication time). Resulting average chromatin size was 200–500 bp as verified by the Bioanalyzer. Cleared Chromatin was diluted 10 times with Dilution buffer (0.01% SDS, 1.1% Triton X-100, 1.2mM EDTA, 16.7mM Tris-HCl, pH 8.1, 167mM NaCl) and was incubated overnight at 4C with 3ug of appropriate antibody. Mixture of protein A and G magnetic beads (Life Techonologies) was preblocked overnight with BSA and tRNA and was added to the chromatin next day for 3 hours. Immune complexes were washed once with Low Salt buffer (0.1% SDS, 1% Triton X-100, 2mM EDTA, 20mM Tris-HCl, pH 8.1, 150mM NaCl), High Salt buffer (0.1% SDS, 1% Triton X-100, 2mM EDTA, 20mM Tris-HCl, pH 8.1, 500mM NaCl), LiCl buffer (0.25M LiCl, 1% NP-40, 1% deoxycholic acid (sodium salt), 1mM EDTA, 10mM Tris, pH 8.1) and TE buffer. Complexes were eluted in 1% SDS, 10mM Sodium bicarbonate buffer at 65C for 10min. Crosslinks were reversed at 65C overnight, proteins were digested using proteinase K and DNA was purified using Zymo Chip DNA Clean and Concentrator kit (Zymo research). Immunoprecipitation was performed using the following antibody: Pbx1/2/3/(sc-888, Santa Cruz). As a negative control in PBX ChIP experiments, the PBX antibody was incubated with PBX blocking peptide (sc-888P, Santa Cruz) at 1:400 molar ratio, and added to the chromatin lysates. ChIP-Seq Libraries were prepared using NEBNext library Prep kit (New England Biolabs) and Illumina standard adaptors and sequenced using a 50bp single end strategy using Illumina HiSeq platform.
Supplementary Material
Acknowledgments
This work was supported by funds from NARSAD to O.G., and Nina Ireland, Weston Havens Foundation, NINDS (NS34661), and NIMH (MH049428 and MH081880) to J.L.R.R. A.V. was supported by NIH grants R01HG003988 and U54HG006997. Research conducted at the E.O. Lawrence Berkeley National Laboratory was performed under Department of Energy Contract DE-AC02-05CH11231, University of California. JLRR is a Founder and Consultant for Neurona; this company has no financial interests related to this paper.
Footnotes
AUTHOR CONTRIBUTIONS
O.G. designed, conducted, and analyzed data for all experiments described in this manuscript. S.L. helped perform and analyze ChIP-Seq experiments, and provided comments on the manuscript. A.N., P.T. and A.V. performed informatic analyses of the ChIP-Seq data. A.Y. performed analysis of Pbx2 mutants. L.S. provided the Pbx mutant mice and information about the PBX antibodies. J.L.R.R. provided funding and laboratory resources, helped guide the project and analyze results. O.G. and J.L.R.R. together prepared the manuscript.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Alfano C, Magrinelli E, Harb K, Hevner RF, Studer M. Postmitotic control of sensory area specification during neocortical development. Nat Commun. 2014;5:5632. doi: 10.1038/ncomms6632. [DOI] [PubMed] [Google Scholar]
- Amaral DG, Schumann CM, Nordahl CW. Neuroanatomy of autism. Trends Neurosci. 2008;31:137–145. doi: 10.1016/j.tins.2007.12.005. [DOI] [PubMed] [Google Scholar]
- Bacon C, Schneider M, Le Magueresse C, Froehlich H, Sticht C, Gluch C, Monyer H, Rappold GA. Brain-specific Foxp1 deletion impairs neuronal development and causes autistic-like behaviour. Mol Psychiatry. 2014 doi: 10.1038/mp.2014.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berthelsen J, Viggiano L, Schulz H, Ferretti E, Consalez GG, Rocchi M, Blasi F. PKNOX1, a gene encoding PREP1, a new regulator of Pbx activity, maps on human chromosome 21q22.3 and murine chromosome 17B/C. Genomics. 1998a;47:323–324. doi: 10.1006/geno.1997.5086. [DOI] [PubMed] [Google Scholar]
- Berthelsen J, Zappavigna V, Ferretti E, Mavilio F, Blasi F. The novel homeoprotein Prep1 modulates Pbx-Hox protein cooperativity. Embo J. 1998b;17:1434–1445. doi: 10.1093/emboj/17.5.1434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berthelsen J, Zappavigna V, Mavilio F, Blasi F. Prep1, a novel functional partner of Pbx proteins. Embo J. 1998c;17:1423–1433. doi: 10.1093/emboj/17.5.1423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bjerke GA, Hyman-Walsh C, Wotton D. Cooperative transcriptional activation by Klf4, Meis2, and Pbx1. Mol Cell Biol. 2011;31:3723–3733. doi: 10.1128/MCB.01456-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolte S, Cordelieres FP. A guided tour into subcellular colocalization analysis in light microscopy. J Microsc. 2006;224:213–232. doi: 10.1111/j.1365-2818.2006.01706.x. [DOI] [PubMed] [Google Scholar]
- Borello U, Madhavan M, Vilinsky I, Faedo A, Pierani A, Rubenstein J, Campbell K. Sp8 and COUP-TF1 reciprocally regulate patterning and Fgf signaling in cortical progenitors. Cereb Cortex. 2014;24:1409–1421. doi: 10.1093/cercor/bhs412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bulchand S, Grove EA, Porter FD, Tole S. LIM-homeodomain gene Lhx2 regulates the formation of the cortical hem. Mech Dev. 2001;100:165–175. doi: 10.1016/s0925-4773(00)00515-3. [DOI] [PubMed] [Google Scholar]
- Burglin TR. Analysis of TALE superclass homeobox genes (MEIS, PBC, KNOX, Iroquois, TGIF) reveals a novel domain conserved between plants and animals. Nucleic Acids Res. 1997;25:4173–4180. doi: 10.1093/nar/25.21.4173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Capellini TD, Di Giacomo G, Salsi V, Brendolan A, Ferretti E, Srivastava D, Zappavigna V, Selleri L. Pbx1/Pbx2 requirement for distal limb patterning is mediated by the hierarchical control of Hox gene spatial distribution and Shh expression. Development. 2006;133:2263–2273. doi: 10.1242/dev.02395. [DOI] [PubMed] [Google Scholar]
- Capellini TD, Handschuh K, Quintana L, Ferretti E, Di Giacomo G, Fantini S, Vaccari G, Clarke SL, Wenger AM, Bejerano G, et al. Control of pelvic girdle development by genes of the Pbx family and Emx2. Dev Dyn. 2011a;240:1173–1189. doi: 10.1002/dvdy.22617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Capellini TD, Zappavigna V, Selleri L. Pbx homeodomain proteins: TALEnted regulators of limb patterning and outgrowth. Dev Dyn. 2011b;240:1063–1086. doi: 10.1002/dvdy.22605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Capellini TD, Zewdu R, Di Giacomo G, Asciutti S, Kugler JE, Di Gregorio A, Selleri L. Pbx1/Pbx2 govern axial skeletal development by controlling Polycomb and Hox in mesoderm and Pax1/Pax9 in sclerotome. Dev Biol. 2008;321:500–514. doi: 10.1016/j.ydbio.2008.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caronia-Brown G, Yoshida M, Gulden F, Assimacopoulos S, Grove EA. The cortical hem regulates the size and patterning of neocortex. Development. 2014;141:2855–2865. doi: 10.1242/dev.106914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cederquist GY, Azim E, Shnider SJ, Padmanabhan H, Macklis JD. Lmo4 establishes rostral motor cortex projection neuron subtype diversity. J Neurosci. 2013;33:6321–6332. doi: 10.1523/JNEUROSCI.5140-12.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cholfin JA, Rubenstein JL. Patterning of frontal cortex subdivisions by Fgf17. Proc Natl Acad Sci U S A. 2007;104:7652–7657. doi: 10.1073/pnas.0702225104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cholfin JA, Rubenstein JL. Frontal cortex subdivision patterning is coordinately regulated by Fgf8, Fgf17, and Emx2. J Comp Neurol. 2008;509:144–155. doi: 10.1002/cne.21709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chou SJ, Perez-Garcia CG, Kroll TT, O’Leary DD. Lhx2 specifies regional fate in Emx1 lineage of telencephalic progenitors generating cerebral cortex. Nat Neurosci. 2009;12:1381–1389. doi: 10.1038/nn.2427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crespo-Facorro B, Kim J, Andreasen NC, O’Leary DS, Bockholt HJ, Magnotta V. Insular cortex abnormalities in schizophrenia: a structural magnetic resonance imaging study of first-episode patients. Schizophr Res. 2000;46:35–43. doi: 10.1016/s0920-9964(00)00028-1. [DOI] [PubMed] [Google Scholar]
- DiMartino JF, Selleri L, Traver D, Firpo MT, Rhee J, Warnke R, O’Gorman S, Weissman IL, Cleary ML. The Hox cofactor and proto-oncogene Pbx1 is required for maintenance of definitive hematopoiesis in the fetal liver. Blood. 2001;98:618–626. doi: 10.1182/blood.v98.3.618. [DOI] [PubMed] [Google Scholar]
- Fernandes M, Gutin G, Alcorn H, McConnell SK, Hebert JM. Mutations in the BMP pathway in mice support the existence of two molecular classes of holoprosencephaly. Development. 2007;134:3789–3794. doi: 10.1242/dev.004325. [DOI] [PubMed] [Google Scholar]
- Ferretti E, Li B, Zewdu R, Wells V, Hebert JM, Karner C, Anderson MJ, Williams T, Dixon J, Dixon MJ, et al. A conserved Pbx-Wnt-p63-Irf6 regulatory module controls face morphogenesis by promoting epithelial apoptosis. Dev Cell. 2011;21:627–641. doi: 10.1016/j.devcel.2011.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ficara F, Murphy MJ, Lin M, Cleary ML. Pbx1 regulates self-renewal of long-term hematopoietic stem cells by maintaining their quiescence. Cell Stem Cell. 2008;2:484–496. doi: 10.1016/j.stem.2008.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fode C, Ma Q, Casarosa S, Ang SL, Anderson DJ, Guillemot F. A role for neural determination genes in specifying the dorsoventral identity of telencephalic neurons. Genes Dev. 2000;14:67–80. [PMC free article] [PubMed] [Google Scholar]
- Fukuchi-Shimogori T, Grove EA. Neocortex patterning by the secreted signaling molecule FGF8. Science. 2001;294:1071–1074. doi: 10.1126/science.1064252. [DOI] [PubMed] [Google Scholar]
- Garel S, Huffman KJ, Rubenstein JL. Molecular regionalization of the neocortex is disrupted in Fgf8 hypomorphic mutants. Development. 2003;130:1903–1914. doi: 10.1242/dev.00416. [DOI] [PubMed] [Google Scholar]
- Goebbels S, Bormuth I, Bode U, Hermanson O, Schwab MH, Nave KA. Genetic targeting of principal neurons in neocortex and hippocampus of NEX-Cre mice. Genesis. 2006;44:611–621. doi: 10.1002/dvg.20256. [DOI] [PubMed] [Google Scholar]
- Gorski JA, Talley T, Qiu M, Puelles L, Rubenstein JL, Jones KR. Cortical excitatory neurons and glia, but not GABAergic neurons, are produced in the Emx1-expressing lineage. J Neurosci. 2002;22:6309–6314. doi: 10.1523/JNEUROSCI.22-15-06309.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goumans MJ, Mummery C. Functional analysis of the TGFbeta receptor/Smad pathway through gene ablation in mice. Int J Dev Biol. 2000;44:253–265. [PubMed] [Google Scholar]
- Gourion D, Gourevitch R, Leprovost JB, Olie HlJP, Krebs MO. Neurodevelopmental hypothesis in schizophrenia. Encephale. 2004;30:109–118. doi: 10.1016/s0013-7006(04)95421-8. [DOI] [PubMed] [Google Scholar]
- Hamasaki T, Leingartner A, Ringstedt T, O’Leary DD. EMX2 regulates sizes and positioning of the primary sensory and motor areas in neocortex by direct specification of cortical progenitors. Neuron. 2004;43:359–372. doi: 10.1016/j.neuron.2004.07.016. [DOI] [PubMed] [Google Scholar]
- Hoerder-Suabedissen A, Wang WZ, Lee S, Davies KE, Goffinet AM, Rakic S, Parnavelas J, Reim K, Nicolic M, Paulsen O, et al. Novel markers reveal subpopulations of subplate neurons in the murine cerebral cortex. Cereb Cortex. 2009;19:1738–1750. doi: 10.1093/cercor/bhn195. [DOI] [PubMed] [Google Scholar]
- Itoh S, Itoh F, Goumans MJ, Ten Dijke P. Signaling of transforming growth factor-beta family members through Smad proteins. Eur J Biochem. 2000;267:6954–6967. doi: 10.1046/j.1432-1327.2000.01828.x. [DOI] [PubMed] [Google Scholar]
- Joshi PS, Molyneaux BJ, Feng L, Xie X, Macklis JD, Gan L. Bhlhb5 regulates the postmitotic acquisition of area identities in layers II-V of the developing neocortex. Neuron. 2008;60:258–272. doi: 10.1016/j.neuron.2008.08.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kikkawa T, Obayashi T, Takahashi M, Fukuzaki-Dohi U, Numayama-Tsuruta K, Osumi N. Dmrta1 regulates proneural gene expression downstream of Pax6 in the mammalian telencephalon. Genes Cells. 2013;18:636–649. doi: 10.1111/gtc.12061. [DOI] [PubMed] [Google Scholar]
- Kitisin K, Saha T, Blake T, Golestaneh N, Deng M, Kim C, Tang Y, Shetty K, Mishra B, Mishra L. Tgf-Beta signaling in development. Sci STKE. 2007;2007:cm1. doi: 10.1126/stke.3992007cm1. [DOI] [PubMed] [Google Scholar]
- Knoepfler PS, Calvo KR, Chen H, Antonarakis SE, Kamps MP. Meis1 and pKnox1 bind DNA cooperatively with Pbx1 utilizing an interaction surface disrupted in oncoprotein E2a-Pbx1. Proc Natl Acad Sci U S A. 1997;94:14553–14558. doi: 10.1073/pnas.94.26.14553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Konno D, Iwashita M, Satoh Y, Momiyama A, Abe T, Kiyonari H, Matsuzaki F. The mammalian DM domain transcription factor Dmrta2 is required for early embryonic development of the cerebral cortex. PLoS One. 2012;7:e46577. doi: 10.1371/journal.pone.0046577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koss M, Bolze A, Brendolan A, Saggese M, Capellini TD, Bojilova E, Boisson B, Prall OW, Elliott DA, Solloway M, et al. Congenital asplenia in mice and humans with mutations in a Pbx/Nkx2-5/p15 module. Dev Cell. 2012;22:913–926. doi: 10.1016/j.devcel.2012.02.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwan KY, Lam MM, Krsnik Z, Kawasawa YI, Lefebvre V, Sestan N. SOX5 postmitotically regulates migration, postmigratory differentiation, and projections of subplate and deep-layer neocortical neurons. Proc Natl Acad Sci U S A. 2008;105:16021–16026. doi: 10.1073/pnas.0806791105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai T, Jabaudon D, Molyneaux BJ, Azim E, Arlotta P, Menezes JR, Macklis JD. SOX5 controls the sequential generation of distinct corticofugal neuron subtypes. Neuron. 2008;57:232–247. doi: 10.1016/j.neuron.2007.12.023. [DOI] [PubMed] [Google Scholar]
- Lee SM, Tole S, Grove E, McMahon AP. A local Wnt-3a signal is required for development of the mammalian hippocampus. Development. 2000;127:457–467. doi: 10.1242/dev.127.3.457. [DOI] [PubMed] [Google Scholar]
- Leone DP, Srinivasan K, Chen B, Alcamo E, McConnell SK. The determination of projection neuron identity in the developing cerebral cortex. Curr Opin Neurobiol. 2008;18:28–35. doi: 10.1016/j.conb.2008.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li G, Adesnik H, Li J, Long J, Nicoll RA, Rubenstein JL, Pleasure SJ. Regional distribution of cortical interneurons and development of inhibitory tone are regulated by Cxcl12/Cxcr4 signaling. J Neurosci. 2008;28:1085–1098. doi: 10.1523/JNEUROSCI.4602-07.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Long JE, Swan C, Liang WS, Cobos I, Potter GB, Rubenstein JL. Dlx1&2 and Mash1 transcription factors control striatal patterning and differentiation through parallel and overlapping pathways. J Comp Neurol. 2009;512:556–572. doi: 10.1002/cne.21854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mallamaci A, Muzio L, Chan CH, Parnavelas J, Boncinelli E. Area identity shifts in the early cerebral cortex of Emx2−/− mutant mice. Nat Neurosci. 2000;3:679–686. doi: 10.1038/76630. [DOI] [PubMed] [Google Scholar]
- Mangale VS, Hirokawa KE, Satyaki PR, Gokulchandran N, Chikbire S, Subramanian L, Shetty AS, Martynoga B, Paul J, Mai MV, et al. Lhx2 selector activity specifies cortical identity and suppresses hippocampal organizer fate. Science. 2008;319:304–309. doi: 10.1126/science.1151695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mattar P, Langevin LM, Markham K, Klenin N, Shivji S, Zinyk D, Schuurmans C. Basic helix-loop-helix transcription factors cooperate to specify a cortical projection neuron identity. Mol Cell Biol. 2008;28:1456–1469. doi: 10.1128/MCB.01510-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCloy RA, Rogers S, Caldon CE, Lorca T, Castro A, Burgess A. Partial inhibition of Cdk1 in G 2 phase overrides the SAC and decouples mitotic events. Cell Cycle. 2014;13:1400–1412. doi: 10.4161/cc.28401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McLean CY, Bristor D, Hiller M, Clarke SL, Schaar BT, Lowe CB, Wenger AM, Bejerano G. GREAT improves functional interpretation of cis-regulatory regions. Nat Biotechnol. 2010;28:495–501. doi: 10.1038/nbt.1630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyashita-Lin EM, Hevner R, Wassarman KM, Martinez S, Rubenstein JL. Early neocortical regionalization in the absence of thalamic innervation. Science. 1999;285:906–909. doi: 10.1126/science.285.5429.906. [DOI] [PubMed] [Google Scholar]
- Molyneaux BJ, Arlotta P, Menezes JR, Macklis JD. Neuronal subtype specification in the cerebral cortex. Nat Rev Neurosci. 2007;8:427–437. doi: 10.1038/nrn2151. [DOI] [PubMed] [Google Scholar]
- Monuki ES, Porter FD, Walsh CA. Patterning of the dorsal telencephalon and cerebral cortex by a roof plate-Lhx2 pathway. Neuron. 2001;32:591–604. doi: 10.1016/s0896-6273(01)00504-9. [DOI] [PubMed] [Google Scholar]
- Muzio L, Mallamaci A. Emx1, emx2 and pax6 in specification, regionalization and arealization of the cerebral cortex. Cereb Cortex. 2003;13:641–647. doi: 10.1093/cercor/13.6.641. [DOI] [PubMed] [Google Scholar]
- Nakagawa Y, Johnson JE, O’Leary DD. Graded and areal expression patterns of regulatory genes and cadherins in embryonic neocortex independent of thalamocortical input. J Neurosci. 1999;19:10877–10885. doi: 10.1523/JNEUROSCI.19-24-10877.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nieto M, Monuki ES, Tang H, Imitola J, Haubst N, Khoury SJ, Cunningham J, Gotz M, Walsh CA. Expression of Cux-1 and Cux-2 in the subventricular zone and upper layers II–IV of the cerebral cortex. J Comp Neurol. 2004;479:168–180. doi: 10.1002/cne.20322. [DOI] [PubMed] [Google Scholar]
- Ogawa M, Miyata T, Nakajima K, Yagyu K, Seike M, Ikenaka K, Yamamoto H, Mikoshiba K. The reeler gene-associated antigen on Cajal-Retzius neurons is a crucial molecule for laminar organization of cortical neurons. Neuron. 1995;14:899–912. doi: 10.1016/0896-6273(95)90329-1. [DOI] [PubMed] [Google Scholar]
- Olson JM, Asakura A, Snider L, Hawkes R, Strand A, Stoeck J, Hallahan A, Pritchard J, Tapscott SJ. NeuroD2 is necessary for development and survival of central nervous system neurons. Dev Biol. 2001;234:174–187. doi: 10.1006/dbio.2001.0245. [DOI] [PubMed] [Google Scholar]
- Phelan ML, Rambaldi I, Featherstone MS. Cooperative interactions between HOX and PBX proteins mediated by a conserved peptide motif. Mol Cell Biol. 1995;15:3989–3997. doi: 10.1128/mcb.15.8.3989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pierani A, Moran-Rivard L, Sunshine MJ, Littman DR, Goulding M, Jessell TM. Control of interneuron fate in the developing spinal cord by the progenitor homeodomain protein Dbx1. Neuron. 2001;29:367–384. doi: 10.1016/s0896-6273(01)00212-4. [DOI] [PubMed] [Google Scholar]
- Piven J, Arndt S, Bailey J, Havercamp S, Andreasen NC, Palmer P. An MRI study of brain size in autism. Am J Psychiatry. 1995;152:1145–1149. doi: 10.1176/ajp.152.8.1145. [DOI] [PubMed] [Google Scholar]
- Sahara S, Kawakami Y, Izpisua Belmonte JC, O’Leary DD. Sp8 exhibits reciprocal induction with Fgf8 but has an opposing effect on anterior-posterior cortical area patterning. Neural Dev. 2007;2:10. doi: 10.1186/1749-8104-2-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaeren-Wiemers N, Andre E, Kapfhammer JP, Becker-Andre M. The expression pattern of the orphan nuclear receptor RORbeta in the developing and adult rat nervous system suggests a role in the processing of sensory information and in circadian rhythm. Eur J Neurosci. 1997;9:2687–2701. doi: 10.1111/j.1460-9568.1997.tb01698.x. [DOI] [PubMed] [Google Scholar]
- Selleri L, Depew MJ, Jacobs Y, Chanda SK, Tsang KY, Cheah KS, Rubenstein JL, O’Gorman S, Cleary ML. Requirement for Pbx1 in skeletal patterning and programming chondrocyte proliferation and differentiation. Development. 2001;128:3543–3557. doi: 10.1242/dev.128.18.3543. [DOI] [PubMed] [Google Scholar]
- Selleri L, DiMartino J, van Deursen J, Brendolan A, Sanyal M, Boon E, Capellini T, Smith KS, Rhee J, Popperl H, et al. The TALE homeodomain protein Pbx2 is not essential for development and long-term survival. Mol Cell Biol. 2004;24:5324–5331. doi: 10.1128/MCB.24.12.5324-5331.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen WF, Chang CP, Rozenfeld S, Sauvageau G, Humphries RK, Lu M, Lawrence HJ, Cleary ML, Largman C. Hox homeodomain proteins exhibit selective complex stabilities with Pbx and DNA. Nucleic Acids Res. 1996;24:898–906. doi: 10.1093/nar/24.5.898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen WF, Montgomery JC, Rozenfeld S, Moskow JJ, Lawrence HJ, Buchberg AM, Largman C. AbdB-like Hox proteins stabilize DNA binding by the Meis1 homeodomain proteins. Mol Cell Biol. 1997;17:6448–6458. doi: 10.1128/mcb.17.11.6448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stoykova A, Treichel D, Hallonet M, Gruss P. Pax6 modulates the dorsoventral patterning of the mammalian telencephalon. J Neurosci. 2000;20:8042–8050. doi: 10.1523/JNEUROSCI.20-21-08042.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun Y, Nadal-Vicens M, Misono S, Lin MZ, Zubiaga A, Hua X, Fan G, Greenberg ME. Neurogenin promotes neurogenesis and inhibits glial differentiation by independent mechanisms. Cell. 2001;104:365–376. doi: 10.1016/s0092-8674(01)00224-0. [DOI] [PubMed] [Google Scholar]
- Swift GH, Liu Y, Rose SD, Bischof LJ, Steelman S, Buchberg AM, Wright CV, MacDonald RJ. An endocrine-exocrine switch in the activity of the pancreatic homeodomain protein PDX1 through formation of a trimeric complex with PBX1b and MRG1 (MEIS2) Mol Cell Biol. 1998;18:5109–5120. doi: 10.1128/mcb.18.9.5109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka S, Kamachi Y, Tanouchi A, Hamada H, Jing N, Kondoh H. Interplay of SOX and POU factors in regulation of the Nestin gene in neural primordial cells. Mol Cell Biol. 2004;24:8834–8846. doi: 10.1128/MCB.24.20.8834-8846.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas JT, Canelos P, Luyten FP, Moos M., Jr Xenopus SMOC-1 Inhibits bone morphogenetic protein signaling downstream of receptor binding and is essential for postgastrulation development in Xenopus. J Biol Chem. 2009;284:18994–19005. doi: 10.1074/jbc.M807759200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas-Chollier M, Herrmann C, Defrance M, Sand O, Thieffry D, van Helden J. RSAT peak-motifs: motif analysis in full-size ChIP-seq datasets. Nucleic Acids Res. 2012;40:e31. doi: 10.1093/nar/gkr1104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toresson H, Parmar M, Campbell K. Expression of Meis and Pbx genes and their protein products in the developing telencephalon: implications for regional differentiation. Mech Dev. 2000;94:183–187. doi: 10.1016/s0925-4773(00)00324-5. [DOI] [PubMed] [Google Scholar]
- Tsui D, Vessey JP, Tomita H, Kaplan DR, Miller FD. FoxP2 regulates neurogenesis during embryonic cortical development. J Neurosci. 2013;33:244–258. doi: 10.1523/JNEUROSCI.1665-12.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Visel A, Blow MJ, Li Z, Zhang T, Akiyama JA, Holt A, Plajzer-Frick I, Shoukry M, Wright C, Chen F, et al. ChIP-seq accurately predicts tissue-specific activity of enhancers. Nature. 2009;457:854–858. doi: 10.1038/nature07730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Visel A, Taher L, Girgis H, May D, Golonzhka O, Hoch RV, McKinsey GL, Pattabiraman K, Silberberg SN, Blow MJ, et al. A high-resolution enhancer atlas of the developing telencephalon. Cell. 2013;152:895–908. doi: 10.1016/j.cell.2012.12.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki R, Shimodaira H. Pvclust: an R package for assessing the uncertainty in hierarchical clustering. Bioinformatics. 2006;22:1540–1542. doi: 10.1093/bioinformatics/btl117. [DOI] [PubMed] [Google Scholar]
- Vitobello A, Ferretti E, Lampe X, Vilain N, Ducret S, Ori M, Spetz JF, Selleri L, Rijli FM. Hox and Pbx factors control retinoic acid synthesis during hindbrain segmentation. Dev Cell. 2011;20:469–482. doi: 10.1016/j.devcel.2011.03.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Li G, Stanco A, Long JE, Crawford D, Potter GB, Pleasure SJ, Behrens T, Rubenstein JL. CXCR4 and CXCR7 have distinct functions in regulating interneuron migration. Neuron. 2011;69:61–76. doi: 10.1016/j.neuron.2010.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamasue H, Iwanami A, Hirayasu Y, Yamada H, Abe O, Kuroki N, Fukuda R, Tsujii K, Aoki S, Ohtomo K, et al. Localized volume reduction in prefrontal, temporolimbic, and paralimbic regions in schizophrenia: an MRI parcellation study. Psychiatry Res. 2004;131:195–207. doi: 10.1016/j.pscychresns.2004.05.004. [DOI] [PubMed] [Google Scholar]
- Yun K, Potter S, Rubenstein JL. Gsh2 and Pax6 play complementary roles in dorsoventral patterning of the mammalian telencephalon. Development. 2001;128:193–205. doi: 10.1242/dev.128.2.193. [DOI] [PubMed] [Google Scholar]
- Zembrzycki A, Perez-Garcia CG, Wang CF, Chou SJ, O’Leary DD. Postmitotic regulation of sensory area patterning in the mammalian neocortex by Lhx2. Proc Natl Acad Sci U S A. 2015;112:6736–6741. doi: 10.1073/pnas.1424440112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou CJ, Borello U, Rubenstein JL, Pleasure SJ. Neuronal production and precursor proliferation defects in the neocortex of mice with loss of function in the canonical Wnt signaling pathway. Neuroscience. 2006;142:1119–1131. doi: 10.1016/j.neuroscience.2006.07.007. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.