Summary
Plants exposed to heavy metals rapidly induce changes in gene expression that activate and enhance detoxification mechanisms, including toxic-metal chelation and the scavenging of reactive oxygen species. However, the mechanisms mediating toxic heavy metal-induced gene expression remain largely unknown. To genetically elucidate cadmium-specific transcriptional responses in Arabidopsis, we designed a genetic screen based on the activation of a cadmium-inducible reporter gene. Microarray studies identified a high-affinity sulfate transporter (SULTR1;2) among the most robust and rapid cadmium-inducible transcripts. The SULTR1;2 promoter (2.2 kb) was fused with the firefly luciferase reporter gene to quantitatively report the transcriptional response of plants exposed to cadmium. Stably transformed luciferase reporter lines were ethyl methanesulfonate (EMS) mutagenized, and stable M2 seedlings were screened for an abnormal luciferase response during exposure to cadmium. The screen identified non-allelic mutant lines that fell into one of three categories: (i) super response to cadmium (SRC) mutants; (ii) constitutive response to cadmium (CRC) mutants; or (iii) non-response and reduced response to cadmium (NRC) mutants. Two nrc mutants, nrc1 and nrc2, were mapped, cloned and further characterized. The nrc1 mutation was mapped to the γ-glutamylcysteine synthetase gene and the nrc2 mutation was identified as the first viable recessive mutant allele in the glutathione synthetase gene. Moreover, genetic, HPLC mass spectrometry, and gene expression analysis of the nrc1 and nrc2 mutants, revealed that intracellular glutathione depletion alone would be insufficient to induce gene expression of sulfate uptake and assimilation mechanisms. Our results modify the glutathione-depletion driven model for sulfate assimilation gene induction during cadmium stress, and suggest that an enhanced oxidative state and depletion of upstream thiols, in addition to glutathione depletion, are necessary to induce the transcription of sulfate assimilation genes during early cadmium stress.
Keywords: glutathione biosynthesis, heavy metal, γ-glutamylcysteine synthetase, metabolite-based cloning, phytochelatins, Arabidopsis thaliana
Introduction
Toxic metals such as lead, cadmium (Cd), mercury and the metalloid arsenic can accumulate in soils and water to levels that are detrimental to human and environmental health. Many human disorders have been attributed to the ingestion of heavy metals, including learning disabilities in children, dementia, impairment of bone metabolism and increased cancer rates (Tong et al., 2000; Allen et al., 2002; Aschner and Walker, 2002; Ohta et al., 2002; Yu et al., 2002; Waisberg et al., 2003; Heck et al., 2009; Satarug et al., 2010). Food crops are a major source of heavy metal intake in humans, which has prompted interest in understanding how plants take up, detoxify and retain heavy metals. In addition, plants hold the potential for the development of a cost-effective approach for the removal and remediation of heavy metalladen soils and water through the use of metal-hyperaccumulating plants (phytoremediation) (Raskin et al., 1994; Dushenkov et al., 1995; Salt et al., 1995, 1998; Clemens, 2006).
Metal trafficking, both within the cell and between different tissues, often requires the use of metal ligand molecules such as citrate, nicotianamine, glutathione (GSH) and phytochelatins (PCs) (Lee et al., 1978; Grill et al., 1985; Howden et al., 1995; Kramer et al., 2000; Sanchez-Fernandez et al., 2001; Klein et al., 2002; Richau et al., 2009; Mendoza-Cozatl et al., 2011). Glutathione is a crucial molecule required for the synthesis of PCs, which detoxify mercury, Cd and the metalloid arsenic. PCs are small glutathione polymers synthesized in the cytosol (Grill et al., 1985; Clemens et al., 1999, 2001; Ha et al., 1999; Vatamaniuk et al., 1999, 2001). PCs bind highly toxic heavy metals and metalloids, and transport them into the vacuoles by ABC transporters (Li et al., 2004; Chen et al., 2006; Mendoza-Cozatl et al., 2010; Song et al., 2010). Glutathione and PCs have been shown to undergo long-distance transport of Cd through the phloem, but the identities of these transporters remain unknown (Gong et al., 2003; Chen et al., 2006; Mendoza-Cozatl et al., 2008). Thus, exposure to heavy metals can rapidly deplete glutathione levels and create an extremely high demand for glutathione.
At the transcriptional level, heavy metal exposure elicits a robust gene expression response in plants (Herbette et al., 2006; Weber et al., 2006). For instance, Cd exposure rapidly depletes cells of GSH, which in turn induces transcripts that encode sulfate uptake, sulfate assimilation and glutathione biosynthesis mechanisms (Lee and Leustek, 1999; Nocito et al., 2006; Davidian and Kopriva, 2010). These findings have led to the development of a metabolite demand-driven model for the regulation of sulfate assimilation and glutathione biosynthesis in which heavy metal-induced GSH depletion induces gene expression (Vauclare et al., 2002; Kopriva, 2006). However, the molecular mechanisms that trigger rapid changes in gene expression following heavy metal exposure in plants remain unknown.
To uncover the molecular and genetic mechanisms that mediate rapid Cd-induced gene expression in Arabidopsis, we have pursued Cd-induced microarray experiments and a forward genetic screen to identify mutants with altered responses to Cd exposure, using a Cd-inducible promoter driving the expression of the firefly luciferase gene. Unexpectedly, two of the mutants showing a dramatically decreased Cd response are impaired in steps upstream of GSH synthesis. HPLC-MS analyses of thiol compounds suggest that upstream thiols and an oxidative redox state functions in the induction of sulfate uptake genes during Cd exposure, and not during GSH depletion alone. Characterization of the transcriptional response to Cd in these mutants revealed a new level of regulation (hierarchical regulation) of sulfur assimilation signaling and glutathione biosynthesis in response to Cd exposure in plants.
Results
Identification of a rapid cadmium-inducible promoter
To identify genes in Arabidopsis that are highly and rapidly induced by Cd, we performed oligonucleotide chip-microarray experiments (Affymetrix, ATH1) on 1-week-old Arabidopsis seedlings exposed to 200 μm CdCl2 for 6 h (Table S1). Cd-inducible transcripts were identified (Table S1) and transcriptional activation following Cd exposure was confirmed for six strongly induced transcripts by RT-PCR (Figure 1a). Nine promoter-luciferase constructs containing 2.2-kb promoter fragments of the cadmium-inducible genes were introduced into Arabidopsis (Col-0), and stable T3 homozygous seedlings were analyzed for cadmium-induced luminescence. Luciferase (LUC) reporter lines carrying the high-affinity sulfate transporter SULTR1;2 promoter (pSULTR1;2) showed a quantitative and highly reproducible luciferase response to Cd, and one line (line A) was chosen for mutagenesis (Figure 1b). This pSULTR1;20∷LUC reporter line will henceforth be referred to as the control reporter line or parental line.
Figure 1.
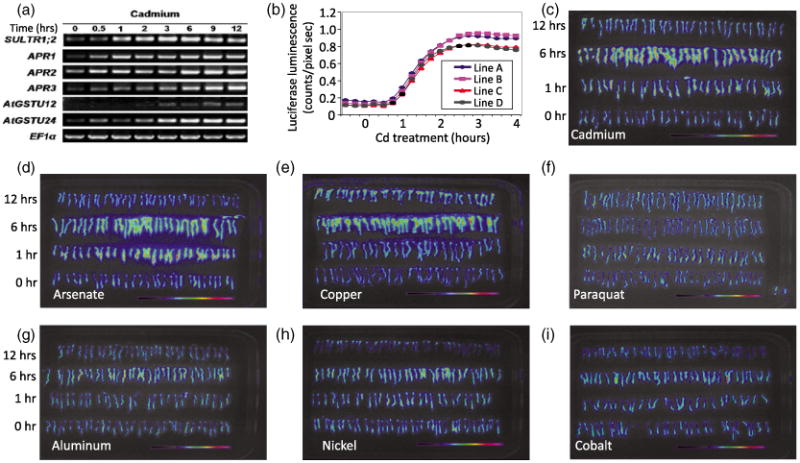
Development of a cadmium-inducible reporter line. (a) Genes identified by microarray analysis as being induced following 200 μm cadmium (Cd) treatment for 12 h were verified by RT-PCR. Tissue was collected every 30 min and RNA was extracted as described in the Experimental Procedures. (b) Quantification of the luciferase activity of pSULTR1;2∷LUC showing the dynamic response of four independent pSULTR1;2∷LUC reporter lines over a 4-h time course. Measurements were taken in 15-min intervals.
(c–h) False color luminescence images after exposure to heavy metals, metalloids and reactive oxygen species (ROS)-generating compounds over a 12-h time course. Seedlings were exposed to 200 μm of the indicted substance and imaged as described in the Experimental Procedures. Specificity of the control reporter line response was determined by luminescence imaging after exposure to (c) cadmium, (d) arsenate, (e) copper, (f) paraquat, (g) aluminum, (h) nickel and (i) cobalt over a 12-h period.
pSULTR1;2∷LUC is induced by cadmium, arsenate and copper, but not by exogenous reactive oxygen species
The dynamic response of 4 reporter lines was analyzed over a 12-h period following Cd exposure (Figure 1b,c). Luciferase activity was highest in roots, and the induction was evident after 1 h of Cd exposure, reaching a maximum after 3 h of exposure (Figure 1b) and decreasing steadily to half of the maximal induction after 12 h of Cd exposure (Figure 1c). To determine if the SULTR1;2 promoter is induced broadly by metals or exogenous reactive oxygen species (ROS), luciferase induction was measured following exposure to arsenate, copper, aluminum, nickel, cobalt and the ROS-inducing agent paraquat (Figure 1d–i). Figure 1 shows that Cd (Figure 1c), arsenate (Figure 1d) and copper (Figure 1e) elicit a strong transcriptional response, whereas the remaining metals and paraquat showed limited or no induction during the 12-h exposure period. These results suggest that SULTR1;2 is not broadly induced by oxidative stress and that the SULTR1;2 induction line is a suitable parental line for a forward genetic screen to identify mutants with an impaired Cd-induced transcriptional response.
Seeds of the control reporter line were ethyl methanesulfonate (EMS) -mutagenized (approximately 6000 seeds), and 60 000 M2 seedlings were screened for altered luciferase induction after 6 h of Cd exposure. Putative mutants were selected and the altered luciferase response was confirmed in M3 seedlings (Figure 2). Mutants were classified into one of three different groups: (i) constitutive response without Cd (CRC) mutants, showing a constitutive luciferase induction (Figure 2b) without being exposed to Cd; (ii) super response to Cd (SRC) mutants, which showed higher luciferase activity compared with the control reporter lines following Cd exposure (Figure 2c); (iii) non-response or reduced response to Cd (NRC) mutants, which failed to induce strong luciferase activity after Cd exposure (Figure 2d,e).
Figure 2.
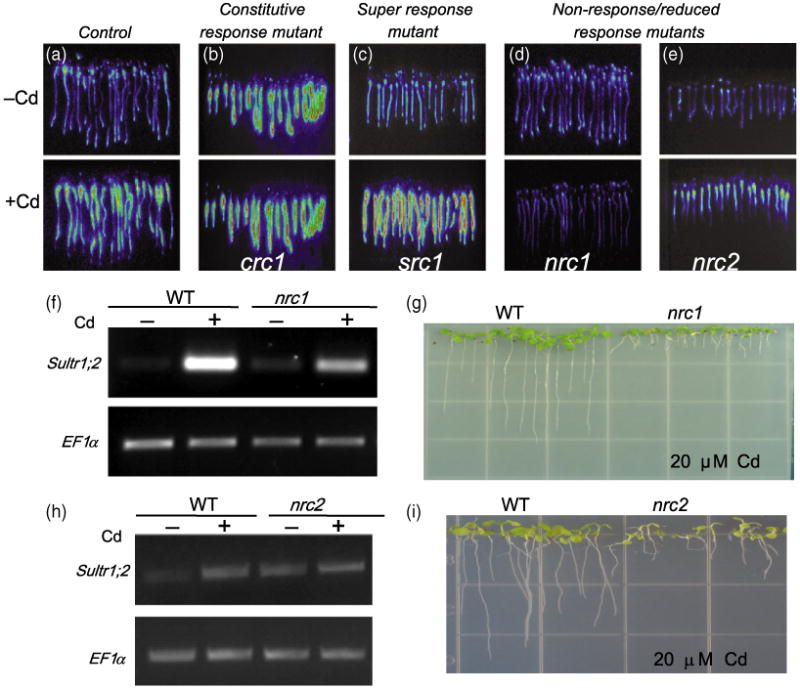
Ethyl methanesulfonate (EMS)-mutagenized pSULTR1;2∷LUC seeds were screened for altered cadmium-induced luciferase activity responses: (a) mutants were classified into three different groups based on luciferase response, compared with the control reporter line; (b) constitutive response to cadmium or CRC mutants (crc1); (c) super response to cadmium or SRC mutants (src1); (d) non-response to cadmium or NRC mutants (nrc1, d; nrc2, e).
(f) RT-PCR of the native SULTR1;2 expression using 35 PCR cycles confirmed the NR reporter phenotype of nrc1.
(g) nrc1 shows a short root phenotype compared with control when grown on 20 μm cadmium for 9 days.
(h) RT-PCR of the native SULTR1;2 transcript using 30 PCR cycles showed a reduced induction in the nrc2 mutant. (i) nrc2 also has a short-root phenotype compared with the control when grown on 20 μm Cd for 14 days.
Glutathione-deficient mutants show reduced luciferase induction during Cd exposure
We focused on characterization of two recessive non-response mutants, designated non-response to cadmium 1 and 2 (nrc1 and nrc2), which are Cd sensitive and have short roots when grown in the presence of Cd. Figure 2(d,e) shows the luciferase phenotype of the nrc1 and nrc2 mutants. To validate the decreased luciferase response of the nrc1 and nrc2 mutants, RT-PCR analysis of the native SULTR1;2 gene was performed in the control and in the nrc1 and nrc2 mutants. Figure 2(f,h) shows that the induction of the SULTR1;2 transcript in nrc1 was severely decreased compared with the control reporter line (Figure 2f), but only moderately decreased in the nrc2 mutant (Figure 2h). The reduced size of nrc2 seedlings probably contributed to the difference between the measured decreases in luciferase response in the nrc2 mutant, with a moderate decrease in Sultr1;2 transcript level in the nrc2 mutant. Thus, the nrc1 mutant is a strong non-response mutant, whereas the nrc2 mutant, which retains some Sultr1;2 induction, is more accurately described as a reduced response mutant. Root elongation experiments on plates containing 20 μm CdCl2 showed that nrc1 and nrc2 seedlings are Cd hypersensitive (Figure 2g,i). Crosses between nrc2 and nrc1 showed that they are non-allelic.
The organic thiols cysteine, γ-glutamylcysteine (γ-EC) and GSH are known to be key metabolites required for the production of PCs that mediate Cd detoxification. Therefore, we analyzed the metabolic thiol profile of the nrc1 and nrc2 mutants by fluorescence HPLC coupled to a mass spectrometer (HPLC-MS) (Figure 3a-f). After exposure to 20 μm Cd for 48 h, GSH levels were decreased in both the nrc1 (44.8 ± 2.31 nmol GSH per g fresh weight) and the nrc2 (94.9 ± 8.22 nmol GSH per g fresh weight) mutants compared with parental controls (126.2 nmol GSH per g fresh weight; Figure 3a-c,f). Conversely, cysteine levels in the nrc1 (66.6 ± 8.19 nmol Cys per g fresh weight) and nrc2 (51.3 ± 8.93 nmol Cys per g fresh weight) mutants in the presence of Cd were elevated compared with parental controls (11.8 ± 0.71 nmol Cys per g fresh weight; Figure 3a-d). Interestingly, γ-EC levels were decreased following Cd exposure in the nrc1 mutant (2.30 ± 0.40 nmol γ-EC per g fresh weight), and were elevated in the nrc2 mutant (431.7 ± 72.7 nmol γ-EC per g fresh weight), compared with parental controls (8.57 ± 0.25 nmol γ-EC per g fresh weight; Figure 3a-c,e).
Figure 3.
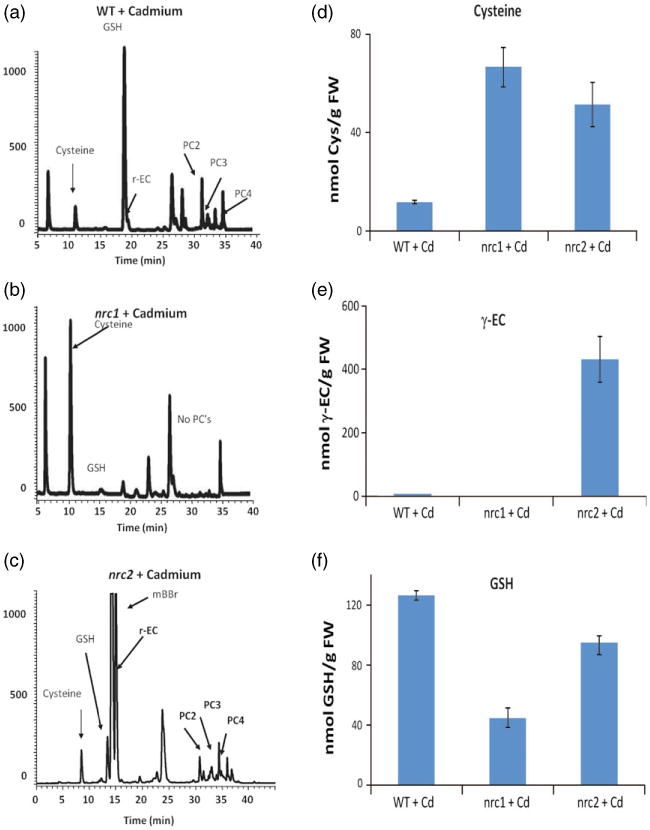
Thiol profile of the nrc1 and nrc2 non-response mutants reveals enhanced cysteine levels in nrc1 and enhanced γ-EC levelsin nrc2. Fluorescence HPLC-MS was used to identify and quantify thiols
(a) in control plants; (b) in the nrc1 mutant; (c) in the nrc2 mutant.
(d) Cysteine quantification using HPLC-MS revealed a five to sixfold accumulation of cysteine in both the nrc1 and nrc2 mutants relative to the control following treatment with cadmium.
(e) Fluorescence HPLC-MS also revealed a approximately 50-fold accumulation of γ-glutamylcysteine in nrc2 relative to the control and a depletion of γ-EC in the nrc1 mutant (approximately 25% of wild-type levels).
(f) Glutathione quantification using HPLC-MS revealed that both the nrc1 and nrc2 mutants are GSH deficient. All bar graphs show the mean of between three and six independent samples. Error bars represent the standard error of the mean (SEM).
Physical mapping and characterization of the nrc1 and nrc2 mutants
Our HPLC-MS findings suggest that nrc1 inefficiently converts cysteine into γ-EC, the precursor of GSH and PCs. Initial rough mapping using an F2 population of a nrc1 × Landsberg erecta (Ler) backcross located the mutation on chromosome 4, between the nga1107 and ciw7 markers (Figure 4a). Based on the thiol profile of nrc1, candidate genes involved in sulfur assimilation and GSH synthesis from this mapping region were PCR-amplified and sequenced. The locus At4g23100 contained a single C→T mutation in the fourth exon causing a Pro →Leu (P214L) change in the amino acid sequence of γ-EC synthetase (γ-ECS), a key enzyme in glutathione biosynthesis (Figure 4a).
Figure 4.
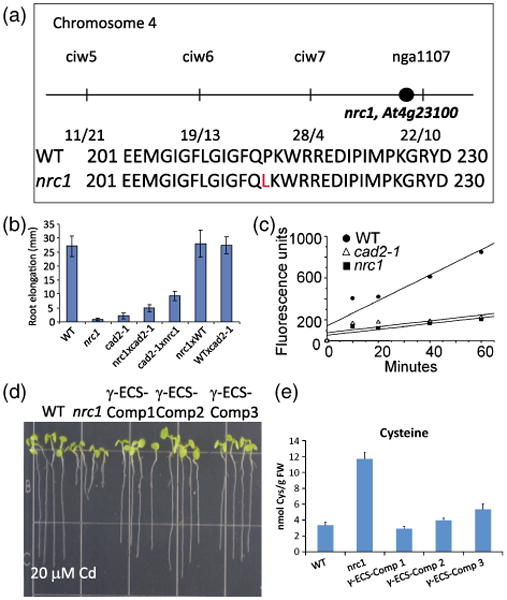
Characterization of the non-response to cadmium mutant nrc1.
(a) Mapping of the nrc1 mutant placed the mutation on chromosome 4 between the ciw7 and nga1107 markers. Candidate gene sequencing identified a point mutation in At4g23100, which causes a Pro → Leu (P214L) change in the γ-glutamylcysteine synthetase protein.
(b) Crosses between nrc1 and cad2-1 show the mutants are allelic. The short-root phenotype of the nrc1 and cad2-1 mutants grown on 20 μm cadmium (Cd) is not rescued in the nrc1 × cad2-1 or the cad2-1 × nrc1 F1 cross, but crossing either mutant into WT restores root elongation in the F1 generation.
(c) nrc1 is deficient in γ-EC synthetase (γ-ECS) activity. An in vitro assay was performed to determine the activity of the γ-ECS protein from wild-type (WT), nrc1 and cad2-1 plants. γ-EC synthetase activity was measured by tracking γ-EC appearance using HPLC-MS in crude extracts (rosette leaves) obtained from wild-type Col-0 (full circles), nrc1 (full squares) and cad2-1 (empty triangles). Extracts obtained from WT plants showed a steady synthesis of γ-EC during the assay, whereas nrc1 and cad2-1 showed only marginal increases in γ-EC concentration during the assay.
(d) Expression of genomic γ-ECS in the nrc1 mutant behind a constitutive promoter restores root elongation on Cd. Three independent T2 transformants (Comp1–Comp3) were used for root elongation experiments on 20 μm Cd.
(e) Seven-day-old complementation lines (γ-ECS Comp1–Comp3) were also used for fluorescence HPLC-MS analyses following 48 h of 100 μm Cd treatment. Cysteine levels in the nrc1 mutant are three to sixfold higher than in the WT (66.6 ± 8.22 nmol Cys per g fresh weight in nrc1 compared with 11.8 ± 0.71 nmol Cys per g fresh weight in WT), whereas all three of the γ-ECS complemented (γ-ECS-Comp) lines show a WT cysteine accumulation. All bar graphs show the mean of between three and six independent samples. Error bars represent the standard error of the mean (SEM).
To further determine whether this mutation in γ-ECS was responsible for the nrc1 phenotype, nrc1 was crossed into the previously characterized γ-ECS allele, cad2-1 (Howden et al., 1995) (Figure 4b). F1 seedlings from the reciprocal crosses between the recessive nrc1 and cad2-1 mutants were Cd hypersensitive, as determined by root elongation assays in the presence of cadmium (Figure 4b). In contrast, in the presence of Cd, seedlings from crosses of wild-type Col-0 (WT) and nrc1 or cad2-1 were not Cd hypersensitive (Figure 4b). These results show that nrc1 is allelic to cad2-1.
To further compare the nrc1allele with the cad2-1allele, we compared the γ-ECS activity in protein extracts obtained from the two mutants versus the activity of γ-ECS in WT extracts. Using HPLC-MS to determine the initial rates of activity of γ-ECS, we determined that WT extracts synthesizes γ-EC at a rate of 74pmoles –SH min)−1 mg protein)−1, whereas nrc1 and cad2-1 synthesize γ-EC at a rate of 13 pmoles –SH min−1 mg protein−1 (17.56% of the WT rate) and 14 pmoles –SH min−1 mg protein−1 (18.91% of the WT rate), respectively (Figure 4c). These results suggest that the point mutation in nrc1is as severe as the 6-bp deletion found in the cad2-1 mutant. To further confirm the causative mutation in the nrc1 mutant, we expressed the genomic γ-ECS gene, beginning with the start codon and excluding the 5ʹ and 3ʹ untranslated regions (UTRs), ectopically behind the CaMV 35S promoter in the nrc1 mutant background. Three independent transformant lines (γ-ECS-Comp1–γ-ECS-Comp3) were selected and T2 seedlings from these lines were used for root elongation studies and fluorescence HPLC-MS. Ectopic expression of γ-ECS in the nrc1 mutant background rescued the Cd-sensitive root growth phenotype of the nrc1 mutant (Figure 4d), and greatly decreased the cysteine accumulation phenotype (Figure 4e). Taken together, these results support the conclusion that the identified mutation in γ-ECS is the causative mutation in the nrc1 mutant.
Our HPLC-MS results also suggest that nrc2 inefficiently converts γ-EC into GSH (Figure 3a,c,f). Genetic mapping using an F2 population of an nrc2 × Landsberg erecta (Ler) backcross located the mutation on chromosome 5, between the T21B4 and F15F15 markers (Figure 5a). Based on the thiol profile of nrc2, candidate genes involved in sulfur assimilation and GSH synthesis from this mapping region were PCR amplified and sequenced. The locus At5g27380 contained a single C → T mutation in the 10th exon, causing an Ala → Val (A404V) change in the amino acid sequence of glutathione synthetase (GS), the final enzyme in glutathione biosynthesis (Figure 5a).
Figure 5.
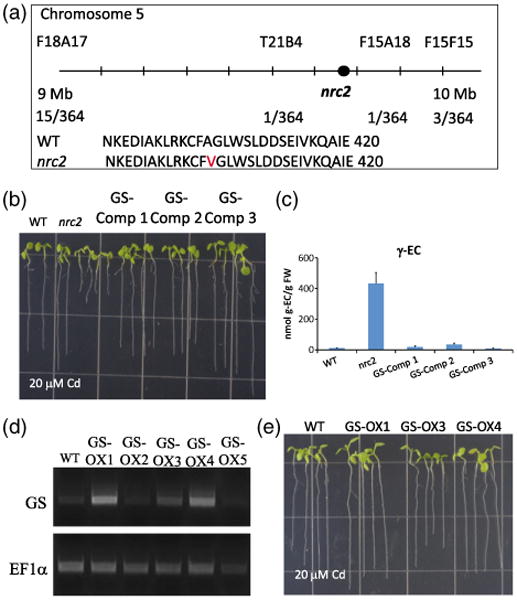
Characterization of the nrc2 non-response to the cadmium mutant as a first recessive viable allele in the Arabidopsis glutathione synthetase gene.
(a) Physical mapping of the nrc2 mutant placed the mutation on chromosome 5, between the F18A17 and F15F15 markers. Candidate gene sequencing identified a point mutation in At5g23780, which causes an Ala → Val (A404V) change in the glutathione synthetase protein.
(b) Expression of genomic GS in the nrc2 mutant behind a constitutive promoter restores root elongation on Cd. Three independent T2 transfor-mants (GS Comp1–Comp3) were used for root elongation experiments on 20 μm Cd.
(c) Seven-day-old complementation lines (GS Comp1–Comp3) were also used for fluorescence HPLC-MS analyses following 48 h of 100 μm Cd treatment. Gamma-glutamylcysteine (γ-EC) levels in the nrc2 mutant are approximately 50-fold higher than in the WT (431.7 ± 72.7 nmol per g fresh weight in nrc2 compared with 8.57 ± 0.25 nmol per g fresh weight in WT), whereas all three of the GS Comp lines show a WT γ-EC accumulation.
(d) The same genomic GS construct was also used to transform WT Col-0 plants to produce GS overexpression lines (GS OX1-OX5). RT-PCR of the GS gene was performed on T2 seedlings to identify lines expressing increased GS transcript levels compared with WT.
(e) T2 GS overexpression lines (GS-OX1, GS-OX3 and GS-OX4) were then used for root elongation experiments to show that extopic GS expression does not increases tolerance to 20 μm Cd. All bar graphs show the mean values from between three and six independent samples. Error bars represent the standard error of the mean (SEM).
No previous viable mutation in the Arabidopsis GS gene has been identified, and GS T-DNA insertion mutants have been shown to be seedling lethal (Pasternak et al., 2008). Therefore, to determine whether the GS point mutation was responsible for the nrc2 mutant phenotype, the genomic GS gene, beginning from the start codon and excluding the 5′ and 3′ UTR regions, was ectopically expressed behind the CaMV 35S promoter in the nrc2 genetic background. Root elongation using T2 transformant seedlings from three independent transformant lines (GS-Comp1–GS-Comp3) grown on 20 μm Cd confirmed that ectopic expression of the GS gene complemented the Cd-sensitive phenotype in the nrc2 mutant (Figure 5b). Furthermore, 21-day-old soil-grown seedlings appeared less chlorotic than the nrc2 mutant (Figure S1). Subsequent HPLC-MS analyses of WT, nrc2 and T2 seedlings treated with 20 μm Cd show that γ-EC levels were drastically decreased in the complemented lines (Figure 5c). To determine whether this increase in Cd tolerance was an artifact of ectopic expression of GS, we also transformed WT Col-0 with the same GS construct, and selected independent T2 transformant lines (GS-OX1–GS-OX5). RT-PCR analysis was performed to select lines showing an increase in GS transcript (Figure 5d) relative to the WT. We then performed root elongation experiments on 20 μm Cd using these overexpression lines and confirmed that ectopic expression of GS does not increase Cd tolerance compared with wild-type lines (Figure 5e). These findings together provide strong evidence that the nrc2 phenotype is caused by the identified recessive point mutation in GS.
Sultr1;2 induction is repressed, even when GSH is depleted
The transcriptional upregulation of sulfate assimilation genes has been described as being part of the plant response to GSH depletion (e.g. PC synthesis during Cd exposure causing GSH depletion; Rouached et al., 2008; Saito, 2004). However, in contrast to this model, the nrc1 and nrc2 mutants showed clear GSH depletion (Figure 3d), but failed to produce a strong induction of the SULTR1;2 promoter-driven luciferase reporter (Figure 2d,e) and the native Sultr1;2 mRNA to wild-type levels after cadmium exposure (Figure 2f,g). These findings point to an alternative hypothesis that the over-accumulation of thiol compounds, either as cysteine (Figure 3d) or γ-EC (Figure 3e), represses the induction of SULTR1;2 gene expression during Cd exposure in these mutants, even though GSH levels are reduced (Figure 3f). A possible mechanism mediating this response may be that the thiol-dependent cellular redox state also contributes to the Cd-induced gene expression of Sultr1;2. To test this hypothesis, we conducted thiol feeding experiments in the presence of cysteine or γ-EC added to the growth medium. Figure 6a shows that the addition of cysteine or γ-EC to the growth media attenuates the induction of SULTR1;2 gene expression in response to Cd. From the above experiments, however, it was unclear whether the addition of cysteine or γ-EC repressed the luciferase activity by extracellular Cd chelation, metabolite repression or altered cellular redox state. Therefore, we analyzed whether the addition of cysteine, GSH, DTT (a non-physiological thiol) and the non-thiol reducing agent, butylated hydroxyanisole (BHA, which is not known to chelate Cd; Gulcin et al., 2003), altered the Cd-dependent induction of SULTR1;2 gene expression. As shown in Figure 6b, feeding the non-physiological reducing agents DTT and BHA repressed Cd-induced luciferase activity (Figure 6b) to a similar degree as the metabolites cysteine or GSH (Figure 6b). These results are consistent with a hypothesis where a reducing cellular environment in the nrc1 and nrc2 mutants, caused by cysteine or γ-EC over-accumulation, represses Cd-induced gene expression despite low GSH levels in these mutants. Thus, a reducing cellular environment would have hierarchical control, repressing SULTR1;2 gene induction (Figure 6c). In this experiment, reducing compounds, including the non-physiological reducing agents DTT and BHA, prevent Cd-induced signal transduction, despite the depletion in GSH levels caused by PC production. Furthermore, our results indicate that an increased level of reducing thiols, including cysteine or γ-EC, inhibits Cd-induced Sultr1;2 gene expression in vivo, even when GSH levels are depleted (Figure 6c).
Figure 6.
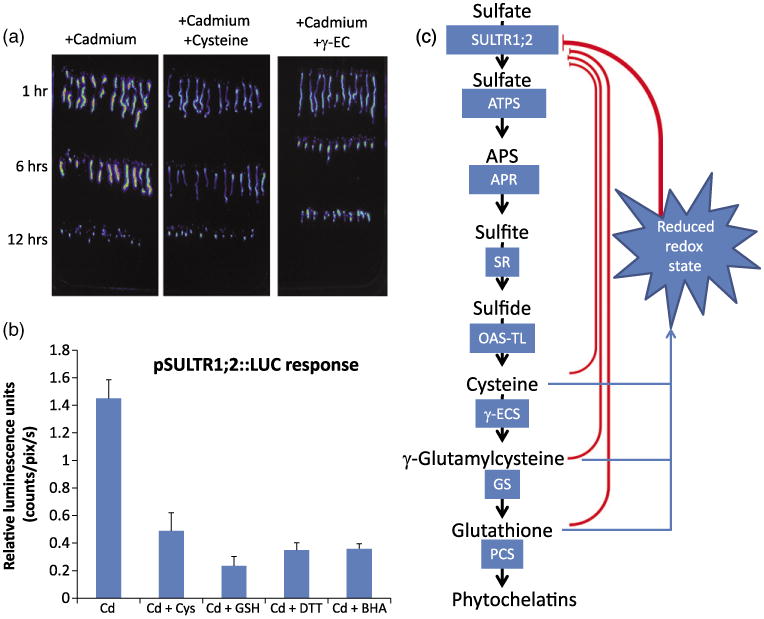
Reducing compounds affect cadmium-dependent induction of the pSULTR1;2∷LUC reporter.
(a) Addition of 1 mm cysteine or γ-EC to the growth media lowers the response of the control pSULTR1;2∷LUC line to cadmium over a 12-h time course.
(b) Cadmium-dependent induction of the luciferase reporter gene is affected by reducing compounds. Exposure to 100 μm CdCl2 induces the transcription of the pSULTR1;2∷LUC reporter gene (Cd). The addition of the reducing compounds cysteine (Cd + 1 mm Cys) or glutathione (Cd + 1 mm GSH) to the growth media strongly represses the cadmium-induced luciferase activity. The non-physiological reducing agents DTT (Cd + 1 mm DTT) and BHA (Cd + 200 μm BHA) also repressed the induction of the reporter gene, suggesting that changes in the cellular redox state is a factor controlling the expression of the SULTR1;2 gene. All images were quantified after 6 h of exposure to the specified compound.
(c) Schematic representation of putative signals involved in the regulation of the high-affinity sulfate transporter, SULTR1;2, during cadmium stress. Glutathione and cysteine are known to repress SULTR1;2 under many conditions, including sulfur starvation and cadmium stress. Characterization of the nrc1 and nrc2 mutants suggests, however, that both GSH demand and changes in cellular redox are required for Cd-dependent transcriptional changes.
Discussion
A luciferase-based genetic screen was devised using Cd-dependent microarray analysis and the Cd-inducible Sultr1;2 promoter. In Arabidopsis the Sultr1;2 transcript is induced by Cd (Rouached et al., 2008) (Figure 1). Microarray analysis (Table S1), RT-PCR analysis (Figures 1a and 2a) and luciferase imaging (Figure 1b,c) demonstrate that the 2.2-kb SULTR1;2 promoter fragment is a rapid and robust reporter of Cd exposure. Furthermore, we show that the SULTR1;2 promoter fragment is induced by a well-defined set of metals and metalloids (arsenic), but is induced less well by ROS-inducing agents, such as paraquat (Figure 1c–i).
Our Cd-inducible reporter screen allowed us to identify mutants with decreased, constitutive and increased activity of the reporter gene (Figure 2). These classes of mutants suggest that sulfate uptake in Arabidopsis is regulated by antagonistic transcriptional activators and repressors. To date, one transcriptional regulator of SULTR1;2, SLIM1, has been reported and is regulated under sulfur limiting conditions (Maruyama-Nakashita et al., 2006). It remains unknown whether SLIM1 functions in the Cd response, and none of our strong nrc mutants mapped to the SLIM1 gene. The components of the Cd-dependent transcriptional signaling pathway in Arabidopsis remain unknown. Isolation and characterization of the Arabidopsis nrc mutants was pursued to advance our understanding of the levels of genetic and mechanistic regulation of the sulfate assimilation pathway that occurs during Cd exposure.
Current model of sulfur homeostasis and GSH depletion during Cd stress
Glutathione is known to be important for mitigating stress as the GSH-deficient mutants cad2-1, pad2, rax1-1 and zir1 have all been identified by their sensitivity to abiotic or biotic stresses (Cobbett et al., 1998; Ball et al., 2004; Parisy et al., 2007; Shanmugam et al., 2011). Another GSH-deficient mutant, rml, was identified as lacking a root meristem and having a severe growth and developmental phenotype (Vernoux et al., 2000). These mutants display GSH depletion of various degrees, with the rml mutant having the least GSH (approximately 3% of wild-type levels) and rax1-1 having the highest GSH levels (approximately 50% of wild-type levels) (Vernoux et al., 2000; Ball et al., 2004; Shanmugam et al., 2011). Whereas the severity of these mutations is typically linked to the degree of GSH depletion, Shanmugam et al. (2011) have recently shown by systematically analyzing the iron-induced zinc tolerance of each of these mutants that a threshold level of GSH is required for some phenotypes. These results suggest that the phenotypes observed in these mutants may not be linearly correlated with GSH levels alone. Furthermore, the nrc1 × cad2-1 F1 plants (Figure 4b) showed slightly longer root growth than either the nrc1 mutant or the cad2-1 mutant alone. This suggests that when the two mutant alleles are expressed together, despite having similar GSH content, the nrc1 × cad2-1 F1 crosses are slightly less sensitive to Cd. One explanation for this observation is that in the nrc1 × cad2-1 cross, the γ-ECS dimer is more functional than in the nrc1 or cad2-1 offspring (Hothorn et al., 2006; Gromes et al., 2008). This would be consistent with recent findings showing that the regulation of γ-ECS in plants is complex and occurs at both the transcriptional and post-transcriptional levels (Hothorn et al., 2006; Gromes et al., 2008).
The current model for sulfur homeostasis in plants proposes that GSH, the most abundant organic thiol in plants, is a strong negative regulator of both sulfate assimilation and cysteine biosynthesis. Glutathione is known to repress the expression and activity of high-affinity sulfate transporters, ATP sulfurylase and APS reductase (Kopriva, 2006). Cellular GSH levels decrease during Cd stress as a result of PC production (Rauser, 1995). This GSH depletion causes an increase in cellular GSH demand, increasing the transcription of sulfate uptake-related genes (i.e. SULTR1;2) (Kopriva and Rennenberg, 2004; Kopriva, 2006) (Figure 1). This model argues that as sulfate assimilation restores thiol levels, GSH represses sulfate uptake and assimilation genes (Kopriva and Rennenberg, 2004; Kopriva, 2006). Tight regulation of GSH synthesis is needed because of the high reactivity yet essential nature of GSH. Feedback regulation allows the rapid activation of sulfate assimilation during a sudden decrease in GSH levels. This model accounts for many observations, but it assumes that GSH is the major regulator of sulfate assimilation during Cd stress. Growing evidence indicates that cysteine and H2S are also potent repressors of sulfate transporters; however, it is unclear if cysteine and H2S repression are direct or mediated through an increase in GSH (Lappartient and Touraine, 1996; Vauclare et al., 2002; Maruyama-Nakashita et al., 2004). Here, by isolating and characterizing mutations that insulate variations in cysteine and γ-glutamylcysteine from changes in GSH concentration, we show the key role of sulfur-containing compounds synthesized prior to GSH production in repressing Cd-induced gene expression (Figure 5c).
nrc mutants reveal cysteine and γ-EC as potent SULTR1;2 repressors
The identification and characterization of the nrc1 and nrc2 mutants suggest that another level of Cd-induced gene expression regulation exists. The nrc1 mutant is GSH deficient but accumulates high levels of cysteine (Figure 3b,d,e), whereas the nrc2 mutant has decreased GSH levels but accumulates high levels of γ-EC (Figure 4c–e). According to the current model, the GSH status of these mutants should induce SULTR1;2, particularly after Cd exposure. Thus, we would expect these mutants to be constitutive or super-response mutants (Figure 2). However, in the nrc1 mutant, Cd-induced SULTR1;2 gene expression was strongly repressed, and in the nrc2 mutant, Cd-induced SULTR1;2 gene expression was decreased (Figure 2d,e).
We hypothesized that the aberrant accumulation of thiol compounds in the nrc1 (cysteine) and nrc2 (γ-EC) mutants also caused a reducing redox environment during Cd exposure, leading to a downregulation of SULTR1;2 (Figure 6c). Our findings indicate that this reducing cellular state may contribute as a repression mechanism of sulfate assimilation genes in response to Cd stress. Evidence to support this was obtained by feeding experiments with physiological (cysteine and GSH) and non-physiological (DTT) reducing agents, as well as reducing agents not known to chelate Cd (BHA) (Figure 6b). Interestingly, feeding cysteine, γ-EC or GSH to the pSULTR∷LUC parental line lowered the Cd-induced luciferase response (Figure 6a,b). Furthermore, feeding non-physiological reducing agents such as DTT and BHA also decreased the Sultr1;2 induction during Cd exposure (Figure 6b). These results, together with our thiol profiling of isolated genetic mutants, are inconsistent with a solely GSH depletion-driven model, and point to a model where GSH depletion, upstream thiol concentrations and an oxidized cellular redox state are required to induce sulfate assimilation in Arabidopsis in response to Cd stress (Figure 6c). The elevated cysteine or γ-EC thiol levels and reducing cellular conditions in the nrc1 and nrc2 mutants are proposed to repress the induction of sulfate assimilation genes, despite the low GSH concentrations in the mutants (Figure 3d). Thus, our results suggest that sulfate assimilation in Arabidopsis is controlled in a hierarchical manner by upstream thiols, the redox state of the cell and the concentration of GSH (Figure 6c). However, oxidative stress alone is not sufficient for mediating Cd-induced SULTR1;2 expression. Indeed, oxidizing agents such as paraquat (Figure 1f) and H2O2 did not induce SULTR1;2 expression as highly or rapidly as Cd (Figure 1), presumably because they do not cause a decrease in organic thiols despite altering the cellular redox state (Figure 6c).
Regulation of sulfate assimilation and glutathione biosynthesis
The Sultr1;2 transcript is induced by several stresses, including Cd, attack by pathogens and sulfur deprivation (Maruyama-Nakashita et al., 2006). The SULTR1;2 promoter was previously used as a reporter gene to screen for mutants unable to induce genes regulated during sulfur starvation (Maruyama-Nakashita et al., 2005). This screen led to the identification of SLIM1 (EIL3), an ethylene insensitive-like transcription factor that regulates the expression of SULTR1;2 and of genes that mediate glucosinolate synthesis (Maruyama-Nakashita et al., 2006). The SLIM1 protein has been proposed to be a transcriptional activator under conditions of sulfur starvation (Segarra et al., 2009). Presently, there is no direct evidence supporting its role as a transcriptional activator of the SULTR1;2 promoter. To date it is not known whether SLIM1 directly regulates the expression of Sultr1;2 or whether its function is independent of O-acetylserine (a precursor of cysteine) and GSH concentrations (Maruyama-Nakashita et al., 2006). In summary, the isolation and characterization of the non-response to Cd mutants nrc1 and nrc2 points to a new model (Figure 6c) for the regulation of gene expression in response to Cd stress, in which several criteria are necessary for Cd-induced gene expression: (i) GSH depletion; (ii) depletion of upstream thiol levels; (iii) an oxidative cellular redox state, which together control the Cd-induced transcription of SULTR1;2.
Experimental procedures
Arabidopsis accessions
The WT Arabidopsis thaliana ecotypes used for mapping were Columbia (Col-0) and Landsberg erecta (Ler-0). The nrc1 and nrc2 mutants are in the Col-0 genetic background, and the transformant line pSULTR1;2∷LUC is also in the Col-0 genetic background.
Plant growth conditions
Seeds were sterilized and plated on plates containing quarter-strength MS standard medium (M5519; Sigma-Aldrich, http:// www.sigmaaldrich.com), 1 mm 2-(N-morpholine)-ethanesulphonic acid (MES), 1% phytoagar (Duchefa, http://www.duchefa.com) and the pH adjusted to 5.6 with 1.0 m KOH (Maser et al., 2002; Lee et al., 2003). Sterilized nylon mesh with a 200-μm pore size (Spectrum Labs, http://www.spectrumlabs.com) was placed on the surface of the media prior to sowing the seeds. The seeds were then stratified with cold treatment at 4°C for 48 h, and grown under growth room conditions for 5 days (300 μmol m−2 s−1,70% Hr, 16-h light at 21°C/8-h dark at 18°C)(Sung et al., 2007). Seedlings were then transferred to quarter-strength MS, 1 mm MES and 1% agar plates containing 20 μm CdCl2.
Construction of cadmium-response luciferase reporter line
Nine Arabidopsis promoters (2.2 kb before the ATG) from genes found to be induced by cadmium were cloned into the pPZPXomegaL+ vector (kindly provided by Dr. Steve Kay) with BamH1 and/or HindIII restriction digestion. The correct orientation of the promoters was confirmed by sequencing the cloned promoter regions in the vector. These nine luciferase constructs were transformed into Arabidopsis and the resulting T3 homozygous transgenic luciferase plant lines were tested for induction of luciferase protein by monitoring bioluminescence for up to 12 h after exposure to Cd. Of the nine transgenic luciferase reporter lines, several reporter lines containing the promoter of a high-affinity sulfate transporter gene (SULTR1;2) showed the most reproducible induction of luciferase, and were hence selected as the reporter lines for the cadmium transcriptional response screening.
Mutant isolation by luciferase luminescence imaging
Homozygous T3 seeds of a pSULTR1;2 reporter line were mutagenized with 0.25% EMS for 14 h. The survival of 50% of the mutagenized seeds was confirmed as an indicator of adequate mutagenesis. M1 seeds were bulk harvested from approximately 6000 M0 plants and approximately 200 000 M1 seeds were screened for altered luminescence patterns in response to cadmium treatment (200 μm for 6 h). This is between two and four times more than the minimum M1 population required to find a mutation in any given G:C pair (Jander et al., 2003). For luciferase imaging, the protocol described by Chinnusamy et al. (2002) was followed with the following modifications (Chinnusamy et al., 2002). Seedlings were grown for 5 days horizontally on 36-μm Nitex mesh (Small Parts, Seattle, WA, USA) before being presprayed with 5 mm luciferin (Promega, http://www.promega.com) 6 h before being transferred to either control or treatment plates in order to minimize non-specific luminescence. After transfer the seedlings were subjected to another spraying of 5 mm luciferin and incubated for a period of time (as indicated in the results section) before being imaged using a BERTHOLD NightOWL LB981 imaging system (EG&G Berthold, http://www.berthold.com). A 2-min exposure time was used for capturing the bioluminescent images. Luminescence was quantified using nightowl.
Total RNA isolation and RT-PCR analysis
Plant materials were flash frozen into liquid nitrogen immediately after treatments. Plant materials were ground using a pre-chilled mortar and pestle. A 100-mg portion of ground plant powder was used to extract total RNA by using a commercial RNA extraction kit (Qiagen, http://www.qiagen.com) (Sunarpi et al., 2005). The quantity and quality of total RNA was recorded using spectrophotometry and gel electrophoresis. A 5-μg portion of total RNA was treated with DNase1 (Ambion, now Invitrogen, http://www.invitrogen.com) to remove DNA contamination from total RNA samples. Prior to the reverse-transcription reaction, DNase-treated total RNA was heated to 65°C for 10 min and immediately cooled down in ice to minimize the secondary structures of total RNAs. A 1-μg portion of total RNA was reverse transcribed with a NotI-d(T)18 primer using a First Strand cDNA kit (GE Healthcare, http:// www.gehealthcare.com) for 60 min at 37°C. Reverse-transcribed cDNA was subjected to PCR to amplify the expression signal of each gene, with the following typical conditions: initial denaturation of cDNA/RNA and inactivation of reverse transcriptase at 95°C for 5 min, then DNA amplification with 25–40 cycles of 95°C for 15 s, 52°C for 15 s, 72°C for 1 min, then a final extension at 72°C for 5 min using an MJ Research PTC 100 Thermal Cycler (GMI, http:// www.gmi-inc.com). As a loading control, elongation factor-1α (EF-1α) mRNA was analyzed. Table S2 contains a complete list of all primers used in this study.
Thiol measurements by fluorescence HPLC
Thiol-containing compounds in plant samples, including cysteine, γ-EC, GSH and PCs, were analyzed using fluorescence detection HPLC, as described by Fahey and Newton (1987). To analyze the levels of thiol compounds produced by plants in response to treatment, plants were grown on minimal growth media plates for 5 days then transferred to fresh media plates containing 200 μm cadmium. In order to minimize the oxidation of thiol compounds during the extraction, plant seedlings were flash-frozen in liquid nitrogen, and then ground and extracted as previously described (Sung et al., 2009). The peaks of thiol compounds were identified by coupled parallel mass spectrometry measurement, as previously described (Chen et al., 2006), and quantified using xcalibur (Thermo Scientific, http:// www.thermoscientific.com). To identify the peptides from plant extracts, PC2, PC3 and PC4 standards were synthesized on a MILLIGEN 9050 PepSynthesizer (Millipore, http://www.millipore. com) using Fmoc-Glu-OtBu (Bachem, http://www.bachem.com). Other thiol standards, such as glutathione, cysteine, γ-EC and NAC, were purchased from Sigma-Aldrich. All reported thiol quantities are means of between three and nine biologically independent samples, and error bars indicate the standard error of the mean (SEM).
Plant growth conditions and cadmium treatment
For plate-based assays, including heavy metal treatments, luciferase luminescence assay and plant growth for RT-PCR analysis, seeds were germinated on nylon mesh (Spectrum Laboratories Inc., http://www.spectrumlabs.com) on quarter-strength MS media and grown for 1 week at 22°C, 75% humidity, with a 16-h light/8-h dark photoperiod regime at approximately 75 μmol m−2 s−1 light intensity in a Conviron growth chamber (Controlled Environments Inc., http://www.conviron.com). Seedlings on nylon mesh were transferred either to treatment or control plates and incubated for up to 12 h, as previously described (Sung et al., 2009).
Feeding experiments
We performed feeding experiments using the control pSULTR1;2∷LUC reporter line (Figure 1). We exposed 5-day-old seedlings to 100 μm Cd, 100 μm Cd + 2500 μm Cys, 100 μm Cd + 2500 μm DTT, 100 μm Cd + 300 μm BHA or 100 μm Cd + 2500 μm GSH for 6 h (Figure 4e), before performing luciferase imaging on the roots and quantifying the relative luciferase luminescence.
nrc1 and cad2-1 enzyme activity experiments
The activity of γ-ECS was measured in protein extracts using HPLC fluorescence and thiol-derivatization with monobromobimane, using a modified version of the protocol described by Hell and Bergmann (1990). Rosette leaves from wild-type (Col-0), nrc1 or cad2-1 were ground in liquid nitrogen and 100-300 mg of ground tissue were mixed 1:1 with extraction buffer (100 mm Tris-HCl, 10 mm MgCl2, 1 mm EDTA, pH 8.0). Unless otherwise stated, all steps were carried out at 4°C and all buffers were saturated with nitrogen prior to being used to minimize the oxidation of thiols during protein extraction and enzymatic activity measurements. The protein extract was centrifuged for 10 min at 10 000 g, and the supernatant was desalted using Sephadex G-25 previously equilibrated with extraction buffer. The enzyme activity assay (500 μl final volume) contained 100 mm Tris (pH 8.0), 50 mm MgCl2, 20 mm glutamate, 5 mm ATP and an ATP regenerating system that consisted of 5 mm phosphoenolpyruvate, 1 mm DTT and 10 U ml−1 of pyruvate kinase. The protein extract was incubated for 10 min at 30°C, and γ-ECS activity was started by adding 1 mm cysteine. At defined time points, 50 μl of the reaction solution were taken and thiols were derivatized by adding 40 μl of monobromobimane (2 mm) and incubated for 20 min at 40°C. Derivatization was stopped by adding 10 μl of PCA30% v/v. Samples were mixed using a vortex and protein was precipitated by centrifugation (10 min at 10 000 g). The supernatant was filtered using 0.45-μm Ultrafree-MC filters (Amicon; Millipore) before being analyzed by HPLC as described previously (Mendoza-Cozatl et al., 2008). The quantity of γ-EC synthesized over time was normalized to the protein content in the plant extract, quantified using the Bradford reagent (Sigma-Aldrich) and BSA as a protein standard.
nrc1 and nrc2 complementation
All primers used for PCR amplification for cloning are listed in Table S2. For constitutive γ-ECS expression in the nrc1 mutant, a γ-ECS genomic DNA fragment was amplified from Col-0 gDNA using the primers TJ199-F and TJ199-R (0-3294 bp). The amplified genomic γ-ECS DNA fragment, which excluded both the 5′ and 3′ untranslated regions (UTRs), was cloned into pENTR/D-TOPO® (Invitrogen), following the manufacturer's instructions. The CaMV 35S:γ-ECS-NOS construct was obtained by recombining the γ-ECS genomic sequence into a Gateway®-compatible pGreenII plasmid (Hellens et al., 2000) containing the 35Spro and the NOS terminator (35Spro:GW-NOSter), using LR Clonase II® (Invitrogen). The pGREE-NII 35S:γ-ECS-NOS construct was transformed using electroporation into Agrobacterium tumefacians strain GV3101.
For constitutive GS expression in the nrc2 mutant, a GS genomic DNA fragment was amplified from Col-0 gDNA using the primers TJ198-R and TJ198-R (0-2702 bp). The amplified genomic GS DNA fragment, which excluded both the 5′ and 3′ UTRs, was cloned into pENTR/D-TOPO® (Invitrogen), following the manufacturer's instructions. The CaMV 35S:GS-NOS construct was obtained by recombining the GS genomic sequence into a Gateway®-compatible pGreenII plasmid (Hellens et al., 2000) containing the 35Spro and the NOS terminator (35Spro:GW-NOSter), using LR Clonase II® (Invitrogen). Note that our attempts to complement nrc2 using a 35S-driven GS cDNA were not successful, whereas the genomic DNA complimented the nrc2 growth and yellowing phenotype (Figure S1) in six independent lines (all isolated transformants). We tested the complementation of the Cd-dependent root growth and thiol accumulation in three of these lines, as shown in the results section. The pGREENII 35S:GS-NOS construct was transformed using electroporation into Agrobacterium tumefacians strain GV3101.
Arabidopsis thaliana was transformed using the floral-dip method (Clough and Bent, 1998) with the GV3101 strains described above. The helper plasmid, pSoup, was used for the pGreenII-carrying strains (Hellens et al., 2000). The pGREENII 35S:GS-NOS construct was used to transform the nrc2 mutant for complementation and Col-0, whereas the pGREENII 35S:γ-ECS-NOS construct was used to transform the nrc1 mutant for complementation. Hygromycin selection of transformants was performed in both the T1 and T2 generations.
Supplementary Material
Figure S1. Visual phenotype of 20-day-old nrc2 mutant plants grown in soil.
Table S1. A list of the 30 genes most influenced by cadmium exposure, ranked in order of decreasing induction.
Table S2. A list of primers used for RT-PCR, promoter cloning, gene sequencing and complementation experiments.
Please note: As a service to our authors and readers, this journal provides supporting information supplied by the authors. Such materials are peer-reviewed and may be re-organized for online delivery, but are not copy-edited or typeset. Technical support issues arising from supporting information (other than missing files) should be addressed to the authors.
Acknowledgments
This research was supported by the National Institute of Environmental Health Sciences (grant no. ES010337; JIS and EAK). The Division of Chemical Sciences, Geosciences, and Biosciences, Office of Basic Energy Sciences of the U.S. Department of Energy Grant DE-FG02-03ER15449 supported the metal-dependent screen. DGMC is the recipient of a PEW Latin American Fellowship. TOJ was supported by the UCSD-Salk IGERT Plant Systems Biology Interdisciplinary Graduate Training Program (grant no. 0504645). Cadmium-dependent microarray data are accessible through GEO Series accession number GSE35869 (http://www.ncbi.nlm.nih.gov/ geo/query/acc.cgi?acc=GSE35869).
Footnotes
Supporting information: Additional Supporting Information may be found in the online version of this article:
References
- Allen JW, Shanker G, Tan KH, Aschner M. The consequences of methylmercury exposure on interactive functions between astrocytes and neurons. Neurotoxicology. 2002;23:755–759. doi: 10.1016/S0161-813X(01)00076-6. [DOI] [PubMed] [Google Scholar]
- Aschner M, Walker SJ. The neuropathogenesis of mercury toxicity. Mol Psychiatry. 2002;7(Suppl 2):S40–41. doi: 10.1038/sj.mp.4001176. [DOI] [PubMed] [Google Scholar]
- Ball L, Accotto GP, Bechtold U, et al. Evidence for a direct link between glutathione biosynthesis and stress defense gene expression in Arabidopsis. Plant Cell. 2004;16:2448–2462. doi: 10.1105/tpc.104.022608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen A, Komives EA, Schroeder JI. An improved grafting technique for mature Arabidopsis plants demonstrates long-distance shoot-to-root transport of phytochelatins in Arabidopsis. Plant Physiol. 2006;141:108–120. doi: 10.1104/pp.105.072637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chinnusamy V, Stevenson B, Lee Bh, Zhu JK. Screening for gene regulation mutants by bioluminescence imaging. Sci STKE. 2002;2002:pl10. doi: 10.1126/stke.2002.140.pl10. [DOI] [PubMed] [Google Scholar]
- Clemens S. Toxic metal accumulation, responses to exposure and mechanisms of tolerance in plants. Biochimie. 2006;88:1707–1719. doi: 10.1016/j.biochi.2006.07.003. [DOI] [PubMed] [Google Scholar]
- Clemens S, Kim EJ, Neumann D, Schroeder JI. Tolerance to toxic metals by a gene family of phytochelatin synthases from plants and yeast. EMBO J. 1999;18:3326–3333. doi: 10.1093/emboj/18.12.3325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clemens S, Schroeder J, Degenkolb T. Caenorhabditis elegans expresses a functional phytochelatin synthase. Eur J Biochem. 2001;268:3640–3643. doi: 10.1046/j.1432-1327.2001.02293.x. [DOI] [PubMed] [Google Scholar]
- Clough SJ, Bent AF. Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 1998;16:735–743. doi: 10.1046/j.1365-313x.1998.00343.x. [DOI] [PubMed] [Google Scholar]
- Cobbett CS, May MJ, Howden R, Rolls B. The glutathione-deficient, cadmium-sensitive mutant, cad2-1, of Arabidopsis thaliana is deficient in gamma-glutamylcysteine synthetase. Plant J. 1998;16:73–78. doi: 10.1046/j.1365-313x.1998.00262.x. [DOI] [PubMed] [Google Scholar]
- Davidian JC, Kopriva S. Regulation of sulfate uptake and assimilation—the same or not the same? Mol Plant. 2010;3:314–325. doi: 10.1093/mp/ssq001. [DOI] [PubMed] [Google Scholar]
- Dushenkov V, Kumar P, Motto H, Raskin I. Rhizofiltration – the use of plants to remove heavy metals from aqueous streams. Environ Sci Tech. 1995;29:1239–1245. doi: 10.1021/es00005a015. [DOI] [PubMed] [Google Scholar]
- Fahey RC, Newton GL. Determination of low-molecular-weight thiols using monobromobimane fluorescent labeling and high-performance liquid chromatography. Methods Enzymol. 1987;143:85–96. doi: 10.1016/0076-6879(87)43016-4. [DOI] [PubMed] [Google Scholar]
- Gong JM, Lee DA, Schroeder JI. Long-distance root-to-shoot transport of phytochelatins and cadmium in Arabidopsis. Proc Natl Acad Sci USA. 2003;100:10118–10123. doi: 10.1073/pnas.1734072100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grill E, Loeffler S, Winnacker EL, Zenk MH. Phytochelatins: the principal heavy-metal complexing peptides of higher plants. Science. 1985;230:674–676. doi: 10.1126/science.230.4726.674. [DOI] [PubMed] [Google Scholar]
- Gromes R, Hothorn M, Lenherr ED, Rybin V, Scheffzek K, Rausch T. The redox switch of γ-glutamylcysteine ligase via a reversible monomer–dimer transition is a mechanism unique to plants. Plant J. 2008;54:1063–1075. doi: 10.1111/j.1365-313X.2008.03477.x. [DOI] [PubMed] [Google Scholar]
- Gulcin I, Buyukokuroglu ME, Kufrevioglu OI. Metal chelating and hydrogen peroxide scavenging effects of melatonin. J Pineal Res. 2003;34:278–281. doi: 10.1034/j.1600-079x.2003.00042.x. [DOI] [PubMed] [Google Scholar]
- Ha SB, Smith AP, Howden R, Dietrich WM, Cobbett C. Phytochelatin synthase genes from Arabidopsis and the yeast Schizosac-charomyces pombe. Plant Cell. 1999;11:1153–1164. doi: 10.1105/tpc.11.6.1153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heck JE, Andrew AS, Onega T, Rigas JR, Jackson BP, Karagas MR, Duell EJ. Lung cancer in a U.S. population with low to moderate arsenic exposure. Environ Health Perspect. 2009;117:1718–1723. doi: 10.1289/ehp.0900566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hell R, Bergmann L. Gamma-glutamylcysteine synthetase in higher-plants - catalytic properties and subcellular-localization. Planta. 1990;180:603–612. doi: 10.1007/BF02411460. [DOI] [PubMed] [Google Scholar]
- Hellens RP, Edwards EA, Leyland NR, Bean S, Mullineaux PM. pGreen: a versatile and flexible binary Ti vector for Agrobacterium-mediated plant transformation. Plant Mol Biol. 2000;42:819–832. doi: 10.1023/a:1006496308160. [DOI] [PubMed] [Google Scholar]
- Herbette S, Taconnat L, Hugouvieux V, et al. Genome-wide transcriptome profiling of the early cadmium response of Arabidopsis roots and shoots. Biochimie. 2006;88:1751–1765. doi: 10.1016/j.biochi.2006.04.018. [DOI] [PubMed] [Google Scholar]
- Hothorn M, Wachter A, Gromes R, Stuwe T, Rausch T, Scheffzek K. Structural basis for the redox control of plant glutamate cysteine ligase. J Biol Chem. 2006;281:27557–27565. doi: 10.1074/jbc.M602770200. [DOI] [PubMed] [Google Scholar]
- Howden R, Andersen CR, Goldsbrough PB, Cobbett CS. A cadmium-sensitive, glutathione-deficient mutant of Arabidopsis thaliana. Plant Physiol. 1995;107:1067–1073. doi: 10.1104/pp.107.4.1067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jander G, Baerson SR, Hudak JA, Gonzalez KA, Gruys KJ, Last RL. Ethylmethanesulfonate saturation mutagenesis in Arabidopsis to determine frequency of herbicide resistance. Plant Physiol. 2003;131:139–146. doi: 10.1104/pp.102.010397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klein M, Mamnum Y, Eggmann T, Schuller C, Wolfger H, Martinoia E, Kuchler K. The ATP-binding cassette (ABC) transporter Bpt1p mediates vacuolar sequestration of glutathione conjugates in yeast. FEBS Lett. 2002;520:63–67. doi: 10.1016/s0014-5793(02)02767-9. [DOI] [PubMed] [Google Scholar]
- Kopriva S. Regulation of sulfate assimilation in Arabidopsis and beyond. Ann Bot. 2006;97:479–495. doi: 10.1093/aob/mcl006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kopriva S, Rennenberg H. Control of sulphate assimilation and glutathione synthesis: interaction with N and C metabolism. J Exp Bot. 2004;55:1831–1842. doi: 10.1093/jxb/erh203. [DOI] [PubMed] [Google Scholar]
- Kramer U, Pickering IJ, Prince RC, Raskin I, Salt DE. Subcellular localization and speciation of nickel in hyperaccumulator and non-accumulator thlaspi species. Plant Physiol. 2000;122:1343–1354. doi: 10.1104/pp.122.4.1343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lappartient AG, Touraine B. Demand-driven control of root ATP sulfurylase activity and SO42 – uptake in intact canola (the role of phloem-translocated glutathione) Plant Physiol. 1996;111:147–157. doi: 10.1104/pp.111.1.147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee S, Leustek T. The affect of cadmium on sulfate assimilation enzymes in Brassica juncea. Plant Sci. 1999;141:201–207. [Google Scholar]
- Lee J, Reeves RD, Brooks RR, Jaffré T. The relation between nickel and citric acid in some nickel-accumulating plants. Phytochemistry. 1978;17:1033–1035. [Google Scholar]
- Lee DA, Chen A, Schroeder JI. Ars1, an Arabidopsis mutant exhibiting increased tolerance to arsenate and increased phosphate uptake. Plant J. 2003;35:637–646. doi: 10.1046/j.1365-313x.2003.01835.x. [DOI] [PubMed] [Google Scholar]
- Li Y, Dhankher O, Carreira L, Lee D, Chen A, Schroeder J, Balish R, Meagher R. Overexpression of phytochelatin synthase in Arabidopsis leads to enhanced arsenic tolerance and cadmium hypersensitivity. Plant Cell Physiol. 2004;45:1787–1797. doi: 10.1093/pcp/pch202. [DOI] [PubMed] [Google Scholar]
- Maruyama-Nakashita A, Nakamura Y, Yamaya T, Takahashi H. Regulation of high-affinity sulphate transporters in plants: towards systematic analysis of sulphur signalling and regulation. J Exp Bot. 2004;55:1843–1849. doi: 10.1093/jxb/erh175. [DOI] [PubMed] [Google Scholar]
- Maruyama-Nakashita A, Nakamura Y, Watanabe-Takahashi A, Inoue E, Yamaya T, Takahashi H. Identification of a novel cis-acting element conferring sulfur deficiency response in Arabidopsis roots. Plant J. 2005;42:305–314. doi: 10.1111/j.1365-313X.2005.02363.x. [DOI] [PubMed] [Google Scholar]
- Maruyama-Nakashita A, Nakamura Y, Tohge T, Saito K, Takahashi H. Arabidopsis SLIM1 is a central transcriptional regulator of plant sulfur response and metabolism. Plant Cell. 2006;18:3235–3251. doi: 10.1105/tpc.106.046458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mäser P, Eckelman B, Vaidyanathan T, et al. Altered shoot/root Na+ distribution and bifurcating salt sensitivity in Arabidopsis by genetic disruption of the Na+ transporter AtHKT1. FEBS Lett. 2002;531:157–533. doi: 10.1016/s0014-5793(02)03488-9. [DOI] [PubMed] [Google Scholar]
- Mendoza-Cozatl DG, Butko E, Springer F, Torpey JW, Komives EA, Kehr J, Schroeder JI. Identification of high levels of phytochelatins, glutathione and cadmium in the phloem sap of Brassica napus. A role for thiol-peptides in the long-distance transport of cadmium and the effect of cadmium on iron translocation. Plant J. 2008;54:249–259. doi: 10.1111/j.1365-313X.2008.03410.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mendoza-Cozatl DG, Zhai Z, Jobe TO, et al. Tonoplast-localized Abc2 transporter mediates phytochelatin accumulation in vacuoles and confers cadmium tolerance. J Biol Chem. 2010;285:40416–40426. doi: 10.1074/jbc.M110.155408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mendoza-Cozatl DG, Jobe TO, Hauser F, Schroeder JI. Long-distance transport, vacuolar sequestration, tolerance, and transcriptional responses induced by cadmium and arsenic. Curr Opin Plant Biol. 2011;14:554–562. doi: 10.1016/j.pbi.2011.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nocito FF, Lancilli C, Crema B, Fourcroy P, Davidian JC, Sacchi GA. Heavy metal stress and sulfate uptake in maize roots. Plant Physiol. 2006;141:1138–1148. doi: 10.1104/pp.105.076240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohta H, Ichikawa M, Seki Y. Effects of cadmium intake on bone metabolism of mothers during pregnancy and lactation. Tohoku J Exp Med. 2002;196:33–42. doi: 10.1620/tjem.196.33. [DOI] [PubMed] [Google Scholar]
- Parisy V, Poinssot B, Owsianowski L, Buchala A, Glazebrook J, Mauch F. Identification of PAD2 as a γ-glutamylcysteine synthetase highlights the importance of glutathione in disease resistance of Arabidopsis. Plant J. 2007;49:159–172. doi: 10.1111/j.1365-313X.2006.02938.x. [DOI] [PubMed] [Google Scholar]
- Pasternak M, Lim B, Wirtz M, Hell R, Cobbett CS, Meyer AJ. Restricting glutathione biosynthesis to the cytosol is sufficient for normal plant development. Plant J. 2008;53:999–1012. doi: 10.1111/j.1365-313X.2007.03389.x. [DOI] [PubMed] [Google Scholar]
- Raskin I, Kumar PBAN, Dushenkov S, Salt DE. Bioconcentration of heavy metals by plants. Curr Opin Biotechnol. 1994;5:285–290. [Google Scholar]
- Rauser WE. Phytochelatins and related peptides-structure, biosynthesis, and function. Plant Physiol. 1995;109:1141–1149. doi: 10.1104/pp.109.4.1141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richau KH, Kozhevnikova AD, Seregin IV, Vooijs R, Koevoets PLM, Smith JAC, Ivanov VB, Schat H. Chelation by histidine inhibits the vacuolar sequestration of nickel in roots of the hyperaccumulator Thlaspi caerulescens. New Phytol. 2009;183:106–116. doi: 10.1111/j.1469-8137.2009.02826.x. [DOI] [PubMed] [Google Scholar]
- Rouached H, Wirtz M, Alary R, Hell R, Arpat AB, Davidian JC, Fourcroy P, Berthomieu P. Differential regulation of the expression of two high-affinity sulfate transporters, SULTR1.1 and SULTR1.2, in Arabidopsis. Plant Physiol. 2008;147:897–911. doi: 10.1104/pp.108.118612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saito K. Sulfur assimilatory metabolism. The long and smelling road. Plant Physiol. 2004;136:2443–2450. doi: 10.1104/pp.104.046755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salt DE, Blaylock M, Kumar NP, Dushenkov V, Raskin I. Phytoremediation – a novel strategy for the removal of toxic metals from the environment using plants. Bio-Technology. 1995;13:468–474. doi: 10.1038/nbt0595-468. [DOI] [PubMed] [Google Scholar]
- Salt DE, Smith RD, Raskin I. Phytoremediation. Annu Rev Plant Physiol Plant Mol Biol. 1998;49:643–668. doi: 10.1146/annurev.arplant.49.1.643. [DOI] [PubMed] [Google Scholar]
- Sanchez-Fernandez R, Davies T, Coleman J, Rea P. The Arabidopsis thaliana ABC protein superfamily, a complete inventory. J Biol Chem. 2001;276:30231–30244. doi: 10.1074/jbc.M103104200. [DOI] [PubMed] [Google Scholar]
- Satarug S, Garrett SH, Sens MA, Sens DA. Cadmium, environmental exposure, and health outcomes. Environ Health Perspect. 2010;118:182–190. doi: 10.1289/ehp.0901234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Segarra G, Van der Ent S, Trillas I, Pieterse CMJ. MYB72, a node of convergence in induced systemic resistance triggered by a fungal and a bacterial beneficial microbe. Plant Biol. 2009;11:90–96. doi: 10.1111/j.1438-8677.2008.00162.x. [DOI] [PubMed] [Google Scholar]
- Shanmugam V, Tsednee M, Yeh KC. ZINC TOLERANCE INDUCED BY IRON 1 reveals the importance of glutathione in the cross-homeostasis between zinc and iron in Arabidopsis thaliana. The Plant Journal. 2012 doi: 10.1111/j.1365-313X.2011.04850.x.\. [DOI] [PubMed] [Google Scholar]
- Song WY, Park J, Mendoza-Co´zatl DG, et al. Arsenic tolerance in Arabidopsis is mediated by two ABCC-type phytochelatin transporters. Proc Nat Acad Sci. 2010;107:21187–21192. doi: 10.1073/pnas.1013964107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sunarpi Horie Y, Motoda J, et al. Enhanced salt tolerance mediated by AtHKT1 transporter-induced Na unloading from xylem vessels to xylem parenchyma cells. Plant J. 2005;44:928–938. doi: 10.1111/j.1365-313X.2005.02595.x. [DOI] [PubMed] [Google Scholar]
- Sung DY, Lee D, Harris H, Raab A, Feldmann J, Meharg A, Kumabe B, Komives EA, Schroeder JI. Identification of an arsenic tolerant double mutant with a thiol-mediated component and increased arsenic tolerance in phyA mutants. Plant J. 2007;49:1064–1075. doi: 10.1111/j.1365-313X.2006.03018.x. [DOI] [PubMed] [Google Scholar]
- Sung DY, Kim TH, Komives EA, Mendoza-Cozatl DG, Schroeder JI. ARS5 is a component of the 26S proteasome complex, and negatively regulates thiol biosynthesis and arsenic tolerance in Arabidopsis. Plant J. 2009;59:802–813. doi: 10.1111/j.1365-313X.2009.03914.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tong S, von Schirnding YE, Prapamontol T. Environmental lead exposure: a public health problem of global dimensions. Bull World Health Organ. 2000;78:1068–1077. [PMC free article] [PubMed] [Google Scholar]
- Vatamaniuk OK, Mari S, Lu YP, Rea PA. AtPCS1, a phytochelatin synthase from Arabidopsis: isolation and in vitro reconstitution. Proc Natl Acad Sci USA. 1999;96:7110–7115. doi: 10.1073/pnas.96.12.7110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vatamaniuk OK, Bucher EA, Ward JT, Rea PA. A new pathway for heavy metal detoxification in animals. Phytochelatin synthase is required for cadmium tolerance in Caenorhabditis elegans. J Biol Chem. 2001;276:20817–20820. doi: 10.1074/jbc.C100152200. [DOI] [PubMed] [Google Scholar]
- Vauclare P, Kopriva S, Fell D, Suter M, Sticher L, Ballmoos Pv, Kraä-henbuühl U, Camp ROd, Brunold C. Flux control of sulphate assimilation in Arabidopsis thaliana: adenosine 5′-phosphosulphate reductase is more susceptible than ATP sulphurylase to negative control by thiols. Plant J. 2002;31:729–740. doi: 10.1046/j.1365-313x.2002.01391.x. [DOI] [PubMed] [Google Scholar]
- Vernoux T, Wilson RC, Seeley KA, et al. The ROOT MERISTEM-LESS1/CADMIUM SENSITIVE2 gene defines a glutathione-dependent pathway involved in initiation and maintenance of cell division during postembryonic root development. Plant Cell. 2000;12:97–110. doi: 10.1105/tpc.12.1.97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waisberg M, Joseph P, Hale B, Beyersmann D. Molecular and cellular mechanisms of cadmium carcinogenesis. Toxicology. 2003;192:95–117. doi: 10.1016/s0300-483x(03)00305-6. [DOI] [PubMed] [Google Scholar]
- Weber M, Trampczynska A, Clemens S. Comparative transcriptome analysis of toxic metal responses in Arabidopsis thaliana and the Cd2+-hypertolerant facultative metallophyte Arabidopsis halleri. Plant Cell Environ. 2006;29:950–963. doi: 10.1111/j.1365-3040.2005.01479.x. [DOI] [PubMed] [Google Scholar]
- Yu HS, Lee CH, Chen GS. Peripheral vascular diseases resulting from chronic arsenical poisoning. J Dermatol. 2002;29:123–130. doi: 10.1111/j.1346-8138.2002.tb00234.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1. Visual phenotype of 20-day-old nrc2 mutant plants grown in soil.
Table S1. A list of the 30 genes most influenced by cadmium exposure, ranked in order of decreasing induction.
Table S2. A list of primers used for RT-PCR, promoter cloning, gene sequencing and complementation experiments.
Please note: As a service to our authors and readers, this journal provides supporting information supplied by the authors. Such materials are peer-reviewed and may be re-organized for online delivery, but are not copy-edited or typeset. Technical support issues arising from supporting information (other than missing files) should be addressed to the authors.


