Abstract
Cells of Flavobacterium johnsoniae, a rod-shaped bacterium about 6 μm long, do not have flagella or pili, yet they move over surfaces at speeds of about 2 μm/s. This motion is called gliding. Recent advances in F johnsoniae research include the discovery of mobile cell-surface adhesins and rotary motors. The puzzle is how rotary motion leads to linear motion. We suggest a possible mechanism, inspired by the snowmobile.
Keywords: Flavobacterium johnsoniae, rotary motor, SprB filament
Introduction
Flavobacterium (phylum bacteroidetes), Myxococcus (phylum proteobacteria) and Mycoplasma (phylum firmicutes) are three bacterial genera that provide model systems for gliding that can be manipulated genetically. Flavobacterium johnsoniae and Myxococcus xanthus contain peptidoglycan and are rod shaped, whereas Mycoplasma mobile is devoid of peptidoglycan and is flask shaped. M. mobile gliding machinery localizes around the neck region of the cell and utilizes ATP hydrolysis to power movement. A protein present at the neck anchored by a cytoskeletal network acts as a ‘leg’ that adheres to a surface and enables gliding by a centipede-like mechanism [1]. The M. mobile model cannot explain gliding of the other bacteria, such as F. johnsoniae and M xanthus, which are powered by a protonmotive force [2,3]. Several models have been proposed to explain gliding of M xanthus [4,5], which occurs at about 1/60th the speed of gliding in F johnsoniae. Of these models, the helical rotor model appears to be the most widely accepted [6]. According to this model, the gliding motor of M. xanthus moves along a helical intracellular track, and this motion somehow causes movement of cell-surface components. In contrast, F johnsoniae has rotary motors that stay in place and power gliding [7]. This solves the ‘peptidoglycan problem’ [5] associated with the helical rotor model for M. xanthus. The F. johnsoniae motor drive shafts pierce the peptidoglycan layer; they don't move laterally through the peptidoglycan. The cell-surface and motor components implicated in F. johnsoniae gliding, discussed below, are genetically unique, different from those identified in other bacteria. The mechanism of F. johnsoniae gliding is different from that of both M. xanthus and M. mobile.
Bacteria of the phylum bacteroidetes are present in diverse ecological niches ranging from aquatic and terrestrial environments to the gut microflora. The family Flavobacteriaceae, which is a part of this phylum, comprises commensal bacteria and opportunistic pathogens. Some of these bacteria infect fish and are causative agents of columnaris disease, bacterial coldwater disease and bacterial gill disease. They also secrete a large array of enzymes that help degrade and utilize extracellular proteins and polysaccharides. They are distinctive in being able to move rapidly over surfaces, even glass, without flagella or pili, organelles that enable other bacteria to swim, swarm, or twitch [8].
For large flying objects, such as birds, airplanes, or humans in gliders, gliding refers to a smooth continuous motion that occurs without the use of engines. A glider falls through the air, buoyed by its wings. It moves at high Reynolds number, carried forward by inertia, resisted by drag. A microscopic organism immersed in water moves at low Reynolds number, overwhelmed by drag (whether on a surface or not). It cannot coast [9]. Thus, bacterial gliding is an active process that requires constant influx of energy and must be caused by movement of proteins that carry cells over a surface. We think the machinery used for bacterial gliding might be analogous to the machinery that powers a snowmobile. How does one build such a machine on a microscopic scale?
Cells of Flavobacterium johnsoniae are amongst the fastest known gliders and have emerged as a powerful model system to study bacteroidetes-specific gliding. They have mobile cell-surface adhesins that move in a looped fashion and interact with another surface on which the bacteria glide. Recent advances in genetic manipulation of F. johnsoniae (reviewed by McBride and Nakane in this issue) led to discovery of proteins that are required for gliding. Some of these proteins form filaments that project from the surface of the cell and move along its length [10,11], while others form motors that can rotate these filaments [7]. How rotation leads to linear displacement is an open question. Are the cell-surface filaments carried by treads driven by rotary motors, in the way that chains are driven by sprockets powered by rotary motors? Clearly, more experimental evidence is required. In any event, the filaments interact with a surface and enable motion of the cell.
The mechanism for propulsion
The movement of cell-surface adhesins was first reported by Pate and Chang [2] for Flavobacterium columnare (previously known as Flexibacter columnaris) and Flavobacterium johnsoniae (previously known as Cytophaga johnsoniae). Movement was detected by attaching polystyrene latex beads to a cell and tracking their displacement. Similar work was done with Cytophaga U67, which is a close relative of F. johnsoniae [12]. Electron microscopy data show that one F. johnsoniae cell has many cell-surface filaments. More recently, it was discovered that the filaments are composed of a protein, SprB. Motion of anti-SprB antibody conjugated polystyrene beads, and of anti-SprB antibodies labeled with a fluorescent dye, was recorded and tracked [11,13]. Some reports suggest that filaments move along looped tracks [12-14] while one report suggests that they move helically [11].
Recording of a cell gliding over a stationary polystyrene bead (Fig.1 and MovieS1) provides useful insights on the motion of a SprB filament. These recordings suggest (i) the presence of a continuous track and (ii) that once a filament is attached to a surface, it does not need to detach for a cell to glide. In Fig.1 and MovieS1, one or more SprB filaments are attached to a bead at their distal ends. This end of the filament does not move relative to the bead and the glass surface to which the bead is adsorbed. The filament however, is loaded onto a component (call it a tread) that moves along a track fixed to the rigid framework of the cell, presumably, the peptidoglycan layer. The filament and tread are in motion relative to the cell-surface, while the track is fixed to that surface. The filament and tread are pushed or pulled along the track, which results in motion of the cell body relative to the end of the filament that is attached to the bead. The track loops around the end of the cell, so when the cell pole reaches the bead, the cell flips over, i.e., the lagging pole becomes the leading pole, while the direction of cell motion in the laboratory frame remains the same. On the other hand, if the distal end of SprB filament is free, it is pulled along the surface of the cell. The motion of SprB can be visualized by attachment of a free anti-sprB coated latex bead or of fluorescent anti-SprB antibody. The identity of the filament, SprB, is known, but the identities of the tread and track are not. Structures visualized by cryo-em tomography as patches that are connected to SprB filaments and are present at the base of the outer membrane [10] might be sections of tread.
Figure 1.
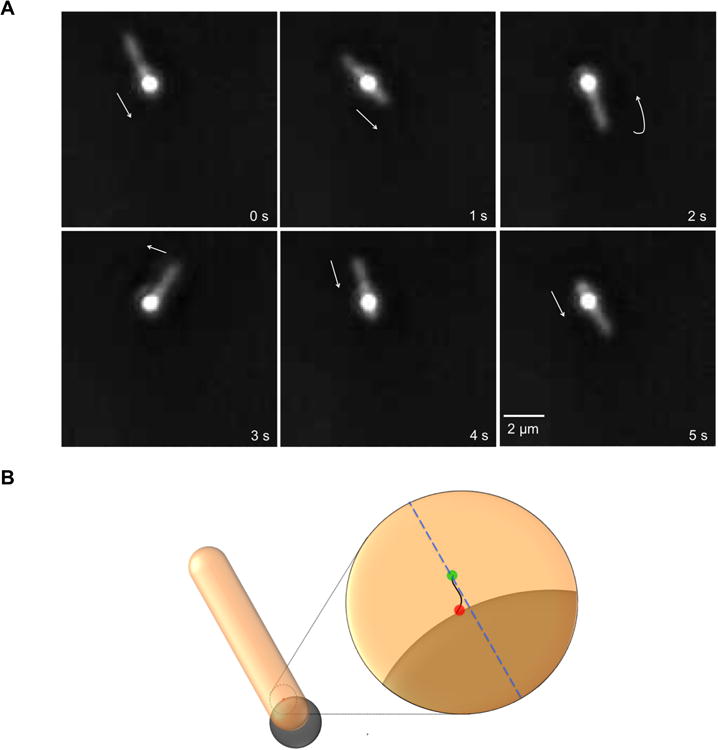
Movement of F. johnsoniae on a stationary polystyrene bead. (A) Images at 1 s time intervals from Movie S1, taken using a phase contrast microscope with a bright-phase objective (Nikon Plan 40× BM), showing a rod-shaped F. johnsoniae cell (slightly out of focus) moving over a spherical bead (0.5 μm). Time is shown at the bottom right of each frame and a scale bar is shown at bottom left of the last frame. Arrows depict direction of motion of the leading pole (0-3 s), which later becomes the lagging pole (4 s). (B) Diagrammatic representation of a cell moving over a stationary bead, with a magnified view in the inset. In the magnified view, a SprB filament (black), which is underneath the cell, is bound to the bead at an attachment site (red dot) that remains fixed. The SprB filament is loaded on a tread guided by a track (blue dotted line), and the SprB filament-to-tread attachment site (green dot) is in motion relative to the bead.
The average length of an F. johnsoniae cell is 6 μm, and the average gliding speed is 2 μm/s. So, one SprB filament can propel a cell for 3 s. After that, if the cell is to continue to glide in the same direction, the filament must detach from the glass surface and another SprB filament must take its place. One F. johnsoniae cell has many filaments that move at the speed of ∼2 μm/s, which is also the speed of a gliding cell. So, the filaments work in parallel rather than in tandem. The F. johnsoniae rotary motor is a constant-speed device [7], which is convenient if different SprB filaments are to move in parallel. As long as all filaments are moving along the same track or on parallel tracks, the number of points of adhesion increases the propulsive force that can be exerted but not the speed of cell movement.
Surface adhesins for gliding
Sticking to a surface is the first step a cell takes towards gliding. So, it is important that the surface filament has regions that are adhesive. F. johnsoniae has two surface adhesins, SprB and RemA. RemA has a lectin domain that binds exo-polysaccharides, and it's over-expression results in cell aggregation [14]. SprB is a protein of molecular weight 669 kDa, four times larger than RemA. Both SprB and RemA are involved in sticking a cell to glass. The adhesive domains of SprB are not known. Flavobacteria are present in most aquatic and terrestrial environments. Filament-like structures were recently observed for Flavobacterium columnare [15]. Sequence alignment of F. johnsoniae and F. columnare SprB shows that while the two proteins have regions of similarity, they also have large regions that are not conserved. Large regions of F. johnsoniae SprB, spread all over the amino-acid sequence, are absent in F. columnare SprB. Despite missing these regions, F. columnare glides. F. columnare is a fish pathogen and is the causative agent of columnaris disease. It has the ability to adhere to fish and form biofilms [16], it is likely that surface-adhesion domains of F. columnare SprB might be required for both surface colonization and gliding.
From rotation to gliding
Our work demonstrated that a rotary motor is involved in F. johnsoniae gliding [7]. One possible mechanism for converting rotation to longitudinal motion is that of a protein tread connecting two rotary motors in a manner similar to that by which a chain connects two sprockets, or by which treads in a snowmobile are linked to a gasoline engine. In F. johnsoniae this chain would be the moving tread carrying SprB. Gld proteins probably form the rotary motor, but the identity of other components is not known. To identify the components, more information about mutants that are defective in gliding is needed. Besides being defective in gliding, many such mutants lack the ability to secrete an array of proteins, which include the cell-surface adhesins SprB and RemA, via the Type IX protein secretion system (T9SS) [17-24]. It is unclear if mutants lacking genes that encode T9SS proteins are defective in gliding because they lack the secretion apparatus or the gliding motor. Differentiating between proteins that play exclusive roles in either gliding or secretion via T9SS appears to be crucial. While most structural information for the Gld proteins is lacking, recently, the preliminary X-ray analysis for the carboxy-terminal fragment of GldM (PorM) was reported [25]. With increasing knowledge about gliding proteins, the next step forward might be to study their interactions and to visualize the structure of the gliding apparatus.
It is instructive to recall how the basal body of the flagellar rotary motor was visualized. The iconic images made by DeRosier [26] were of motors dissolved out of Salmonella with neutral detergents, by the method of Aizawa et al. (1985) [27], who followed the procedure of DePamphilis and Adler (1971) [28] but worked at higher pH. Cell walls were weakened by treatment with lysozyme-EDTA and the spheroplasts were dissolved with Triton X-100. The viscosity of the lysate was lowered by treatment with DNase, and insoluble components were separated by differential centrifugation. The basal bodies could be found because they were at the proximal ends of flagellar filaments. Might one covalently link beads to SprB filaments and fish for components isolated by a similar procedure? More recent work by cryo-em tomography [29,30] began with spirochetes, because their cell bodies were thin [31]. Might cryo-em be used to find gliding motor components in F. johnsoniae minicells?
Supplementary Material
Movie S1: Movie depicts motion of a F. johnsoniae cell over a 0.5 μm diameter polystyrene bead. The bead is stationary with reference to the glass surface.
Highlights.
Bacterial gliding, i.e. movement over a surface, of bacteria that have neither flagella nor pili, requires constant propulsion.
Gliding Flavobacterium have a rotary motor and mobile cell-surface filaments.
The machinery required for gliding could be analogous to the components that power and drive a snowmobile.
Acknowledgments
This work is supported by National Institutes of Health Grant AI016478.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References and recommended reading
Papers of particular interest, published within the period of review, have been highlighted as:
* of special interest
** of outstanding interest
- 1.Miyata M. Unique centipede mechanism of Mycoplasma gliding. Annu Rev Microbiol. 2010;64:519–537. doi: 10.1146/annurev.micro.112408.134116. [DOI] [PubMed] [Google Scholar]
- 2.Pate JL, Chang LYE. Evidence that gliding motility in prokaryotic cells is driven by rotary assemblies in the cell envelopes. Curr Microbiol. 1979;2:59–64. [Google Scholar]
- 3.Nan B, Chen J, Neu JC, Berry RM, Oster G, Zusman DR. Myxobacteria gliding motility requires cytoskeleton rotation powered by proton motive force. Proceedings of the National Academy of Sciences of the United States of America. 2011;108:2498–2503. doi: 10.1073/pnas.1018556108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Mauriello EM, Mignot T, Yang Z, Zusman DR. Gliding motility revisited: how do the myxobacteria move without flagella? Microbiology and molecular biology reviews : MMBR. 2010;74:229–249. doi: 10.1128/MMBR.00043-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Nan B, McBride MJ, Chen J, Zusman DR, Oster G. Bacteria that glide with helical tracks. Curr Biol. 2014;24:R169–173. doi: 10.1016/j.cub.2013.12.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Nan B, Bandaria JN, Moghtaderi A, Sun IH, Yildiz A, Zusman DR. Flagella stator homologs function as motors for myxobacterial gliding motility by moving in helical trajectories. Proc Natl Acad Sci U S A. 2013;110:E1508–1513. doi: 10.1073/pnas.1219982110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7**.Shrivastava A, Lele PP, Berg HC. A rotary motor drives flavobacterium gliding. Curr Biol. 2015;25:338–341. doi: 10.1016/j.cub.2014.11.045. This study shows that a rotary motor that generates high torque is involved in gliding. The torque changes with load, while the speed remains constant. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Jarrell KF, McBride MJ. The surprisingly diverse ways that prokaryotes move. Nat Rev Microbiol. 2008;6:466–476. doi: 10.1038/nrmicro1900. [DOI] [PubMed] [Google Scholar]
- 9.Berg HC. Random walks in biology. Princeton University Press; 1993. [Google Scholar]
- 10.Liu J, McBride MJ, Subramaniam S. Cell-surface filaments of the gliding bacterium Flavobacterium johnsoniae revealed by cryo-electron tomography. J Bacteriol. 2007;189:7503–7506. doi: 10.1128/JB.00957-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11*.Nakane D, Sato K, Wada H, McBride MJ, Nakayama K. Helical flow of surface protein required for bacterial gliding motility. Proc Natl Acad Sci U S A. 2013;110:11145–11150. doi: 10.1073/pnas.1219753110. In this study SprB filaments were visualized by electron microscopy. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lapidus IR, Berg HC. Gliding motility of Cytophaga sp. strain U67. J Bacteriol. 1982;151:384–398. doi: 10.1128/jb.151.1.384-398.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Nelson SS, Bollampalli S, McBride MJ. SprB is a cell surface component of the Flavobacterium johnsoniae gliding motility machinery. J Bacteriol. 2008;190:2851–2857. doi: 10.1128/JB.01904-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Shrivastava A, Rhodes RG, Pochiraju S, Nakane D, McBride MJ. Flavobacterium johnsoniae RemA is a mobile cell surface lectin involved in gliding. Journal of bacteriology. 2012;194:3678–3688. doi: 10.1128/JB.00588-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Laanto E, Penttinen RK, Bamford JK, Sundberg LR. Comparing the different morphotypes of a fish pathogen--implications for key virulence factors in Flavobacterium columnare. BMC Microbiol. 2014;14:170. doi: 10.1186/1471-2180-14-170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cai W, De La Fuente L, Arias CR. Biofilm formation by the fish pathogen Flavobacterium columnare: development and parameters affecting surface attachment. Appl Environ Microbiol. 2013;79:5633–5642. doi: 10.1128/AEM.01192-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sato K, Naito M, Yukitake H, Hirakawa H, Shoji M, McBride MJ, Rhodes RG, Nakayama K. A protein secretion system linked to bacteroidete gliding motility and pathogenesis. Proc Natl Acad Sci USA. 2010;107:276–281. doi: 10.1073/pnas.0912010107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18*.Shrivastava A, Johnston JJ, van Baaren JM, McBride MJ. Flavobacterium johnsoniae GldK, GldL, GldM, and SprA are required for secretion of the cell surface gliding motility adhesins SprB and RemA. Journal of bacteriology. 2013;195:3201–3212. doi: 10.1128/JB.00333-13. This study identifies the cellular localization of core T9SS proteins GldK, GldL and GldM. It shows that T9SS proteins are required for cell-surface localization of motility adhesins. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kharade SS, McBride MJ. Flavobacterium johnsoniae PorV is required for secretion of a subset of proteins targeted to the type IX secretion system. J Bacteriol. 2015;197:147–158. doi: 10.1128/JB.02085-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20*.Kharade SS, McBride MJ. Flavobacterium johnsoniae chitinase ChiA is required for chitin utilization and is secreted by the type IX secretion system. J Bacteriol. 2014;196:961–970. doi: 10.1128/JB.01170-13. This study shows that Gld proteins (T9SS) are required for secretion of a novel chitinase. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21**.McBride MJ, Zhu Y. Gliding motility and Por secretion system genes are widespread among members of the phylum bacteroidetes. J Bacteriol. 2013;195:270–278. doi: 10.1128/JB.01962-12. This study surveys the gliding of bacteria of the phylum bacteroidetes. It reports that five bacteria previously recorded as non-motile, exhibit gliding. It defines a core set of genes required for gliding and shows that all gliding members of this phylum have genes that encode components of the T9SS. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rhodes RG, Samarasam MN, Shrivastava A, van Baaren JM, Pochiraju S, Bollampalli S, McBride MJ. Flavobacterium johnsoniae gldN and gldO Are Partially Redundant Genes Required for Gliding Motility and Surface Localization of SprB. Journal of Bacteriology. 2010;192:1201–1211. doi: 10.1128/JB.01495-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wang Y, Wang Z, Cao J, Guan Z, Lu X. FLP-FRT-based method to obtain unmarked deletions of CHU_3237 (porU) and large genomic fragments of Cytophaga hutchinsonii. Appl Environ Microbiol. 2014;80:6037–6045. doi: 10.1128/AEM.01785-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24*.Zhu Y, McBride MJ. Deletion of the Cytophaga hutchinsonii type IX secretion system gene sprP results in defects in gliding motility and cellulose utilization. Appl Microbiol Biotechnol. 2014;98:763–775. doi: 10.1007/s00253-013-5355-2. This study shows that gliding motility and T9SS are linked in Cytophaga hutchinsonii, which is a member of the phylum bacteroidetes. [DOI] [PubMed] [Google Scholar]
- 25.Stathopulos J, Cambillau C, Cascales E, Roussel A, Leone P. Crystallization and preliminary X-ray analysis of the C-terminal fragment of PorM, a subunit of the Porphyromonas gingivalis type IX secretion system. Acta Crystallogr F Struct Biol Commun. 2015;71:71–74. doi: 10.1107/S2053230X1402559X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Francis NR, Sosinsky GE, Thomas D, DeRosier DJ. Isolation, characterization and structure of bacterial flagellar motors containing the switch complex. J Mol Biol. 1994;235:1261–1270. doi: 10.1006/jmbi.1994.1079. [DOI] [PubMed] [Google Scholar]
- 27.Aizawa SI, Dean GE, Jones CJ, Macnab RM, Yamaguchi S. Purification and characterization of the flagellar hook-basal body complex of Salmonella typhimurium. J Bacteriol. 1985;161:836–849. doi: 10.1128/jb.161.3.836-849.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.DePamphilis ML, Adler J. Fine structure and isolation of the hook-basal body complex of flagella from Escherichia coli and Bacillus subtilis. J Bacteriol. 1971;105:384–395. doi: 10.1128/jb.105.1.384-395.1971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chen S, Beeby M, Murphy GE, Leadbetter JR, Hendrixson DR, Briegel A, Li Z, Shi J, Tocheva EI, Muller A, et al. Structural diversity of bacterial flagellar motors. EMBO J. 2011;30:2972–2981. doi: 10.1038/emboj.2011.186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhao X, Norris SJ, Liu J. Molecular architecture of the bacterial flagellar motor in cells. Biochemistry. 2014;53:4323–4333. doi: 10.1021/bi500059y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Murphy GE, Leadbetter JR, Jensen GJ. In situ structure of the complete Treponema primitia flagellar motor. Nature. 2006;442:1062–1064. doi: 10.1038/nature05015. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Movie S1: Movie depicts motion of a F. johnsoniae cell over a 0.5 μm diameter polystyrene bead. The bead is stationary with reference to the glass surface.


