SUMMARY
We report the development of a next-generation sequencing-based technology that entails construction of a DNA library comprising up to at least 47 (~16,000) bar-coded sequences, production of RNA transcripts, and analysis of transcript ends and transcript yields ("massively systematic transcript end readout," MASTER). Using MASTER, we define full inventories of transcription start sites ("TSSomes") of Escherichia coli RNA polymerase for initiation at a consensus core promoter in vitro and in vivo, we define the TSS-region DNA-sequence determinants for TSS selection, reiterative initiation ("slippage synthesis"), and transcript yield, and we define effects of DNA topology and NTP concentration. The results reveal that slippage synthesis occurs from the majority of TSS-region DNA sequences and that TSS-region DNA sequences have profound, up to 100-fold, effects on transcript yield. The results further reveal that TSSomes depend on DNA topology, consistent with the proposal that TSS selection involves transcription-bubble expansion ("scrunching") and transcription-bubble contraction ("anti-scrunching").
INTRODUCTION
Defining the mechanism of any process involving protein-nucleic acid interactions requires a full understanding of the relationship between nucleic acid sequence and functional output. Often such studies require the construction and analysis of many individual variants of a given DNA sequence. However, for processes involving extensive protein-nucleic acid interactions, a comprehensive analysis of the functional output derived from all sequence variants is not feasible with conventional approaches that test individual sequence variants on a one-by-one basis. Instead, such analyses require approaches that facilitate the parallel interrogation of thousands of individual sequence variants en masse (Melnikov et al., 2012; Patwardhan et al., 2012; Heyduk and Heyduk, 2014; Heyduk and Heyduk, 2015).
The process of transcription involves extensive interactions between RNA polymerase (RNAP) and a double-stranded DNA template. Each stage of transcription, and each reaction carried out by RNAP during transcription, is affected by the sequence context of the DNA template (Larson et al., 2011; Wang and Greene, 2011; Dangkulwanich et al., 2014; Ruff et al., 2015; Washburn and Gottesman, 2015). While structural studies have revealed some of the RNAP-nucleic acid interactions that modulate transcription (Murakami, 2015), a quantitative understanding of how DNA sequence influences transcription output requires comprehensive knowledge of RNAP activity in all sequence contexts. Toward achievement of this goal, we report the development of a technology that facilitates the comprehensive analysis of the relationship between nucleic acid sequence and functional output during transcription for Escherichia coli RNAP on a scale previously inaccessible. In particular, we report the development of an experimental platform that enables the parallel functional analysis of at least 47 (~16,000), and potentially at least 410 (~1,000,000), distinct DNA template sequence variants in vitro and in vivo ("massively systematic transcript end readout," MASTER). We apply MASTER to the analysis of the determinants and mechanism of transcription initiation. Specifically, by use of MASTER we (i) perform a comprehensive analysis of the DNA-sequence determinants for transcription start site (TSS) selection, (ii) perform a comprehensive analysis of DNA-sequence determinants for reiterative initiation (a non-standard form of transcription initiation also termed “slippage synthesis”), (iii) perform a comprehensive analysis of the relationship between TSS-region sequence and transcript yield, and (iv) define the effects of DNA topology and alterations in nucleoside triphosphate (NTP) concentrations on TSS-selection, slippage synthesis, and transcript yield. Our results provide a full description of the relationship between TSS-region sequence and transcriptional output from a consensus core promoter for E. coli RNAP both in vitro and in vivo. Our results further demonstrate the broad utility of MASTER for analysis of biological processes that involve extensive protein-nucleic acid interactions.
DESIGN
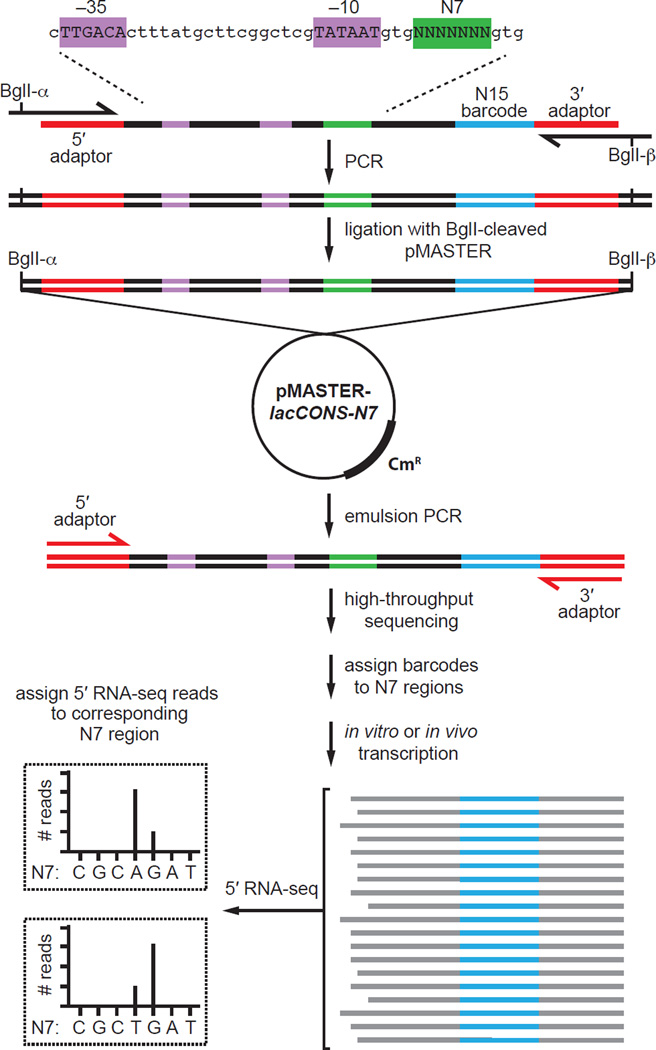
To overcome limitations of conventional approaches for the analysis of effects of sequence variation on transcription, we sought to develop a technology that would enable a comprehensive analysis of the relationship between sequence and transcription output. Thus, we developed a next-generation sequencing-based approach (Figure 1) that entails construction of a template library that contains up to 47 (~16,000), and potentially up to 410 (~1,000,000), barcoded sequences Figure S1A,B), production of RNA transcripts from the template library in vitro or in vivo, and analysis of transcript ends and transcript yields. To generate a template library, we synthesize an oligodeoxyribonucleotide containing a first randomized sequence spanning the region of interest (Figure 1, top, green) and a second, longer randomized sequence that serves as a barcode (Figure 1, top, blue). The oligodeoxyribonucleotide is converted to double-stranded DNA by use of PCR, the double-stranded DNA is ligated with a vector backbone, the resulting recombinant plasmid DNA is used to transform cells, and recombinant plasmid DNA is isolated from 106-107 transformants (Figure 1, middle). The resulting library is amplified by emulsion PCR and sequenced to define each unique sequence and thereby to associate each first randomized sequence with its corresponding second randomized sequence that serves as its barcode. This procedure provides a “self-assembling barcode,” in which for each DNA molecule in the library, the first randomized sequence in the library is associated with a known corresponding second randomized sequence. RNA produced from the library, either in an in vitro transcription reaction or in vivo, is isolated and subjected to deep sequencing in a manner that enables the identification of the sequence of transcript ends and the sequence of the barcode within each transcript (Figure 1, bottom). We term this approach “massively systematic transcript end readout, MASTER.”
Figure 1. Massively systematic transcript end readout (MASTER).
Top: generation of pMASTER-lacCONS-N7 library. An oligodeoxyribonucleotide carrying the lacCONS-N7 promoter and 15-nt barcode sequence (blue) is used as template in a PCR reaction using primers that introduce BglI sites. The BglI digested PCR product is cloned into BglI digested plasmid pSG289 (Figure S1A) to generate plasmid pMASTER-lacCONS-N7, which contains 47 (~16,000) sequences at positions 4–10 bps downstream of the lacCONS −10 element (green). Middle: product generated by emulsion PCR is used for high-throughput sequencing analysis to assign barcodes to TSS-sequence variants. PCR primers shown in red (5′ and 3′ adaptor) carry sequences that facilitate analysis using an Illumina HiSeq. Bottom: 5′ RNA-seq analysis of RNA produced from the library in vitro and in vivo. The sequence of the barcode is used to assign the RNA to a TSS-region, the sequence of the 5′ end is used to define the TSS, and the number of reads is used to measure transcript yield from each TSS-region sequence. (See Figure S1)
RESULTS
Application of MASTER for analysis of transcription initiation
During transcription initiation, the RNAP holoenzyme binds to promoter DNA, unwinds a turn of promoter DNA to form an RNAP-promoter open complex containing an unwound “transcription bubble,” and selects a transcription start site (TSS) (Ruff et al., 2015). Some progress has been made to define the DNA sequence determinants for TSS selection (Aoyama and Takanami, 1985; Sorensen et al., 1993; Jeong and Kang, 1994; Liu and Turnbough, 1994; Walker and Osuna, 2002; Lewis and Adhya, 2004). However, a complete description of how the sequence of the TSS-region of a given promoter determines TSS selection, and the impact of the sequence of the TSS-region on yields of full-length transcripts, has not been provided. We therefore sought to employ MASTER to perform a comprehensive analysis of the effects of TSS-region sequences on functional output during transcription initiation for E. coli RNAP.
We constructed a MASTER library containing 47 (~16,000) sequences at the TSS-region, i.e. positions 4–10 base pairs (bps) downstream of the −10 element, of a consensus E. coli σ70-dependent promoter (Figures 1, S1C). We generated RNA from the library in transcription reactions performed in vitro using non-supercoiled DNA templates, transcription reactions performed in vitro using negatively supercoiled DNA templates, and transcription of negatively supercoiled DNA templates in vivo. The RNA transcripts were analyzed by high-throughput sequencing of RNA 5′ ends (5′ RNA-seq) to identify the sequence of the RNA 5′ end and to identify the sequence of the barcode, which defined the identity of the template that produced the RNA. The sequences of RNA 5′ ends generated from a given template in the library were used to define the TSS position(s) from the template and the total number of sequencing reads generated from the template were used to define transcript yields from the template (Figure 1, bottom).
MASTER analysis of sequence determinants of TSS selection
To analyze TSS selection we considered only sequencing reads with 5′ end sequences that precisely matched the sequence of the TSS region from which the transcript was generated “matched RNAs”. For each TSS-region sequence we calculated the number of matched RNAs emanating from each position 4–10 bps downstream of the −10 element (hereafter termed positions 4, 5, 6, 7, 8, 9 and 10). Next, we calculated the percentage of reads derived from matched RNAs emanating from each position (X) within the TSS region [%TSSX = 100 * (# reads at position X) / (# reads at all positions 4–10)], and used the %TSSX values to calculate a mean TSS for each sequence variant. The results define the full TSS inventories ("TSSomes") from a consensus core promoter for non-supercoiled templates in vitro, negatively supercoiled templates in vitro, and negatively supercoiled templates in vivo (Tables S1–S7).
MASTER analysis of TSS selection using non-supercoiled linear DNA
Averaging the %TSS at positions 4–10 observed for all TSS-sequence variants in reactions performed using non-supercoiled linear DNA templates (Table S1) shows, consistent with previous analyses of individual promoters (Aoyama and Takanami, 1985; Sorensen et al., 1993; Jeong and Kang, 1994; Liu and Turnbough, 1994; Walker and Osuna, 2002; Lewis and Adhya, 2004), that TSS selection is determined by position (Figure 2A) (range = positions 6–10; mean = 7.36 bp downstream of −10 element; mode = 7 bp downstream of −10 element). The results further show the order of preference for TSS selection for non-supercoiled linear templates in vitro is position 7 > 8 > 6 ~ 9 > 10 (Figures 2A,B, S2A; p < 0.001).
Figure 2. TSS selection on a non-supercoiled linear DNA template in vitro.
A. TSS-distribution histogram. Average %TSS at positions 4–10 for TSS-regions with ≥25 matched RNA reads (Table S1).
B. Sequence determinants for TSS selection. Table lists the amount of the total %TSS at positions 6–10 derived from TSS-regions carrying (i) R or Y at the indicated TSS position, (ii) A, G, C, or T at the indicated TSS position, or (iii) YTSS-1RTSS or RTSS-1RTSS at the indicated TSS position.
C. Sequence preferences for TSS selection. Sequence logo for the 162 TSS-region sequences (top 1%) with the highest %TSS at positions 6–10. Red bases indicate the TSS.
(See Figure S3)
To investigate sequence determinants for TSS selection we sorted TSS-region sequences on the basis of the identities of the bases at positions 6–10. As prior analyses of TSS selection have noted a bias for purine at the TSS position (Maitra and Hurwitz, 1965; Jorgensen et al., 1969; Hawley and McClure, 1983), we first determined the %TSS for promoter variants that specify use of a purine (R) TSS or pyrimidine (Y) TSS at positions 6–10. The results show a strong preference for R over Y at each TSS position (Figures 2B, S2B; p < 0.001). Further analysis of TSS region variants carrying an A, G, C, or T at each TSS position revealed the order of preference as G > A > C ~ T (Figures 2B, S2C; p < 0.001). Prior analyses of TSS selection also noted a preference for initiation at YTSS-1RTSS sequence motifs (Shultzaberger et al., 2007). To investigate the influence of YTSS-1RTSS sequences on TSS selection we determined the %TSS for promoter variants carrying YTSS-1RTSS or RTSS-1RTSS. The results show that YTSS-1RTSS is preferred over RTSS-1RTSS at each TSS position (Figures 2B, S2D; p < 0.001).
We next identified TSS-region sequences that yielded the highest %TSS at each of the five positions within the TSS range (Figure 2C). For each TSS position, sequences that favor the highest %TSS exhibit a strong preference for YTSS-1RTSS. In addition, for TSS position 9 there is a strong preference for TTSS-1, while for TSS position 10, the sequences with the highest %TSS show a preference for a TTSS-2TTSS-1.
MASTER analysis of TSS selection using negatively supercoiled DNA
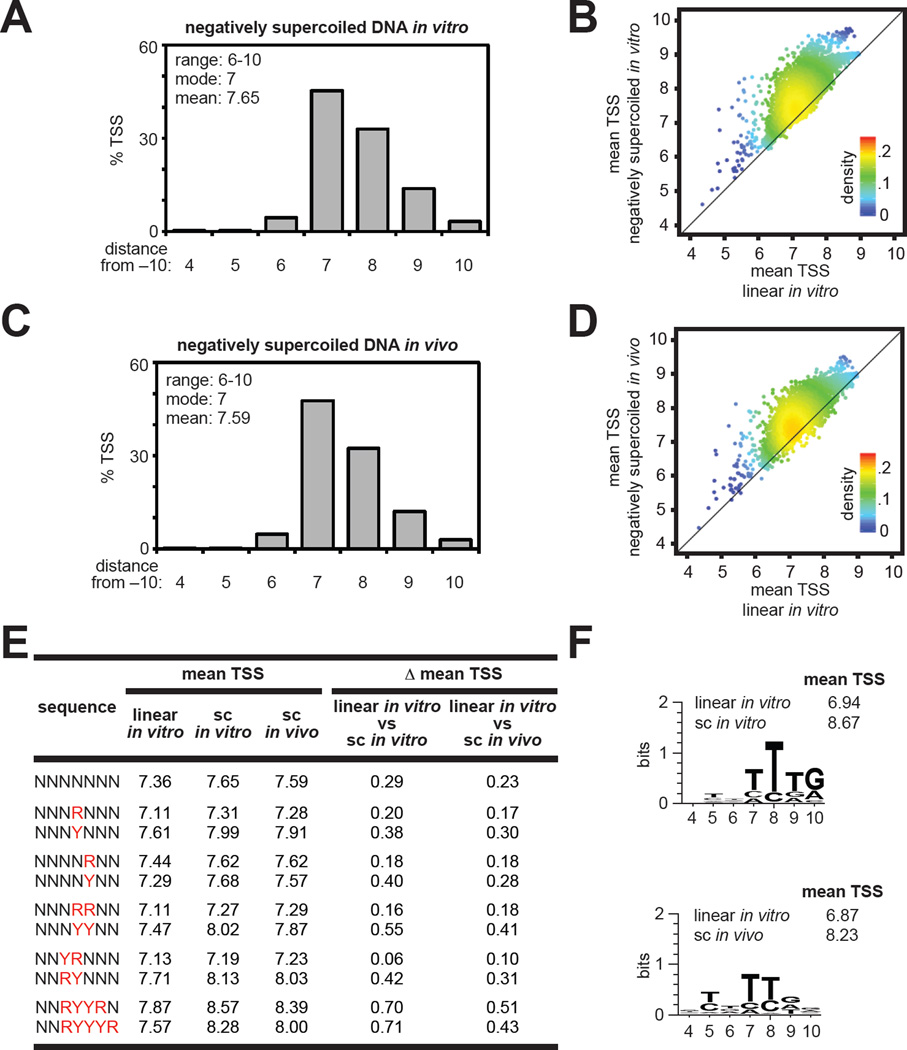
For analysis of the effects of DNA topology on TSS selection we compared results obtained in vitro using either a non-supercoiled linear DNA template (Table S1) or a relaxed circular DNA template (Table S6) with results obtained using a negatively supercoiled DNA template in vitro (Tables S2, S7) and in vivo (Table S3). Analysis of the %TSS at positions 4–10 observed for all TSS-sequence variants in reactions performed using negatively supercoiled DNA templates (Figure 3A,C) shows that the range of TSS positions (positions 6–10) and modal TSS (position 7) was identical to the range of TSS positions and modal TSS observed using a non-supercoiled DNA template (Figure 2A).
Figure 3. TSS selection on negatively supercoiled DNA templates.
A. TSS-distribution histogram for experiments performed in vitro. Average %TSS at positions 4–10 for TSS-regions with ≥25 matched RNA reads (Table S2).
B. Plot of the mean TSS with negatively supercoiled DNA in vitro versus the mean TSS with non-supercoiled linear DNA in vitro for individual TSS-region sequences.
C. TSS-distribution histogram for experiments performed in vivo. Average %TSS at positions 4–10 for TSS-regions with ≥25 matched RNA reads (Table S3).
D. Plot of the mean TSS with negatively supercoiled DNA in vivo versus the mean TSS with non-supercoiled linear DNA in vitro for individual TSS-region sequences.
E. Average of the mean TSS values for the indicated TSS-region sequences. (Δ mean TSS; differences between values observed on linear and supercoiled templates).
F. Sequence preferences for topology dependent effects on TSS selection. Sequence logo and average mean TSS values for 162 TSS-region sequences (top 1%) with the highest values of Δ mean TSS.
(See Figure S4)
In addition, the sequence determinants for TSS selection observed using negatively supercoiled DNA templates (Figure S3) were identical to those observed using non-supercoiled DNA templates (Figure 2B,C). In particular, R is favored over Y at each TSS position, the order of TSS preference for each base is G > A > C ~ T, and YTSS-1RTSS is preferred over RTSS-1RTSS at each TSS position (Figure S2A–D; p < 0.001). Furthermore, the TSS-region sequences that yielded the highest %TSS at each of the five TSS positions within the TSS range were similar on negatively supercoiled DNA templates and non-supercoiled DNA templates (Figures 2C, S3).
However, there were also notable differences between results obtained using negatively supercoiled DNA templates and non-supercoiled DNA templates. First, while the %TSS observed at position 6 and position 9 were similar with non-supercoiled DNA (Figures 2A, 2B, S4A), the %TSS at position 9 was ~3-times larger than the %TSS at position 6 with negatively supercoiled DNA (Figures 3A, 3C, S3, S4B). Second, comparison of the mean TSS observed with negatively supercoiled DNA (Figure 3A,C) with the mean TSS observed using non-supercoiled DNA (Figure 2A) reveals TSS distributions with negatively supercoiled DNA (mean = 7.65 bp downstream of −10 element with negatively supercoiled DNA in vitro; mean = 7.59 bp downstream of −10 element with negatively supercoiled DNA in vivo) are shifted downstream relative to TSS distribution with non-supercoiled DNA (mean = 7.36 bp downstream of −10 element). In addition, ~95% of TSS-region sequence variants exhibited an increase in the mean TSS with negatively supercoiled DNA templates compared with a non-supercoiled DNA template (Figures 3B, 3D, S4C).
Analysis of the effects of TSS-region sequence on sensitivity to topology revealed that TSS-region sequences carrying R at positions 7 and 8 are less susceptible to topology effects, while TSS-region sequences carrying a Y at positions 7 and 8 are more susceptible to topology effects (Figures 3E, S2E, S4D; p < 0.001). In addition, TSS-region sequences that contained YR at positions 6/7 are less susceptible to topology effects, while TSS-region sequences carrying a RY at positions 6/7 are more susceptible to topology effects (Figures 3E, S2E, S4D; p < 0.001). Furthermore, identification of the TSS-region sequences that exhibited the highest difference in mean TSS in a comparison of negatively supercoiled DNA and non-supercoiled DNA revealed that TSS-region sequences that contained RYYR at positions 6–9 or RYYYR at positions 6–10 were highly susceptible to topology dependent changes in TSS selection (Figures 3E, 3F, S2E, S4D, S4E; p < 0.001).
The results obtained from our MASTER analysis of TSS selection on non-supercoiled and negatively supercoiled DNA indicate that DNA topology is a determinant of TSS selection and reveal TSS-region sequence determinants that confer sensitivity or resistance to topology-dependent changes in TSS selection for a consensus core promoter. In addition, a comparison of the results obtained using negatively supercoiled DNA in vitro (Figures 3A, S3A, S4B) with results obtained using negatively supercoiled DNA in vivo (Figures 3C, S3B) show that TSS selection with negatively supercoiled DNA in vitro accurately recapitulates TSS selection with negatively supercoiled DNA in vivo, suggesting that all determinants for TSS selection for a consensus core promoter are contained within an in vitro transcription reaction (i.e., RNAP, DNA, and NTPs) and that no other determinants or factors have major effects on global TSS distributions in vivo (Figure 3C) or on global sequence determinants for TSS selection in vivo (Figure S3B).
MASTER analysis of effects of NTP concentrations on TSS selection
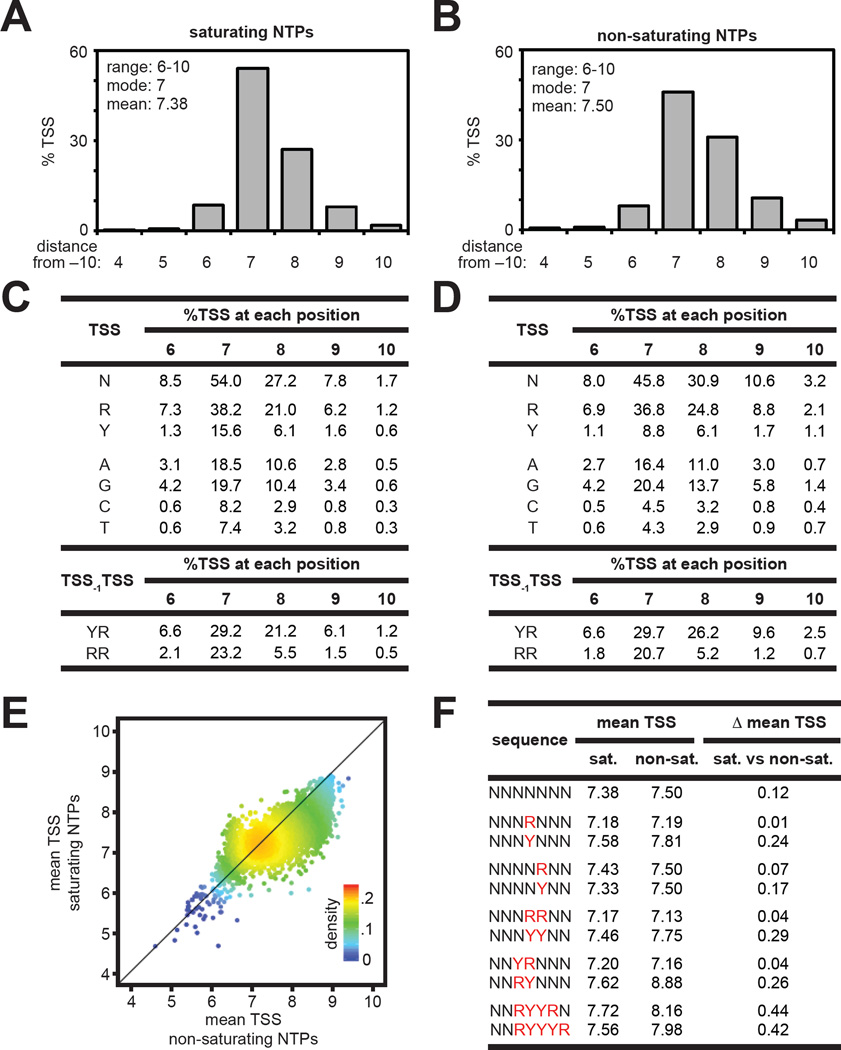
To analyze the effects of NTP concentrations on TSS selection we compared results obtained from the analysis of in vitro reactions performed in the presence of saturating NTPs (2.5 mM NTPs:Mg2+) (Table S4) with results obtained from the analysis of in vitro reactions performed in the presence of non-saturating NTPs (0.1 mM) (Table S5). Results show that the range of TSS positions (6–10) and modal TSS (position 7) are identical at saturating and non-saturating NTP concentrations (Figure 4A,B) and identical to the range and mode observed in the analysis of non-supercoiled and supercoiled DNA templates in vitro at 1 mM NTPs and the analysis of supercoiled DNA templates in vivo (Figures 2A, 3A, 3C, S4A, S4B). Comparison of the sequence determinants for TSS selection at saturating and non-saturating NTP concentrations revealed that preference for an R TSS over a Y TSS increases at non-saturating NTP concentrations, preference for a G TSS increases at non-saturating NTP concentrations, and preference for YTSS-1RTSS over RTSS-1RTSS increases at non-saturating NTP concentrations (Figures 4C, 4D, S2A–D).
Figure 4. Effects of NTP concentrations on TSS selection in vitro.
A. and B. TSS-distribution histograms at saturating (A) and non-saturating (B) NTP concentrations in vitro. Average %TSS at positions 4–10 for TSS-regions with ≥25 matched RNA reads (Tables S4 and S5). Experiments were performed at 2.5 mM NTPs:Mg2+ (saturating) or 0.1 mM NTPs (non-saturating) using a non-supercoiled linear DNA template.
C. and D. Sequence determinants for TSS selection. (C, saturating NTPs; D, non-saturating NTPs)
E. Plot of the mean TSS at saturating NTP concentrations versus non-saturating NTP concentrations for individual TSS-region sequences.
F. Average of the mean TSS values observed for the indicated TSS-region sequences at saturating (sat.) and non-saturating (non-sat.) NTP concentrations. (Δ mean TSS; differences between values observed at saturating and non-saturating NTP concentrations)
Comparison of the mean TSS observed at saturating and non-saturating NTP concentrations (Figure 4A,B) reveals the overall TSS distribution at non-saturating NTP concentrations (mean = 7.50 bp downstream of −10 element) is shifted slightly downstream relative to the overall TSS distribution at saturating NTP concentrations (mean = 7.38 bp downstream of −10 element). Comparison of individual TSS-sequence variants revealed that a majority of TSS-sequence variants exhibited a change in the mean TSS at non-saturating NTP concentrations compared with saturating NTP concentrations (Figure 4E). TSS-region sequences carrying an R at position 7 and position 8 are less susceptible to alterations in TSS selection in response to alterations in NTP concentrations, while TSS-region sequences carrying a Y at position 7 and position 8 are more susceptible to alterations in TSS selection in response to alterations in NTP concentrations (Figures 4F, S2E; p < 0.001). In addition, TSS-region sequences that contained YR at positions 6/7 are less susceptible to effects of alterations in NTP concentrations, while TSS-region sequences carrying a RY at positions 6/7 are more susceptible to effects of alterations in NTP concentrations (Figures 4F, S2E; p < 0.001). Furthermore, TSS-region sequences that contained RYYR at positions 6–9 or RYYYR at positions 6–10 were highly susceptible to NTP-concentration dependent changes in TSS selection (Figures 4F, S2E; p < 0.001). Strikingly, the patterns observed in the comparison of sequences that are more or less susceptible to NTP-concentration dependent changes in TSS selection (Figure 4F) were similar to the patterns observed in the comparison of sequences that are more or less susceptible to topology dependent changes in TSS selection (Figures 3E, S4D).
MASTER analysis defines the extent of productive slippage synthesis during transcription initiation
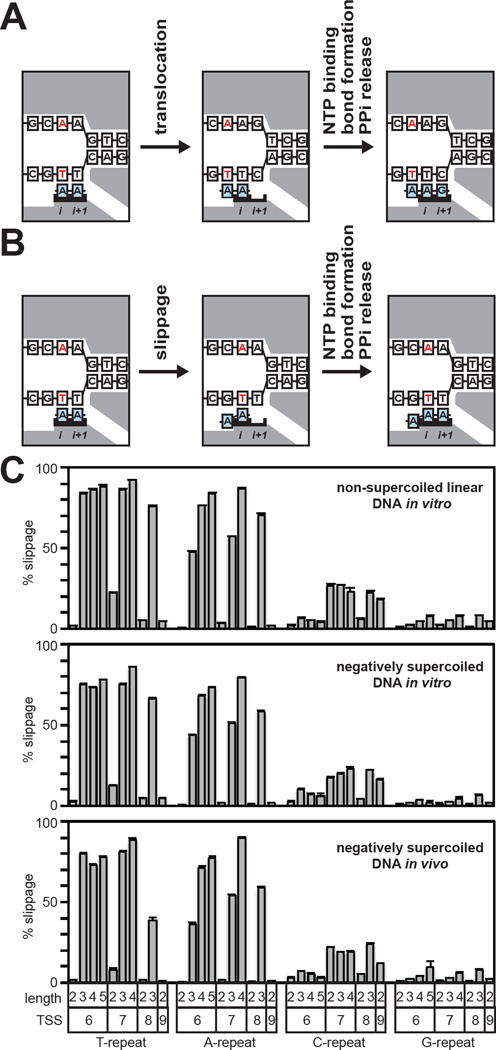
During the standard pathway of transcription initiation, in each nucleotide addition step, RNAP translocates relative to the DNA and RNA, and the DNA template strand and the 3′ end of the RNA product remain in register (Figure 5A). However, during initial transcription RNAP can enter into an alternative pathway of transcription termed "slippage synthesis" (Jacques and Kolakofsky, 1991; Turnbough and Switzer, 2008; Turnbough, 2011). In slippage synthesis, RNAP does not translocate relative to the DNA and RNA, and, instead, the RNA product slips upstream relative to the DNA template strand, establishing a new register of DNA and RNA (Figure 5B). Slippage synthesis can occur in multiple cycles, including very large numbers of cycles. Accordingly, slippage synthesis is also referred to as "reiterative transcription initiation."
Figure 5. Comprehensive analysis of productive slippage synthesis.
A. Nucleotide addition cycle for the standard pathway of transcription initiation. Left: initial transcribing complex with a 2-nt RNA in a pre-translocated state. Middle: initial transcribing complex with a 2-nt RNA in a post-translocated state. Right: 3-nt product complex in a pre-translocated state. The RNA and DNA template strand remain in lock-step register and the sequence of the RNA is fully complementary to the template strand. White boxes, DNA; blue boxes, RNA; gray shading, RNAP; red, TSS bases; i and i+1, RNAP active-center i and i+1 sites.
B. Nucleotide addition cycle for the slippage pathway. Left: initial transcribing complex with a 2-nt RNA in a pre-translocated state. Middle: RNA has moved backward relative to the DNA template by one base. Right: 3-nt product complex in a pre-translocated state. The 5′ end of the RNA carries an RNA/DNA difference and is not complementary to the template strand.
C. Analysis of productive slippage synthesis. Graphs show % slippage (mean + SEM) for TSS-region sequences containing 5′ end homopolymeric repeat sequences of the indicated length that begin at the indicated position (TSS).
(See Figure S5)
RNAs produced as a consequence of slippage synthesis either can be released from the initial transcribing complex ("non-productive slippage synthesis") or can be extended to yield full-length RNA products ("productive slippage synthesis"). Full-length RNA products generated by productive slippage synthesis typically contain at least one 5' nucleotide that does not match the sequence of the DNA template ("RNA/DNA difference"; Figure 5B). Therefore, to identify products of productive slippage synthesis in our analysis of transcription output, we enumerated reads that carried at least one RNA/DNA difference (Tables S1–S7) and, since slippage synthesis can occur in multiple cycles, we also enumerated reads with 5′ ends that were up to five bases longer than position 4, i.e. the first position of the randomized TSS-region ("L reads" in Tables S1–S7).
To assess the extent of productive slippage synthesis, we analyzed the total transcription output from each TSS-region sequence variant for transcripts predicted to be generated by productive slippage synthesis.
First, we assessed the extent of productive slippage synthesis at promoters that contain homopolymeric repeat sequences that start at the TSS (i.e., Tn, An, Gn, and Cn, where n > 1 and where the sequence starts at the TSS). Such sequences potentially are expected to facilitate productive slippage synthesis, in increments of one base pair, since homopolymeric repeat sequences allow slippage to occur, in increments of one base pair, with the net loss of only one RNA-DNA base pair (Figure 5B). Products of standard synthesis and productive slippage synthesis from homopolymeric repeat sequences were distinguished by the absence or presence, respectively, of at least one RNA/DNA difference. For example, for the TSS-region sequence AAT, which carries an A2 homopolymeric repeat starting at the underlined TSS base, RNAs generated by standard synthesis would have the fully templated 5′-end sequence AAU-, whereas potential RNAs generated by productive slippage synthesis would have 5′-end sequences AAAU-, AA AAU-, AAAAAU-, etc. that carry one or more 5′-RNA/DNA difference. We calculated the percentage of RNAs from the homopolymeric tract that are produced by slippage [% slippage = 100(#slippage reads) / (#slippage reads + #standard reads)]. The results indicate that: (i) slippage occurs at promoters that contain Tn, An, Cn, and Gn homopolymeric repeats at TSS positions 6, 7, 8, and 9 (Figure 5C); (ii) % slippage is especially high for Tn and An homopolymeric repeats (up to >80%; Figure 5C); (iii) % slippage increases in all cases as the length of a Tn, An and Gn homopolymeric repeat increases and increases in many cases as the length of a Cn homopolymeric repeat increases (Figure 5C); (iv) the number of nucleotides added to the RNA 5′ end by repeated cycles of slippage increases as the length of a Tn, An and Gn homopolymeric repeat increases (Tables S1–S7); and (v) the number of nucleotides added to the 5′ end by repeated cycles of slippage can be strikingly long (up to at least 8 for Tn and An homopolymeric repeats; Tables S1–S7).
Next, we assessed the extent of productive slippage synthesis at promoters that contain homopolymeric repeat sequences that start one base downstream of the TSS. Such sequences also potentially are expected to facilitate productive slippage synthesis, in increments of one nucleotide, since such sequences also allow slippage to occur, in increments of one nucleotide, with the net loss of only one RNA-DNA base pair. Products of standard synthesis and potential products of productive slippage synthesis again were distinguished by the absence or presence, respectively, of at least one RNA/DNA difference. The results indicate that (i) slippage occurs at promoters that contain Tn, An, Cn, and Gn homopolymeric repeat sequences that start one base downstream of a TSS at positions 6, 7, and 8 (Figure S5A); (ii) % slippage is especially high for Tn and An homopolymeric repeats (Figure S5A); (iii) % slippage increases in all cases as the length of a Tn, An and Gn homopolymeric repeat increases and increases in many cases as the length of a Cn homopolymeric repeat increases (Figure S5A); (iv) the number of nucleotides added to the RNA 5′ end by repeated cycles of slippage increases as the length of a Tn, An and Gn homopolymeric repeat increases (Tables S1–S7); and (v) the number of nucleotides added to the 5′ end by repeated cycles of slippage can be strikingly long (up to at least 8 for Tn and An homopolymeric repeats; Tables S1–S7).
Next, we assessed the extent of productive slippage synthesis at promoters that do not contain homopolymeric repeat sequences starting at the TSS. Such sequences potentially may allow productive slippage synthesis in increments of two nucleotides. Products of standard synthesis and potential products of productive slippage synthesis were distinguished, in this case, by extension of the RNA 5′-end by increments of two nucleotides. For example, for the TSS-region sequence AGAT, RNAs generated by standard synthesis would have the fully templated 5′-end sequence AGAU-, whereas potential RNAs generated by productive slippage synthesis in increments of two nucleotides would have the 5′-end sequences AGAGAU-, AGAGAGAU-, AGAGAGAGAU-, etc. The results indicate that slippage in increments of two nucleotides occurs for promoters that lack homopolymeric repeat sequences at the TSS (~0.1% to ~1% slippage in increments of two base pairs; Figure S5C). The results further indicate that slippage in increments of two base pairs is especially high for the TSS-region dinucleotide sequences TG, TA, CG, and AG (Figure S5C), and that the local sequence context of the TSS-region dinucleotide sequence affects % slippage. Thus, for most cases, when the nucleotide preceding the TSS-region dinucleotide sequence matches the second nucleotide of the TSS-region dinucleotide sequence, or when the nucleotide following the TSS-region dinucleotide sequence matches the first nucleotide of the TSS-region dinucleotide sequence, the % slippage increases (Figure S5C). More broadly, when sequences preceding or following the TSS-region dinucleotide sequence (ab) create a dinucleotide repeat pattern of the form b-ab, ab-ab, ab-a, b-ab-a, or ab-ab-a, % slippage is especially higher (Figure S5C). We infer that slippage in increments of two nucleotides occurs, is widespread, and is facilitated by upstream or downstream extensions of the TSS region dinucleotide sequence that increase complementarity between the RNA product and the DNA template strand upon slippage in increments of two nucleotides.
Prompted by the above results, we re-analyzed published RNA-seq data obtained in vivo in E. coli (Thomason et al, 2015), assessing levels of productive slippage synthesis for chromosomal promoters in vivo in E. coli (Figure S5B,C). We find extents of slippage for chromosomal promoters containing homopolymeric repeats at the TSS (Figure S5B, top) or one base pair downstream of the TSS (Figure S5B, bottom) similar to those observed for a consensus core promoter by MASTER (Figures 5C, S5A). In addition, we find slippage in increments of two nucleotides at chromosomal promoters containing non-repeat TSS-region dinucleotide sequences (Figure S5C).
Finally, we assessed the contribution of productive slippage synthesis to the total transcription output for each TSS-region sequence variant. We divided the total number of sequencing reads derived from productive slippage synthesis by the total number of sequencing reads, combining data for all analyzed experimental conditions. The results indicate, surprisingly, that productive slippage synthesis is detectable (≥0.5%) for the majority of TSS-region sequences (~52%; 8,590 of 16,384), occurs at moderate levels (≥5%) for ~18% of TSS-region sequences (3,007 of 16,384), and accounts for the majority of the total transcription output (≥50%), for a large number of TSS-region sequences (3.1%; 509 out of 16,384).
MASTER analysis reveals effects of TSS-region sequences on transcript yield
To document effects of TSS-region sequence on relative transcript yield, we divided the read count representing the total transcription output for each TSS-region sequence variant by the relative number of DNA templates that carried the sequence variant. We refer to the value obtained as the “relative expression” of each TSS-region sequence variant (Tables S1–S7). A comparison of the relative expression values allowed us to determine the influence of sequence variation in the TSS-region on the range of expression observed for a given experimental condition (Figures 6A, 6B, S6).
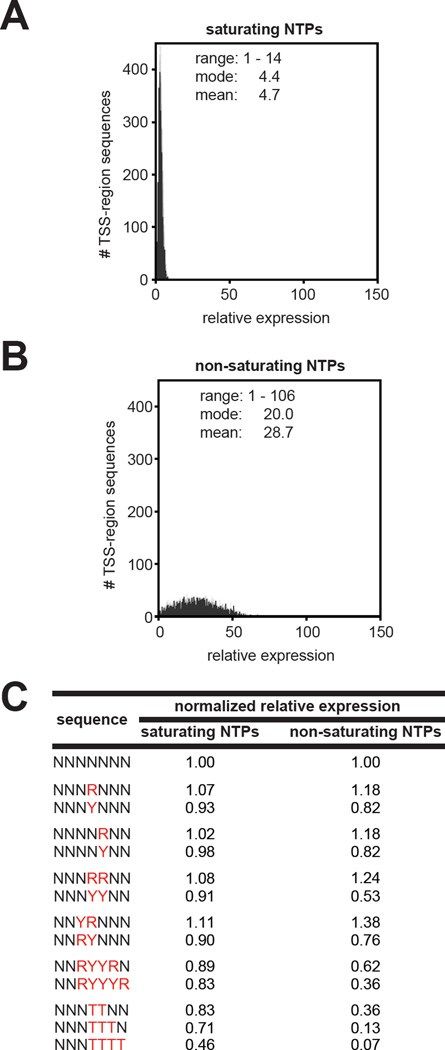
Figure 6. Effects of NTP concentrations on transcript yields in vitro.
A. and B. Relative expression histograms for experiments performed at saturating NTP (A) and non-saturating (B) NTP concentrations using a non-supercoiled linear DNA template in vitro. Relative expression for TSS-region sequences with ≥ 25 total RNA reads for which the number of DNA templates was not in the top or bottom 10% (Tables S4 and S5). For each experimental condition the lowest value of relative expression was normalized to 1.
C. Normalized relative expression for the indicated TSS-region sequences. Values were calculated by dividing the average relative expression for the indicated TSS-region sequence by the relative expression observed for all TSS-region sequences.
(See Figure S6)
For experiments performed in vitro using non-supercoiled linear DNA or negatively supercoiled DNA (at 1 mM NTPs) TSS-region sequence variation led to a ~40-fold range of relative expression (Figure S6A,B). For experiments performed with negatively supercoiled DNA in vivo we found that TSS-region sequence variation led to a > 100-fold range of relative expression (Figure S6C). However, interpreting the effects of TSS-region sequences on transcript yields in vivo is complicated by the potential contribution of sequence variation at the RNA 5′ end to the stability of full-length transcripts. Thus, we infer that the analysis of effects of TSS-region sequence variation on relative transcript yield in vitro provides a more accurate measure of the true impact of TSS-region sequence variation on transcription output. We therefore conclude that DNA topology does not exert a significant global impact on the relationship between TSS-region sequence and the range of expression.
We next compared the effect of NTP concentrations on the range of expression. For assays performed in vitro at saturating NTP concentrations, TSS-region sequence variation led to a ~14-fold range of relative expression (Figure 6A). In contrast, for assays performed in vitro at non-saturating NTP concentrations TSS-region sequence variation led to a ~100-fold range of relative expression (Figure 6B). Thus, the results show that limiting the concentrations of NTPs significantly enhances effects of TSS-region sequence variation on expression. Results further show that the magnitude of the difference in relative expression of TSS-region sequences carrying an R at TSS positions 7 and 8 relative to TSS-region sequence variants carrying a Y at TSS positions 7 and 8 increased at non-saturating NTP concentrations (Figure 6C). In addition, the magnitude of the difference in relative expression observed from TSS-region sequences carrying YR versus RY at positions 6/7 also increased at non-saturating NTP concentrations (Figure 6C). Furthermore, TSS-region sequences carrying 2–4 consecutive T bases, which exhibit high % slippage (Figure 5), exhibit a large decrease in relative expression at non-saturating NTP concentrations compared with saturating NTP concentrations (Figure 6C). Thus, we propose that this decrease in relative expression occurs, at least in part, due to a decrease in productive slippage synthesis and concomitant increase in non-productive slippage synthesis (undetectable by MASTER), as a consequence of performing reactions at non-saturating NTP concentrations.
Our analysis of the effects of TSS-region sequence on transcript yield show that sequence variation in the TSS-region can impact overall transcript yields (i.e. promoter strength) by at least two orders of magnitude. We conclude that the TSS-region sequence is a key determinant of promoter strength.
MASTER analysis reveals a correlation between precision of TSS-selection and transcript yield
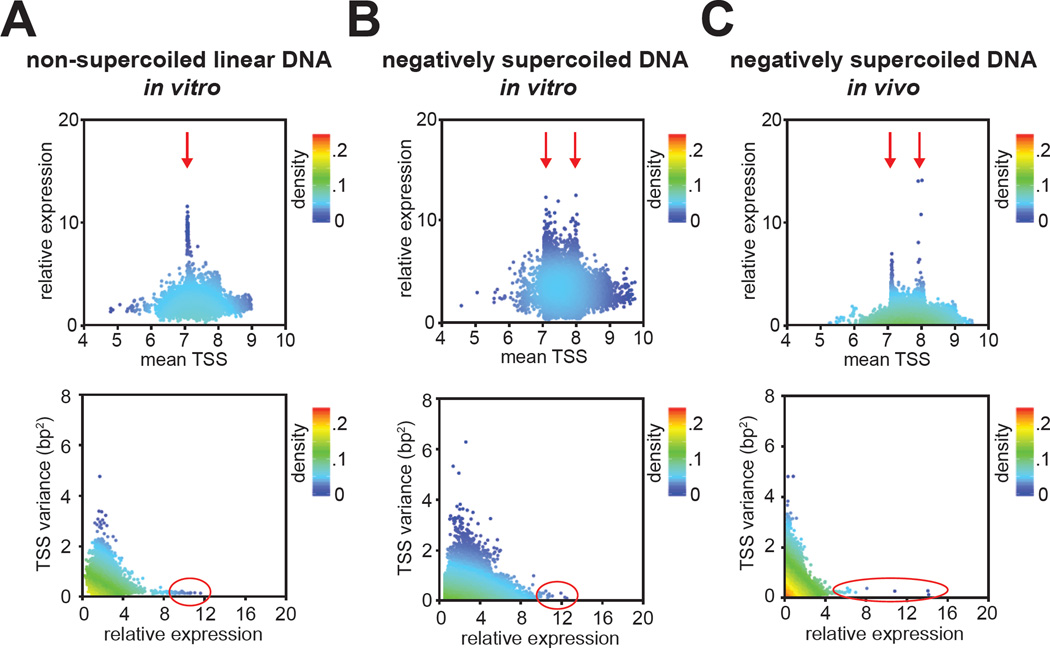
We next analyzed the relationship between TSS selection and transcript yields by comparing the mean TSS value with the relative expression value for each TSS-region sequence variant (Figure 7). This analysis revealed that sequence variants with the highest relative expression had mean TSS values extremely close to one of the two preferred TSS positions, position 7 and position 8 (Figure 7A–C, red arrows).
Figure 7. Precision of TSS selection is a determinant of transcript yield.
Top: plot of relative expression versus mean TSS. Bottom: plot of TSS variance versus relative expression.
Next, we compared the variance from the mean TSS versus the relative expression for each sequence variant (Figure 7A–C, bottom). Results show that TSS-region sequences with the highest relative expression exhibited very low variance from the mean TSS (Figure 7, red circles; Figure S2F; p < 0.001). Thus, our MASTER analysis reveals that TSS-region sequences that exhibit the highest expression under a particular experimental condition also exhibit “precise” TSS selection at either position 7 or position 8. We conclude that precision of TSS selection is a previously undocumented contributor to promoter strength.
DISCUSSION
Here we report the development and application of MASTER, a technology leveraging the capabilities of high-throughput sequencing to enable the comprehensive analysis of the relationship between up to at least 47 (~16,000), and potentially at least 410 (~1,000,000), DNA sequences and transcriptional output. MASTER provides many orders of magnitude higher throughput than conventional methods of interrogating the effects of DNA sequence on transcription output, can be carried out within a reasonable timescale, and can be performed at reasonable cost. We demonstrate the utility of MASTER by applying this method to the analysis of transcription initiation. Our results provide a complete description of the relationship between TSS-region sequence and TSS selection, slippage synthesis, and transcript yields for initiation at a consensus core promoter by E. coli RNAP in vitro and in vivo.
MASTER analysis provides a comprehensive description of transcription initiation
We documented three measurable outputs of transcription initiation: TSS position, productive slippage synthesis, and yields of full-length transcripts, for a library comprising 47 (~16,000) sequence variants of a consensus E. coli promoter. The results define full inventories of transcription start sites ("TSSomes") of E. coli RNAP in vitro and in vivo (Tables S1–S7) and full inventories of transcripts generated by productive slippage synthesis ("slippomes") of E. coli RNAP in vitro and in vivo (Tables S1–S7). Furthermore, our analyses of productive slippage synthesis provide, to our knowledge, the first comprehensive description of the extent of slippage in vitro and in vivo for any RNAP. Results indicate that slippage synthesis occurs from the majority of TSS-region DNA sequences and reveal slippage by increments of two nucleotides occurs at surprisingly high levels (Figures 5, S5, Tables S1–S7).
Our analysis of the effect of DNA topology on TSS-selection provides mechanistic insight into this process. In particular, single-molecule FRET experiments indicate that transcription-bubble size in the RNAP-promoter open complex (RPo) can vary (Robb et al., 2013). Based on the flexibility of RPo, it has been proposed that variability in the position of the TSS relative to core promoter elements is mediated by changes in the size of the unwound transcription bubble in RPo. Specifically, it has been proposed that TSS selection at downstream sites is mediated by transcription-bubble expansion ("scrunching"), and TSS selection at upstream sites is mediated by transcription-bubble contraction ("anti-scrunching") (Robb et al., 2013). Our MASTER results indicate that negative supercoiling, which can provide a driving force for transcription-bubble expansion, favors TSS selection at downstream positions (Figures 3, S4), consistent with the hypothesis that TSS-selection involves scrunching and anti-scrunching.
MASTER analysis of the effects of TSS-region sequences on yields of full-length transcripts show that TSS-region DNA sequences can have profound, up to 100-fold, effects on transcript yield (Figures 6, S6, Tables S1–S7). Furthermore the impact of TSS-region sequence on the range of expression varies in response to changes in NTP concentrations. Our findings that promoter TSS-region sequences can dictate a wide-range of expression levels suggests that these sequences serve as a reservoir of expression level diversity. Moreover, this diversity should easily be accessible to mutational processes and natural selection for tuning or altering promoter output.
MASTER analysis comparing TSS selection and transcript yield revealed a previously unknown relationship between precision of TSS selection and promoter strength (Figure 7). In particular, we found that sequences with the highest expression exhibit precise TSS selection at positions 7 or 8 (Figure 7, Tables S1–S7). Our findings indicate that optimal expression is provided, in part, by TSS-region sequences that promote efficient start site selection at a single position. The identification of the precision of TSS selection as a previously unidentified contributor to promoter strength exemplifies the utility of MASTER for illuminating aspects of transcription that would be difficult, if not impossible, to discover through conventional approaches.
MASTER as a new approach for comprehensive analysis of the relationship between nucleic-acid sequence and functional output
Analyses of the behavior of chromosomally encoded promoters using high-throughput approaches (e.g. RNA-seq, ChIP-seq) have provided a wealth of information regarding mechanisms employed by cells to regulate gene expression. However, every promoter is an evolved, unique sequence, which constrains the ability of researchers using such approaches to infer universal properties based on aggregate behavior. Furthermore, for sequence regions of more than a few bases, the total sequence diversity represented by all promoters in a genome is significantly less than the maximum theoretical diversity contained in an equivalent length of randomized DNA sequence. MASTER overcomes the inherent limitations imposed by analysis of chromosomally encoded promoters by enabling the comprehensive measurement of transcription output for all possible sequence variants of a given region of a transcription unit. In addition, MASTER allows the analysis of the behavior of all possible sequence variants in a region of interest to be performed over diverse conditions in vitro and in vivo.
Furthermore, although for sequence-specific promoter elements one can predict the effect of mutations on the overall affinity of RNAP for the promoter element, correlating the effect on transcription output is not straightforward. Thus, predicting how a given promoter will respond to alterations in conditions, and identifying the sequence determinants that dictate the response, represents an immense challenge. MASTER overcomes these limitations by enabling systematic variation of sequence attributes in a controlled fashion. Thus, we anticipate that results obtained from these and future studies using MASTER will enable more accurate predictions of the behavior of chromosomally encoded promoters, and will inform the design of synthetic promoters for use in artificial biological circuits. In addition, MASTER can be readily adapted for comprehensive analysis of sequence determinants for transcription elongation, transcription termination, or any other biological process that involves complex nucleic-acid interactions.
Limitations
In this work, we have considered effects of TSS-region sequences on transcriptional output within the context of a consensus core promoter. However, in principle, the sequence of other promoter sequences (i.e. the UP-element, −35 element, the spacer region, the extended −10 element, the −10 element, and the discriminator element) also may impact transcription start site selection. The potential roles of other promoter sequences can be addressed simply by including additional regions of the promoter within the randomized segments. Currently, the method enables up to at least 7 bp, and potentially up to at least 10 bp, to be included in the randomized segment (Figures 1, S1). As sequencing technologies improve, the method will provide comprehensive coverage of even larger segments.
In addition, the in vivo experiments reported in this work are performed in the context of a promoter that is plasmid-borne. Thus, a priority for future work will be to modify the MASTER procedure in order to analyze the behavior of promoter libraries in vivo in the context of the bacterial chromosome.
EXPERIMENTAL PROCEDURES
Construction of MASTER plasmid libraries
To generate library pMASTER-lacCONS-N7 (Figures 1, S1A), a PCR product containing the lacCONS promoter with a 7-nt randomized region positioned 4–10 nt downstream of the −10 element and a 15-nt randomized “barcode” region positioned 27–41 nt downstream of the −10 element was subcloned into pSG289 (see Supplemental Experimental Procedures and Figure S1A). Sequencing analysis of the pMASTER-lacCONS-N7 library indicated that it contains 16,295 (~99.5%) of a possible 16,384 sequences in the TSS-region (Figure S1C). Library pMASTER-lacCONS-N10 was generated using a similar procedure and contains 1,019,505 (~97.2%) of a possible 1,048,576 sequences carrying a 10-bp randomized region (Figure S1B).
Generation of RNAs from the pMASTER-lacCONS-N7 library in vitro
10 nM of template DNA (see Supplemental Experimental Procedures) was mixed with 50 nM RNAP holoenzyme in transcription buffer (50 mM Tris HCl pH 8.0, 10 mM MgCl2, 0.01 mg/ml BSA, 100 mM KCl, 5% glycerol, 10 mM DTT, 0.4U/µl RNase OUT) and incubated at 37°C for 10 min. Transcription was initiated by adding NTPs (to a final concentration of 10 mM, 1 mM or 0.1 mM each NTP) and heparin (to 0.1 mg/ml). (We note that the Mg2+ concentration of 10 mM is limiting. Accordingly, the effective NTP:Mg2+ concentration in reactions performed in the presence of 10 mM NTPs is 2.5 mM for each NTP.) Reactions were stopped after 15 min by addition of EDTA to 10 mM. RNAs were analyzed by 5′ RNA-seq as described in the Supplemental Experimental Procedures.
Generation of RNAs from the pMASTER-lacCONS-N7 library in vivo
DH10B–T1R cells carrying pMASTER-lacCONS-N7 were grown at 37°C in 50 ml of LB containing chloramphenicol (25 µg/ml) in a 250 ml flask shaken at 210 RPM. RNA was isolated at OD600 ~0.5 and used for 5′ RNA-seq analysis as described in the Supplemental Experimental Procedures. Plasmid DNA was also isolated from cells and used as a template in emulsion PCR reactions to generate a product that was sequenced to assign barcodes (see Supplemental Experimental Procedures).
High-throughput sequencing of RNA 5’ ends (5’ RNA-seq)
5′ RNA-seq was done using procedures described in (Vvedenskaya et al., 2015) and in the Supplemental Experimental Procedures. cDNA libraries were sequenced using an Illumina HiSeq 2500.
Data analysis
Sequencing of template DNA was used to associate the 7-bp randomized sequence in the region of interest with a corresponding second 15-bp randomized sequence that serves as its barcode. For 5′ RNA-seq analysis we considered only those reads that contained a perfect match to the sequence downstream of position 10 and a perfect match to the 5 bases downstream of the 15-bp barcode. The identity of the 15-bp barcode was used to determine the identity of bases at positions 4–10 of the lacCONS template. Sequences derived from the RNA 5′ end of reads that were perfect matches to the sequence of the template were used for analysis of TSS selection. Sequences derived from RNA 5′ ends that carried one or more mismatches from the DNA template and/or extended up to 5 bases upstream of position 4, were not considered in the analysis of TSS position, but were used for analysis of productive slippage synthesis and for analysis of transcript yields (for further details see Supplemental Experimental Procedures). All of the reads used for our data our analyses are provided in Tables S1–S7.
Source code and documentation for analysis of DNA templates and 5′ RNA-seq libraries are provided at: https://github.com/NickelsLabRutgers/MASTER-Data-Analysis.
Data deposition
Raw reads have been deposited in the NIH/NCBI Sequence Read Archive under the study accession number SRP057850.
Supplementary Material
ACKNOWLEDGEMENTS
We thank Craig Kaplan for discussion, Jeremy Bird and Maria Ciaramella for assistance with preparation of template libraries, Jared Knoblauch for preliminary data analysis, and Hanif Vahedian Movahed for RNAP. Work was supported by FIRB-Futuro in Ricerca RBFR12OO1G_002 “Nematic” (AV and VV) and NIH grants GM041376 (RHE), GM088343 (BEN), GM096454 (BEN), and GM115910 (BEN).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
AUTHOR CONTRIBUTIONS
IOV, SRG, AV and VV performed experiments. YZ, DT, RHE and BEN analyzed data. RHE and BEN designed experiments and wrote the paper.
REFERENCES
- Aoyama T, Takanami M. Essential structure of E. coli promoter II. Effect of the sequences around the RNA start point on promoter function. Nucleic Acids Res. 1985;13:4085–4096. doi: 10.1093/nar/13.11.4085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dangkulwanich M, Ishibashi T, Bintu L, Bustamante C. Molecular mechanisms of transcription through single-molecule experiments. Chem. Rev. 2014;114:3203–3223. doi: 10.1021/cr400730x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hawley DK, McClure WR. Compilation and analysis of Escherichia coli promoter DNA sequences. Nucleic Acids Res. 1983;11:2237–2255. doi: 10.1093/nar/11.8.2237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heyduk E, Heyduk T. Next generation sequencing-based parallel analysis of melting kinetics of 4096 variants of a bacterial promoter. Biochemistry. 2014;53:282–292. doi: 10.1021/bi401277w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heyduk T, Heyduk E. Next Generation Sequencing-based analysis of RNA polymerase functions. Methods. 2015 doi: 10.1016/j.ymeth.2015.04.030. [DOI] [PubMed] [Google Scholar]
- Jacques JP, Kolakofsky D. Pseudo-templated transcription in prokaryotic and eukaryotic organisms. Gene Dev. 1991;5:707–713. doi: 10.1101/gad.5.5.707. [DOI] [PubMed] [Google Scholar]
- Jeong W, Kang C. Start site selection at lacUV5 promoter affected by the sequence context around the initiation sites. Nucleic Acids Res. 1994;22:4667–4672. doi: 10.1093/nar/22.22.4667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jorgensen SE, Buch LB, Nierlich DP. Nucleoside triphosphate termini from RNA synthesized in vivo by Escherichia coli . Science. 1969;164:1067–1070. doi: 10.1126/science.164.3883.1067. [DOI] [PubMed] [Google Scholar]
- Larson MH, Landick R, Block SM. Single-molecule studies of RNA polymerase. Mol. Cell. 2011;41:249–262. doi: 10.1016/j.molcel.2011.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis DE, Adhya S. Axiom of determining transcription start points by RNA polymerase in Escherichia coli. Mol. Micro. 2004;54:692–701. doi: 10.1111/j.1365-2958.2004.04318.x. [DOI] [PubMed] [Google Scholar]
- Liu J, Turnbough CL. Effects of transcriptional start site sequence and position on nucleotide-sensitive selection of alternative start sites at the pyrC promoter in Escherichia coli . J. Bacteriol. 1994;176:2938–2945. doi: 10.1128/jb.176.10.2938-2945.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maitra U, Hurwitz H. The role of DNA in RNA synthesis, IX. Nucleoside triphosphate termini in RNA polymerase products. Proc. Natl. Acad. Sci. U.S.A. 1965;54:815–822. doi: 10.1073/pnas.54.3.815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melnikov A, Murugan A, Zhang X, Tesileanu T, Wang L, Rogov P, Feizi S, Gnirke A, Callan CG, Jr, Kinney JB, et al. Systematic dissection and optimization of inducible enhancers in human cells using a massively parallel reporter assay. Nat. Biotechnol. 2012;30:271–277. doi: 10.1038/nbt.2137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murakami KS. Structural biology of bacterial RNA polymerase. Biomolecules. 2015;5:848–864. doi: 10.3390/biom5020848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patwardhan RP, Hiatt JB, Witten DM, Kim MJ, Smith RP, May D, Lee C, Andrie JM, Lee SI, Cooper GM, et al. Massively parallel functional dissection of mammalian enhancers in vivo. Nat. Biotechnol. 2012;30:265–270. doi: 10.1038/nbt.2136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robb NC, Cordes T, Hwang LC, Gryte K, Duchi D, Craggs TD, Santoso Y, Weiss S, Ebright RH, Kapanidis AN. The transcription bubble of the RNA polymerase-promoter open complex exhibits conformational heterogeneity and millisecond-scale dynamics: implications for transcription start-site selection. J. Mol. Biol. 2013;425:875–885. doi: 10.1016/j.jmb.2012.12.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruff EF, Record MT, Jr, Artsimovitch I. Initial events in bacterial transcription initiation. Biomolecules. 2015;5:1035–1062. doi: 10.3390/biom5021035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shultzaberger RK, Chen Z, Lewis KA, Schneider TD. Anatomy of Escherichia coli σ70 promoters. Nucleic Acids Res. 2007;35:771–788. doi: 10.1093/nar/gkl956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sorensen KI, Baker KE, Kelln RA, Neuhard J. Nucleotide pool-sensitive selection of the transcriptional start site in vivo at the Salmonella typhimurium pyrC and pyrD promoters. J. Bacteriol. 1993;175:4137–4144. doi: 10.1128/jb.175.13.4137-4144.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomason MK, Bischler T, Eisenbart SK, Forstner KU, Zhang A, Herbig A, Nieselt K, Sharma CM, Storz G. Global transcriptional start site mapping using differential RNA sequencing reveals novel antisense RNAs in Escherichia coli. J. Bacteriol. 2015;197:18–28. doi: 10.1128/JB.02096-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turnbough CL. Regulation of gene expression by reiterative transcription. Curr. Opin. Microbiol. 2011;14:142–147. doi: 10.1016/j.mib.2011.01.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turnbough CL, Switzer RL. Regulation of pyrimidine biosynthetic gene expression in bacteria. Microbiol. Mol. Biol. Rev. 2008;72:266–300. doi: 10.1128/MMBR.00001-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vvedenskaya IO, Goldman SR, Nickels BE. Preparation of cDNA libraries for high-throughput RNA sequencing analysis of RNA 5' ends. Methods Mol. Biol. 2015;1276:211–228. doi: 10.1007/978-1-4939-2392-2_12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker KA, Osuna R. Factors affecting start site selection at the Escherichia coli fis promoter. J. Bacteriol. 2002;184:4783–4791. doi: 10.1128/JB.184.17.4783-4791.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang F, Greene EC. Single-molecule studies of transcription: from one RNA polymerase at a time to the gene expression profile of a cell. J. Mol. Biol. 2011;412:814–831. doi: 10.1016/j.jmb.2011.01.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Washburn RS, Gottesman ME. Regulation of transcription elongation and termination. Biomolecules. 2015;5:1063–1078. doi: 10.3390/biom5021063. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.