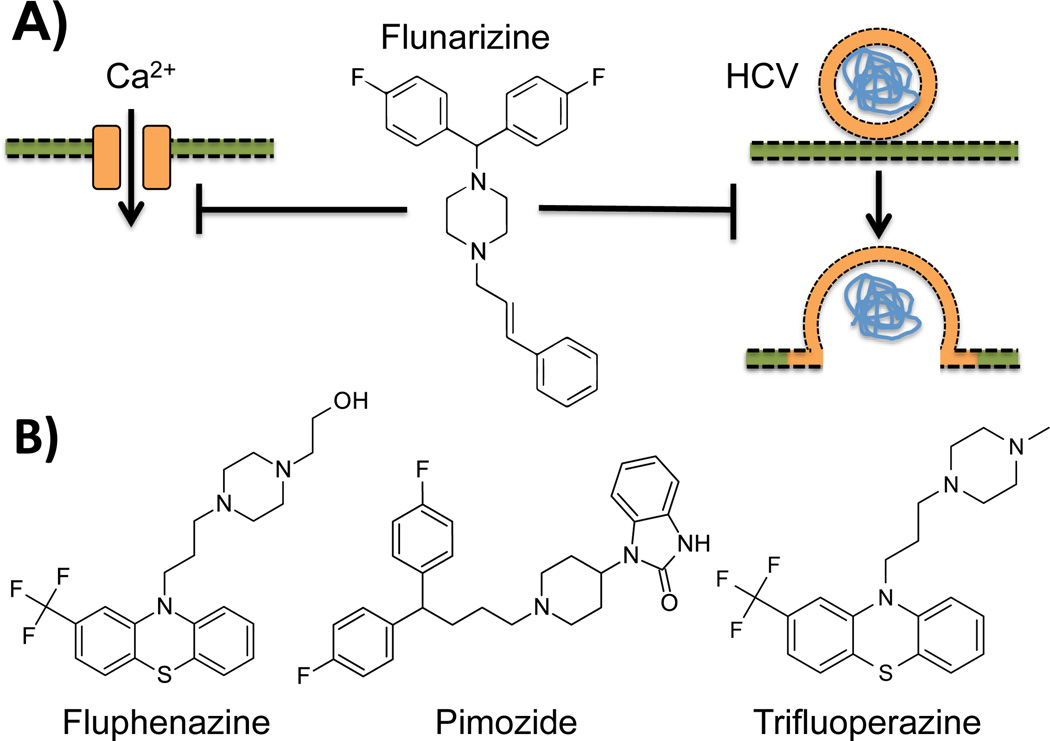
Hepatitis C virus (HCV) is a blood-borne pathogen that causes widespread chronic liver disease (1). In an article in the current issue of Hepatology, Perin et al. identify flunarizine, a clinically approved drug for treating migraine, as an inhibitor of specific HCV genotypes (Fig. 1A). Leveraging several experimental approaches, the authors demonstrate that fusion of viral and host-cell membranes during viral entry is the step that is susceptible to flunarizine inhibition. Determinants of susceptibility to this and related drugs including fluphenazine, pimozide, and trifluoperizine (Fig. 1B) reside in the E1 and E2 envelope glycoproteins of HCV. These findings suggest a cost-efficient potential anti-HCV therapeutic strategy and shed light on the mechanism of HCV cell entry.
Fig. 1.
The chemical structure and activity of flunarizine. (A) Flunarizine is a large hydrophobic fluorinated piperazine derivative used in the prophylaxis of migraine. Its activity is based on blocking calcium influx into vascular smooth muscle. In an article in the current issue of Hepatology, Perin et al. report that flunarizine also blocks the HCV lifecycle at the membrane fusion step. (B) Fluphenazine, pimozide, and trifluoperazine form a subset of related calcium antagonists that show similar ability to inhibit HCV entry as flunarizine and are likely to share the inhibitory mechanism of flunarizine.
HCV is an enveloped positive-sense single-stranded RNA virus from the hepacivirus genus in the Flaviviridae family. In the bloodstream of infected patients it travels in association with lipoprotein particles. HCV is highly hepatotropic and follows a complex entry pathway reliant on multiple host entry factors (2). Following attachment and trafficking to tight junctions in polarized hepatocytes, the virus is internalized and fusion of the viral and endosomal membranes proceeds by mechanism dependent on the reduced pH of the endosomal lumen. Once HCV genome is delivered to the cytoplasm, a single polyprotein is translated and processed into three structural and seven non-structural proteins. Genome replication and egress of newly formed virions complete the lifecycle. Several recently approved anti-HCV drugs target viral polyprotein processing and genome replication (3).
HCV E1 and E2 are the two envelope glycoproteins required for cellular attachment and membrane fusion. E1 and E2 form stable heterodimers and are anchored in the viral membrane by their C-terminal transmembrane domains. Recent structural studies revealed that HCV E1 and E2 proteins have molecular architectures distinct from the envelope proteins of other viruses from other genera within the Flaviviridae family (4–6). E2 contains the determinants of attachment while E1 has been proposed to contain the determinants of fusion. However, the molecular trajectory leading to membrane fusion is not fully understood due in part to the lack of a complete three-dimensional structure of the E1/E2 heterodimer.
More than a decade before the discovery of HCV, flunarizine was developed as a calcium antagonist with potential applications in treatment of vascular disorders (7). In the 1980s it found widespread use as an effective anti-migraine therapeutic and is still used in this capacity in Canada and continental Europe today. While the structural basis of flunarizine binding and inhibition of voltage-gated calcium channels is not known, flunarizine acts to inhibit calcium influx into smooth muscles cells preventing constriction of blood vessels and thus helps improve blood flow to the brain to combat the pathophysiology of migraine.
Perin et al. identified flunarizine as a submicromolar HCV inhibitor in a whole lifecycle screen of a library of clinically approved drugs. They confirmed this finding in assays using primary human hepatocytes, humanized reporter mice, and in vitro grown hepatic organoids. Using chimeric viruses they showed that susceptibility to flunarizine and related compounds varies widely between HCV subtypes and is encoded in the E1 and E2 proteins. Isolation of flunarizine-resistant mutants allowed determination of specific residues necessary for inhibiting the viral lifecycle. These included M267V and Q289H in E1, M405T in E2 and I757T in p7. Interestingly, M267, a key determinant of flunarizine susceptibility, is limited to the susceptible genotype, genotype 2a – a result that neatly dovetails observations obtained with chimeric viruses.
In elegant time of addition experiments Perin et al. demonstrated that flunarizine inhibition occurs at the membrane fusion step of cell entry. In these experiments individual steps of the entry pathway were controlled by temperature and pH shifts. Flunarizine showed a unique ability to block viral lifecycle (assayed using a luciferase reporter, an indirect readout) when added during the low-pH incubation that triggers fusion. This result was strengthened by the direct observations obtained by infecting hepatic organoids with viruses carrying fluorescently labeled membranes. While HCV association with tight junction markers and endocytic pathway markers was unaffected by flunarizine, the increase in fluorescence that follows mixing of viral and host membranes was strongly inhibited.
Molecular mechanism underlying flunarizine fusion inhibition remains unclear. Lack of effect of calcium chelators and unrelated calcium channel inhibitors, such as nickel (Ni2+) ions, suggests that calcium flux inhibition is not required for fusion arrest. Likewise, membrane stiffness and cholesterol content appeared unaffected by flunarizine and the related compounds. Additionally, similar genotype specificities and shared resistance mutants of the flunarizine-like compounds argue against an indirect mode of action. Instead, it is possible that genotype 2 viruses use a unique host cofactor during entry and that flunarizine inhibits that interaction. Alternatively, flunarizine may directly stabilize the genotype 2 envelope proteins in a fusion incompetent conformation. Given the hydrophobic nature of these compounds, it is possible that they share a common binding site on the E1/E2 dimer despite their different chemical structures. Further investigation into the mechanism of inhibition will shed light on both the clinical applicability of these scaffolds and on the HCV entry pathway.
The findings reported by Perin et al. are significant because targeting of viral fusion step has been underexplored for its therapeutic potential. So far, mechanism-based fusion inhibitors have been developed for class I fusion proteins, such as those of HIV (8), and several approaches for inhibiting class II fusion proteins have been advanced including broad specificity drugs affecting membrane stiffness (9). Discovery of the antiviral activity of flunarizine adds membrane fusion as a new target to the anti-HCV repertoire. Although flunarizine is only effective against HCV genotype 2, it may offer a cost-effective complement to the currently available treatments, which will remain unaffordable to a significant fraction of the infected population for some time. Moreover, flunarizine may serve as an invaluable molecular platform for the design of more broadly active and more potent HCV entry inhibitors. Such design efforts would be greatly aided if the structure of the flunarizine binding pocket on HCV E1/E2 could be determined.
Acknowledgments
Written with support from a Senior Research Fellowship from the Wellcome Trust (grant no.: 101908/Z/13/Z) to Y.M. and from grant R01 GM102869 from the National Institutes of Health to Y.M.
Abbreviations
- HCV
hepatitis C virus
Footnotes
Potential conflict of interest: Nothing to report.
References
- 1.Lindenbach BD, Murray CL, Thiel HJ, Rice CM. Flaviviridae. In: Knipe DM, Howley PM, editors. Fields Virology. Sixth ed. Vol. 1. Philadelphia: Lippincott Williams & Wilkins; 2013. pp. 712–746. [Google Scholar]
- 2.Lindenbach BD, Rice CM. The ins and outs of hepatitis C virus entry and assembly. Nat Rev Microbiol. 2013;11:688–700. doi: 10.1038/nrmicro3098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.European Association for the Study of the Liver. EASL Recommendations on Treatment of Hepatitis C 2015. J Hepatol. 2015;63:199–236. doi: 10.1016/j.jhep.2022.10.006. [DOI] [PubMed] [Google Scholar]
- 4.Kong L, Giang E, Nieusma T, Kadam RU, Cogburn KE, Hua Y, Dai X, et al. Hepatitis C virus E2 envelope glycoprotein core structure. Science. 2013;342:1090–1094. doi: 10.1126/science.1243876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Khan AG, Whidby J, Miller MT, Scarborough H, Zatorski AV, Cygan A, Price AA, et al. Structure of the core ectodomain of the hepatitis C virus envelope glycoprotein 2. Nature. 2014;509:381–384. doi: 10.1038/nature13117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.El Omari K, Iourin O, Kadlec J, Sutton G, Harlos K, Grimes JM, Stuart DI. Unexpected structure for the N-terminal domain of hepatitis C virus envelope glycoprotein E1. Nat Commun. 2014;5:4874. doi: 10.1038/ncomms5874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Van Nueten JM, Janssen PA. Comparative study of the effects of flunarizine and cinnarizine on smooth muscles and cardiac tissues. Arch Int Pharmacodyn Ther. 1973;204:37–55. [PubMed] [Google Scholar]
- 8.Didigu CA, Doms RW. Novel approaches to inhibit HIV entry. Viruses. 2012;4:309–324. doi: 10.3390/v4020309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Pierson TC, Kielian M. Flaviviruses: braking the entering. Curr Opin Virol. 2013;3:3–12. doi: 10.1016/j.coviro.2012.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]