Abstract
Maintenance of the balance of DNA methylation and demethylation is fundamental for normal cellular development and function. Members of the Ten-Eleven-Translocation (TET) family proteins are Fe(II)- and 2-oxoglutarate-dependent dioxygenases that catalyze sequential oxidation of 5-methylcytosine (5mC) to 5-hydroxymethylcytosine (5hmC) and subsequent oxidized derivatives in DNA. In addition to their roles as intermediates in DNA demethylation, these oxidized methylcytosines are novel epigenetic modifications of DNA. DNA methylation and hydroxymethylation profiles are markedly disrupted in a wide range of cancers but how these changes are related to the pathogenesis of cancers is still ambiguous. In this review, we discuss the current understanding of TET protein functions in normal and malignant hematopoietic development and the ongoing questions to be resolved.
Introduction
In mammalian cells, cytosines in DNA undergo a series of covalent modifications by DNA methyltransferases (DNMTs) and TET family enzymes [1,2]. During the early stages of embryogenesis, the de novo DNA methyltransferases DNMT3A and DNMT3B add a methyl group to the 5-position of cytosine to create 5-methylcytosine (5mC), which mostly occurs as a symmetrical modification in the context of CpG dinucleotides. During DNA replication, the DNA methylation pattern is faithfully inherited by the daughter cells through the action of the maintenance DNA methyltransferase DNMT1, which is recruited to newly replicated hemi-methylated DNAs by its partner protein UHRF1 and restores symmetrical methylation by adding methyl groups to the newly synthesized DNA strands (Figure 1A).
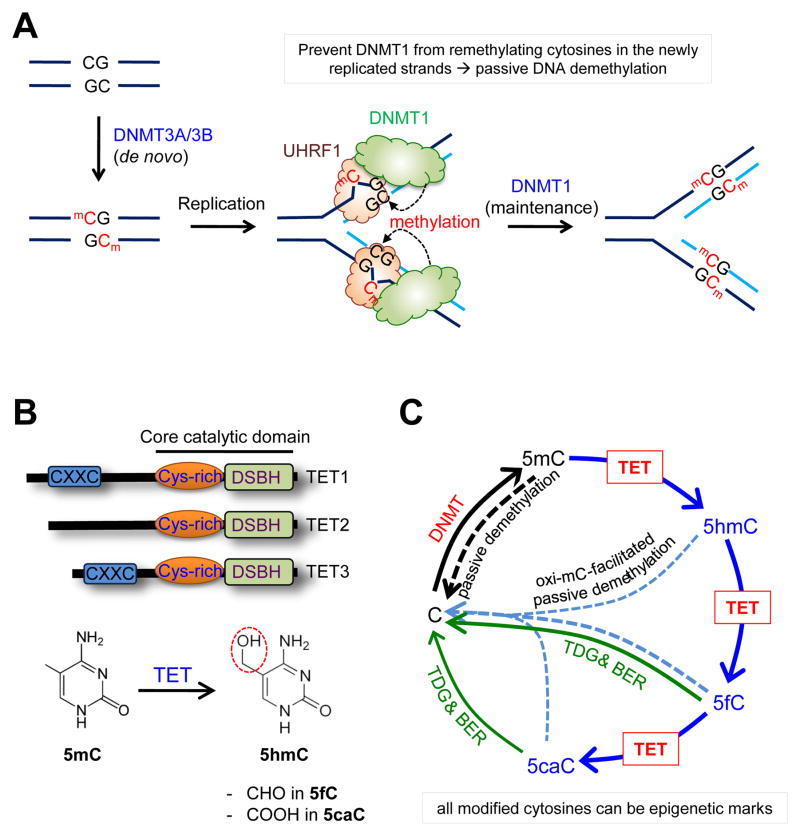
Figure 1. Interplay between DNMTs and TETs in DNA demethylation pathway.
A, DNA methylation and passive DNA demethylation pathway. De novo DNA methyltransferases DNMT3a and DNMT3b create 5-methylcytosines in the context of CpG dinucleotides. After DNA replication, DNMT1 is recruited to the hemimethylated regions via interaction with its critical partner UHRF1, then restores symmetrical methylation pattern in the newly synthesized DNA strands. When DNMT1 is lost or inactivated, 5mC undergoes passive dilution after replication, a process termed passive DNA demethylation. B, Domain structure and function of TET proteins. All three TET family members (TET1, TET2 and TET3) contain a core catalytic domain that is composed of a cysteine-rich domain (Cys-rich) and double stranded β-helix domain (DSBH). TET1 and TET3, but not TET2, additionally contain an Zn finger type DNA-binding domain named CXXC domain at the N-terminal region, which preferentially binds chromatin regions containing unmethylated CpG sequences. C, Control of DNA methylation-demethylation dynamics by TET proteins. TET proteins successively oxidize 5-methylcytosine to produce 5-hydroxymethylcytosine (5hmC), 5-formylcytosine (5fC) and 5-carboxylcytosine (5caC) in DNA. All these oxi-mCs inhibit DNMT1, promoting passive DNA demethylaton. 5fC and 5caC are directly excised by thymine DNA glycosylase (TDG) to generate abasic sites, which triggers base excision repair to replace the abasic sites with unmodified cytosines.
TET family dioxygenases (TET1, TET2 and TET3) sequentially oxidize the methyl group of 5mC (Figure 1B). They first add a hydroxyl group to form 5-hydroxymethylcytosine (5hmC) [3], after which they can catalyze further stepwise oxidation of 5hmC to generate 5-formylcytosine (5fC) and 5-carboxylcytosine (5caC) [4,5]. The three oxidized methylcytosines (oxi-mCs) are implicated in both passive and active DNA demethylation (reviewed in [2,6]) and can also behave as stable epigenetic marks (Figure 1C) [7–10]. TET proteins may also affect chromatin architecture and gene expression independently of their catalytic activity, through physical interactions with various cellular proteins [11,12].
It is well documented that loss of TET function is closely associated with diverse types of cancers including hematological and non-hematological cancers (reviewed in [13,14]). However, whether impaired TET function is directly implicated in driving cancer development remains to be determined. Furthermore, inactivation of DNMTs is also known to contribute to oncogenesis in certain contexts [15–17]. Because DNMTs generate 5mC that TET proteins consume as substrate, it is unclear whether DNMT loss-of-function results in malignancy via loss of 5mCs, oxi-mCs or both. Elucidation of the molecular links between TET/DNMT dysregulation and oncogenic transformation may enable us to develop ways to restore a normal methylation and hydroxymethylation landscape in cancers, and reveal novel pathways that can be clinically targeted for the prevention or treatment of cancers.
The potential mechanisms by which TET protein expression or function is regulated, and possible ways to manipulate TET activity in cancers for the development of novel epigenetic therapies, have already been discussed in detail elsewhere [13,14]. Therefore, in this review, we discuss our current understanding of the function of TET loss-of-function in normal and oncogenic cellular development, with an emphasis on several open questions related to hematopoietic transformation that need to be resolved in future studies.
Aberrant DNA methylation in normal and oncogenic hematopoietic development
HSC repopulating capacity
Studies using murine models show that DNA methylation is critical for homeostasis and differentiation of hematopoietic stem cells (HSCs). Interestingly, maintenance and de novo DNA methyltransferases seem to play distinct roles in regulating the repopulating capacity of HSCs. In serial transplantation assays under both competitive and noncompetitive conditions, disruption or partial reduction of Dnmt1 activity in hematopoietic system results in severe impairment of trilineage engraftment [18,19]. In contrast, loss of Dnmt3a strongly augments hematopoietic reconstitution over serial transplantation [20] whereas that of Dnmt3b exhibited a milder phenotype consistent with the fact that Dnmt3b highly expressed in HSCs corresponds to a catalytically inactive isoform [21]. Interestingly, combined deficiency of Dnmt3a and Dnmt3b synergistically increased the repopulating capacity of HSCs [21].
HSC self-renewal
Early reports suggested that both de novo (Dnmt3a and Dnmt3b) and maintenance (Dnmt1) methyltransferases are necessary for HSC self-renewal [18,19,22]. However, in these studies, instead of directly assessing the ability of HSCs to self-renew, the extent of peripheral blood chimerism after serial transplantation was used as a marker for HSC self-renewal. This assay does not faithfully reflect HSC self-renewal because the outcome could be affected by multiple factors such as proliferation or apoptosis of mature cells and/or their differentiating precursors. More recently, by directly quantifying HSCs that are amplified after transplantation, Challen et al. showed that Dnmt3a-deficient HSCs indeed display augmented self-renewal [20]. Ablation of Dnmt3b only weakly affected HSC self-renewal while simultaneous deletion of both enzymes led to a dramatic increase in HSC self-renewal capability [21]. Mechanistically, loss of Dnmt3a was suggested to increase self-renewal by upregulating multipotency genes crucial for HSC functions (Gata3, Runx1, Pbx1 and Cdkn1a) and downregulating differentiation-related genes (Flk2, Sfpi1, and Mef2c), but this pattern of altered gene expression poorly correlated with methylation changes induced by Dnmt3a loss [20], suggesting that there might be unidentified mechanisms by which loss of Dnmt3a controls gene expression, including mechanisms independent of catalytic activity or coordinated regulation of multiple genes through effects on master regulators. Furthermore, in most studies, the effect of Dnmt loss on DNA methylation status was assessed by bisulfite sequencing of genomic DNAs, which does not distinguish 5mC from 5hmC or C from 5fC and 5caC [2,23]. Thus, it has yet to be clearly defined how loss of Dnmt activity affects DNA methylation status in target gene loci and how changes in DNA methylation in turn affect gene transcription.
Fate decisions and differentiation
DNA methylation also seems to be implicated in fate decisions during hematopoiesis at the very early stage of HSCs as well as subsequent differentiating progenitors [24,25]. Lineage-associated genes are weakly expressed in HSCs prior to lineage commitment, a process termed lineage priming [26], and their expression is differentially up- or down-regulated depending on the differentiation pathway involved. In HSCs from Dnmt1 hypomorphic mice, lymphoid, but not myeloerythroid, genes were strikingly suppressed, resulting in developmental skewing toward myeloerythroid lineages with impaired B lymphopoiesis [19]. Consistently, the expression of CD48, an early marker whose expression is rapidly induced upon differentiation of HSCs, is also upregulated in Dnmt1-deficient HSCs [19], suggesting that HSCs utilize DNA methylation to prevent premature induction of differentiation programs. In contrast, Dnmt3a-disrupted HSCs strongly favor self-renewal over differentiation toward mature cells, a phenomenon exacerbated by combined deletion of Dnmt3b [20,21].
Analyses of dynamic DNA methylation patterns during hematopoietic differentiation suggest that DNA methylation can also affect lineage choice at later stages [24,25]. The general idea is that in early progenitors undergoing differentiation towards a specific lineage, the regulatory regions of lineage-related genes are demethylated and expressed, whereas the regulatory regions of genes that specify alternative lineages are silenced by robust methylation [25]. Compared with myeloid progenitors, lymphoid progenitors seem to rely more on DNA methylation for efficient suppression of alternate-lineage (i.e. myeloerythroid) genes [24,25]. However, the precise mechanisms by which DNA methylation differentially controls lineage gene expression in HSCs and subsequent differentiating progenitors remains to be established.
Hematopoietic transformation
The connection between aberrant DNA methylation and cancer was first experimentally demonstrated by the development of aggressive T cell lymphoma in Dnmt1 hypomorphic mice [27]. The molecular basis of cancer development in these mice is not clear but increased genome instability was considered an important contributing factor [27]. Exome-sequencing showed that genetic aberrations at DNMT1 locus are very rare in hematopoietic cancers, so DNMT1 function might be controlled in ways other than through simple coding region mutations.
It was recently discovered that somatic DNMT3A mutations are recurrent in myeloid and lymphoid malignancies including acute myeloid leukemias (AMLs; 20~30%), myelodysplastic syndromes (MDS; 10~15%) and myeloproliferative neoplasms (MPN; ~8%), and they are generally associated with poor prognosis [28]. Interestingly, DNMT3A mutations are typically heterozygous and most frequently occur at the Arginine 882 residue within the methyltransferase domain. More than 60% of DNMT3A mutations in AML target this residue [28,29], and the resulting DNMT3AR882H mutant proteins exert a dominant negative effect on the remaining wild-type DNMT3A [30]. Consistently, mice reconstituted with hematopoietic stem/progenitor cells (HSPCs) overexpressing DNMT3AR882H mutant succumb to chronic myelomonocytic leukemia (CMML)-like disease [15], and transfer of Dnmt3a-deficient HSCs or bone marrow cells also predisposes wild-type mice to diverse types of myeloid and lymphoid malignancies resembling MDS, AML and acute lymphoid leukemia (ALL) albeit with long latency [20,21]. Some Dnmt3a-deficient leukemia samples acquired second mutations such as c-Kit, Notch1, Kras and Npm1 mutations [20,21], implying that Dnmt3a loss-of-function cooperates with other mutations to drive oncogenic transformation, shortening latency and affecting the spectrum of diseases that develop. Again, most of these studies are descriptive and the underlying molecular mechanisms remained to be clarified.
Recently, extended (> 3.5 kb) regions with low CpG methylation, termed ‘canyons’, were identified as distinct from CpG islands in the genome [31] and this novel genomic feature was associated with genes that are highly conserved and developmentally regulated. Despite low level of DNA methylation, all canyon-associated genes are not highly expressed. In murine HSCs, about half of canyons were marked only with H3K4me3 marks whereas 45.9% contained both H3K4me3 and H3K27me3 marks, with the remaining 6% exhibiting only H3K27me3. The genes associated with H3K4me3-marked canyons were highly expressed whereas those associated with H3K27me3-marked canyons displayed no or low expression, irrespective of the presence of H3K4me3 marks, implying that H3K27me3 marks may be the determining factor for gene expression. Interestingly, the methylation/hydroxymethylation status of canyon borders seems to be dynamically regulated by cooperation between Dnmt3a and Tet proteins, which is important for normal and malignant hematopoietic development. Some canyon-associated genes that are active in HSCs are significantly enriched for genes whose expression is dysregulated in leukemias. However, how histone modifications are differentially regulated in canyons and whether canyon size alterations are related to gene expression, differentiation and transformation have not been directly assessed.
Contrasting roles of TET1 in myeloid and lymphoid cancers
TET1, the founding member of the TET family, was originally identified as a fusion partner of the mixed-lineage leukemia gene (MLL; also known as KMT2A) in a handful of acute myeloid and lymphocytic leukemias bearing the chromosomal translocation t(10;11)(q22;q23) [32,33]. TET1 and TET3 mutations have garnered relatively less attention compared to TET2 mutations, because their coding region mutations are rarely identified in hematological malignancies. However, recent studies indicate that TET1 also retains a crucial regulatory role in the oncogenic transformation of hematopoietic cells. Intriguingly, TET1 has contrasting roles in myeloid and lymphoid transformation. In MLL-rearranged leukemia, it functions as an oncogene [34]. TET1 expression was significantly upregulated by various MLL fusion proteins, leading to an increase in the expression of oncogenic target genes such as Hoxa9, Meis1 and Pbx3 and thus promoting leukemogenesis [34]. In contrast, TET1 also has tumor suppressor function: a recent study reported that loss of Tet1 predisposes mice to B cell lymphoma that resembles follicular lymphoma and diffuse large B cell lymphoma [35]. TET1 transcription was frequently downregulated in non-Hodgkin B cell lymphoma (B-NHL) through promoter CpG methylation. In mice, Tet1 loss resulted in aberrant hematopoiesis characterized by augmented repopulating capacity of HSCs, skewed differentiation toward B cell lineage and increased frequency of lymphoid-primed multipotent progenitors (LMPPs). Tet1-deficient pro-B cells also displayed enhanced serial replating capacity and accumulation of DNA damage, potentially through downregulation of genes encoding components of the DNA repair machinery. Whole-exome sequencing of Tet1-deficient cancers identified multiple mutations that are also frequently observed in human B-NHL, suggesting that TET1 deficiency impairs genomic integrity and renders cells more susceptible to secondary mutations to drive full malignancy. However, it still remains unclear how TET1 exerts distinct functions in different cancers, how deficiency of Tet1 results in altered gene expression, differentiation bias and genomic instability, and how these alterations are connected to oncogenic transformation.
TET2 mutations in hematological cancers
The potential connection between TET2 mutation and myeloid transformation was suggested by early observations that the chromosomal region 4q24 that harbors the TET2 gene shows recurrent microdeletions and copy number-neutral loss-of-heterozygosity (also termed uniparental disomy) in myeloid cancers [36,37]. Subsequently, extensive exome-sequencing studies revealed various types of somatic TET2 mutations in large cohorts of patients with myeloid malignancies such as MDS (~20%), CMML (~50%), AML (~20%) and secondary AML (reviewed in [13,14]). Although TET2 mutations are prevalent throughout the TET2 coding regions, missense mutations tend to be relatively clustered in regions encoding the core catalytic domain of TET2 protein [13], and most of these impair the normal catalytic activity of TET2 to cause impaired 5mC oxidation and aberrant DNA methylation in genome [38–40].
In addition to myeloid cancers, TET2 mutations are also frequently found in various lymphoid malignancies. Compared to B cell lymphoma, peripheral T cell lymphoma (PTCL) including angioimmunoblastic T cell lymphoma (AITL; about 33~76%) and PTCL, not otherwise specified (PTCL-NOS; about 20~38%) display highest frequency of TET2 mutations [41–43]. In AITL, TET2 mutations co-exist with RHOAG17V and DNMT3A mutations at very high frequency [43–46]. Unlike patients with myeloid malignancies where TET2 and IDH mutations are mutually exclusive [47], TET2 and IDH2 mutations co-occur very frequently (~88.2%) in patients with PTCL [42–44], suggesting that the mode and outcome of the functional interplay between TET2 and IDH mutations could be divergent in different cancers. Whether and how TET2 mutations cooperate with these mutations to facilitate the development of lymphomas remains to be determined.
Unlike TET1, TET2 loss-of-function commonly induces a developmental bias toward the myeloid lineage. HSPCs isolated from MPN patients harboring TET2 mutations [36] or murine progenitor cells with diminished expression of Tet2 preferentially differentiate toward the myeloid lineage at the expense of other lineages in vitro [47,48]. In most of the murine models reported so far, ablation of Tet2 results in an accumulation of the HSPC population in the bone marrow. Tet2-deficient HSPCs seem to more efficiently replate serially in methylcellulose, compared to wild-type controls [41,49,50]. Transplantation assays showed that Tet2-deficient bone marrow or fetal liver cells acquired a significant advantage over wild-type cells for reconstitution into all hematopoietic lineages [41,48–53]. The effect of Tet2 on self-renewal of HSCs was not directly assessed but serial transplantation assays using wild-type or Tet2-disrupted CD150+ LSK cells suggested that Tet2 deficiency augments HSC self-renewal in a cell-autonomous manner [54]. Thus, Tet2 normally restricts the expansion and function of HSCs in the fetus as well as in adults. Notably, some strains of Tet2-disrupted mice are predisposed to develop CMML-like myeloid malignancies with advanced age, displaying a characteristic increase in myelopoiesis, splenomegaly, hepatomegaly, extramedullary hematopoiesis and elevated white blood cell counts associated with monocytosis and neutrophilia [41,49,50]. As commonly observed in various mouse models of myeloid malignancies, normal splenic architecture in these mice was disrupted by massive infiltration of myeloid or erythroid cells. In some mice, MDS with expansion of erythroid progenitors and highly transplantable MPN-like myeloid leukemias were also induced upon Tet2 deficiency [41,49]. Consistent with the frequent occurrence of heterozygous TET2 mutations in patient samples, Tet2 haploinsufficiency sufficed to alter HSC properties and induce MPN- or CMML-like myeloid leukemias [41,49,50].
Although descriptive, these studies suggest that there is a causal relation between loss of TET2 function and myeloid transformation. However, the sporadic incidence of full–blown leukemias at a very late time point hinders our understanding of the exact pathogenic function of TET2. For unknown reasons, no malignancies were observed in certain strains of Tet2-deficient mice [41]. Furthermore, despite frequent TET2 mutations in lymphoid malignancies, lymphoma formation is rare in murine models. In addition, some elderly people have loss-of-function mutations in TET2 that are similar to those observed in myeloid leukemias but they do not always have overt hematopoietic cancers [55]. Taken together, it seems clear that TET2 mutations alone do not strongly induce malignancy: rather, TET2 loss-of-function may contribute to the initiation of oncogenic transformation by inducing a precancerous condition, after which cells acquire additional genetic aberrations (‘second hits’) to develop full-blown malignancy. Depending on the timing and combination of cooperating mutations, the rate and fate of transformation might be determined. Indeed, numerous mutations were shown to co-exist with TET2 mutations in MDS patients [56,57] and studies using various murine models demonstrated that these mutations synergize with Tet2 deficiency to accelerate development of a more advanced disease phenotype with markedly reduced latency [51,58–62].
Potential redundancy between TET2 and TET3
TET proteins and their immediate catalytic product, 5hmC, are ubiquitously expressed throughout tissues. Tet2 and Tet3 are particularly highly expressed in the hematopoietic system, and their expression levels are dynamically regulated during hematopoietic differentiation [38,49,50]. What are the roles of Tet3 in normal and malignant hematopoiesis? Ablation of either Tet2 or Tet3 led to a modest decrease in 5hmC levels [14,48] but their combined deficiency was associated with a dramatic loss of genomic 5hmC, implying that Tet2 and Tet3 are the major 5mC oxidases in the hematopoietic system. As with Tet2 deficiency, Tet3 deficiency neither strongly disturbs normal hematopoiesis nor induces hematopoietic malignancy. Thus, Tet2 and Tet3 play redundant roles in controlling homeostasis, function and differentiation of HSCs, as well as in suppressing transformation. To understand more precisely the role of TET proteins in gene expression, lineage specification and malignant transformation, it will be necessary to assess whether combined deficiency of Tet2 and Tet3 facilitates or accelerates the development of myeloid malignancies.
As mentioned earlier, TET1 and TET3 coding region mutations are very rare in hematopoietic cancers. However, this does not necessarily imply that TET1 and TET3 do not retain function in these cancers; low 5hmC levels in these cancers suggest that these TET proteins could be inactivated in many other ways (Figure 2) [13,14]. For instance, mutations in regulatory sequences (such as promoters, enhancers or splice sites), promoter methylation and/or upregulation of TET-targeting E3 ligases or microRNAs could result in reduced expression of TET mRNAs and proteins [35,63–67]. As reported in glioblastomas and myeloid leukemias, increased levels of 2-hydroxyglutarate by gain-of-function mutations in IDH genes could inactivate TET enzymes [68,69]. Moreover, dysregulation of interaction with critical binding partners of TET proteins such as WT1, IDAX and HDAC1/2 could also result in loss of TET expression/activities or aberrant chromatin targeting of TET proteins [11,12,70,71]. Like other dioxygenases, TET enzymes require molecular oxygen, 2-oxoglutarate and ferrous iron as substrates or essential cofactors [72], and therefore would be sensitive to hypoxia, altered metabolism and altered redox function in cancer. Finally, at least two studies have shown that TET3 can translocate between the cytoplasm and the nucleus [73,74], and therefore is likely subject to regulation by signals from the tumor microenvironment. Further studies are necessary to determine how the expression and activity of TET3 are controlled in cancers.
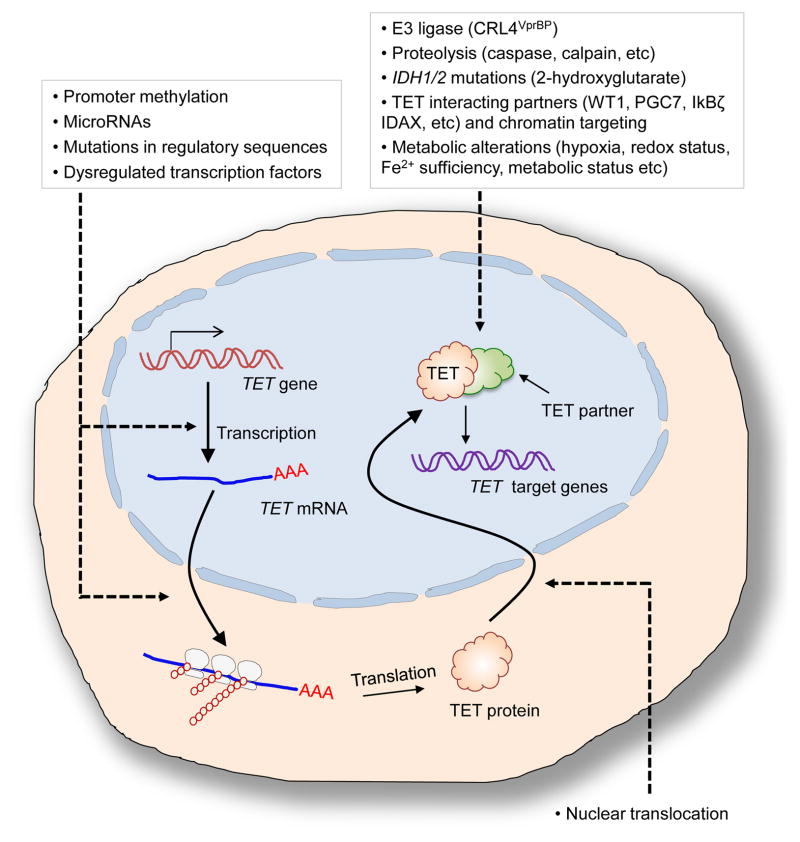
Figure 2. Modulation of TET protein expression and function.
In addition to the coding region mutations in TET genes, TET protein expression and activity can be modulated in a variety of ways. TET transcription or translation could be inhibited by promoter methylation, microRNAs, mutations in regulatory sequences (i.e. promoters, enhancers or splice sites) or altered transcription factor expression or function. TET protein levels could be regulated by proteolysis mediated by caspase or calpain. Ubiquitylation of TET2 protein by E3 ligases CRL4VprBP and interactions with other proteins such as WT1, IDAX and PGC7 that are important for recruiting TET proteins to specific chromatin regions are known to regulate its DNA binding activity. TET function can also be modulated by IDH mutations leading to the production of 2-hydroxyglutarate or alterations in cellular metabolic status (i.e. hypoxia, Fe2+ levels or redox state).
Proposed mechanisms by which TET loss-of-function could contribute to cancer
5mCs produced by DNMTs are essential substrates for TET enzymes, and it is clear that TET loss-of-function mutations impair the consumption of 5mC at certain genomic loci, leading to accumulation of 5mC [47]. However, recent whole-genome 5mC mapping studies in embryonic stem cells show that TET deficiency leads to alterations of genomic 5mC levels in both directions: hypermethylation and hypomethylation [75–77]. Importantly, these changes in DNA methylation do not directly correlate with alterations in gene expression. These data suggest that the epigenetic impact of TET dysregulation on gene expression and lineage specification may not critically depend on changes in DNA methylation. In this respect, there is substantial evidence that changes in DNA methylation are a long-term “consequence” of altered gene transcription rather than an immediate cause (reviewed in [78,79]).
What might be the mechanisms that underlie the gene expression changes induced by TET dysregulation, if alterations of DNA methylation are not the primary cause? The most relevant factor might be the profound loss of oxi-mCs produced by TET proteins in TET-deficient cells (Figure 3). In support of this point, loss of Dnmt3a results in similar hematopoietic phenotypes as Tet2 loss-of-function such as expansion, augmented repopulation and self-renewal of HSCs, myeloid skewing, and ultimately myeloid leukemia, as discussed in detail above. In terms of 5mC accumulation, loss of Dnmt3a and Tet2 proteins have antagonistic effects as the former would result in decreased 5mC but the latter contributes to increase of 5mCs; however, in terms of oxi-mC loss, they exhibit the same effects, since levels of oxi-mCs would decrease in both conditions (Figure 3).
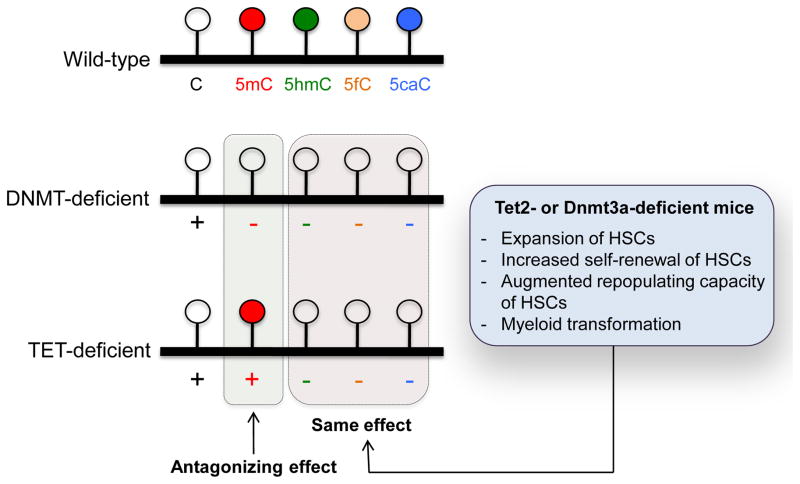
Figure 3. Significance of loss of oxi-mCs in the pathogenesis of cancers.
In terms of 5mC, loss of TET function results in increased 5mC and so is antagonistic to loss of DNMT function, which results in decreased 5mC. However, in terms of oxi-mC, loss of TET and DNMT function have the the same effect, a decrease of oxi-mC. Given that ablation of Tet2 or Dnmt3a in HSCs displays similar effects such as expansion, increased repopulating capacity and myeloid transformation of HSCs in murine models, loss of oxi-mCs seems more likely to be relevant to malignant transformation in hematopoietic malignancies than alterations in DNA methylation.
Considering that 5hmC is enriched at high levels at active enhancers and the gene bodies of actively transcribing genes, both TET and DNMT inactivation would be expected to decrease oxi-mC at a wide range of functional regions in the genome that are involved in gene transcription. However, 5hmC is a stable modification that is lost mainly through cell proliferation [9], and the modified base generated on a newly synthesized strand immediately after DNA replication is 5mC generated by DNMT, which is only subsequently acted upon by TET proteins to produce 5hmC and other oxi-mC. Therefore, the effect of 5hmC loss in TET- or DNMT-deficient cells on gene transcription would be observed only after several replication cycles and moreover would be most apparent in dividing cells. Future studies to assess whether combined deficiency of Dnmt3a and Tet2 accelerates hematopoietic transformation would be of great help to clarify the molecular mechanisms by which TET loss-of-function drive the development of hematopoietic cancers.
5fC and 5caC were shown to directly affect, albeit quite mildly, the processivity of RNA polymerase during transcriptional elongation [80]. This finding suggests that oxi-mC levels in gene bodies does not simply correlate with, but rather could be used to predict, the level of gene transcription. If this hypothesis is correct, oxi-mC would be most heavily lost, and gene transcription would be most prominently decreased, at genes that encode key signaling proteins and transcriptional regulators that govern cell lineage commitment and cell-type-specific function, as observed in early embryos that are deficient of Tet1 and Tet3 [81].
As an alternative but not mutually exclusive hypothesis, the tumor suppressor function of TET proteins may possibly be mediated by functions that are independent of their catalytic activities. It has been recently shown that WT1 physically associates with TET proteins, which is critical for chromatin targeting of TET enzymes and their target gene expression [70,71]. Tet2 interacts with HDAC1/2 in innate immune cells to suppress genes that are important for regulating inflammatory response such as IL-6 [12]. Therefore, the absence of intact TET proteins themselves would likely alter expression of a subset of master regulators of lineage specification, genomic integrity or tumor suppression; in consequence, the downstream target genes might be subject to a coordinate dysregulation that hastens oncogenesis. Further studies will be required to resolve this issue.
Conclusions
TET loss-of-function is strongly associated with hematologic malignancies of lymphoid and myeloid origin as well as diverse solid cancers. Previous studies have uncovered a role for TET proteins in regulating gene expression, lineage specification and genomic integrity to facilitate oncogenic transformation. However, we still lack a detailed understanding of the molecular mechanisms by which TET proteins control these processes. Further mechanistic studies on the role of TET dysregulation during oncogenesis and identification of ways to target TET activity in cancer cells will facilitate the development of novel epigenetic therapy of a wide range of cancers.
Acknowledgments
This work was supported by the research fund (1.150084) of Ulsan National Institute of Science and Technology (to M.K), and by NIH R01 grant CA151535 and grant 6464-15 from the Leukemia and Lymphoma Society Translational Research Program (to A.R).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References and recommended reading
Papers of particular interest, published within the period of review, have been highlighted as:
* of special interest
** of outstanding interest
- 1.Ooi SK, O’Donnell AH, Bestor TH. Mammalian cytosine methylation at a glance. J Cell Sci. 2009;122:2787–2791. doi: 10.1242/jcs.015123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Pastor WA, Aravind L, Rao A. TETonic shift: biological roles of TET proteins in DNA demethylation and transcription. Nature reviews Molecular cell biology. 2013;14:341–356. doi: 10.1038/nrm3589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Tahiliani M, Koh KP, Shen Y, Pastor WA, Bandukwala H, Brudno Y, Agarwal S, Iyer LM, Liu DR, Aravind L, et al. Conversion of 5-methylcytosine to 5-hydroxymethylcytosine in mammalian DNA by MLL partner TET1. Science. 2009;324:930–935. doi: 10.1126/science.1170116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ito S, Shen L, Dai Q, Wu SC, Collins LB, Swenberg JA, He C, Zhang Y. Tet proteins can convert 5-methylcytosine to 5-formylcytosine and 5-carboxylcytosine. Science. 2011;333:1300–1303. doi: 10.1126/science.1210597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.He YF, Li BZ, Li Z, Liu P, Wang Y, Tang Q, Ding J, Jia Y, Chen Z, Li L, et al. Tet-mediated formation of 5-carboxylcytosine and its excision by TDG in mammalian DNA. Science. 2011;333:1303–1307. doi: 10.1126/science.1210944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Shen L, Song CX, He C, Zhang Y. Mechanism and function of oxidative reversal of DNA and RNA methylation. Annual review of biochemistry. 2014;83:585–614. doi: 10.1146/annurev-biochem-060713-035513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Spruijt CG, Gnerlich F, Smits AH, Pfaffeneder T, Jansen PW, Bauer C, Munzel M, Wagner M, Muller M, Khan F, et al. Dynamic readers for 5-(hydroxy)methylcytosine and its oxidized derivatives. Cell. 2013;152:1146–1159. doi: 10.1016/j.cell.2013.02.004. [DOI] [PubMed] [Google Scholar]
- 8.Bachman M, Uribe-Lewis S, Yang X, Burgess HE, Iurlaro M, Reik W, Murrell A, Balasubramanian S. 5-Formylcytosine can be a stable DNA modification in mammals. Nature chemical biology. 2015;11:555–557. doi: 10.1038/nchembio.1848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- *9.Bachman M, Uribe-Lewis S, Yang X, Williams M, Murrell A, Balasubramanian S. 5-Hydroxymethylcytosine is a predominantly stable DNA modification. Nature chemistry. 2014;6:1049–1055. doi: 10.1038/nchem.2064. This study demonstrated that 5hmC is a stable modification. 5hmC loss primarily results from DNA replication, suggesting that 5hmC has a regulatory role as an epigenetic mark per se. Reference 8 shows that 5-formylcytosine is also a stable mark. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Raiber EA, Murat P, Chirgadze DY, Beraldi D, Luisi BF, Balasubramanian S. 5-Formylcytosine alters the structure of the DNA double helix. Nature structural & molecular biology. 2015;22:44–49. doi: 10.1038/nsmb.2936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- *11.Ko M, An J, Bandukwala HS, Chavez L, Aijo T, Pastor WA, Segal MF, Li H, Koh KP, Lahdesmaki H, et al. Modulation of TET2 expression and 5-methylcytosine oxidation by the CXXC domain protein IDAX. Nature. 2013;497:122–126. doi: 10.1038/nature12052. This paper shows dual functions for IDAX in regulating TET2 expression and function. By physically associating with TET2, IDAX may control chromatin targeting of TET2. However, when expressed at high levels, IDAX activates caspase pathway to degrade TET2 proteins, restricting further production and/or oxidation of ox-mCs. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhang Q, Zhao K, Shen Q, Han Y, Gu Y, Li X, Zhao D, Liu Y, Wang C, Zhang X, et al. Tet2 is required to resolve inflammation by recruiting Hdac2 to specifically repress IL-6. Nature. 2015;525:389–393. doi: 10.1038/nature15252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Huang Y, Rao A. Connections between TET proteins and aberrant DNA modification in cancer. Trends in genetics : TIG. 2014;30:464–474. doi: 10.1016/j.tig.2014.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ko M, An J, Pastor WA, Koralov SB, Rajewsky K, Rao A. TET proteins and 5-methylcytosine oxidation in hematological cancers. Immunological reviews. 2015;263:6–21. doi: 10.1111/imr.12239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Xu J, Wang YY, Dai YJ, Zhang W, Zhang WN, Xiong SM, Gu ZH, Wang KK, Zeng R, Chen Z, et al. DNMT3A Arg882 mutation drives chronic myelomonocytic leukemia through disturbing gene expression/DNA methylation in hematopoietic cells. Proceedings of the National Academy of Sciences of the United States of America. 2014;111:2620–2625. doi: 10.1073/pnas.1400150111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- **16.Mayle A, Yang L, Rodriguez B, Zhou T, Chang E, Curry CV, Challen GA, Li W, Wheeler D, Rebel VI, et al. Dnmt3a loss predisposes murine hematopoietic stem cells to malignant transformation. Blood. 2015;125:629–638. doi: 10.1182/blood-2014-08-594648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ***17.Celik H, Mallaney C, Kothari A, Ostrander EL, Eultgen E, Martens A, Miller CA, Hundal J, Klco JM, Challen GA. Enforced differentiation of Dnmt3a-null bone marrow leads to failure with c-Kit mutations driving leukemic transformation. Blood. 2015;125:619–628. doi: 10.1182/blood-2014-08-594564. References 16 and 17 show that transplantation of Dnmt3a-disrupted HSCs or bone marrow cells under noncompetitive condition results in diverse types of hematopoietic malignancies in myeloid or lymphoid cells. Dnmt3a-deficient malignant cells showed distinct methylation profiles and acquired additional mutations. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Trowbridge JJ, Snow JW, Kim J, Orkin SH. DNA methyltransferase 1 is essential for and uniquely regulates hematopoietic stem and progenitor cells. Cell Stem Cell. 2009;5:442–449. doi: 10.1016/j.stem.2009.08.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Broske AM, Vockentanz L, Kharazi S, Huska MR, Mancini E, Scheller M, Kuhl C, Enns A, Prinz M, Jaenisch R, et al. DNA methylation protects hematopoietic stem cell multipotency from myeloerythroid restriction. Nat Genet. 2009;41:1207–1215. doi: 10.1038/ng.463. [DOI] [PubMed] [Google Scholar]
- 20.Challen GA, Sun D, Jeong M, Luo M, Jelinek J, Berg JS, Bock C, Vasanthakumar A, Gu H, Xi Y, et al. Dnmt3a is essential for hematopoietic stem cell differentiation. Nature genetics. 2012;44:23–31. doi: 10.1038/ng.1009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- *21.Challen GA, Sun D, Mayle A, Jeong M, Luo M, Rodriguez B, Mallaney C, Celik H, Yang L, Xia Z, et al. Dnmt3a and Dnmt3b have overlapping and distinct functions in hematopoietic stem cells. Cell stem cell. 2014;15:350–364. doi: 10.1016/j.stem.2014.06.018. This paper shows that combined deficiency of Dnmt3a and Dnmt3b results in a dramatic increase in HSC self-renewal and a profound differentiation block by activating β-catenin signaling. In reference 20, the same authors demonstrated that loss of Dnmt3a augments self-renewal of HSCs and strongly impairs differentiation over serial transplantation. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tadokoro Y, Ema H, Okano M, Li E, Nakauchi H. De novo DNA methyltransferase is essential for self-renewal, but not for differentiation, in hematopoietic stem cells. J Exp Med. 2007;204:715–722. doi: 10.1084/jem.20060750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Huang YPW, Shen Y, Tahiliani M, Liu DR, Rao A. The behaviour of 5-hydroxymethylcytosine in bisulfite sequencing. PLoS One. 2009;5:e8888. doi: 10.1371/journal.pone.0008888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ji H, Ehrlich LI, Seita J, Murakami P, Doi A, Lindau P, Lee H, Aryee MJ, Irizarry RA, Kim K, et al. Comprehensive methylome map of lineage commitment from haematopoietic progenitors. Nature. 2010;467:338–342. doi: 10.1038/nature09367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bock C, Beerman I, Lien WH, Smith ZD, Gu H, Boyle P, Gnirke A, Fuchs E, Rossi DJ, Meissner A. DNA methylation dynamics during in vivo differentiation of blood and skin stem cells. Molecular cell. 2012;47:633–647. doi: 10.1016/j.molcel.2012.06.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hu M, Krause D, Greaves M, Sharkis S, Dexter M, Heyworth C, Enver T. Multilineage gene expression precedes commitment in the hemopoietic system. Genes & development. 1997;11:774–785. doi: 10.1101/gad.11.6.774. [DOI] [PubMed] [Google Scholar]
- 27.Gaudet F, Hodgson JG, Eden A, Jackson-Grusby L, Dausman J, Gray JW, Leonhardt H, Jaenisch R. Induction of tumors in mice by genomic hypomethylation. Science. 2003;300:489–492. doi: 10.1126/science.1083558. [DOI] [PubMed] [Google Scholar]
- 28.Yang L, Rau R, Goodell MA. DNMT3A in haematological malignancies. Nature reviews Cancer. 2015;15:152–165. doi: 10.1038/nrc3895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ley TJ, Ding L, Walter MJ, McLellan MD, Lamprecht T, Larson DE, Kandoth C, Payton JE, Baty J, Welch J, et al. DNMT3A mutations in acute myeloid leukemia. The New England journal of medicine. 2010;363:2424–2433. doi: 10.1056/NEJMoa1005143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Russler-Germain DA, Spencer DH, Young MA, Lamprecht TL, Miller CA, Fulton R, Meyer MR, Erdmann-Gilmore P, Townsend RR, Wilson RK, et al. The R882H DNMT3A mutation associated with AML dominantly inhibits wild-type DNMT3A by blocking its ability to form active tetramers. Cancer cell. 2014;25:442–454. doi: 10.1016/j.ccr.2014.02.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- **31.Jeong M, Sun D, Luo M, Huang Y, Challen GA, Rodriguez B, Zhang X, Chavez L, Wang H, Hannah R, et al. Large conserved domains of low DNA methylation maintained by Dnmt3a. Nature genetics. 2014;46:17–23. doi: 10.1038/ng.2836. By performing whole-genome bisulfite sequencing of HSCs, this study identified ‘canyons’, extended (> 3.5 kb) regions with low CpG methylation in the genome. The authors suggest that the methylation and hydroxymethylation status of canyon borders is dynamically regulated by the interplay between Dnmt3a and Tet proteins. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ono R, Taki T, Taketani T, Taniwaki M, Kobayashi H, Hayashi Y. LCX, leukemia-associated protein with a CXXC domain, is fused to MLL in acute myeloid leukemia with trilineage dysplasia having t(10;11)(q22;q23) Cancer Res. 2002;62:4075–4080. [PubMed] [Google Scholar]
- 33.Lorsbach RB, Moore J, Mathew S, Raimondi SC, Mukatira ST, Downing JR. TET1, a member of a novel protein family, is fused to MLL in acute myeloid leukemia containing the t(10;11)(q22;q23) Leukemia. 2003;17:637–641. doi: 10.1038/sj.leu.2402834. [DOI] [PubMed] [Google Scholar]
- 34.Huang H, Jiang X, Li Z, Li Y, Song CX, He C, Sun M, Chen P, Gurbuxani S, Wang J, et al. TET1 plays an essential oncogenic role in MLL-rearranged leukemia. Proceedings of the National Academy of Sciences of the United States of America. 2013;110:11994–11999. doi: 10.1073/pnas.1310656110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- **35.Cimmino L, Dawlaty MM, Ndiaye-Lobry D, Yap YS, Bakogianni S, Yu Y, Bhattacharyya S, Shaknovich R, Geng H, Lobry C, et al. TET1 is a tumor suppressor of hematopoietic malignancy. Nature immunology. 2015;16:653–662. doi: 10.1038/ni.3148. This study shows that ablation of Tet1 results in the development of B cell lymphoma in mice at advanced ages. Tet1 deficiency induced developmental skewing toward the B cell lineage and accumulation of DNA damage. Tet1-deficient murine tumors harbored a number of mutations that are also present in non-Hodgkin B cell lymphoma in humans. TET1 transcription was diminished in these cancers by promoter methylation. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Delhommeau F, Dupont S, Della Valle V, James C, Trannoy S, Masse A, Kosmider O, Le Couedic JP, Robert F, Alberdi A, et al. Mutation in TET2 in myeloid cancers. N Engl J Med. 2009;360:2289–2301. doi: 10.1056/NEJMoa0810069. [DOI] [PubMed] [Google Scholar]
- 37.Langemeijer SM, Kuiper RP, Berends M, Knops R, Aslanyan MG, Massop M, Stevens-Linders E, van Hoogen P, van Kessel AG, Raymakers RA, et al. Acquired mutations in TET2 are common in myelodysplastic syndromes. Nat Genet. 2009;41:838–842. doi: 10.1038/ng.391. [DOI] [PubMed] [Google Scholar]
- 38.Ko M, Huang Y, Jankowska AM, Pape UJ, Tahiliani M, Bandukwala HS, An J, Lamperti ED, Koh KP, Ganetzky R, et al. Impaired hydroxylation of 5-methylcytosine in myeloid cancers with mutant TET2. Nature. 2010;468:839–843. doi: 10.1038/nature09586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Pronier E, Almire C, Mokrani H, Vasanthakumar A, Simon A, da Costa Reis Monte Mor B, Masse A, Le Couedic JP, Pendino F, Carbonne B, et al. Inhibition of TET2-mediated conversion of 5-methylcytosine to 5-hydroxymethylcytosine disturbs erythroid and granulomonocytic differentiation of human hematopoietic progenitors. Blood. 2011;118:2551–2555. doi: 10.1182/blood-2010-12-324707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Chang YI, Damnernsawad A, Allen LK, Yang D, Ranheim EA, Young KH, Zhang J, Kong G, Wang J, Liu Y, et al. Evaluation of allelic strength of human TET2 mutations and cooperation between Tet2 knockdown and oncogenic Nras mutation. British journal of haematology. 2014;166:461–465. doi: 10.1111/bjh.12871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- *41.Quivoron C, Couronne L, Della Valle V, Lopez CK, Plo I, Wagner-Ballon O, Do Cruzeiro M, Delhommeau F, Arnulf B, Stern MH, et al. TET2 inactivation results in pleiotropic hematopoietic abnormalities in mouse and is a recurrent event during human lymphomagenesis. Cancer Cell. 2011;20:25–38. doi: 10.1016/j.ccr.2011.06.003. [DOI] [PubMed] [Google Scholar]
- *42.Lemonnier F, Couronne L, Parrens M, Jais JP, Travert M, Lamant L, Tournillac O, Rousset T, Fabiani B, Cairns RA, et al. Recurrent TET2 mutations in peripheral T-cell lymphomas correlate with TFH-like features and adverse clinical parameters. Blood. 2012;120:1466–1469. doi: 10.1182/blood-2012-02-408542. [DOI] [PubMed] [Google Scholar]
- *43.Odejide O, Weigert O, Lane AA, Toscano D, Lunning MA, Kopp N, Kim S, van Bodegom D, Bolla S, Schatz JH, et al. A targeted mutational landscape of angioimmunoblastic T-cell lymphoma. Blood. 2014;123:1293–1296. doi: 10.1182/blood-2013-10-531509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- *44.Sakata-Yanagimoto M, Enami T, Yoshida K, Shiraishi Y, Ishii R, Miyake Y, Muto H, Tsuyama N, Sato-Otsubo A, Okuno Y, et al. Somatic RHOA mutation in angioimmunoblastic T cell lymphoma. Nature genetics. 2014;46:171–175. doi: 10.1038/ng.2872. [DOI] [PubMed] [Google Scholar]
- *45.Palomero T, Couronne L, Khiabanian H, Kim MY, Ambesi-Impiombato A, Perez-Garcia A, Carpenter Z, Abate F, Allegretta M, Haydu JE, et al. Recurrent mutations in epigenetic regulators, RHOA and FYN kinase in peripheral T cell lymphomas. Nature genetics. 2014;46:166–170. doi: 10.1038/ng.2873. References 41–45 identified recurrent TET2 mutations in peripheral T cell lymphoma and their striking association with DNMT3A, RHOA and IDH2 mutations. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Couronne L, Bastard C, Bernard OA. TET2 and DNMT3A mutations in human T-cell lymphoma. The New England journal of medicine. 2012;366:95–96. doi: 10.1056/NEJMc1111708. [DOI] [PubMed] [Google Scholar]
- 47.Figueroa ME, Abdel-Wahab O, Lu C, Ward PS, Patel J, Shih A, Li Y, Bhagwat N, Vasanthakumar A, Fernandez HF, et al. Leukemic IDH1 and IDH2 mutations result in a hypermethylation phenotype, disrupt TET2 function, and impair hematopoietic differentiation. Cancer Cell. 2010;18:553–567. doi: 10.1016/j.ccr.2010.11.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ko M, Bandukwala HS, An J, Lamperti ED, Thompson EC, Hastie R, Tsangaratou A, Rajewsky K, Koralov SB, Rao A. Ten-Eleven-Translocation 2 (TET2) negatively regulates homeostasis and differentiation of hematopoietic stem cells in mice. Proceedings of the National Academy of Sciences of the United States of America. 2011;108:14566–14571. doi: 10.1073/pnas.1112317108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Li Z, Cai X, Cai CL, Wang J, Zhang W, Petersen BE, Yang FC, Xu M. Deletion of Tet2 in mice leads to dysregulated hematopoietic stem cells and subsequent development of myeloid malignancies. Blood. 2011;118:4509–4518. doi: 10.1182/blood-2010-12-325241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Moran-Crusio K, Reavie L, Shih A, Abdel-Wahab O, Ndiaye-Lobry D, Lobry C, Figueroa ME, Vasanthakumar A, Patel J, Zhao X, et al. Tet2 loss leads to increased hematopoietic stem cell self-renewal and myeloid transformation. Cancer Cell. 2011;20:11–24. doi: 10.1016/j.ccr.2011.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Muto T, Sashida G, Oshima M, Wendt GR, Mochizuki-Kashio M, Nagata Y, Sanada M, Miyagi S, Saraya A, Kamio A, et al. Concurrent loss of Ezh2 and Tet2 cooperates in the pathogenesis of myelodysplastic disorders. The Journal of experimental medicine. 2013;210:2627–2639. doi: 10.1084/jem.20131144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Shide K, Kameda T, Shimoda H, Yamaji T, Abe H, Kamiunten A, Sekine M, Hidaka T, Katayose K, Kubuki Y, et al. TET2 is essential for survival and hematopoietic stem cell homeostasis. Leukemia. 2012;26:2216–2223. doi: 10.1038/leu.2012.94. [DOI] [PubMed] [Google Scholar]
- 53.Kunimoto H, Fukuchi Y, Sakurai M, Sadahira K, Ikeda Y, Okamoto S, Nakajima H. Tet2 disruption leads to enhanced self-renewal and altered differentiation of fetal liver hematopoietic stem cells. Scientific reports. 2012;2:273. doi: 10.1038/srep00273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Kunimoto H, Fukuchi Y, Sakurai M, Takubo K, Okamoto S, Nakajima H. Tet2-mutated myeloid progenitors possess aberrant in vitro self-renewal capacity. Blood. 2014;123:2897–2899. doi: 10.1182/blood-2014-01-552471. [DOI] [PubMed] [Google Scholar]
- 55.Busque L, Patel JP, Figueroa ME, Vasanthakumar A, Provost S, Hamilou Z, Mollica L, Li J, Viale A, Heguy A, et al. Recurrent somatic TET2 mutations in normal elderly individuals with clonal hematopoiesis. Nature genetics. 2012;44:1179–1181. doi: 10.1038/ng.2413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Haferlach T, Nagata Y, Grossmann V, Okuno Y, Bacher U, Nagae G, Schnittger S, Sanada M, Kon A, Alpermann T, et al. Landscape of genetic lesions in 944 patients with myelodysplastic syndromes. Leukemia. 2014;28:241–247. doi: 10.1038/leu.2013.336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Papaemmanuil E, Gerstung M, Malcovati L, Tauro S, Gundem G, Van Loo P, Yoon CJ, Ellis P, Wedge DC, Pellagatti A, et al. Clinical and biological implications of driver mutations in myelodysplastic syndromes. Blood. 2013;122:3616–3627. doi: 10.1182/blood-2013-08-518886. quiz 3699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Abdel-Wahab O, Gao J, Adli M, Dey A, Trimarchi T, Chung YR, Kuscu C, Hricik T, Ndiaye-Lobry D, Lafave LM, et al. Deletion of Asxl1 results in myelodysplasia and severe developmental defects in vivo. The Journal of experimental medicine. 2013;210:2641–2659. doi: 10.1084/jem.20131141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Lobry C, Ntziachristos P, Ndiaye-Lobry D, Oh P, Cimmino L, Zhu N, Araldi E, Hu W, Freund J, Abdel-Wahab O, et al. Notch pathway activation targets AML-initiating cell homeostasis and differentiation. The Journal of experimental medicine. 2013;210:301–319. doi: 10.1084/jem.20121484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.De Vita S, Schneider RK, Garcia M, Wood J, Gavillet M, Ebert BL, Gerbaulet A, Roers A, Levine RL, Mullally A, et al. Loss of function of TET2 cooperates with constitutively active KIT in murine and human models of mastocytosis. PloS one. 2014;9:e96209. doi: 10.1371/journal.pone.0096209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Soucie E, Hanssens K, Mercher T, Georgin-Lavialle S, Damaj G, Livideanu C, Chandesris MO, Acin Y, Letard S, de Sepulveda P, et al. In aggressive forms of mastocytosis, TET2 loss cooperates with c-KITD816V to transform mast cells. Blood. 2012;120:4846–4849. doi: 10.1182/blood-2011-12-397588. [DOI] [PubMed] [Google Scholar]
- 62.Shih AH, Jiang Y, Meydan C, Shank K, Pandey S, Barreyro L, Antony-Debre I, Viale A, Socci N, Sun Y, et al. Mutational cooperativity linked to combinatorial epigenetic gain of function in acute myeloid leukemia. Cancer cell. 2015;27:502–515. doi: 10.1016/j.ccell.2015.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- *63.Nakagawa T, Lv L, Nakagawa M, Yu Y, Yu C, D’Alessio AC, Nakayama K, Fan HY, Chen X, Xiong Y. CRL4(VprBP) E3 ligase promotes monoubiquitylation and chromatin binding of TET dioxygenases. Molecular cell. 2015;57:247–260. doi: 10.1016/j.molcel.2014.12.002. This article shows that TET2 is monoubiquitinated by the CRL4VprBP complex, and that this modification promotes the DNA binding of TET2 proteins. Several leukemia-associated TET2 mutations impair VprBP-mediated monoubiquitylation. [DOI] [PMC free article] [PubMed] [Google Scholar]
- *64.Song SJ, Ito K, Ala U, Kats L, Webster K, Sun SM, Jongen-Lavrencic M, Manova-Todorova K, Teruya-Feldstein J, Avigan DE, et al. The oncogenic microRNA miR-22 targets the TET2 tumor suppressor to promote hematopoietic stem cell self-renewal and transformation. Cell stem cell. 2013;13:87–101. doi: 10.1016/j.stem.2013.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- *65.Song SJ, Poliseno L, Song MS, Ala U, Webster K, Ng C, Beringer G, Brikbak NJ, Yuan X, Cantley LC, et al. MicroRNA-antagonism regulates breast cancer stemness and metastasis via TET-family-dependent chromatin remodeling. Cell. 2013;154:311–324. doi: 10.1016/j.cell.2013.06.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- *66.Cheng J, Guo S, Chen S, Mastriano SJ, Liu C, D’Alessio AC, Hysolli E, Guo Y, Yao H, Megyola CM, et al. An extensive network of TET2-targeting MicroRNAs regulates malignant hematopoiesis. Cell reports. 2013;5:471–481. doi: 10.1016/j.celrep.2013.08.050. References 64–66 demonstrate that TET2 expression is controlled by numerous microRNAs. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Kim YH, Pierscianek D, Mittelbronn M, Vital A, Mariani L, Hasselblatt M, Ohgaki H. TET2 promoter methylation in low-grade diffuse gliomas lacking IDH1/2 mutations. Journal of clinical pathology. 2011;64:850–852. doi: 10.1136/jclinpath-2011-200133. [DOI] [PubMed] [Google Scholar]
- 68.Dang L, Jin S, Su SM. IDH mutations in glioma and acute myeloid leukemia. Trends in molecular medicine. 2010;16:387–397. doi: 10.1016/j.molmed.2010.07.002. [DOI] [PubMed] [Google Scholar]
- 69.Xu W, Yang H, Liu Y, Yang Y, Wang P, Kim SH, Ito S, Yang C, Wang P, Xiao MT, et al. Oncometabolite 2-hydroxyglutarate is a competitive inhibitor of alpha-ketoglutarate-dependent dioxygenases. Cancer cell. 2011;19:17–30. doi: 10.1016/j.ccr.2010.12.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- *70.Rampal R, Alkalin A, Madzo J, Vasanthakumar A, Pronier E, Patel J, Li Y, Ahn J, Abdel-Wahab O, Shih A, et al. DNA hydroxymethylation profiling reveals that WT1 mutations result in loss of TET2 function in acute myeloid leukemia. Cell reports. 2014;9:1841–1855. doi: 10.1016/j.celrep.2014.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- *71.Wang Y, Xiao M, Chen X, Chen L, Xu Y, Lv L, Wang P, Yang H, Ma S, Lin H, et al. WT1 recruits TET2 to regulate its target gene expression and suppress leukemia cell proliferation. Molecular cell. 2015;57:662–673. doi: 10.1016/j.molcel.2014.12.023. References 70 and 71 show that WT1 mutations are mutually exclusive with mutations in TET2, and IDH genes, and that WT1 recruits TET2 to specific chromatin regions, particulary promoters, to activate TET2 target genes. WT1 was necessary for TET2 to suppress leukemic cell growth and colony formation. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Loenarz C, Schofield CJ. Expanding chemical biology of 2-oxoglutarate oxygenases. Nature chemical biology. 2008;4:152–156. doi: 10.1038/nchembio0308-152. [DOI] [PubMed] [Google Scholar]
- 73.Arioka Y, Watanabe A, Saito K, Yamada Y. Activation-induced cytidine deaminase alters the subcellular localization of Tet family proteins. PloS one. 2012;7:e45031. doi: 10.1371/journal.pone.0045031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Zhang Q, Liu X, Gao W, Li P, Hou J, Li J, Wong J. Differential regulation of the ten-eleven translocation (TET) family of dioxygenases by O-linked beta-N-acetylglucosamine transferase (OGT) The Journal of biological chemistry. 2014;289:5986–5996. doi: 10.1074/jbc.M113.524140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Lu F, Liu Y, Jiang L, Yamaguchi S, Zhang Y. Role of Tet proteins in enhancer activity and telomere elongation. Genes & development. 2014;28:2103–2119. doi: 10.1101/gad.248005.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Huang Y, Chavez L, Chang X, Wang X, Pastor WA, Kang J, Zepeda-Martinez JA, Pape UJ, Jacobsen SE, Peters B, et al. Distinct roles of the methylcytosine oxidases Tet1 and Tet2 in mouse embryonic stem cells. Proceedings of the National Academy of Sciences of the United States of America. 2014;111:1361–1366. doi: 10.1073/pnas.1322921111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- **77.Hon GC, Song CX, Du T, Jin F, Selvaraj S, Lee AY, Yen CA, Ye Z, Mao SQ, Wang BA, et al. 5mC oxidation by Tet2 modulates enhancer activity and timing of transcriptome reprogramming during differentiation. Molecular cell. 2014;56:286–297. doi: 10.1016/j.molcel.2014.08.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- **78.Bestor TH, Edwards JR, Boulard M. Notes on the role of dynamic DNA methylation in mammalian development. Proceedings of the National Academy of Sciences of the United States of America. 2015;112:6796–6799. doi: 10.1073/pnas.1415301111. References 77 and 78 point out the lack of experimental evidence supporting the prevailing view that DNA methylation controls gene expression, and propose the converse hypothesis, that altered gene transcription may cause changes of DNA methylation. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Schubeler D. Function and information content of DNA methylation. Nature. 2015;517:321–326. doi: 10.1038/nature14192. [DOI] [PubMed] [Google Scholar]
- 80.Wang L, Zhou Y, Xu L, Xiao R, Lu X, Chen L, Chong J, Li H, He C, Fu XD, et al. Molecular basis for 5-carboxycytosine recognition by RNA polymerase II elongation complex. Nature. 2015 doi: 10.1038/nature14482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Kang J, Lienhard M, Pastor WA, Chawla A, Novotny M, Tsagaratou A, Lasken RS, Thompson EC, Surani MA, Koralov SB, et al. Simultaneous deletion of the methylcytosine oxidases Tet1 and Tet3 increases transcriptome variability in early embryogenesis. Proceedings of the National Academy of Sciences of the United States of America. 2015;112:E4236–4245. doi: 10.1073/pnas.1510510112. [DOI] [PMC free article] [PubMed] [Google Scholar]