Abstract
The bacterial populations in the human intestine impact host physiological functions through their metabolic activity. In addition to performing essential catabolic and biotransformation functions, the gut microbiota produces bioactive small molecules that mediate interactions with the host and contribute to the neurohumoral axes connecting the intestine with other parts of the body. This review discusses recent progress in characterizing the metabolic products of the gut microbiota and their biological functions, focusing on studies that investigate the responsible bacterial pathways and cognate host receptors. Several key areas are highlighted for future development: context-based analysis targeting pathways; integration of analytical approaches; metabolic modeling; and synthetic systems for in vivo manipulation of microbiota functions. Prospectively, these developments could further our mechanistic understanding of host-microbiota interactions.
Keywords: Microbiota metabolites, pathways, receptor ligands, gnotobiotic mice, metagenomics, metabolomics, metabolic model
Graphical abstract
Introduction
Colonized at birth, the adult human gastrointestinal (GI) tract harbors ~1014 bacteria belonging to at least several hundred species [1]. Collectively termed the gut microbiota, these bacterial populations impact an array of physiological functions in the GI tract, including digestion and immune response to foodborne pathogens. A well-known digestive function of the microbiota is the fermentation of complex carbohydrates to short chain fatty acids (SCFAs). Recent metabolomic studies have detected SCFAs and many other bioactive microbiota-derived metabolites in systemic circulation [2], supporting the view that these molecules constitute part of the neurohumoral communication axes that link the intestine with other organs such as the liver and brain.
Significant alteration in the microbial populations, or dysbiosis, correlates with not only GI diseases, but also other chronic diseases such as diabetes, cancer, and asthma. While it is difficult to establish causation, the case for dysbiosis as a contributing mechanism for some diseases has become increasingly compelling. Seminal work by the Gordon laboratory identified a pattern of dysbiosis in obesity that is characterized by a greater capacity for energy harvest [3]. Psychiatric disorders such as autism spectrum disorder (ASD) are frequently accompanied by changes to the intestinal microbiota composition [4,5]. Using a murine model of ASD, Hsiao et al. showed that alterations to the serum metabolite profile resulting from dysbiosis correlate with behavioral abnormalities; administration of a probiotic attenuated these abnormalities while reducing the serum levels of microbiota metabolites elevated in ASD mice [6].
These studies provide firm evidence that dysbiosis dramatically impacts host physiology; moreover, the functional consequences reflect alterations in the profile of microbiota-derived metabolic products. Many of these metabolites show activity as signaling molecules [7], and engage various host receptors and regulatory molecules in vitro and in vivo [8,9]; however, specific functions have been identified for only a small subset of these metabolites [10]. Aided by advances in sequencing and data analysis pipelines [11], the catalog of annotated genomes for bacteria found in humans [12,13] and model organisms [14] has rapidly expanded over the last several years. These developments present an exciting opportunity to integrate compositional data with measurements on the functional outputs of the microbiota to address fundamental questions regarding molecular mechanisms mediating host-microbiota interactions.
In this review, we discuss recent progress in characterizing the metabolic products of the gut microbiota and their biological functions in the context of host physiology. We focus on studies that investigate the responsible enzymatic pathways and bacterial groups. We also discuss representative methods and models, including metabolic models that could facilitate the integration of different types of data on microbiota composition and functional readouts.
Metabolic functions and outputs of the gut microbiota
Substrates metabolized by the gut microbiota include dietary residues, mucosal macromolecules (e.g. mucins), endogenous metabolites (notably bile acids), and xenobiotic chemicals. The major classes of dietary substrates (Fig. 1) comprise carbohydrates, amino acids, certain lipids (e.g., polyunsaturated fatty acids [15]), and phytochemicals [16].
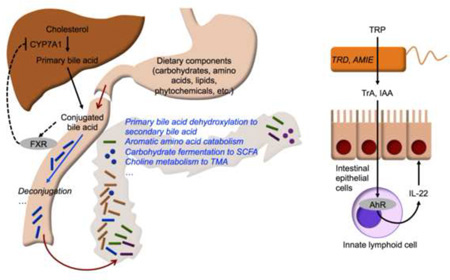
Figure 1.
Representative dietary inputs and metabolic functions of intestinal microbiota. Metabolic functions are indicated in italics. Closed and open arrows indicate flow of specific metabolites and dietary residues, respectively. Dotted lines illustrate negative feedback of hepatic bile acid synthesis under the regulation of intestinal farnesoid X receptor (FXR), which is antagonized by conjugated bile acids.
Short-chain fatty acids (SCFAs)
The most abundant SCFAs in the intestine are acetate, propionate, and butyrate, which are primarily derived from carbohydrates [17]. Some fermentation of amino acids also occurs, generating alkyl carboxylic acids such as valerate and caproate [18]. Acetate and propionate are found throughout the small and large intestines, while butyrate is found mainly in the cecum and colon.
The main route for propionate synthesis proceeds through succinate, with methylmalonyl-CoA decarboxylase catalyzing a key step that results in the formation of propanoyl-CoA [19] (Fig. 2). An alternative pathway involves hydrolysis of propane-1,2-diol into propanal. This pathway is present in several major phylogenetic groups, especially Lachnospiraceae, and likely plays an important role in fermenting deoxy sugar residues of host-derived glycans [19]. Butyrate production can also occur through several pathways, utilizing acetyl-CoA, glutarate, 4-aminobutyrate, or lysine as the major substrate (Fig. 2). A recent metagenomic analysis of stool samples from healthy human subjects found that the acetyl- CoA pathway is the most prevalent, followed by the lysine pathway [20]. The same study identified Firmicutes strains as the major butyrate producers, with the vast majority of these strains harboring genes for the acetyl-CoA pathway. Interestingly, the lysine and acetyl-CoA pathways often occur together, suggesting that the latter pathway could be cross-fed with protein degradation pathways, thereby providing adaptability to a protein-rich diet [20].
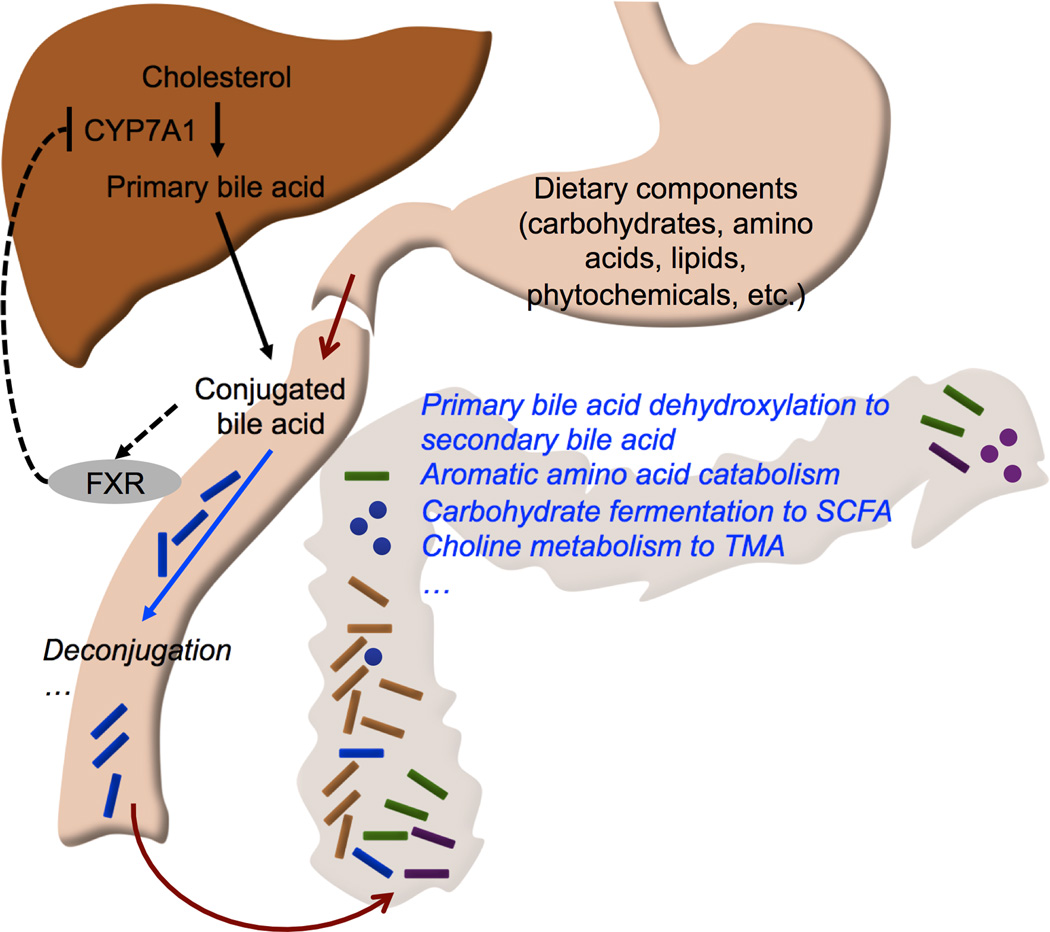
Figure 2.
Major pathways of propionate (green arrows) and butyrate (blue arrows) metabolism in gut bacteria. For clarity, only a subset of the intermediates and reactions are shown. Enzyme name abbreviations are shown in bold italics. Dotted and solid arrows indicate multiple and single reaction steps, respectively. PDU: propionaldehyde dehydrogenase; MMD: methylmalonyl-CoA decarboxylase; GCD: glutaconyl-CoA decarboxylase; CRO: crotonase; 4HBT: 4-hydroxybutyrate CoA-transferase; KAL: 3-aminobutyryl-CoA ammonia-lyase; BCD: butyryl-CoA dehydrogenase.
Utilized locally as an energy source by colonic epithelial cells, SCFAs are critical for maintaining intestinal barrier integrity by regulating expression of tight junction proteins. The best characterized host receptors for SCFAs are G protein-coupled receptors 41 (GPR41) and 43 (GPR43). GPR41 is widely expressed in a variety of tissues and cell types, and plays an important role in regulating whole body energy balance through its interaction with the gut microbiota [21]. GPR43 is most highly expressed in immune cells. Targeted gene knockout studies have shown that SCFAs activate cytokines and chemokines in vitro and in vivo, and that this response is GPR43 dependent [22].
Aromatic amino acid (AAA) derivatives
A major group of immunomodulatory microbiota metabolites derives from catabolism of AAAs (Fig. 3a). In both humans and mice, a principal route for tryptophan (TRP) catabolism is the kynurenine pathway. Depending on the cell type, the rate-limiting first step is catalyzed by tryptophan 2,3-dioxygenase or indoleamine 2,3-dioxygenase (IDO1). Kynurenine is an endogenous ligand for the arylhydrocarbon receptor (AhR), which regulates immune responses and inflammation through its effects on cytokine production and Treg cell development [23]. Because kynurenine production competes with other TRP pathways, IDO1 activity in immune cells of the intestine modulates the TRP pool available for microbiota metabolism [24]. Major products of TRP catabolism include indole-3-acetamide, tryptamine (TrA), and indole, formed via tryptophan 2-monooxygenase, tryptophan decarboxylase, and tryptophanase A (TNA), respectively [24]. These products can be further transformed into other indole-containing molecules such as indole-3-acetate (IAA) and indole-3-acetaldehyde (IAAld). The genes encoding the TRP reactions are found in many different phylogenic groups of the gut microbiota [25] as well as in human and mouse genomes; of the aforementioned enzymes, only TNA is strictly bacterial. Nevertheless, metabolomic analyses of intestinal and fecal samples from GF and gnotobiotic mice indicate that the gut microbiota is a major source of these and other AAA derivatives (e.g. phenethylamine from phenylalanine and phenol from tyrosine) [2,25]. Like kynurenine, TrA, IAA, and IAld are AhR ligands, and can modulate the host’s immune reactivity [24] (Fig. 3b). Indole itself is a weak AhR agonist [26], but dose-dependently increases tight junction resistance of intestinal epithelial cells and attenuates indicators of inflammation [27], suggesting that microbiota-derived TRP metabolites may interact with host pathways through other receptors.
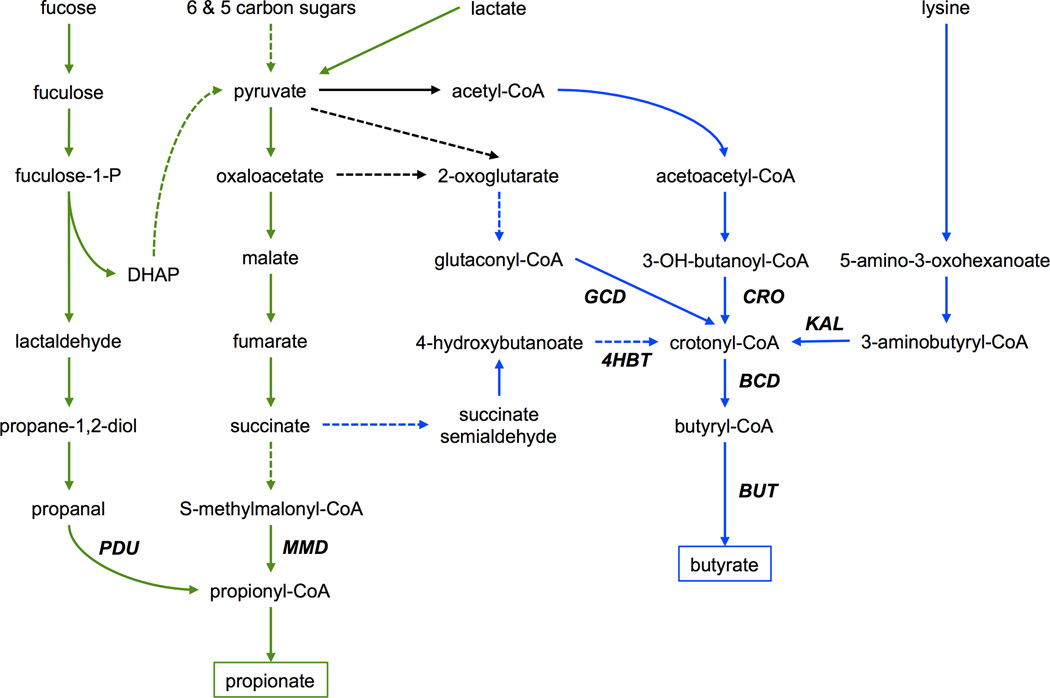
Figure 3.
(a) Tryptophan (TRP) co-metabolism by host and microbiota. Enzyme name abbreviations are shown in bold italics. Blue and red fonts indicate strictly bacterial and host enzymes, respectively. ARAT: aromatic amino acid transferase; IPD: indolepyruvate decarboxylase; TRD: tryptophan decarboxylase; MAO: monoamine oxidase; ALDH: IAAld dehydrogenase; AMI: amidase; TMO: tryptophan 2-monooxygenase. See text for additional definitions of abbreviations. (b) TRP catabolism in gut bacteria produces TrA and IAA, which can translocate across intestinal epithelial cells (IECs) to activate the arylhydrocarbon receptor (AhR) expressed in innate lymphoid cells. This in turn induces secretion of interleukin-22 (IL-22), which triggers an immune response against pathogens, e.g. resulting in production of antimicrobial peptides by Paneth cells.
Metabolism of AAAs illustrates another emerging function of the gut microbiota, modulation of the neuroendocrine communication between the digestive and nervous systems. The serum concentration of serotonin is significantly reduced in GF mice, but restored when their GI tract is colonized with Clostridium species [28]. A similar study on catecholamines found that their levels were similar in GF and conventionally raised (CONV-R) mice; however, the catecholamines in GF mice were mostly in biologically less active conjugated form [29]. Colonizing the GF mice with CONV-R fecal isolates dramatically increased the levels of free catecholamines, suggesting that the gut microbiota could modulate systemic catecholamine levels through deconjugation activity.
Bile acids
Microbial deconjugation also plays an important role in regulating bile acids. These cholesterol-derived metabolites facilitate intestinal absorption of dietary lipids and fat-soluble vitamins, while their rate of synthesis influences systemic cholesterol levels. Synthesis of primary bile acids, cholate (CA) and chenodeoxycholate (CDCA), occurs in the liver. Before entering the intestine, the primary bile acids undergo conjugation with either glycine or taurine, which enhances the bile acids’ surfactant function. More than 95% of the bile acids are reabsorbed in the distal ileum and returned to the liver as part of the enterohepatic circulation (Fig. 1). The remainder enters the cecum and colon, where they can be transformed into secondary bile acids. Major secondary bile acids include deoxycholate (DCA) and lithocholate, generated through 7α-dehydroxylation of CA and CDCA, respectively (Fig. 4a). The enzyme system catalyzing this dehydroxylation step has been found in several Firmicutes species, notably Clostridium scindens [30]; however, the general distribution of this system in colonic microbiota remains poorly characterized. In contrast, the distribution of bile salt hydrolases (BSHs), which regenerate primary bile acids through deconjugation, has been extensively characterized [31]. Phylogenic groups encoding BSH include microbiota genera belonging to Firmicutes (Lactobacillus, Clostridium), Actinobacteria (Bifidobcterium), and Bacteroidetes (Bacteroides). Together, the deconjugation and dehydoxylation reactions greatly diversify the bile acids found in the intestine [32].
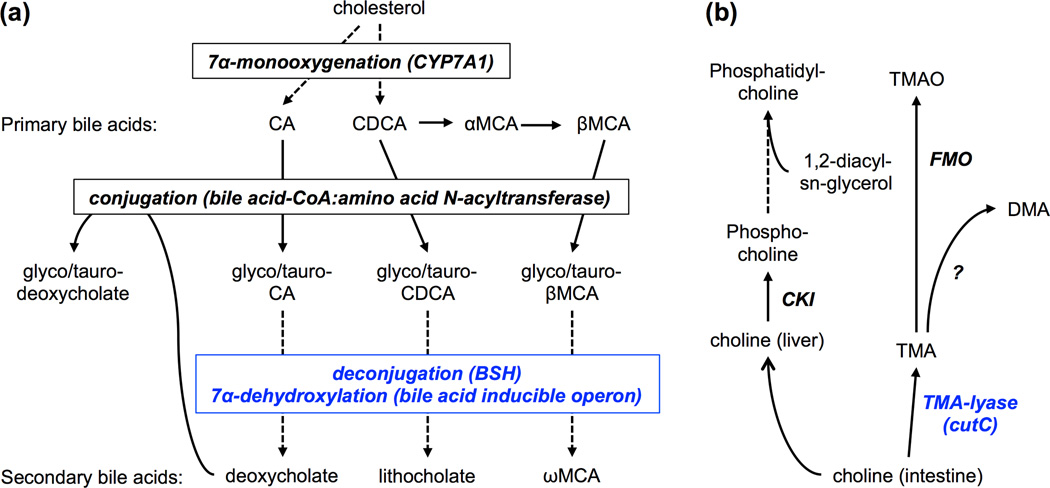
Figure 4.
(a) Key steps in host-microbiota co-metabolism of bile acids. Enzymatic steps are shown in bold italics. Blue font indicates bacterial reactions taking place in the intestine. Note that deoxycholate, a secondary bile acid formed by the microbiota, can enter the liver through the enteroheaptic circulation and then conjugated with glycine or taurine to a bile salt. See text for definitions of abbreviations. (b) Choline co-metabolism by the host and microbiota. Dietary choline is metabolized in the intestine by bacterial trimethylamine (TMA)-lyase to yield TMA, ethanol, and acetate. Blue font indicates bacterial enzyme. In the liver, TMA is further metabolized into trimethylamine N-oxide (TMAO) by a flavin-containing monooxygenase (FMO). Additional metabolic products of TMA include dimethylamine (DMA); however, the responsible enzyme remains to be elucidated. Choline can also enter the liver, phosphorylated by choline kinase (CKI), and activated to form a substrate for phosphatidylcholine synthesis.
A major component of the host’s bile acid signaling system is the farnesoid X receptor (FXR), a transcription factor expressed in both the liver and intestine. Liver bile acid synthesis is under negative feedback regulation by intestinal FXR through a fibroblast growth factor (FGF15)-dependent mechanism. In mice, FXR is antagonized by tauro-conjugated muricholate (TβMCA; MCA is a primary bile acid in mice). The levels of MCA and TβMCA in the distal small intestine are significantly higher in GF compared to CONV-R mice [32]. As a result, FXR-dependent inhibition of bile acid synthesis is impaired in GF mice, consequently increasing cytochrome P450 7A1 (CYP7A1)-catalyzed cholesterol metabolism in the liver [33]. Using intestine specific FXR−/− mice, Li et al. showed that inhibition of FXR signaling correlates with depletion of Lactobacillus and Clostridium species and with decreases in BSH and 7α-dehydroxylase activities [34]. Similar correlations were observed when bacterial BSH variants were overexpressed in the intestines of GF or CONV-R mice [35], demonstrating that even a subtle shift in BSH activity impacts the intestinal bile acid profile and host lipid metabolism.
Another role for microbiota metabolism of bile acids is to inhibit colonization by gut pathogens. In its vegetative state, C. difficile is susceptible to secondary bile acid (e.g. DCA) toxicity. Employing mouse models, clinical studies, metagenomic analyses, and mathematical modeling, Buffie et al. found that resistance to C. difficile infection strongly correlates with presence of C. scindens, and showed that C. scindens inhibits C. difficile growth in a bile salt dependent manner [36].
Choline
In humans, dietary uptake of choline is necessary to maintain a sufficient level for biosynthesis of phospholipids. Metabolomic data indicate that certain species of the gut microbiota carry out anaerobic choline metabolism to produce trimethylamine (TMA), acetate and ethanol (Fig. 4b). Dysbiosis leading to aberrant choline metabolism has been proposed as potential contributing factor in non-alcoholic fatty liver disease, and increased TMA in circulation has been identified as a risk factor for cardiovascular disease and colon cancer. The specific microorganisms responsible for choline metabolism remained largely unidentified until recently, when the choline utilization (cut) gene cluster was discovered in sulfatereducing bacteria. Functional characterization of this gene cluster in human gut bacteria showed that the gene encoding TMA-lyase (cutC) was widely distributed across different phyla, but the distribution did not correlate with phylogeny, suggesting that the pathway may have been acquired in some strains via horizontal gene transfer [37]. The study also found that every human gut isolate experimentally confirmed to metabolize choline possesses cutC, demonstrating the utility of the gene as a functional marker of choline metabolizing capacity while also underscoring the need to complement genomic and metagenomic analyses with molecular and biochemical characterization.
Methods and Models
Manipulation of gut microbial populations
Due to ethical concerns and practical limitations of manipulating the gut microbiota in human subjects, GF and gnotobiotic mice have featured prominently as experimental model systems. Administration of broad-spectrum antibiotics via drinking water has also been used, but the results can be difficult to reproduce. While gavage feeding of the antibiotics improves reproducibility [38], this can severely stress the animal, presenting difficulties for long-term studies. Another strategy for characterizing the metabolism of specific gut microbes is to conduct in vitro cultures, which affords experimental control over substrates and environmental parameters (e.g. pH). Utilizing anaerobic cultures in conjunction with gnotobiotic mice, Goodman et al. showed that the taxonomic groups dominant in human fecal microbiota are represented in its readily cultured members [39]. An intriguing extension of this approach is to introduce culture-expanded isolates into gnotobiotic mice to study the interaction between specific bacterial groups and diet. Additionally, GF mice can be colonized with isolates from individuals with different health status, e.g. obese vs. lean, to study the effects of gut bacteria on the host’s metabolic phenotype [40]. Limitations remain, however, with respect to the ability to recapitulate the spatial distribution of microorganisms (site specific clustering [41]) along different parts of the digestive tract. Conventional culture systems grow the cells in a mixed environment, and thus cannot provide this feature. In this regard, the development of staged bioreactor systems [42] or engineered intestinal tissue models supporting stable colonization by bacteria would be immensely useful.
An alternative to isolating specific bacterial subgroups is to characterize the metabolite profile of bodily fluids or intestinal samples, and integrate these profiles with metagenomic data to investigate the microbial origin of the detected metabolites. Untargeted analyses using high-resolution mass spectrometry paired with comparisons using GF or gnotobiotic mice have been especially useful in identifying metabolites whose levels depend on the microbiota and thus reflect bacterial metabolic activity [43]. Due to the high dimensionality of these data sets, multivariate statistical methods are often employed to select discriminatory metabolites that characterize significant differences between samples. However, challenges remain in determining whether a metabolite is the product of host or microbiota metabolism, as many metabolites can be produced in both mammalian and bacterial cells. Moreover, the metabolites present in the intestine could result from co-metabolism involving reactions from multiple organisms [44,45], including the host.
Metabolic models
Analysis of metabolomic data can be facilitated by computational methods that utilize metabolic models. Metabolites identified from an untargeted analysis could be mapped to known pathways via reaction definitions available in databases, which in turn provides a link to enzymes, genes, and genomes. Using pathway analysis, routes could be traced through a metabolic model of the gut microbiota to determine the organism(s) harboring the necessary genes and thus capable of producing the metabolites of interest. Current modeling efforts can be broadly grouped into two approaches. In the first approach, genome-scale metabolic models (GEMs) are built for specific microorganisms, and analyzed using simulation frameworks such as constrained optimization to characterize the organism’s metabolic capacities. For example, Heinken et al. utilized a GEM of B. thetaiotaomicron in conjunction with flux balance analysis to find that co-metabolism with the bacteria could rescue a potentially lethal loss of enzymatic function in the host [46]. More recently, Shoaie et al. utilized GEMs to simulate metabolic interactions across three representative species of gut bacteria (Eubacterium rectale, Methanobrevibacter smithii, and B. thetaiotaomicron) under varying nutrient settings [47]. In the second approach, the gut microbiota is modeled as a single “super” organism that represents the metabolic capability of an entire microbial community. This approach has the drawback that interactions between particular species cannot be examined detail. However, the community-level models can be constructed in a site-specific manner, comprising a subset of microorganism residing in the same region of the intestine. Moreover, these models can be directly mapped to metagenomic data to comprehensively analyze the diversity of metabolic functions encoded by the intestinal microbiome [43,48]. Recently, Greenblum et al. assembled a community-level model of human gut microbiota to find significant alterations in the functional organization of the microbiota metabolic network in obesity and IBD [49]. In our recent work, we adopted a similar approach to model the gut microbiota and its host as two intersecting reaction networks, and applied a probabilistic search method to identify pathways of AAA catabolism, while discriminating between microbiota and host reactions [25].
Conclusions
Microbiome research continues to advance at a rapid pace, and significant progress has been achieved towards understanding the metabolic functions performed by the gut microbiota. However, many questions remain regarding the enzymatic pathways and species responsible for the metabolites that mediate host-microbiota interactions. Moreover, it is likely that the metabolites characterized to date represent only a fraction of the bioactive chemicals produced by the microbiota. A recent analysis by the Fischbach laboratory identified more than 3,000 small-molecule biosynthetic gene clusters in bacterial genomes associated with humans [50], highlighting the enormous potential to discover novel bioactive molecules in the metabolome of the gut microbiota.
Several promising directions for future research emerge from the examples discussed above. First, functional characterization of microbiota pathways will benefit from workflows that integrate multiple analytical approaches, including not only omics strategies, but also molecular assays to confirm biochemical function. Second, context-based analyses, e.g. targeting a set of genes that constitute a pathway, will likely yield more robust results than searching for a single gene. This also applies to analysis of metabolomic data, as detection of multiple intermediates of a given pathway, rather than only the end product, will improve confidence that the pathway is active. Third, both data integration and context-based analysis will benefit from metabolic models that comprehensively describe the diversity of pathways supported by a microbial community while also capturing species-specific contributions. Here, the ability to reflect compositional changes in simulations of metabolic outputs would be particularly useful, as this would enable in silico predictions on microbiota structure-function relationships leading to experimentally testable hypotheses. Finally, methods and systems will be needed to modulate the activities of specific microbiota pathways in vivo, e.g. using genome edited synthetic probiotic organisms, thereby establishing a route to manipulate a metabolic function of interest. These developments as well as others not discussed here will provide new insights into the molecular mechanisms of host-microbiota interactions, while helping to pave the way for rational design of pre-, pro-, and postbiotics beneficial for human health.
Highlights.
Intestinal microbiota performs diverse metabolic functions essential for host physiology.
Small molecule products of microbiota metabolism act as nutrients and signaling molecules.
Microbiota products modulate lipid metabolism, immune cell reactivity, and neuroendocrine pathways.
Functional characterization of microbiota metabolites will benefit from pathway-oriented analyses and integration of analytical approaches.
Metabolic models could facilitate data integration to relate microbiota composition and function.
Acknowledgements
This work was in part supported by grants from the National Institutes of Health (R21 GM106251) and National Science Foundation (1264502).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
• Recent papers of special interest
••: Recent papers of outstanding interest
- 1.Human Microbiome Project C. Structure, function and diversity of the healthy human microbiome. Nature. 2012;486:207–214. doi: 10.1038/nature11234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wikoff WR, Anfora AT, Liu J, Schultz PG, Lesley SA, Peters EC, Siuzdak G. Metabolomics analysis reveals large effects of gut microflora on mammalian blood metabolites. Proc Natl Acad Sci U S A. 2009;106:3698–3703. doi: 10.1073/pnas.0812874106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Turnbaugh PJ, Ley RE, Mahowald MA, Magrini V, Mardis ER, Gordon JI. An obesity-associated gut microbiome with increased capacity for energy harvest. Nature. 2006;444:1027–1031. doi: 10.1038/nature05414. [DOI] [PubMed] [Google Scholar]
- 4.De Angelis M, Piccolo M, Vannini L, Siragusa S, De Giacomo A, Serrazzanetti DI, Cristofori F, Guerzoni ME, Gobbetti M, Francavilla R. Fecal microbiota and metabolome of children with autism and pervasive developmental disorder not otherwise specified. PLoS One. 2013;8:e76993. doi: 10.1371/journal.pone.0076993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Krajmalnik-Brown R, Lozupone C, Kang DW, Adams JB. Gut bacteria in children with autism spectrum disorders: challenges and promise of studying how a complex community influences a complex disease. Microb Ecol Health Dis. 2015;26:26914. doi: 10.3402/mehd.v26.26914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Hsiao EY, McBride SW, Hsien S, Sharon G, Hyde ER, McCue T, Codelli JA, Chow J, Reisman SE, Petrosino JF, et al. Microbiota modulate behavioral and physiological abnormalities associated with neurodevelopmental disorders. Cell. 2013;155:1451–1463. doi: 10.1016/j.cell.2013.11.024. ••: Demonstrates that dysbiosis associated behavioral abnormalities correlate with accumulation of microbiota metabolites in serum, which can be mitigated with probiotic administration.
- 7.Nicholson JK, Holmes E, Kinross J, Burcelin R, Gibson G, Jia W, Pettersson S. Host-gut microbiota metabolic interactions. Science. 2012;336:1262–1267. doi: 10.1126/science.1223813. [DOI] [PubMed] [Google Scholar]
- 8.Laparra JM, Sanz Y. Interactions of gut microbiota with functional food components and nutraceuticals. Pharmacol Res. 2010;61:219–225. doi: 10.1016/j.phrs.2009.11.001. [DOI] [PubMed] [Google Scholar]
- 9.Sanz Y, De Palma G. Gut microbiota and probiotics in modulation of epithelium and gut-associated lymphoid tissue function. Int Rev Immunol. 2009;28:397–413. doi: 10.3109/08830180903215613. [DOI] [PubMed] [Google Scholar]
- 10.Vogt SL, Pena-Diaz J, Finlay BB. Chemical communication in the gut: Effects of microbiota-generated metabolites on gastrointestinal bacterial pathogens. Anaerobe. 2015;34:106–115. doi: 10.1016/j.anaerobe.2015.05.002. [DOI] [PubMed] [Google Scholar]
- 11.Segata N, Boernigen D, Tickle TL, Morgan XC, Garrett WS, Huttenhower C. Computational meta'omics for microbial community studies. Mol Syst Biol. 2013;9:666. doi: 10.1038/msb.2013.22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Qin J, Li R, Raes J, Arumugam M, Burgdorf KS, Manichanh C, Nielsen T, Pons N, Levenez F, Yamada T, et al. A human gut microbial gene catalogue established by metagenomic sequencing. Nature. 2010;464:59–65. doi: 10.1038/nature08821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Li J, Jia H, Cai X, Zhong H, Feng Q, Sunagawa S, Arumugam M, Kultima JR, Prifti E, Nielsen T, et al. An integrated catalog of reference genes in the human gut microbiome. Nat Biotechnol. 2014;32:834–841. doi: 10.1038/nbt.2942. [DOI] [PubMed] [Google Scholar]
- 14.Turnbaugh PJ, Ridaura VK, Faith JJ, Rey FE, Knight R, Gordon JI. The effect of diet on the human gut microbiome: a metagenomic analysis in humanized gnotobiotic mice. Sci Transl Med. 2009;1:6ra14. doi: 10.1126/scitranslmed.3000322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Druart C, Bindels LB, Schmaltz R, Neyrinck AM, Cani PD, Walter J, Ramer-Tait AE, Delzenne NM. Ability of the gut microbiota to produce PUFA-derived bacterial metabolites: Proof of concept in germ-free versus conventionalized mice. Mol Nutr Food Res. 2015;59:1603–1613. doi: 10.1002/mnfr.201500014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.van Duynhoven J, Vaughan EE, Jacobs DM, Kemperman RA, van Velzen EJ, Gross G, Roger LC, Possemiers S, Smilde AK, Doré J, et al. Metabolic fate of polyphenols in the human superorganism. Proc Natl Acad Sci USA. 2011;108:4531–4538. doi: 10.1073/pnas.1000098107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Macfarlane GT, Macfarlane S. Fermentation in the human large intestine: its physiologic consequences and the potential contribution of prebiotics. J Clin Gastroenterol. 2011;45:S120–S127. doi: 10.1097/MCG.0b013e31822fecfe. [DOI] [PubMed] [Google Scholar]
- 18.De Preter V, Machiels K, Joossens M, Arijs I, Matthys C, Vermeire S, Rutgeerts P, Verbeke K. Faecal metabolite profiling identifies medium-chain fatty acids as discriminating compounds in IBD. Gut. 2015;64:447–458. doi: 10.1136/gutjnl-2013-306423. [DOI] [PubMed] [Google Scholar]
- 19. Reichardt N, Duncan SH, Young P, Belenguer A, McWilliam Leitch C, Scott KP, Flint HJ, Louis P. Phylogenetic distribution of three pathways for propionate production within the human gut microbiota. ISME J. 2014;8:1323–1335. doi: 10.1038/ismej.2014.14. •: Illustrates use of biochemical assays and culture experiments in conjunction with genomic analyses to characterize distribution and confirm metabolic function.
- 20. Vital M, Howe AC, Tiedje JM. Revealing the bacterial butyrate synthesis pathways by analyzing (meta)genomic data. MBio. 2014;5:e00889. doi: 10.1128/mBio.00889-14. •: Provides a complete database of genes from major known butyrate-producing pathways.
- 21.Samuel BS, Shaito A, Motoike T, Rey FE, Backhed F, Manchester JK, Hammer RE, Williams SC, Crowley J, Yanagisawa M, et al. Effects of the gut microbiota on host adiposity are modulated by the short-chain fatty-acid binding G protein-coupled receptor, Gpr41. Proc Natl Acad Sci U S A. 2008;105:16767–16772. doi: 10.1073/pnas.0808567105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kim MH, Kang SG, Park JH, Yanagisawa M, Kim CH. Short-chain fatty acids activate GPR41 and GPR43 on intestinal epithelial cells to promote inflammatory responses in mice. Gastroenterology. 2013;145:396–406. e391–e310. doi: 10.1053/j.gastro.2013.04.056. [DOI] [PubMed] [Google Scholar]
- 23.Nguyen NT, Kimura A, Nakahama T, Chinen I, Masuda K, Nohara K, Fujii-Kuriyama Y, Kishimoto T. Aryl hydrocarbon receptor negatively regulates dendritic cell immunogenicity via a kynurenine-dependent mechanism. Proc Natl Acad Sci U S A. 2010;107:19961–19966. doi: 10.1073/pnas.1014465107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Zelante T, Iannitti RG, Cunha C, De Luca A, Giovannini G, Pieraccini G, Zecchi R, D'Angelo C, Massi-Benedetti C, Fallarino F, et al. Tryptophan catabolites from microbiota engage aryl hydrocarbon receptor and balance mucosal reactivity via interleukin-22. Immunity. 2013;39:372–385. doi: 10.1016/j.immuni.2013.08.003. ••: Provides evidence for a model whereby host and microbiota pathways competing for the same aromatic amino acid interact to modulate mucosal immune reactivity.
- 25. Sridharan GV, Choi K, Klemashevich C, Wu C, Prabakaran D, Pan LB, Steinmeyer S, Mueller C, Yousofshahi M, Alaniz RC, et al. Prediction and quantification of bioactive microbiota metabolites in the mouse gut. Nat Commun. 2014;5:5492. doi: 10.1038/ncomms6492. ••: Combines metabolic modeling, pathway analysis, and metabolomics to identify gut microbiota-dependent metabolites that are AhR ligands.
- 26.Cheng Y, Jin UH, Allred CD, Jayraman A, Chapkin RS, Safe S. Aryl Hydrocarbon Receptor Activity of Tryptophan Metabolites in Young Adult Mouse Colonocytes. Drug Metab Dispos. 2015 doi: 10.1124/dmd.115.063677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bansal T, Alaniz RC, Wood TK, Jayaraman A. The bacterial signal indole increases epithelial-cell tight-junction resistance and attenuates indicators of inflammation. Proc Natl Acad Sci USA. 2010;107:228–233. doi: 10.1073/pnas.0906112107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Yano JM, Yu K, Donaldson GP, Shastri GG, Ann P, Ma L, Nagler CR, Ismagilov RF, Mazmanian SK, Hsiao EY. Indigenous bacteria from the gut microbiota regulate host serotonin biosynthesis. Cell. 2015;161:264–276. doi: 10.1016/j.cell.2015.02.047. •: Findings demonstrate that the gut microbiota directly impacts neuroendocrine hormone synthesis in the intestine.
- 29.Asano Y, Hiramoto T, Nishino R, Aiba Y, Kimura T, Yoshihara K, Koga Y, Sudo N. Critical role of gut microbiota in the production of biologically active, free catecholamines in the gut lumen of mice. Am J Physiol Gastrointest Liver Physiol. 2012;303:G1288–G1295. doi: 10.1152/ajpgi.00341.2012. [DOI] [PubMed] [Google Scholar]
- 30.Ridlon JM, Hylemon PB. Identification and characterization of two bile acid coenzyme A transferases from Clostridium scindens, a bile acid 7alpha-dehydroxylating intestinal bacterium. J Lipid Res. 2012;53:66–76. doi: 10.1194/jlr.M020313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Jones BV, Begley M, Hill C, Gahan CG, Marchesi JR. Functional and comparative metagenomic analysis of bile salt hydrolase activity in the human gut microbiome. Proc Natl Acad Sci U S A. 2008;105:13580–13585. doi: 10.1073/pnas.0804437105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sayin SI, Wahlstrom A, Felin J, Jantti S, Marschall HU, Bamberg K, Angelin B, Hyotylainen T, Oresic M, Backhed F. Gut microbiota regulates bile acid metabolism by reducing the levels of tauro-beta-muricholic acid, a naturally occurring FXR antagonist. Cell Metab. 2013;17:225–235. doi: 10.1016/j.cmet.2013.01.003. [DOI] [PubMed] [Google Scholar]
- 33.Chiang JY. Bile acids: regulation of synthesis. J Lipid Res. 2009;50:1955–1966. doi: 10.1194/jlr.R900010-JLR200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Li F, Jiang C, Krausz KW, Li Y, Albert I, Hao H, Fabre KM, Mitchell JB, Patterson AD, Gonzalez FJ. Microbiome remodelling leads to inhibition of intestinal farnesoid X receptor signalling and decreased obesity. Nat Commun. 2013;4:2384. doi: 10.1038/ncomms3384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Joyce SA, MacSharry J, Casey PG, Kinsella M, Murphy EF, Shanahan F, Hill C, Gahan CG. Regulation of host weight gain and lipid metabolism by bacterial bile acid modification in the gut. Proc Natl Acad Sci U S A. 2014;111:7421–7426. doi: 10.1073/pnas.1323599111. •: Demonstrates that altering the profile of bile acids via overexpression of bacterial bile salt hydrolases profoundly impacts host lipid metabolis.
- 36.Buffie CG, Bucci V, Stein RR, McKenney PT, Ling L, Gobourne A, No D, Liu H, Kinnebrew M, Viale A, et al. Precision microbiome reconstitution restores bile acid mediated resistance to Clostridium difficile. Nature. 2015;517:205–208. doi: 10.1038/nature13828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Martinez-del Campo A, Bodea S, Hamer HA, Marks JA, Haiser HJ, Turnbaugh PJ, Balskus EP. Characterization and detection of a widely distributed gene cluster that predicts anaerobic choline utilization by human gut bacteria. MBio. 2015;6 doi: 10.1128/mBio.00042-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Reikvam DH, Erofeev A, Sandvik A, Grcic V, Jahnsen FL, Gaustad P, McCoy KD, Macpherson AJ, Meza-Zepeda LA, Johansen FE. Depletion of murine intestinal microbiota: effects on gut mucosa and epithelial gene expression. PLoS One. 2011;6:e17996. doi: 10.1371/journal.pone.0017996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Goodman AL, Kallstrom G, Faith JJ, Reyes A, Moore A, Dantas G, Gordon JI. Extensive personal human gut microbiota culture collections characterized and manipulated in gnotobiotic mice. Proc Natl Acad Sci U S A. 2011;108:6252–6257. doi: 10.1073/pnas.1102938108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ridaura VK, Faith JJ, Rey FE, Cheng J, Duncan AE, Kau AL, Griffin NW, Lombard V, Henrissat B, Bain JR, et al. Gut microbiota from twins discordant for obesity modulate metabolism in mice. Science. 2013;341:1241214. doi: 10.1126/science.1241214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Stearns JC, Lynch MD, Senadheera DB, Tenenbaum HC, Goldberg MB, Cvitkovitch DG, Croitoru K, Moreno-Hagelsieb G, Neufeld JD. Bacterial biogeography of the human digestive tract. Sci Rep. 2011;1:170. doi: 10.1038/srep00170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Feria-Gervasio D, Tottey W, Gaci N, Alric M, Cardot JM, Peyret P, Martin JF, Pujos E, Sebedio JL, Brugere JF. Three-stage continuous culture system with a self-generated anaerobia to study the regionalized metabolism of the human gut microbiota. J Microbiol Methods. 2014;96:111–118. doi: 10.1016/j.mimet.2013.11.015. [DOI] [PubMed] [Google Scholar]
- 43.Marcobal A, Kashyap PC, Nelson TA, Aronov PA, Donia MS, Spormann A, Fischbach MA, Sonnenburg JL. A metabolomic view of how the human gut microbiota impacts the host metabolome using humanized and gnotobiotic mice. ISME J. 2013;7:1933–1943. doi: 10.1038/ismej.2013.89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Fischbach MA, Sonnenburg JL. Eating for two: how metabolism establishes interspecies interactions in the gut. Cell Host Microbe. 2011;10:336–347. doi: 10.1016/j.chom.2011.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Magnusdottir S, Ravcheev D, de Crecy-Lagard V, Thiele I. Systematic genome assessment of B-vitamin biosynthesis suggests co-operation among gut microbes. Front Genet. 2015;6:148. doi: 10.3389/fgene.2015.00148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Heinken A, Sahoo S, Fleming RM, Thiele I. Systems-level characterization of a host-microbe metabolic symbiosis in the mammalian gut. Gut Microbes. 2013;4:28–40. doi: 10.4161/gmic.22370. •: Uses a GEM of a representative gut bacterial species to investigate host dependence on bacterial metabolis.
- 47. Shoaie S, Karlsson F, Mardinoglu A, Nookaew I, Bordel S, Nielsen J. Understanding the interactions between bacteria in the human gut through metabolic modeling. Sci Rep. 2013;3:2532. doi: 10.1038/srep02532. •: One of the first reports of GEMs utilized to simulate interactions between different species of the microbiota; illustrates the use of GEMs to reconcile metabolite measurements and transcriptional dat.
- 48.Jiao D, Ye Y, Tang H. Probabilistic inference of biochemical reactions in microbial communities from metagenomic sequences. PLoS Comput Biol. 2013;9:e1002981. doi: 10.1371/journal.pcbi.1002981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Greenblum S, Turnbaugh PJ, Borenstein E. Metagenomic systems biology of the human gut microbiome reveals topological shifts associated with obesity and inflammatory bowel disease. Proc Natl Acad Sci U S A. 2012;109:594–599. doi: 10.1073/pnas.1116053109. ••: Uses metagenomic data to construct a community-level metabolic network, linking shifts in network topology to alterations in microbiota composition as reflected in the abundance of genes encoding the enzyme.
- 50. Donia MS, Cimermancic P, Schulze CJ, Wieland Brown LC, Martin J, Mitreva M, Clardy J, Linington RG, Fischbach MA. A systematic analysis of biosynthetic gene clusters in the human microbiome reveals a common family of antibiotics. Cell. 2014;158:1402–1414. doi: 10.1016/j.cell.2014.08.032. •: Demonstrates biochemical diversity of the gut microbiota metabolome, and highlights the potential to mine this metabolome for novel natural product.