Abstract
The process of base excision repair has been completely reconstituted in vitro and structural and biochemical properties of the component enzymes thoroughly studied on naked DNA templates. More recent work in this field aims to understand how BER operates on the natural substrate, chromatin [1, 2]. Toward this end, a number of researchers, including the Smerdon group, have focused attention to understand how individual enzymes and reconstituted BER operate on nucleosome substrates. While nucleosomes were once thought to completely restrict access of DNA-dependent factors, the surprising finding from these studies suggests that at least some BER components can utilize target DNA bound within nucleosomes as substrates for their enzymatic processes. This data correlates well with both structural studies of these enzymes and our developing understanding of nucleosome conformation and dynamics. While more needs to be learned, these studies highlight the utility of reconstituted BER and chromatin systems to inform our understanding of in vivo biological processes.
Keywords: Base Excision Repair, long-patch (LP) BER, short-patch (SP) BER, Nucleosomes, Chromatin, Histones
1. Introduction
DNA in the cell is constantly damaged from both exogenous and endogenous sources, which can result in chemical modification of DNA, leading to stable mutations [3–5]. The cell employs a variety of mechanisms to repair DNA damage or moderate its mutagenic effects and maintain genomic integrity. Base excision repair (BER), a front-line defense in the repair of damaged bases, excises chemically modified DNA bases that generally do not cause large distortions to the DNA helix [6, 7]. BER corrects an estimated 10,000 lesions/cell/day and can be reconstituted in simplest form by just four enzymes [8, 9]. BER typically is initiated by a DNA glycosylase that recognizes a specific type of damaged, misincorporated, or missing (depurinated) base (Table 1). For example, Family 1 uracil DNA glycosylases (UDGs) specifically remove uracil residues that were misincorporated during DNA replication leading to A-U matches, or generated by deamination of cytosine leading to G-U mismatches. Aberrant uracil in DNA is estimated to occur hundreds of times per cell per day and can result in G:C to A:T transitions, cytotoxic/mutagenic abasic (AP) sites, or changes to transcription due to inhibition of DNA methylation [10, 11]. UDGs have very low activity on uracil in RNA, or cytosine or thymine in DNA [12, 13]. In addition, UDGs play a critical role in generating DNA strand breaks during immunoglobulin gene rearrangement and maturation after enzymatic cystosine deamination to produce G:U mismatches by activation-induced deaminases in B cells [14, 15].
Table 1.
Human DNA Glycosylases
| Glycosylase | Substrate | Mono/Bifunctional | Activity on Nucleosme |
|---|---|---|---|
| AAG/MPG | N-methylpurines, Ethenoadenine | Monofunctional | ND |
| UNG | Uracil, 5-hydroxyuracil | Monofunctional | Yes [34] |
| SMUG1 | Uracil, 5-hydroxymethyluracil | Monofunctional | Yes [34] |
| TDG | Uracil, thymine, 5-hydroxymethyluracil, 5-formylcytosine, 5-carboxylcytosine | Monofunctional | ND |
| MBD4 | Uracil, cytosine, ethenocytosine | Monofunctional | Yes [41] |
| MUTYH | Adenine | Monofunctional | Yes |
| OGG1 | 8-oxoG, FapyG | Bifunctional | Yes [42] |
| NTH1 | Tg, oxidized pyrimidines, formamidopyrimidines | Bifunctional | Yes [43] |
| NEIL1 | Tg, oxidized pyrimidines, formamidopyrimidines | Bifunctional | ND |
| NEIL2 | Tg, oxidized pyrimidines, formamidopyrimidines | Bifunctional | ND |
| NEIL3 | Formamidopyrimides | Bifunctional | ND |
8-oxoG: 8-oxoguanine; FapyG: 2,6-diamino-4-hydroxy-5-formamidopyrimidine; Tg: tymine glycol; ND: not determined
After recognition of a chemically altered or aberrant base, the specific glycosylase, catalyzes a nucleophilic attack by a water molecule at the glycosidic bond, resulting in cleavage of the base - sugar bond, creating an apurinic/apyrimidinic (AP) site in the DNA and releasing the damaged/aberrant base [7, 16, 17]. The backbone at the AP site is either then cleaved by a separate AP endonuclease (APE) activity, or is first cleaved by a glycosylase possessing bifunctional activity, followed by APE cleavage to remove the resulting aldehyde residue. Cleavage results in a 3′-hydroxyl end and a 5′-deoxyribose phosphate (dRP), which can then undergo either short-patch (SP-), or long-patch base excision repair (LP-BER). In short-patch repair, a DNA polymerase with an associated dRP lyase activity inserts a single base, while in long-patch repair a DNA polymerase adds 2–10 nucleotides and displaces a ssDNA flap containing the 5′-dRP, which is subsequently cleaved off by flap endonuclease 1 (FEN1). Both pathways result in a nick that is sealed by DNA ligase [7, 8]. The BER pathway has been entirely reconstituted with purified components in vitro on free DNA substrates. In this review we will consider characterizations of the activity of the major enzymes involved in BER on reconstituted chromatin substrates in vitro.
In vitro characterizations of the activities of the component enzymes involved in BER on well-defined chromatin complexes reconstituted from purified components has been critical in understanding how BER occurs on chromatin substrates in vivo. The basic repeating subunit of chromatin is the nucleosome, consisting of ~147 bp of DNA wrapped ~1.7 times around a protein spool consisting of two copies each of the four core histones H2A, H2B, H3, and H4 [18, 19]. The nucleosome unit also includes 10–80 bps of linker DNA that links cores together to form chromosome-sized oligonucleosome arrays that fold and condense into chromatin fibers and higher order chromatin structures. Typically each nucleosome repeat is bound by one linker histone (e.g. H1) and non-histone chromosomal proteins (NHCPs) that modulate the folding and condensation of nucleosomes into higher order structures present in the cell nucleus [18].
Nuclease digestion studies indicate that the core region is relatively resistant to cleavage due to tight association with the core histones, while the linker DNA is relatively accessible [18]. Likewise, accessibility of nucleosome core DNA is highly restrictive to most DNA-binding factors, including those involved in DNA repair. For example, the activities of factors involved in nucleotide excision repair (NER) are greatly inhibited by chromatin, and thus ATP-dependent chromatin remodeling activities are required for efficient NER [20–24]. However, remodeling processes impose energetic costs to the cell and must be targeted to specific damage locations or at specific times within the cell cycle when access to the DNA is required [20, 25]. Conversely, some BER-associated enzymes appear to exhibit significant activity on nucleosome substrates in vitro, suggesting that some BER events in cells do not require chromatin remodeling. This review will focus on investigations of the activities of the components of the BER pathway on model chromatin substrates.
2. DNA Glycosylases
2.1 Nucleotide excision by DNA glycosylases
The recognition and excision of damaged bases by DNA glycosylases is a critical first step in the base excision repair pathway. Perhaps the best characterized is Uracil DNA glycosylase (UDG), which is conserved from bacteria to humans and serves as a prototypical example of this type of enzyme [16]. UDG has been extensively characterized by X-ray crystallographic studies and biochemical analyses [16, 26–30]. Early UDG crystal structures showed a conically shaped basic channel along one surface with a pocket at the end, suggested to be the damaged duplex DNA-binding site and the active site, respectively [26, 27]. Since uracil was observed to specifically bind within the pocket, substrate recognition was suggested to occur via a base “flipping out” mechanism, similar to that observed for DNA methylases, requiring disruption of canonical base-pairing in order for the uracil to adopt an extrahelical orientation [26, 27]. Later crystal structure studies with UDG in complex with DNA substrates and products substantiated this mechanism, showing that UDG undergoes an open-to-closed structural transition upon binding its target. It was also shown that conserved residues within UDG probe for uracil by binding to primarily the damaged DNA strand and pinching the DNA backbone at successive phosphates, causing disruption of intra-helical uracil base-pairing [16, 29]. In addition, the insertion of a leucine side chain that helps push the uracil base toward the uracil-binding pocket and intercalates into the void, along with the attraction of the highly specific uracil binding pocket, leads to the “flipped out” conformation and hydrolysis of the glycosidic bond. This mechanism appears to be conserved in other DNA glycosylases that use ‘helix-invading’ residues to distort the DNA helix, destabilize damaged bases, and flip them into extra-helical orientations for excision [31, 32].
In most cases binding of DNA glycosylases to target DNA causes a bending of the DNA sequence away from the body of the enzyme. For example, bacterial UDG bends DNA in a 45° angle tangentially away from the enzyme, while the enzyme hOGG1, a glycosylase that initiates the repair of 7,8-dihydro-8-oxoguanine (oxoG) in DNA, causes a sharp, 70° bend in the target DNA directly away from the enzyme [29, 33]. Considering this feature, and the fact that DNA in eukaryotic cells is highly bent around nucleosomes, researchers wondered if these enzymes are able to excise damaged bases in a nucleosome context. Indeed, several groups have shown that nucleosomes may act as substrates for DNA glycosylases, with varying degrees of reduction in the activity of the enzyme compared to that observed for naked DNA.
2.2 Uracil DNA glycosylases
In an initial work, Nilsen and co-workers showed that the two major human uracil-DNA glycosylases, UNG2 and SMUG1 (the latter in combination with APE1), could excise uracil from nucleosome substrates at rates 3-9-fold reduced from that observed for naked DNA, and found little relationship to the position of the U:A base pair within the nucleosome or the rotational orientation of the damage [34]. Smerdon’s group showed that UDG/APE can excise uracil from G:U mismatches within a nucleosome at rates ~10-fold slower than in naked DNA, but found a ~3-fold difference in the excision rates dependent on whether the backbone position of the uracil base was oriented toward or away from the core histone octamer [35]. Differences in the measured rates might be attributed to the use of a template based on the Lytechinus variegatus 5S rRNA positioning element in the former study [34] or the TG-positioning sequence in the latter [35]. Interestingly, both groups demonstrated that at high UDG/APE concentrations, the nucleosome substrates could be digested to completion. These results suggested that UDG/APE identifies uracil within DNA wrapped around nucleosomes and carry out their enzymatic function.
A question arising from these studies was whether these enzymes required transient release and exposure of the uracil-containing DNA away from the confines of the nucleosome, and whether this exposure was in some way facilitated by the enzymes. Widom and colleagues have shown that DNA can spontaneously and transiently unwrap from the nucleosome surface, with the probability of unwrapping being greater closer to the edge of the nucleosome core [36, 37]. Unwrapping allows competition for binding to nucleosome DNA by trans-acting factors on biological timescales, with the extent of unwrapping dependent on histone acetylation and flexibility of the DNA sequence [38, 39]. Moreover, the unwrapped state might be promoted by the cooperative binding of repair factors or other chromatin proteins [40]. Thus, spontaneous unwrapping provides one mechanism whereby BER factors might access and process damage within nucleosome DNA, albeit at rates significantly reduced compared to that on free DNA. A second, distinct mechanism considers the fact that the DNA backbone within nucleosomes is exposed to bulk solvent once every ~10 bp. Given that certain transcription factors and nucleases can productively bind this DNA with affinities near that observed for naked DNA, it is possible that certain sites internal to the nucleosome might be accessible to BER factors without dependency on spontaneous DNA unwrapping. A related possibility is that damage sites within nucleosomes may not be accessible to glycosylases as envisioned in static models from structural studies of nucleosome cores [19], but may dynamically populate conformations that allow the enzymatic processes to proceed without wholesale nucleosome disruption.
A systematic examination of UDG activity throughout a nucleosome substrate provided strong evidence to support the idea that glycosylases can act directly on nucleosome DNA, and that this activity strongly depends on the rotational orientation of the lesion with respect to the histone surface. Cole et al. designed DNA templates based on a well-characterized nucleosome-positioning element (NPE) and quantified rates of UDG activity for U:A base pairs placed at eight locations, from the center to the edge of a nucleosome [41]. At high (single-turnover) concentrations, UDG efficiently removed uracil from sites where the DNA backbone was oriented away from the surface of the histone octamer, with rates that were only a few-fold reduced from that observed for the naked DNA template [41]. Given the modest reduction in UDG cleavage rate, and the much slower rate of spontaneous DNA unwrapping from the nucleosome as shown by Widom [36], these authors concluded that UDG recognized and cleaved its substrate without significant disruption of histone-DNA interactions. In contrast, they found that uracils oriented toward the histone octamer were excised at rates several thousand times slower, consistent with a mechanism in which UDG excision at such sites first requires spontaneous unwrapping of DNA from the histone surface. Also consistent with this mechanism, rates of excision for the ‘in’ sites were additionally dependent on distance from the edge of the nucleosome, being progressively slower for sites closer the nucleosome dyad. In contrast to the nucleosome core, UDG activity on linker DNA was found to be similar to that of naked DNA. However, association of linker histone H1 reduced activity of UDG at selected sites near where the globular domain of H1 is proposed to bind to the nucleosome as well as within the linker DNA [41]. These results are consistent with the idea that nucleosome remodeling is required for efficient DNA glycosylase excision at inward-facing sites [42, 43], and suggests that outward-facing sites within nucleosomes and in linker DNA regions represent hot spots for repair that could influence critical biological processes [41].
A similar observation was made by Smerdon’s group who measured rates of UDG cleavage for outward-facing lesions in nucleosomes to be similar to those measured in naked DNA. Conversely, significant reduction in the efficiency of cleavage was measured for intermediates and inward-facing lesions [44, 45]. Importantly, in formaldehyde crosslinked nucleosomes, in which nucleosome conformational excursions are likely suppressed, these authors found a reduction in the rates of cleavage of an inward site, but a stimulation in the rate of cleavage at an outward-facing site. These data suggest that the nucleosome DNA equilibrates between accessible and non-accessible conformations (with crosslinking biasing the distribution) and that UDG activity traps the accessible conformation, when available [45]. In general, these data support the idea that DNA orientation is of primary importance in defining the rate of UDG activity but that histone-DNA dynamics also plays a significant role in populating enzyme-accessible states throughout the nucleosome [45].
A recent study provides additional molecular insights into how UDG accesses uracils within nucleosomes and brings to light the surprising finding that assembly of DNA into nucleosomes can actually enhance UDG activity at certain sites compared to naked DNA [46]. Examination of the rates of UDG activity within nucleosomes containing the 601 nucleosome-positioning DNA showed that two sites exhibited equal or a 2-fold increase in activity over the naked DNA substrate, respectively. Comparisons with a recent high-resolution X-ray crystal structure of this nucleosome [47] indicates that this position exhibits nucleosome-dependent perturbations in base pair parameters such as twist, roll, and buckle, and a wide minor groove that would facilitate flipping the uracil out from the helix stack and intercalation of Leu272. Moreover the authors provide evidence that less ideal orientations of uracil within the nucleosome DNA undergo conformational excursions to states that are accessible to the enzyme, suggesting that local structural features, in addition to rotational orientation of the lesion contribute to UDG activity [46].
2.3 Base mismatches and base oxidation products
Several groups have investigated whether glycosylases that target base mismatches or base oxidation products can locate their targets within nucleosomes, and whether activity depends on rotational orientation and/or location within the nucleosome. The activity of the glycosylase MBD4 was found to be diminished but not abolished for T/G mismatches within a reconstituted nucleosome in a manner apparently not dependent on position with respect to the dyad, while histone hyperacetylation increased the efficiency with which the bases were excised [48]. Menoni et al. showed that the glycosylase OGG1 was significantly inhibited ~100-fold at a site 10 bps from the nucleosome dyad, but stimulated by ATP-dependent chromatin remodeling in nucleosomes containing major histone variants [49]. Prasad and co-workers investigated the ability of NTH1, a bifunctional DNA glycosylase from humans that targets oxidized pyrimidines, to recognize and excise thymine glycol (Tg) lesions within nucleosomes based on the L. variegatus nucleosome positioning sequence [50]. They found that under substrate-excess conditions NTH1 processed an outward-facing thymine glycol (Tg) lesion, located about 50 bps from the nucleosome dyad, nearly as efficiently as naked DNA. However, a nearby site oriented toward the histone octamer or sites closer to the dyad were removed at ~10-fold reduced efficiency compared to naked DNA. Interestingly, these researchers determined that the lyase activity of bifunctional NTH1 is rate limiting compared to the glycosylase activity [50]. As with the studies on UDG mentioned above, at high concentrations of NTH1, even nucleosomes with inward-oriented Tg residues could be processed to completion. Nucleoprotein gels of processed nucleosomes indicated that NTH1 activity did not induce or cause disruption of the nucleosome structure. These studies suggest that NTH1 takes advantage of transient DNA unwrapping from the nucleosome surface to recognize, excise and cleave inward-facing Tgs, but can directly access outward-facing targets within the nucleosome without significant steric hindrance by the nucleosome structure.
In summary, numerous studies show that glycosylases are able to access lesions within nucleosomes at rates only a few-fold reduced if the target backbone is oriented away from the histone octamer. Hence, they indicate that glycosylases are able to recognize and bind to nucleosome DNA, perhaps because they interact primarily with the damage-containing strand, and bend DNA away from the enzyme in a manner that is compatible with DNA bending in nucleosomes. Moreover, glycosylases recognize outward-facing lesions and carry out their enzymatic function at rates that indicate little or no disruption of histone-DNA interactions. Consequently, data indicate the role of DNA motility within the nucleosome structure, which populate conformations required for enzymatic processing of outward-facing or near outward-facing sites. In contrast, inward-facing sites exhibit rates of cleavage that indicate spontaneous DNA unwrapping in nucleosomes, which is rate-limiting in the enzymatic process [41, 51]. Finally, careful analyses indicate that while spontaneous unwrapping of nucleosomal DNA may contribute to the first step of BER in vivo, it may not be sufficient to account for efficient repair of all oxidative lesions within the time frames of BER established in vivo [51], consistent with recent work showing a contribution of the RSC remodeling complex to overall BER [42, 43].
3. AP Endonuclease
After DNA glycosylase excision of the base, apurinic/apyrimidinic endonuclease (APE) incises the DNA backbone 5′ to the abasic site generating a nick flanked by a 3′-OH and a 5′-dRP. The crystal structure shows that human APE1 has a rigid conformation that does not undergo any major changes as it binds to DNA containing the AP site [52]. Upon binding to the lesion, APE1 stabilizes the AP site external to the helix and induces a kink in DNA, similar to UDG [52]. Importantly, APE1 induces a 35° bend in the DNA helix away from the body of the protein. Also, similar to UDG, APE1 interactions are primarily made with one strand, with nearly 1/3 of the buried solvent-accessible surface area of APE1 centered on the flipped-out abasic site. Thus, APE1 interacts with its substrate in a way that might be conducive to binding and carrying out its enzymatic function on accessible sites within nucleosomes.
While the phosphodiesterase activity of AP endonucleases has not been studied on isolated nucleosomes, APE activity was assayed in the presence of glycosylases to generate AP sites (see above). This is due, in part, to the fact that AP sites are unstable and preparations of AP-containing nucleosomes (with continuous DNA backbones) are problematic. Smerdon’s group examined APE1 activity in nucleosomes, with AP sites generated by both bacterial and human UDG (also known as hUNG2) [44]. By employing limiting concentrations of enzyme, these authors determined that APE1 can efficiently process AP sites within nucleosomes as they arise [44]. Moreover, it is known that UDG binds tightly to its product such that AP site release and enzyme turn-over limit the rate of reactions. Thus, under appropriate substrate-excess conditions, the high affinity of APE1 for AP sites can actually stimulate the rate of UDG activity. This feature is thought to limit the exposure of deleterious AP sites produced by UDG, and to ensure a ‘hand-off’ of an AP site to APE. Although APE1 can stimulate UDG activity on outward-facing lesions within nucleosomes, this stimulation is reduced compared to that observed on naked DNA [44]. In summary, APE1 exhibits relatively robust activity in nucleosomes, and when APE1 and DNA glycosylase are coupled, the overall reaction rate is similar to that of the glycosylase enzyme alone [44, 45]. Interestingly both outward- and inward-facing DNA lesions have similar overall repair rates, suggesting that once UDG traps and cleaves its substrate in nucleosomes, it retains the AP site in an accessible conformation, then ‘hands off’ the substrate to APE.
The bifunctional glycosylase hNTH1 was used to generate the APE substrate DNA 3′-phospho-α, β-unsaturated aldehyde (3′-PUA) in DNA fragments containing a thymine glycol residue. The 3′-PUA-containing DNA was then assembled into nucleosomes in either an outward- or inward-facing orientation. APE was able to digest the outward-facing substrate with rates commensurate to those measured for glycosylase activity, but APE processed the inward-facing lesion at a rate that was about half of that for the outward-facing site [53]. These data demonstrate that APE digests the product of a bifunctional glycosylase and AP sites generated by the mono-functional UDG at similar rates. Moreover, these authors showed that hNTH1 binds to its product, even in nucleosomes, and results in a supershifted nucleosome on a nucleoprotein gel [50]. Addition of excess APE1 displaces hNTH1 from the ternary complex, consistent with the ‘hand-off’ mechanism mentioned above [50]. In summary, these studies indicate that APE appears to process sites within nucleosomes with a facility similar to that of DNA glycosylases, and can do so in conjunction with specific glycosylase activity without significant disruption of nucleosome structure.
4. Flap Endonuclease I
FEN1 was first identified as an enzyme involved in processing of Okazaki fragments and only later was it found to recognize and excise flap-DNA structures, an activity important in LP-BER [54]. Hosfield et al. [55] determined the structure of Pyrococcus furiosus FEN1 to 2.0Å resolution and found it contains a single α/β domain oriented in a saddle-like structure. The center of this structure has a large groove of positive charges forming a DNA binding region, while the bottom of the groove has two regions of conserved aspartate and glutamate residues that coordinate Mg2+. These authors suggest a mechanism in which FEN1 binds to ssDNA flaps with an open clamp structure and tracks in a 5′ to 3′ direction along the single-stranded DNA flap to the flap junction, where the clamp closes as it binds to duplex DNA. Interestingly, the 5′-end of the flap must be unobstructed for FEN1 to bind, as structures like DNA bubbles inhibit FEN1 cleavage [56]. In addition, short oligonucleotides or proteins bound to 5′-flaps also impede FEN1 activity [57, 58]. However, some modifications on the 5′-flap can be accommodated by the enzyme, such as thymine dimers that do not inhibit FEN1 activity [59]. Harrington et al. [60] determined that FEN1 preferentially binds DNA containing 5′-flaps and that the duplex DNA upstream of the 5′-flap is important for enhancing FEN1 binding. Additionally, FEN1 was found to be optimally active on ‘double-flap’ structures in which, in addition to the 5′-flap, the 3′-end of the upstream strand contains a 1-nt flap [61]. Furthermore, FEN1 activation increases upon direct association of PCNA to the DNA flap, as it promotes a higher association rate between FEN1 and DNA [62].
Similar to other factors, the function of FEN1 in LP-BER was well characterized on DNA substrates before its activity was investigated on nucleosomes. The Hayes group assessed FEN1 activity in nucleosomes positioned on the Xenopus laevis 5S rDNA sequence. After establishing that the nucleosome was not affected by the presence of a DNA flap placed one turn from the dyad, they found that at limiting enzyme concentrations FEN1 preferentially cleaved the flap in nucleosomes over those in naked DNA [63]. Interestingly, removing the core histone tails abolished FEN1 preferential cleavage of flaps in the nucleosome, suggesting the tails may play some role in promoting FEN1 binding to nucleosomes. Strikingly, FEN1 was not only able to locate and cleave standard (5 nt) flaps and double-flap structures within the nucleosome, but also cleaved a flap consisting of a single abasic site constituting the flap, representing an intermediate in the BER process [63]. Importantly, inclusion of a restriction endonuclease in the reaction, that cuts the nucleosome DNA near the flap, showed that FEN1 did not induce or require dissociation of the nucleosome DNA [63]. Of note, the restriction enzyme would only detect DNA detached from the nucleosome surface, thus it is possible that FEN1 may require, or induce, local DNA excursions, similar to those suggested for UDG (see above).
Further evidence supporting the idea that FEN1 requires DNA excursions within nucleosomes was provided by the dependence on flap orientation for FEN1 cleavage. Nucleosomes containing DNA flap structures within 5S DNA templates were reconstituted such that the flaps were positioned near the dyad and oriented either toward the histone octamer (flap-in), or away from the histone octamer (flap-out) [64]. Note that in this study, the ‘flaps’ consisted of a ‘double-flap’, with a 1 nt 3′ flap as well as a 5 nt 5′ flap for optimal FEN1 activity (see above). While both flap-containing templates efficiently reconstituted into nucleosomes, hydroxyl radical footprinting, Exo III digestion and site-directed crosslinking indicated subtle structural differences between the flap-in and flap-out nucleosomes suggesting that while, on average, the rotational orientation of DNA is maintained in the presence of the inward-facing flap, the DNA of the flap-in nucleosome undergoes greater conformational excursions on the histone surface. However, FEN1 cleaved both substrates with similar efficiencies, suggesting, again, that DNA conformational motility allowed accessibility to both flap-in and flap-out substrates. In contrast, FEN1 was not able to process flaps within nucleosomes based on the 601 nucleosome positioning sequence, which binds to the core histone octamer with an affinity hundreds of times greater than the 5S rDNA sequence [64]. This data suggests that DNA excursions required for FEN1 activity are greatly suppressed in the tightly bound DNA of the 601 nucleosome. Since the vast majority of nucleosomes in vivo have DNA affinities well below that of the 5S rDNA [65], they should exhibit the conformational motility needed for efficient FEN1 activity. Of note, these studies were not performed with PCNA, thus it would be interesting to determine if PCNA is able to stimulate FEN1 activity on nucleosome substrates.
5. DNA Polymerase β
DNA polymerase β (Pol β) is responsible for filling DNA gaps generated by DNA glycosylases and AP endonuclease, performing DNA synthesis, and 5′-dRP excision activity in both SP- and LP-BER pathways (see elsewhere in this issue). Pol β is a 39-kDa protein made up of an 8-kDa N-terminal domain (residues 1–87) responsible for dRP excision activity, and a 31-kDa C-terminal domain possessing thumb, palm, and finger sub-domains responsible for nucleotidyl transferase activity [8]. The X-ray crystal structure of Pol β shows it is able to induce a 90° kink in DNA allowing for exposure of the gapped or nicked DNA to the enzyme [66]. The crystal structure also indicates that Pol β, in complex with intermediates of the BER pathway, can undergo a conformational change between the thumb and the 8-kDa N-terminal domain: interactions between these domains create a closed conformation and a catalytically active enzyme, while loss of interactions create an open conformation and a catalytically inactive enzyme.
The basic 8-kDa N-terminal domain of Pol β is able to recognize APE incised substrates containing a 3′-OH and 5′-dRP on either side of the excised nucleotide positions. Matsumoto et al. observed purified Pol β successfully repaired AP sites and efficiently excised 5′-dRPs generated by APE1, indicating that in addition to gap-filling DNA synthesis activity, Pol β also possessed a BER rate-limiting dRP-lyase activity [67, 68]. This activity was localized to the 8-kDa N-terminal domain and described a β-elimination excision mechanism [69]. Notably, dRP-lyase activity of Pol β is dependent on the pre-incision by APE1, and reduced AP sites cannot act as a substrate for Pol β lyase activity. This domain also primarily binds to the template DNA strand and upstream of the gap, although some interactions were detected on DNA downstream of the gap as well [70]. The interaction with the downstream DNA is important for gap-filling DNA synthesis, in which Pol β synthesizes up to 6-nt in a processive mechanism, dependent on the presence of a 5′-PO4. DNA templates containing a 5′-OH were poor substrates for Pol β activity and showed incomplete gap-filling synthesis. Gaps longer than 6-nt are also filled by Pol β, but in a distributive manner [71]. In addition, mutational analysis and photochemical crosslinking demonstrate that the 8-kDa N-terminal domain is important for ssDNA-binding and 5′-PO4 recognition [66].
To proceed with SP-BER, Pol β cleaves the 5′-dRP and fills the 1-nt gap with the complementary residue, while in LP-BER, Pol β incorporates 2- or more nucleotides. The long patch base excision repair pathway occurs if the AP site has been generated through oxidative or alkylating damage such that Pol β cannot carry out β-elimination of the 5′-dRP, preventing its excision [72]. Pol β still maintains functionality in this pathway by carrying out gap-filling and strand displacement synthesis, generating a 5′-flap substrate. As discussed previously, inclusion of FEN1 stimulates Pol β strand displacement synthesis activity and is required for excision of the 5′-flap substrate before DNA ligase I can seal the nick. Although Pol β is the main polymerase involved in LP-BER [73], polymerases δ/ε also have activity in this pathway. Loading of the DNA polymerases depends on PCNA/RFC and on FEN1 for stimulation of gap-filling, strand displacement synthesis, and flap cleavage [8]. It is still unclear how the cell differentiates between Pol β LP-BER and polδ/ε LP-BER.
In general, Pol β activity is significantly reduced on nucleosome substrates. In initial studies using a reconstituted BER pathway, Nilsen et al. [34] examined Pol β activity on nucleosomes in the presence of UDG and APE1 at sites about 20 and 50 bp from the nucleosome dyad, employing a 146 bp DNA template based on the Lytechinus variegatus 5S rRNA nucleosome positioning element. While significant one-nucleotide extension was observed at the site near the edge of the nucleosome, extension was severely inhibited at the more internal site [34]. In contrast, Beard et al. found that Pol β was incapable of incorporating nucleotides on nucleosome substrates generated by UDG/APE1 activity, at both outward- and inward-facing positions near the dyad in a nucleosome based on the TG positioning element [35]. Thus, it appears that Pol β activity may depend on both the location and specific features of the surrounding DNA sequence in the nucleosome that may influence the conformational rigidity of the target site.
Pederson and colleagues investigated the activity of Polβ in the midst of sequentially added BER enzymes, starting with hNTH1/APE targeted to thymine glycol placed with outward and inward orientations, about 4.5 and 5 turns from the nucleosome dyad in the 5S nucleosome, and a similar inward orientation was placed in the 601 nucleosome [53]. A robust Polβ activity could be observed in all three cases, but the amounts of polymerase were adjusted to achieve roughly equivalent activities in each case. For example, the outward lesion in the 5S nucleosome required approximately 2.5-fold the amount of enzyme to achieve an activity comparable to that observed in a naked DNA template. The inward lesion in both the 5S and the 601 nucleosomes required 165-fold more enzyme compared to naked DNA [53]. Thus, high polymerase concentrations are required to effectively compete with histones for sites with suboptimal orientations but an outward-facing site requires only a few-fold more enzyme to show approximately equivalent activity as measured on naked DNA. Hence, DNA polymerization can occur on the nucleosome surface at an appropriately oriented site.
To further explore these issues, Rodriguez et al. specifically assayed for Pol β activity at inward- and outward-oriented sites containing single nucleotide gaps within a nucleosome based on the 601 positioning sequence [45]. In contrast to Pederson’s result, they found very limited gap-filling activity, with the greatest reductions for outward-facing gaps, assumed to be due to the greater ease of the enzyme accessing the template strand at inward-facing gaps. Importantly, polymerization at inward-facing gaps was inhibited to a much greater extent by formaldehyde crosslinking than at outward-facing gaps, consistent with a requirement for flexibility of nucleosome DNA for Pol β activity at inward-facing gaps [45]. Together, these data suggest that Pol β has limited gap-filling and strand displacement activity on nucleosome substrates and this may be attributed to the fact that it interacts with both strands of DNA and also needs to create a 90° bend in the DNA backbone to expose the gapped or nicked region. Clearly more detailed investigations are needed to fully elucidate the structural requirements for Pol β activity within nucleosomes.
6. DNA Ligases
DNA ligases are required to complete nucleotide excision repair, repair of double and single stranded DNA breaks, and the processing of Okazaki fragments during replication. During repair, DNA ligases seal nicks in the DNA backbone through generation of phosphodiester bonds between 3′-hydroxyl and 5′-phosphate groups. DNA ligases have a conserved catalytic core formed by the adenylation domain (AdD), the OB-fold domain (OBD) and the DNA-binding domain (DBD) [74]. The interactions between these domains and nicked DNA occur such that the enzyme completely encircles DNA and forms important contacts with the DNA minor groove. The DBD interacts with both the AdD and OBD domains facilitating the end-joining activity of the enzyme, while the OBD domain binds near the nicked DNA and widens both the major and minor grooves through alteration of the DNA backbone [74]. In Eukaryotes, nick-sealing is an ATP-dependent process as it requires adenylation of a conserved lysine residue within the enzyme active site. The AMP group of the adenylated enzyme is transferred to the 5′-phosphate of the DNA nick, where attack of the 3′-hydroxyl results in formation of a phosphodiester bond [74]. Both, DNA ligase I and the DNA ligase IIIα/XRCC1 complex are involved in BER.
While DNA ligase I and DNA ligase IIIα have different interacting factors that specify their functions in LP- or SP-BER, homologies within their catalytic domains suggest that they can compensate for each other. In order to address this question, Sleeth and co-workers generated whole cell extracts immunodepleted for XRCC1-DNA ligase IIIα and found repair of AP sites by SP-BER, suggesting that DNA ligase I is able to compensate for lack of XRCC1-DNA ligase IIIα [75]. Conversely, when whole cell extracts were immunodepleted for DNA ligase I, XRCC1-DNA ligase IIIα was not able to repair reduced AP sites to promote LP-BER. Therefore, while DNA ligase I is able to function in both SP- and LP-BER, XRCC1-DNA ligase IIIα activity is exclusive to SP-BER [75].
6.1 DNA Ligase I
DNA ligase I is a 125-kDa enzyme [76] that is primarily responsible for sealing nicks in LP-BER. In addition to the functional domains shared between ligases (see above), the N-terminal domain of DNA ligase I possesses a nuclear localization signal and a proliferating cell nuclear antigen (PCNA) binding domain. PCNA helps recruit DNA ligase I to sites of DNA damage, promoting efficient ligation of nicks [77]. However, PCNA must completely encircle DNA in order to stimulate efficient ligation of nicked DNA substrates [77], thus the ability of PCNA to enhance DNA ligase I activity in nucleosomes is unclear. In addition to PCNA, replication protein A (RPA) also stimulates DNA ligase I activity in the LP-BER pathway [78].
While much work has been done to elucidate the structure of DNA ligase I, and its mechanism, there is little information on how this enzyme functions in chromatin. The Hayes group addressed this question using nucleosomes reconstituted with the 5S rDNA sequence, containing DNA nicks located at three positions: near the nucleosome dyad, at the edge of the nucleosome, and within the linker DNA region [79]. Nicks in linker DNA were ligated with rates comparable to naked DNA, while nicks positioned near the dyad and near the nucleosome edge were ligated 10- and 6-fold more slowly, respectively. DNA ligase I activity at the dyad of a nucleosome was only reduced 10-fold, suggesting that the enzyme did not disrupt histone-DNA interactions. However, nucleosome core particles (lacking linker DNA) containing a nick at the dyad inhibited the activity of DNA ligase I about 5,000-fold. This was attributed to the increased binding of histone tails to core DNA in the absence of linker DNA binding sites [79, 80]. In fact, removal of the core histone tails via light trypsin digestion restored ligase activity similarly to that observed in nucleosomes containing linker DNA. It was suggested that overcrowding of histone tails and their interactions with core DNA could inhibit the ability of DNA ligase I to seal nicks [79]. Remarkably, incubation of flap-containing nucleosomes with both hFEN1 and hDNA ligase I resulted in the generation of ligated DNA fragments [79]. Inclusion of a restriction enzyme in the reaction indicated that the coupled activity of both enzymes can occur on the surface of the nucleosome, without disruption of the nucleosome structure sufficient to allow restriction enzyme digestion of the DNA template.
6.2 DNA Ligase IIIα
In addition to DNA ligase I, DNA ligase IIIα also plays a role in SP-BER. DNA ligase IIIα contains a catalytic domain homologous to DNA ligase I. Additionally, it has a zinc-finger domain in the N-terminal region that enhances DNA binding [81], and a BRCT II domain in the C-terminal region that is important for interaction with XRCC1 [82]. Caldecott et al. first reported the interaction between DNA ligase IIIα and XRCC1 and showed this interaction is important for DNA ligase IIIα activity [83, 84] as deletion of XRCC1 results in unligated products in SP-BER. Interestingly, XRCC1 is able to interact with poly(ADP-ribose) polymerase (PARP) via a second BRCT domain, suggesting that PARP attracts DNA ligase IIIα/XRCC1 to DNA strand breaks via XRCC1 [85]. In addition, deletion analysis has shown DNA ligase IIIα and PARP-1 directly interact through the N-terminal region of DNA ligase IIIα and that this interaction is responsible for recruitment of DNA ligase IIIα-XRCC1 to ssDNA breaks [85–87].
The activity of DNA ligase IIIα-XRCC1 was investigated on 5S rDNA and 601 nucleosomes that contained DNA nicks [53]. The ligase had limited activity on 5S rDNA nucleosomes, regardless of orientation of the nick. Furthermore, no ligation activity was observed on 601 nucleosomes, consistent with previous reports suggesting that conformational motility within 601 nucleosomes is severely restricted, and that such motility is required for DNA ligase IIIα accessibility and function. In contrast, activity of DNA ligase IIIα/XRCC1 can be observed at higher concentrations, due to its strong interaction with DNA compared to other BER enzymes, which allows effective competition with the core histones for binding the DNA [53]. Increasing the concentration of DNA ligase IIIα-XRCC1 resulted in greater efficiency in ligation, but also disruption of the nucleosome and DNA release. Nucleosome disruption was also noted on gap-containing DNA and may play a role in promoting more efficient extension by Pol β [53].
7. Concluding remarks and future directions
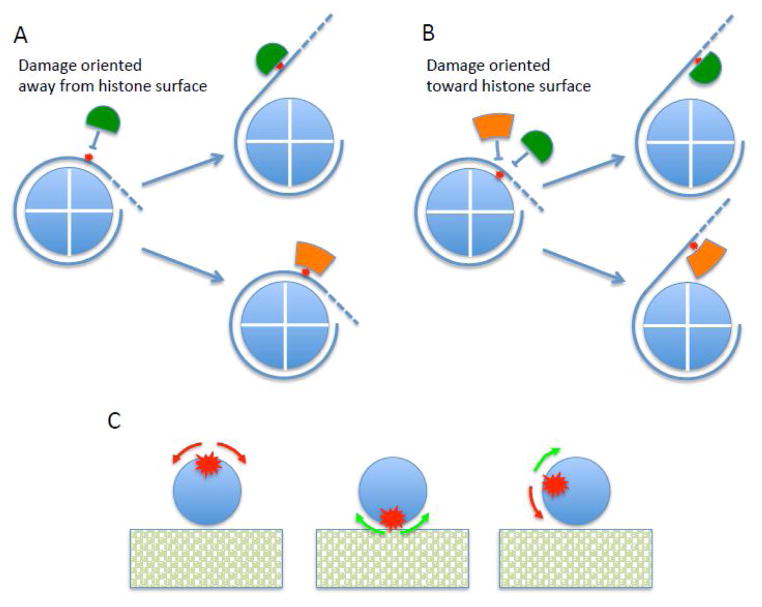
The ability to install specific BER targets in DNA and the advent of well-defined nucleosome positioning sequences has been of great use in investigations of the activity of BER factors on model chromatin substrates. It is clear from these studies that there are two general mechanisms that allow BER activities to function on nucleosome DNA, without influence or aid from external chromatin remodeling factors. First, several activities, including all glycosylases tested, APE, FEN1 and DNA ligase I can directly carry out their enzymatic function so long as the substrate is appropriately rotationally positioned in the nucleosome (Fig. 1A, orange factor). This mechanism does not involve disruption of histone-DNA interactions but for many instances does appear to rely on natural conformational motility within the structure to access states appropriate for catalysis (Fig. 1C). Three aspects of the manner in which the majority of these BER enzymes contact their DNA substrates appear to be contributing factors to the ability to utilize nucleosomes as substrates: in almost all cases, these enzymes i) interact primarily with one of the two strands of the DNA double helix, and in particular, with the strand of the DNA containing the target residue, ii) bind to only one side of the DNA double helix, interacting with less than one helical turn, and iii) bend the DNA in a direction generally away from the main body of the enzyme. Thus, these enzymes seem particularly well-suited for activity on the highly bent, and partially occluded DNA within nucleosomes.
Fig. 1. Schematic showing mechanisms by which BER enzymes access targets within nucleosomes.
A. DNA damage sites (red star) on the surface of the nucleosome can be accessed by spontaneous unwrapping of DNA (blue line) from the core histone octamer surface (blue circle) for factors for which access is impeded by presence of the histones or that require DNA conformations not achievable within the nucleosome (green factors). Factors for which the DNA conformation and accessibilty found within the nucleosome are compatible with enzyme activity, can process targets without the requirement for DNA unwrapping (orange factor).
B. For damage that is that is oriented toward the core histone octamer, or otherwise inaccessible, DNA unwrapping may be required to allow access to both types of factors.
C. DNA motility within the nucleosome influences activity of BER enzymes. This schematic depicts a cut-away view down the helical axis of DNA (blue circle) on the surface of the core histone octamer (rectangle). Left: DNA damage is orientated away from the core histone octamer, and is nominally compatible with BER activities. In this case rotational excursions of the DNA may actually reduce activity of BER enzymes (red arrows). Middle: Damage is oriented directly toward the core histone octamer surface, maximally sub-optimal with respect to accessibility of BER factors. Rotational excursions will generally increase accessibilty (green arrows) Right: Rotational orientations in between maximally optimal and sub-optimal such that excursions may increase or decrease activity of factors.
In contrast, for rotational orientations that are not readily accessibly by BER enzymes, or for enzymes for which substrate binding and catalysis are incompatible with nucleosome DNA, repair-related processes can proceed via a second mechanism, wherein the BER factor thermodynamically competes for binding to the DNA with the core histone octamer (Fig. 1 A and B). This mechanism relies on the natural, spontaneous and facile unwrapping of the nucleosome DNA from the histone surface and, the high affinity of the BER factor for the specific damage/repair site. Indeed, Prasad and co-workers have shown that the concentration of hNTH1 normally found in cell nuclei is more than sufficient to compete with the core histones and allow activity. Moreover, a similar mechanism may be employed by enzymes such as Polβ and the DNA ligase IIIα /XRCC1 complex, which do not appear to have robust activity within nucleosome substrates (Fig. 1 A and B, green factor). This is likely due to the more extensive contacts to DNA and greater distortion in the substrate required for these enzymes, which precludes activity without total displacement of the DNA from the histone surface. In addition, ATP-dependent chromatin remodeling enzymes may be important in providing access to inappropriately positioned damage or to enzymes that cannot function in the nucleosome context [42, 43, 49]. Clearly, more work with well-defined model nucleosome substrates and purified BER components needs to be done to elucidate the mechanism(s) by which chromatin remodeling factors enhance repair in chromatin [2].
Prologue.
I first met Mick Smerdon in the mid 1990’s when he and Ray Reeves invited me for a seminar, not long after I began my independent position at Rochester. On the long trip to Pullman I re-read through my large file of Smerdon papers and was reminded of the breadth and quality of Mick’s work, both in vitro and in vivo, from studies of chromatin repair nucleosome dynamics in UV irradiated cells to investigations of how the wrapping of DNA around nucleosomes influenced the formation of specific types of damage. Mick’s exacting quantitative and physical approach in biological systems has provided a seminal roadmap for many young scientists focusing on factors and mechanisms in the DNA damage and repair field. Since then, it has always been a great pleasure to discuss science with Mick, whether in Pullman, where I have visited numerous times, in Rochester, where Mick generously provided the Plenary Lecture at our Epigenetics and Genome Stability retreat, at many meetings, or by phone. In addition, I have especially appreciated Mick’s friendship, valuable advice, and support through the years - and to hope to continue to enjoy these for many years to come!
Highlights.
BER is a mechanism for repair of a variety of small modifications on DNA bases
The initial step in BER involves recognition and excision of a damaged or aberrant base by a specific DNA glycosylase, with remaining steps performed by common enzymes.
While some BER may be coupled to ATP-dependent remodeling in vivo, numerous studies indicate that may of the BER enzymes can process substrates in nucleosomes.
Both orientation of the damaged base with respect to the histone surface and sequence-specific motility of the DNA within the nucleosome dictate accessibility to BER enzymes.
Acknowledgments
This work supported by NIH Grants R01GM052426 (to JJH) and T32 GM068411.
Footnotes
Conflict of interest statement
None
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Odell ID, Wallace SS, Pederson DS. Rules of engagement for base excision repair in chromatin. J Cell Physiol. 2013;228:258–266. doi: 10.1002/jcp.24134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Rodriguez Y, Hinz JM, Smerdon MJ. Accessing DNA damage in chromatin: Preparing the chromatin landscape for base excision repair. DNA Repair (Amst) 2015 doi: 10.1016/j.dnarep.2015.04.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Friedberg EC. Out of the shadows and into the light: the emergence of DNA repair. Trends Biochem Sci. 1995;20:381. doi: 10.1016/s0968-0004(00)89082-9. [DOI] [PubMed] [Google Scholar]
- 4.Hoeijmakers JJ. Genome maintenance mechanisms for preventing cancer. Nature. 2001;411:366–374. doi: 10.1038/35077232. [DOI] [PubMed] [Google Scholar]
- 5.Lindahl T, Karran P, Wood RD. DNA excision repair pathways. Curr Opin Genet Dev. 1997;7:158–169. doi: 10.1016/s0959-437x(97)80124-4. [DOI] [PubMed] [Google Scholar]
- 6.Parikh SS, Mol CD, Hosfield DJ, Tainer JA. Envisioning the molecular choreography of DNA base excision repair. Curr Opin Struct Biol. 1999;9:37–47. doi: 10.1016/s0959-440x(99)80006-2. [DOI] [PubMed] [Google Scholar]
- 7.Memisoglu A, Samson L. Base excision repair in yeast and mammals. Mutat Res. 2000;451:39–51. doi: 10.1016/s0027-5107(00)00039-7. [DOI] [PubMed] [Google Scholar]
- 8.Wallace SS. Base excision repair: a critical player in many games. DNA Repair (Amst) 2014;19:14–26. doi: 10.1016/j.dnarep.2014.03.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Rodriguez Y, Hinz JM, Smerdon MJ. Accessing DNA damage in chromatin: Preparing the chromatin landscape for base excision repair. DNA Repair. doi: 10.1016/j.dnarep.2015.04.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mosbaugh DW, Bennett SE. Uracil-Excision DNA Repair. Progress in Nucleic Acid Research and Molecular Biology. 1994;48:315–370. doi: 10.1016/s0079-6603(08)60859-4. [DOI] [PubMed] [Google Scholar]
- 11.Krokan HE, Standal R, Slupphaug G. DNA glycosylases in the base excision repair of DNA. Biochemistry Journal. 1997;325:1–16. doi: 10.1042/bj3250001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Pearl LH. Structure and function in the uracil-DNA glycosylase superfamily. Mutation Research. 2000;460:165–181. doi: 10.1016/s0921-8777(00)00025-2. [DOI] [PubMed] [Google Scholar]
- 13.Liu P, Burdzy A, Sowers LC. Substrate Recognition by a Family of Uracil-DNA Glycosylases: UNG, MUG, and TDG. Chemical Research in Toxicology. 2002;15:1001–1009. doi: 10.1021/tx020030a. [DOI] [PubMed] [Google Scholar]
- 14.Krokan HE, Nilsen H, Skorpen F, Otterlei M, Slupphaug G. Base excision repair of DNA in mammalian cells. FEBS Lett. 2000;476:73–77. doi: 10.1016/s0014-5793(00)01674-4. [DOI] [PubMed] [Google Scholar]
- 15.Kavli B, Otterlei M, Slupphaug G, Krokan HE. Uracil in DNA--general mutagen, but normal intermediate in acquired immunity. DNA Repair (Amst) 2007;6:505–516. doi: 10.1016/j.dnarep.2006.10.014. [DOI] [PubMed] [Google Scholar]
- 16.Parikh SS, Putnam CD, Tainer JA. Lessons learned from structural results on uracil-DNA glycosylase. Mutat Res. 2000;460:183–199. doi: 10.1016/s0921-8777(00)00026-4. [DOI] [PubMed] [Google Scholar]
- 17.Stivers J, Drohat AC. Uracil DNA Glycosylases: Insights from a Master Catalyst. Archives of Biochemistry and Biophysics. 2001;396:1–9. doi: 10.1006/abbi.2001.2605. [DOI] [PubMed] [Google Scholar]
- 18.van Holde KE. Chromatin. Springer Verlag; New York: 1989. [Google Scholar]
- 19.Luger K, Mader AW, Richmond RK, Sargent DF, Richmond TJ. Crystal structure of the nucleosome core particle at 2.8 A resolution. Nature. 1997;389:251–260. doi: 10.1038/38444. [DOI] [PubMed] [Google Scholar]
- 20.Ura K, Hayes JJ. Nucleotide excision repair and chromatin remodeling. Eur J Biochem. 2002;269:2288–2293. doi: 10.1046/j.1432-1033.2002.02888.x. [DOI] [PubMed] [Google Scholar]
- 21.Golding A, Chandler S, Ballestar E, Wolffe AP, Schlissel MS. Nucleosome structure completely inhibits in vitro cleavage by the V(D)J recombinase. EMBO J. 1999;18:3712–3723. doi: 10.1093/emboj/18.13.3712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Citterio E, Van Den Boom V, Schnitzler G, Kanaar R, Bonte E, Kingston RE, Hoeijmakers JH, Vermeulen W. ATP-dependent chromatin remodeling by the Cockayne syndrome B DNA repair-transcription-coupling factor. Mol Cell Biol. 2000;20:7643–7653. doi: 10.1128/mcb.20.20.7643-7653.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gong F, Fahy D, Liu H, Wang W, Smerdon MJ. Role of the mammalian SWI/SNF chromatin remodeling complex in the cellular response to UV damage. Cell Cycle. 2008;7:1067–1074. doi: 10.4161/cc.7.8.5647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gong F, Fahy D, Smerdon MJ. Rad4-Rad23 interaction with SWI/SNF links ATP-dependent chromatin remodeling with nucleotide excision repair. Nat Struct Mol Biol. 2006;13:902–907. doi: 10.1038/nsmb1152. [DOI] [PubMed] [Google Scholar]
- 25.Peterson CL, Cote J. Cellular machineries for chromosomal DNA repair. Genes Dev. 2004;18:602–616. doi: 10.1101/gad.1182704. [DOI] [PubMed] [Google Scholar]
- 26.Mol CD, Arvai AS, Slupphaug G, Kavli B, Alseth I, Krokan HE, Tainer JA. Crystal structure and mutational analysis of human uracil-DNA glycosylase: structural basis for specificity and catalysis. Cell. 1995;80:869–878. doi: 10.1016/0092-8674(95)90290-2. [DOI] [PubMed] [Google Scholar]
- 27.Savva R, McAuley-Hetch K, Brown T, Pearl L. The structural basis of specific base-excision repair by uracil-DNA glycosylase. Nature. 1995;373:487–493. doi: 10.1038/373487a0. [DOI] [PubMed] [Google Scholar]
- 28.Slupphaug G, Mol CD, Kavli B, Arvai AS, Krokan HE, Tainer JA. A nucleotide-flipping mechanism from the structure of human uracil-DNA glycosylase bound to DNA. Nature. 1996;384:87–92. doi: 10.1038/384087a0. [DOI] [PubMed] [Google Scholar]
- 29.Parikh SS, Mol CD, Slupphaug G, Bharati S, Krokan HE, Tainer JA. Base excision repair initiation revealed by crystal structures and binding kinetics of human uracil-DNA glycosylase with DNA. EMBO J. 1998;17:5214–5226. doi: 10.1093/emboj/17.17.5214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kosaka H, Hoseki J, Nakagawa N, Kuramitsu S, Masui R. Crystal structure of family 5 uracil-DNA glycosylase bound to DNA. J Mol Biol. 2007;373:839–850. doi: 10.1016/j.jmb.2007.08.022. [DOI] [PubMed] [Google Scholar]
- 31.Sung RJ, Zhang M, Qi Y, Verdine GL. Structural and biochemical analysis of DNA helix invasion by the bacterial 8-oxoguanine DNA glycosylase MutM. J Biol Chem. 2013;288:10012–10023. doi: 10.1074/jbc.M112.415612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Nelson SR, Dunn AR, Kathe SD, Warshaw DM, Wallace SS. Two glycosylase families diffusively scan DNA using a wedge residue to probe for and identify oxidatively damaged bases. Proc Natl Acad Sci U S A. 2014;111:E2091–2099. doi: 10.1073/pnas.1400386111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bruner SD, Norman DP, Verdine GL. Structural basis for recognition and repair of the endogenous mutagen 8-oxoguanine in DNA. Nature. 2000;403:859–866. doi: 10.1038/35002510. [DOI] [PubMed] [Google Scholar]
- 34.Nilsen H, Lindahl T, Verreault A. DNA base excision repair of uracil residues in reconstituted nucleosome core particles. The EMBO Journal. 2002;21:5943–5952. doi: 10.1093/emboj/cdf581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Beard BC, Wilson SH, Smerdon MJ. Suppressed catalytic activity of base excision repair enzymes on rotationally positioned uracil in nucleosomes. PNAS. 2003;100:7465–7470. doi: 10.1073/pnas.1330328100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Polach KJ, Widom J. Mechanism of protein access to specific DNA sequences in chromatin: a dynamic equilibrium model for gene regulation. J Mol Biol. 1995;254:130–149. doi: 10.1006/jmbi.1995.0606. [DOI] [PubMed] [Google Scholar]
- 37.Anderson JD, Widom J. Sequence and position-dependence of the equilibrium accessibility of nucleosomal DNA target sites. J Mol Biol. 2000;296:979–987. doi: 10.1006/jmbi.2000.3531. [DOI] [PubMed] [Google Scholar]
- 38.Anderson JD, Widom J. Poly(dA-dT) promoter elements increase the equilibrium accessibility of nucleosomal DNA target sites. Mol Cell Biol. 2001;21:3830–3839. doi: 10.1128/MCB.21.11.3830-3839.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ngo TT, Zhang Q, Zhou R, Yodh JG, Ha T. Asymmetric Unwrapping of Nucleosomes under Tension Directed by DNA Local Flexibility. Cell. 2015;160:1135–1144. doi: 10.1016/j.cell.2015.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Polach KJ, Widom J. A model for the cooperative binding of eukaryotic regulatory proteins to nucleosomal target sites. J Mol Biol. 1996;258:800–812. doi: 10.1006/jmbi.1996.0288. [DOI] [PubMed] [Google Scholar]
- 41.Cole HA, Tabor-Godwin JM, Hayes JJ. Uracil DNA glycosylase activity on nucleosomal DNA depends on rotational orientation of targets. J Biol Chem. 2010;285:2876–2885. doi: 10.1074/jbc.M109.073544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Czaja W, Mao P, Smerdon MJ. Chromatin remodelling complex RSC promotes base excision repair in chromatin of Saccharomyces cerevisiae. DNA Repair (Amst) 2014;16:35–43. doi: 10.1016/j.dnarep.2014.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Czaja W, Mao P, Smerdon MJ. The Emerging Roles of ATP-Dependent Chromatin Remodeling Enzymes in Nucleotide Excision Repair. International journal of molecular sciences. 2012;13:11954–11973. doi: 10.3390/ijms130911954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hinz JM, Rodriguez Y, Smerdon MJ. Rotational dynamics of DNA on the nucleosome surface markedly impact accessibility to a DNA repair enzyme. Proc Natl Acad Sci U S A. 2010;107:4646–4651. doi: 10.1073/pnas.0914443107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Rodriguez Y, Smerdon MJ. The structural location of DNA lesions in nucleosome core particles determines accessibility by base excision repair enzymes. J Biol Chem. 2013;288:13863–13875. doi: 10.1074/jbc.M112.441444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ye Y, Stahley MR, Xu J, Friedman JI, Sun Y, McKnight JN, Gray JJ, Bowman GD, Stivers JT. Enzymatic excision of uracil residues in nucleosomes depends on the local DNA structure and dynamics. Biochemistry. 2012;51:6028–6038. doi: 10.1021/bi3006412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Makde RD, England JR, Yennawar HP, Tan S. Structure of RCC1 chromatin factor bound to the nucleosome core particle. Nature. 2010;467:562–566. doi: 10.1038/nature09321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ishibashi T, So K, Cupples CG, Ausio J. MBD4-mediated glycosylase activity on a chromatin template is enhanced by acetylation. Mol Cell Biol. 2008;28:4734–4744. doi: 10.1128/MCB.00588-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Menoni H, Gasparutto D, Hamiche A, Cadet J, Dimitrov S, Bouvet P, Angelov D. ATP-dependent chromatin remodeling is required for base excision repair in conventional but not in variant H2A.Bbd nucleosomes. Mol Cell Biol. 2007;27:5949–5956. doi: 10.1128/MCB.00376-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Prasad A, Wallace SS, Pederson DS. Initiation of base excision repair of oxidative lesions in nucleosomes by the human, bifunctional DNA glycosylase NTH1. Mol Cell Biol. 2007;27:8442–8453. doi: 10.1128/MCB.00791-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Maher RL, Prasad A, Rizvanova O, Wallace SS, Pederson DS. Contribution of DNA unwrapping from histone octamers to the repair of oxidatively damaged DNA in nucleosomes. DNA Repair (Amst) 2013;12:964–971. doi: 10.1016/j.dnarep.2013.08.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Mol CD, Izumi T, Mitra S, Tainer JA. DNA-bound structures and mutants reveal abasic DNA binding by APE1 and DNA repair coordination [corrected] Nature. 2000;403:451–456. doi: 10.1038/35000249. [DOI] [PubMed] [Google Scholar]
- 53.Odell ID, Barbour JE, Murphy DL, Della-Maria JA, Sweasy JB, Tomkinson AE, Wallace SS, Pederson DS. Nucleosome disruption by DNA ligase III-XRCC1 promotes efficient base excision repair. Mol Cell Biol. 2011;31:4623–4632. doi: 10.1128/MCB.05715-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Bambara RA, Murante RS, Henricksen LA. Enzymes and reactions at the eukaryotic DNA replication fork. J Biol Chem. 1997;272:4647–4650. doi: 10.1074/jbc.272.8.4647. [DOI] [PubMed] [Google Scholar]
- 55.Hosfield DJ, Mol CD, Shen B, Tainer JA. Structure of the DNA repair and replication endonuclease and exonuclease FEN-1: coupling DNA and PCNA binding to FEN-1 activity. Cell. 1998;95:135–146. doi: 10.1016/s0092-8674(00)81789-4. [DOI] [PubMed] [Google Scholar]
- 56.Robins P, Pappin DJ, Wood RD, Lindahl T. Structural and functional homology between mammalian DNase IV and the 5′-nuclease domain of Escherichia coli DNA polymerase I. J Biol Chem. 1994;269:28535–28538. [PubMed] [Google Scholar]
- 57.Murante RS, Rust L, Bambara RA. Calf 5′ to 3′ exo/endonuclease must slide from a 5′ end of the substrate to perform structure-specific cleavage. J Biol Chem. 1995;270:30377–30383. doi: 10.1074/jbc.270.51.30377. [DOI] [PubMed] [Google Scholar]
- 58.Barnes CJ, Wahl AF, Shen B, Park MS, Bambara RA. Mechanism of tracking and cleavage of adduct-damaged DNA substrates by the mammalian 5′- to 3′-exonuclease/endonuclease RAD2 homologue 1 or flap endonuclease 1. J Biol Chem. 1996;271:29624–29631. doi: 10.1074/jbc.271.47.29624. [DOI] [PubMed] [Google Scholar]
- 59.Bornarth CJ, Ranalli TA, Henricksen LA, Wahl AF, Bambara RA. Effect of flap modifications on human FEN1 cleavage. Biochemistry. 1999;38:13347–13354. doi: 10.1021/bi991321u. [DOI] [PubMed] [Google Scholar]
- 60.Harrington JJ, Lieber MR. DNA structural elements required for FEN-1 binding. J Biol Chem. 1995;270:4503–4508. doi: 10.1074/jbc.270.9.4503. [DOI] [PubMed] [Google Scholar]
- 61.Kao HI, Henricksen LA, Liu Y, Bambara RA. Cleavage specificity of Saccharomyces cerevisiae flap endonuclease 1 suggests a double-flap structure as the cellular substrate. J Biol Chem. 2002;277:14379–14389. doi: 10.1074/jbc.M110662200. [DOI] [PubMed] [Google Scholar]
- 62.Warbrick E, Lane DP, Glover DM, Cox LS. Homologous regions of Fen1 and p21Cip1 compete for binding to the same site on PCNA: a potential mechanism to co-ordinate DNA replication and repair. Oncogene. 1997;14:2313–2321. doi: 10.1038/sj.onc.1201072. [DOI] [PubMed] [Google Scholar]
- 63.Huggins CF, Chafin DR, Aoyagi S, Henricksen LA, Bambara RA, Hayes JJ. Flap Endonuclease 1 Efficiently Cleaves Base Excision Repair and DNA Replication Intermediates Assembled into Nucleosomes. Mol Cell. 2002;10:1201–1211. doi: 10.1016/s1097-2765(02)00736-0. [DOI] [PubMed] [Google Scholar]
- 64.Jagannathan I, Pepenella S, Hayes JJ. Activity of FEN1 endonuclease on nucleosome substrates is dependent upon DNA sequence but not flap orientation. J Biol Chem. 2011;286:17521–17529. doi: 10.1074/jbc.M111.229658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Thastrom A, Lowary PT, Widlund HR, Cao H, Kubista M, Widom J. Sequence motifs and free energies of selected natural and non-natural nucleosome positioning DNA sequences. J Mol Biol. 1999;288:213–229. doi: 10.1006/jmbi.1999.2686. [DOI] [PubMed] [Google Scholar]
- 66.Sawaya MR, Prasad R, Wilson SH, Kraut J, Pelletier H. Crystal structures of human DNA polymerase beta complexed with gapped and nicked DNA: evidence for an induced fit mechanism. Biochemistry. 1997;36:11205–11215. doi: 10.1021/bi9703812. [DOI] [PubMed] [Google Scholar]
- 67.Matsumoto Y, Kim K, Hurwitz J, Gary R, Levin DS, Tomkinson AE, Park MS. Reconstitution of proliferating cell nuclear antigen-dependent repair of apurinic/apyrimidinic sites with purified human proteins. J Biol Chem. 1999;274:33703–33708. doi: 10.1074/jbc.274.47.33703. [DOI] [PubMed] [Google Scholar]
- 68.Srivastava DK, Berg BJ, Prasad R, Molina JT, Beard WA, Tomkinson AE, Wilson SH. Mammalian abasic site base excision repair. Identification of the reaction sequence and rate-determining steps. J Biol Chem. 1998;273:21203–21209. doi: 10.1074/jbc.273.33.21203. [DOI] [PubMed] [Google Scholar]
- 69.Matsumoto Y, Kim K. Excision of deoxyribose phosphate residues by DNA polymerase beta during DNA repair. Science. 1995;269:699–702. doi: 10.1126/science.7624801. [DOI] [PubMed] [Google Scholar]
- 70.Prasad R, Beard WA, Wilson SH. Studies of gapped DNA substrate binding by mammalian DNA polymerase beta. Dependence on 5′-phosphate group. J Biol Chem. 1994;269:18096–18101. [PubMed] [Google Scholar]
- 71.Singhal RK, Wilson SH. Short gap-filling synthesis by DNA polymerase beta is processive. J Biol Chem. 1993;268:15906–15911. [PubMed] [Google Scholar]
- 72.Mol CD, Hosfield DJ, Tainer JA. Abasic site recognition by two apurinic/apyrimidinic endonuclease families in DNA base excision repair: the 3′ ends justify the means. Mutat Res. 2000;460:211–229. doi: 10.1016/s0921-8777(00)00028-8. [DOI] [PubMed] [Google Scholar]
- 73.Dianov GL, Prasad R, Wilson SH, Bohr VA. Role of DNA polymerase beta in the excision step of long patch mammalian base excision repair. J Biol Chem. 1999;274:13741–13743. doi: 10.1074/jbc.274.20.13741. [DOI] [PubMed] [Google Scholar]
- 74.Pascal JM, O’Brien PJ, Tomkinson AE, Ellenberger T. Human DNA ligase I completely encircles and partially unwinds nicked DNA. Nature. 2004;432:473–478. doi: 10.1038/nature03082. [DOI] [PubMed] [Google Scholar]
- 75.Sleeth KM, Robson RL, Dianov GL. Exchangeability of mammalian DNA ligases between base excision repair pathways. Biochemistry. 2004;43:12924–12930. doi: 10.1021/bi0492612. [DOI] [PubMed] [Google Scholar]
- 76.Tomkinson AE, Lasko DD, Daly G, Lindahl T. Mammalian DNA ligases. Catalytic domain and size of DNA ligase I. J Biol Chem. 1990;265:12611–12617. [PubMed] [Google Scholar]
- 77.Tom S, Henricksen LA, Park MS, Bambara RA. DNA ligase I and proliferating cell nuclear antigen form a functional complex. J Biol Chem. 2001;276:24817–24825. doi: 10.1074/jbc.M101673200. [DOI] [PubMed] [Google Scholar]
- 78.DeMott MS, Zigman S, Bambara RA. Replication protein A stimulates long patch DNA base excision repair. J Biol Chem. 1998;273:27492–27498. doi: 10.1074/jbc.273.42.27492. [DOI] [PubMed] [Google Scholar]
- 79.Chafin DR, Vitolo JM, Henricksen LA, Bambara RA, Hayes JJ. Human DNA ligase I efficiently seals nicks in nucleosomes. EMBO J. 2000;19:5492–5501. doi: 10.1093/emboj/19.20.5492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Angelov D, Vitolo JM, Mutskov V, Dimitrov S, Hayes JJ. Preferential interaction of the core histone tail domains with linker DNA. Proc Natl Acad Sci USA. 2001;98:6599–6604. doi: 10.1073/pnas.121171498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Taylor RM, Whitehouse CJ, Caldecott KW. The DNA ligase III zinc finger stimulates binding to DNA secondary structure and promotes end joining. Nucleic Acids Res. 2000;28:3558–3563. doi: 10.1093/nar/28.18.3558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Taylor RM, Moore DJ, Whitehouse J, Johnson P, Caldecott KW. A cell cycle-specific requirement for the XRCC1 BRCT II domain during mammalian DNA strand break repair. Mol Cell Biol. 2000;20:735–740. doi: 10.1128/mcb.20.2.735-740.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Caldecott KW, McKeown CK, Tucker JD, Ljungquist S, Thompson LH. An interaction between the mammalian DNA repair protein XRCC1 and DNA ligase III. Mol Cell Biol. 1994;14:68–76. doi: 10.1128/mcb.14.1.68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Caldecott KW, Tucker JD, Stanker LH, Thompson LH. Characterization of the XRCC1-DNA ligase III complex in vitro and its absence from mutant hamster cells. Nucleic Acids Res. 1995;23:4836–4843. doi: 10.1093/nar/23.23.4836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Masson M, Niedergang C, Schreiber V, Muller S, Menissier-de Murcia J, de Murcia G. XRCC1 is specifically associated with poly(ADP-ribose) polymerase and negatively regulates its activity following DNA damage. Mol Cell Biol. 1998;18:3563–3571. doi: 10.1128/mcb.18.6.3563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Leppard JB, Dong Z, Mackey ZB, Tomkinson AE. Physical and functional interaction between DNA ligase IIIalpha and poly(ADP-Ribose) polymerase 1 in DNA single-strand break repair. Mol Cell Biol. 2003;23:5919–5927. doi: 10.1128/MCB.23.16.5919-5927.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Schreiber V, Ame JC, Dolle P, Schultz I, Rinaldi B, Fraulob V, Menissier-de Murcia J, de Murcia G. Poly(ADP-ribose) polymerase-2 (PARP-2) is required for efficient base excision DNA repair in association with PARP-1 and XRCC1. J Biol Chem. 2002;277:23028–23036. doi: 10.1074/jbc.M202390200. [DOI] [PubMed] [Google Scholar]