Abstract
Objective
To assess the association between the single-nucleotide polymorphism (SNP) rs58542926 in the transmembrane 6 superfamily member 2 (TM6SF2) gene and fatty liver disease in obese youth.
Research and Methods
We genotyped the TM6SF2 rs58542926 SNP in a multiethnic cohort of 957 obese children and adolescents (42% Caucasians, 28% African Americans, 30% Hispanics). All underwent an oral glucose tolerance test, a liver panel and a lipid profile. Of them, 454 children underwent a MRI study to assess hepatic fat content (HFF%), 11 children underwent liver biopsy to assess the degree of disease severity.
Results
The minor allele of the rs58542926 SNP was associated with high HFF% in Caucasians and African Americans (all P<0.05), with high ALT levels in Hispanics (P<0.05) and with a more favorable lipoprotein profile (lower LDL, small dense LDL and very small LDL) in Caucasians and Hispanics (all P<0.05). The liver biopsy showed a higher prevalence of fibrosis (P=0.04) and a higher NAFLD Activity Score (P=0.05) in subjects carrying the minor allele than in those homozygous for the common allele. Moreover, we observed a joint effect among the TM6SF2 rs58542926, the PNPLA3 rs738409 and the GCKR rs1260326 SNPs in determining intrahepatic fat accumulation (P<0.05).
Conclusions
The rs58542926 SNP in the TM6SF2 gene is associated with pediatric NAFLD, but may confer protection against cardiovascular risk.
Keywords: obesity, youth, GCKR, PNPLA3, LDL
Non-alcoholic fatty liver disease (NAFLD) is the most common form of chronic liver disease in pediatrics affecting between 3% and 11% of the pediatric population reaching the rate of 46% among overweight and obese children and adolescents (1).
Recently, an exome-wide association study in a multi-ancestry adult population has identified a non-synonymous single nucleotide polymorphism (SNP) in the Transmembrane 6 Superfamily Member 2 (TM6SF2) gene, to be associated with fatty liver disease, but a clinically favorable lipid profile (low LDL and cholesterol levels) (2, 3). This variant is characterized by a C-to-T substitution in the nucleotide 499, encoding a glutamate with lysine change at the codon 167 (E167K) (2). In vitro studies have shown that the E167K variant form is misfolded and undergoes accelerated intracellular degradation (2), causing an increased intra hepatic fat accumulation and a defect in VLDL secretion (2). Subsequent clinical studies have shown that the molecular defect translates into 1) a higher risk of the progression to NAFLD/NASH (4), and into 2) a lower incidence of cardiovascular events due to the low levels of circulating LDL (5).
Very little is known about the potential effect of the TM6SF2 rs58542926 variant in the pathogenesis of fatty liver in US obese youth. Therefore, in this study, we aimed at determining the role of TM6SF2 rs58542926 in the development of NAFLD in a multiethnic group of obese youth and at verifying whether there is a joint effect on the fatty liver phenotype of the TM6SF2 rs58542926, PNPLA3 rs738409 and GCKR rs1260326 variants, the latter two previously shown to predispose to fatty liver (6, 7); and (4) to explore the effect of the rs58542926 variant on plasma lipoproteins subclasses.
Materials and Methods
We studied 957 obese children and adolescents (42% Caucasians, 28% African Americans, and 30% Hispanics), recruited from the Yale Pediatric Obesity Clinic (Table 1). Our patients were followed by our clinical staff, who monitored glucose, liver function and lipid metabolism and gave standard nutrition counseling and physical activity recommendations. A fast gradient magnetic resonance imaging to assess hepatic fat content (HFF%) and abdominal fat distribution was performed in the subjects who gave their consent to participate to the imaging study. To be eligible for this study, subjects could not be on medications known to affect liver function or alter glucose or lipid metabolism. Information relating to alcohol consumption was obtained in all subjects using our clinical lifestyle questionnaire. Autoimmune hepatitis, Wilson disease, alpha-1-antitrypsin deficiency, hepatitis B and C, and iron overload were excluded with appropriate tests in subjects with persistent elevation in alanine aminotransferase (>6 months). Written informed consent was obtained from the parents and assent from the children and adolescents. The study was approved by the Yale University Human Investigation Committee.
Table 1. Anthropometric characteristics of the study population stratified by ethnicity and TM6SF2 Genotype.
| Total Population | Caucasians | African Americans | Hispanics | |||||||
|---|---|---|---|---|---|---|---|---|---|---|
| N=957 | CC (n=353) | CT (n=49) | P | CC (n= 249) | CT/TT (n=17) | P | CC (n=263) | CT/TT (n=26) | P | |
| Age (years) | 13.338 ± 3.43 | 13.796 ± 3.65 | 13.657 ± 2.68 | 0.797 | 13.353 ± 3.43 | 13.235 ± 3.57 | 0.891 | 12.776 ± 3.701 | 13.342 ± 2.857 | 0.447 |
| Gender (M/F) % | 40/60 | 39/61 | 37/63 | 0.445 | 39/61 | 41/59 | 0.51 | 42/58 | 50/50 | 0.287 |
| Glucose tolerance (NGT/IGT/T2D) % | 73/22/5 | 74/22/4 | 67/27/6 | 0.548 | 70/22/8 | 88/12/0 | 0.39 | 76/20/4 | 61/31/8 | 0.285 |
| BMI (kg/m2) | 32.959 ± 7.601 | 32.744 ± 7.40 | 32.35 ± 8.03 | 0.733 | 34.417 ± 7.97 | 34.199 ± 8.44 | 0.914 | 31.872 ± 7.12 | 31.787 ± 6.97 | 0.954 |
| Z-score BMI | 2.158 ± 0.69 | 2.126 ± 0.66 | 1.954 ± 0.89 | 0.117 | 2.284 ± 0.56 | 2.203 ± 0.88 | 0.595 | 2.124 ± 0.77 | 2.058 ± 0.70 | 0.683 |
| Body Fat (%) | 42.397 ± 10.21 | 42.365 ± 10.07 | 39.198 ± 10.03 | 0.048 | 44.137 ± 10.43 | 44.421 ± 9.91 | 0.921 | 41.737 ± 9.9 | 40.044 ± 10.43 | 0.419 |
| Metabolic Syndrome % | 61/39 | 67/33 | 70/30 | 0.412 | 76/24 | 60/40 | 0.153 | 23/77 | 33/67 | 0.196 |
| Body Fat Composition | ||||||||||
| N=454 | CC (n=155) | CT (n=28) | P | CC (n=116) | CT/TT (n=5) | P | CC (n=134) | CT (n=16) | P | |
| Visceral (cm2) | 53.65 ± 28.53 | 57.414 ± 28.71 | 53.454 ± 30.75 | 0.504 | 43.55 ± 24.16 | 56.62 ± 24.17 | 0.552 | 57.706 ± 29.39 | 56.963 ± 29.62 | 0.727 |
| Deep Subcutaneous (cm2) | 151.35 ± 75.84 | 148.01 ± 72.99 | 131.63 ± 78.01 | 0.057 | 155.808 ±83.25 | 179.98 ± 64.37 | 0.839 | 157.37 ±76.32 | 144.731 ±78.62 | 0.950 |
| Subcutaneous (cm2) | 455.39 ±214.99 | 456.16±206.32 | 389.725±198.87 | 0.003 | 472.804±231.9 | 600.48 ± 170.10 | 0.36 | 452.789±209.3 | 434.431±250.1 | 0.953 |
| Superficial Subcutaneous (cm2) | 129.146 ± 65.19 | 130.369±61.55 | 109.725 ± 57.11 | 0.003 | 141.896±70.84 | 186.88 ± 58.00 | 0.325 | 121.779±63.91 | 111.475±63.64 | 0.699 |
| Deep/Superficial Subcutaneous | 1.233 ± 0.525 | 1.187 ± 0.47 | 1.238 ± 0.51 | 0.802 | 1.144 ± 0.55 | 1.011 ± 0.44 | 0.51 | 1.371 ± 0.56 | 1.348 ± 0.50 | 0.806 |
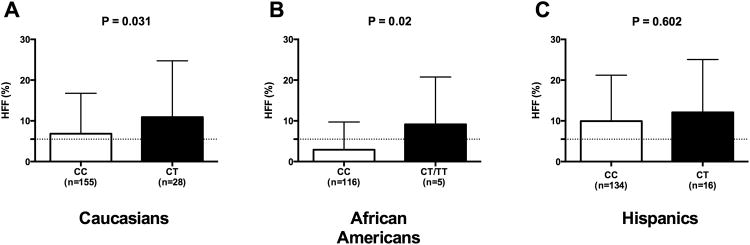
| Hepatic Fat Content (%) | 7.17 ± 10.43 | 6.679 ± 9.71 | 10.911 ± 13.83 | 0.031 | 2.9 ± 6.811 | 9.14 ± 11.62 | 0.02 | 9.931 ± 11.27 | 12.081 ± 12.98 | 0.602 |
Genotyping
Genomic DNA was extracted from peripheral blood leukocytes. To genotype the rs58542926 SNP, the following pair of primers was used: forward = 5′-AAA GTT CAG GCA CAT TGG GAC AAG GC-3′ and reverse = 5′-AGC CAA GAT TGC ATC ACT GCA C-3′. Polymerase chain reaction (PCR) was carried out using the annealing temperature of 65.0°C. PCR products were analyzed by automated sequencing through the Yale W.M. Keck facility.
Metabolic Studies
All metabolic studies were done at the Yale Clinical Center Investigation at 8:00 AM following a 10-hour to 12-hour overnight fast. Subjects were studied at the Hospital Research Unit (HRU) of the Yale New Haven Hospital.
Oral Glucose Tolerance Test
A standard OGTT (1.75 g/kg body weight, up to 75 g) was conducted to determine glucose tolerance (8). Blood samples for determination of glucose, insulin and C-peptide were drawn at −15, 0, 30, 60, 90, 120 and 180 min. The Matsuda index was used to calculate insulin sensitivity (9) (whole-body insulin sensitivity index [WBISI]).
Imaging Studies
Abdominal Magnetic Resonance Imaging
Magnetic resonance imaging (MRI) studies were performed on a GE or Siemens Sonata 1.5 Tesla system (10). The measurement of the liver fat content was performed by MRI using the two-point Dixon (2PD) method as modified by Fishbein et al. Using the MRIcro software program, five regions of interest were drawn on each image and the mean pixel signal intensity level was recorded (11). The HFF was calculated in duplicate from the mean pixel signal intensity data using the formula: [(Sin − Sout)/(2 × Sin)] × 100. The imaging parameters were: matrix size = 128 × 256, flip angle (α) = 30°, TR = 18 ms, TEs = 2.38/4.76 ms out-of-phase and in-phase, respectively, bandwidth = 420 Hz/pixel, six averages, slice thickness = 10 mm, one slice, 2.3 seconds/slice (for 2 points), scan time = 14 seconds in a single breath-hold (11).
Liver Biopsy
Liver Biopsy was performed in 11 subjects (5F) because of persistent elevation in Alanine Aminotransferase (ALT) (mean ALT 130; 95%CI 80-213). Biopsies were formalin-fixed, paraffin embedded, stained with Hematoxyllin & Eosin, trichrome and Gordon's reticulin techniques. All biopsies were 2 cm or more in length and were reviewed by a pediatric pathologist according to the Brunt approach. In detail, steatosis was assessed as the percentage of hepatocytes involved within a lobule (0%–100%, steatosis score) and by using a 4-grade classification modified from Kleiner et al (12): 0, absent; 1, <5%; 2, 5%–33%; 3, 33%–66%; 4, >66%. Staging and grading were performed according to Brunt et al (13). The NAFLD activity score (NAS) was calculated according to Kleiner et al (12). The score is defined as the sum of the scores for steatosis (0, <5%; 1, 5%–33%; 2, 33%–66%; 3, >66%), lobular inflammation (0, none; 1, <2 foci/×200 magnification field; 2, 2–4 foci/×200 magnification field; 3, >4 foci/×200 magnification field), and ballooning (0, none; 1, few; 2, many).
Biochemical Analyses
Plasma glucose was determined using a glucose analyzer by the glucose oxidase method (Beckman Instruments, Brea, CA). Plasma insulin was measured by the Linco RIA, lipid levels were determined with an Auto-Analyzer (model 747-200), and liver enzymes were measured, using standard automated kinetic enzymatic assays. All assays were performed in duplicate, and the absorbance was determined using a microplate reader (molecular Devices M2, Sunnyvale, CA).
Statistical Analyses
The chi-square test was used to assess whether the genotypes were in Hardy Weinberg equilibrium and to test differences in genotype distribution among different ethnic groups. Prior to data analysis, all variables were tested for normality, with non-normally distributed variables log transformed to be better approximated by normality, except for HFF% for which a square root transformation was used. Within each ethnic group, the association between the genotypes and quantitative traits was evaluated by coding the genotype with an additive model of inheritance, i.e. the genotype is coded with 0, 1, or 2 corresponding to the number of minor alleles carried by each individual; age, sex, and BMI z-score were used as covariates when appropriate. To assess whether the effect of the genotype on the degree of liver injury and on lipoprotein profile might be influenced by the presence of the intra-hepatic fat content, we divided the subjects who underwent a fast-MRI in two groups according to the degree of liver fat (<5.5% and >5.5%) and assessed the interaction between the rs58542926 and NAFLD in modulating the levels of ALT, triglycerides, HDL, LDL and total cholesterol in each ethnic group.
The genetic score among the rs58542926 in the TM6SF2, the rs738409 in the PNPLA3 and the rs1260326 in the GCKR was calculated by risk allele counting. In each ethnic group, the association between the genetic score and the main outcome (HFF%) was assessed by a general linear model using an additive model, and age, sex, and BMI z-score were used as covariates. Data are shown as mean and standard deviation.
Results
The minor allele (T) frequency was 0.061 in Caucasians, 0.033 in African Americans and 0.089 in Hispanics; these frequencies were similar to those observed previously (2). The genotype distribution was in Hardy Weinberg equilibrium in all the ethnic groups (all P>0.05).
In all ethnicity the genotype groups had similar age, gender, BMI and glucose tolerance (all P>0.05) (Table 1). Moreover, the two groups showed similar fasting glucose, fasting insulin, HbA1c and WBISI (all P>0.05) (Table 2).
Table 2. Biochemical characteristics of the study population stratified by ethnicity and TM6SF2 Genotype.
| Total Population | Caucasians | African Americans | Hispanics | |||||||
|---|---|---|---|---|---|---|---|---|---|---|
| N=957 | CC (n=353) | CT (n=49) | P | CC (n= 249) | CT/TT (n=17) | P | CC (n=263) | CT/TT (n=26) | P | |
| Glucose Metabolism | ||||||||||
| Fasting glucose (mg/dL) | 90.000 ± 7.66 | 90.1 ± 8.113 | 87.89 ± 6.571 | 0.132 | 89.34 ± 7.53 | 93.44 ± 4.989 | 0.15 | 90.41 ± 6.84 | 92.94 ± 8.4 | 0.18 |
| Fasting insulin (mcU/ml) | 33.300 ± 22.24 | 32.88 ± 18.476 | 29 ± 20.797 | 0.198 | 32.97 ± 22.498 | 48.94 ± 26.135 | 0.098 | 32.493±25.624 | 38.059 ±19.165 | 0.159 |
| 2h glucose (mg/dL) | 123.960 ± 30.79 | 124.06 ±33.282 | 130.42 ± 33.918 | 0.327 | 124.58 ±32.883 | 121.82 ± 23.699 | 0.901 | 121.49 ± 25.45 | 133.24 ±31.053 | 0.029 |
| HbA1C (%) | 5.477 ± 0.35 | 5.41 ± 0.338 | 5.427 ± 0.303 | 0.625 | 5.59 ± 0.385 | 5.721 ± 0.503 | 0.222 | 5.462 ± 0.321 | 5.529 ± 0.297 | 0.368 |
| WBISI | 1.882 ± 1.19 | 1.909 ± 1.176 | 2.209 ± 1.448 | 0.246 | 1.845 ± 1.274 | 2.012 ± 1.247 | 0.703 | 1.821 ± 1.097 | 1.798 ± 1.151 | 0.973 |
| Lipid Profile | ||||||||||
| Cholesterol (mg/dL) | 155.190 ± 33.10 | 159.07 ±35.098 | 145.13 ± 32.043 | 0.013 | 153.72 ±26.132 | 144.47 ± 28.392 | 0.162 | 155.29±36.071 | 134.96 ±32.357 | 0.002 |
| HDL (mg/dL) | 43.750 ± 11.064 | 43.79 ± 11.99 | 42.96 ± 8.844 | 0.598 | 46.06 ± 10.672 | 42.13 ± 11.25 | 0.09 | 41.91 ± 10.043 | 39.08 ± 8.351 | 0.185 |
| LDL (mg/dL) | 90.440 ± 27.62 | 92.03 ± 27.87 | 80.98 ± 28.848 | 0.009 | 91.37 ± 22.553 | 88 ± 25.844 | 0.526 | 91.26 ± 31.242 | 71.21 ± 25.524 | <0.001 |
| Triglyceride (mg/dL) | 106.230 ± 68.74 | 120.17 ± 78.99 | 109.96 ±72.468 | 0.286 | 80.1 ± 52.456 | 71.87 ± 33.73 | 0.532 | 110.5 ± 58.204 | 123.42 ±91.826 | 0.749 |
| VLDL (mg/dL) | 49.461 ± 24.63 | 54.826 ±23.985 | 51.472 ± 26.138 | 0.329 | 37.822 ±21.996 | 29.991 ± 19.568 | 0.172 | 55.397±23.833 | 34.202 ±22.385 | <0.001 |
| Liver Function | ||||||||||
| ALT (U/L) | 25.500 ± 27.41 | 26.27 ± 24.111 | 35.981 ± 58.981 | 0.094 | 16.89 ± 8.653 | 14.21 ± 5.673 | 0.374 | 26.37 ± 21.491 | 47.14 ± 60.113 | 0.029 |
| AST (U/L) | 24.500 ± 15.06 | 24.18 ± 11.625 | 29.36 ± 35.963 | 0.195 | 21.43 ± 6.294 | 20.79 ± 5.74 | 0.749 | 24.71 ± 9.363 | 38.45 ± 42.022 | 0.009 |
| Alkaline Phospatase (U/L) | 183.900 ± 104.61 | 165.69 ±95.947 | 151.45 ± 83.093 | 0.168 | 197.74 ±111.24 | 144.5 ± 92.47 | 0.022 | 202.77±105.286 | 199.9 ±117.505 | 0.761 |
Association of the TM6SF2 rs58542926 with hepatic fat content and liver injury
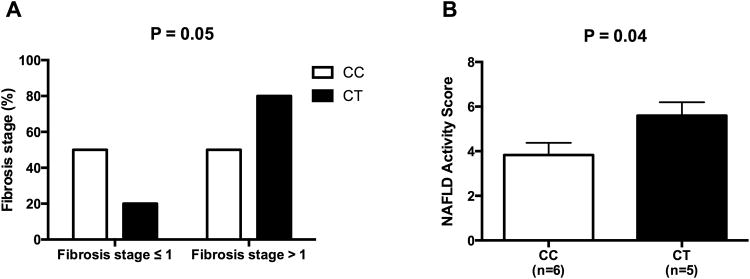
The data in according to the genotype in the whole population are shown in the supplemental tables 1 and 2. Subjects carrying the minor allele of the rs58542926 SNP showed a higher hepatic fat content in Caucasian (P=0.02) and in African American patients (P=0.02), although not statistically significant, a similar trend was observed in Hispanics (P=0.602). (Figure 1). The ALT and AST levels were higher in subjects carrying the T allele in Hispanics (P=0.029 and P=0.009, respectively) and a similar trend was seen in Caucasians (P=0.094 and P=0.195, respectively) (Table 1). In the subgroup of patients who underwent a liver biopsy, 6 carried the CC genotype and 5 the CT genotype. Fibrosis stage > 1 was present in 20% of the CC and 80% of the CT (P=0.05); (Figure 2). Moreover, the two groups of genotype differed for the NAFLD activity score, with the CT genotype showing a significantly higher NAS compared to the CC patients (P=0.04).
Figure 1. Association of the TM6SF2 rs58542926 with hepatic fat content.
Association between TM6SF2 rs58542926 SNP and hepatic fat content (%HFF) in (A) Caucasians, (B) African Americans, and (C) Hispanics. White bars represent the CC and black bars represent CT/TT. P values are adjusted for age, sex, and BMI. Data are shown as mean with SD.
Figure 2. Association between TM6SF2 rs58542926 SNP and liver histology.
The TM6SF2 rs58542926 minor allele is associated with liver fibrosis (panel A) and NAFLD Activity score (NAS) (panel B). Data of NAS are shown as mean with SD.
We did not observe any interaction between the rs58542926 SNP and NAFLD in modulating the levels of ALT, total cholesterol, LDL, HDL and triglycerides (all P>0.05). The associations between the rs58542926 SNP and these outcomes according to the presence of NAFLD and the interactions between the rs58542926 SNP and NAFLD are shown in the supplemental tables 3A, 3B, 3C and 4.
Joint effect of PNPLA3 and GCKR on increased HFF and ALT
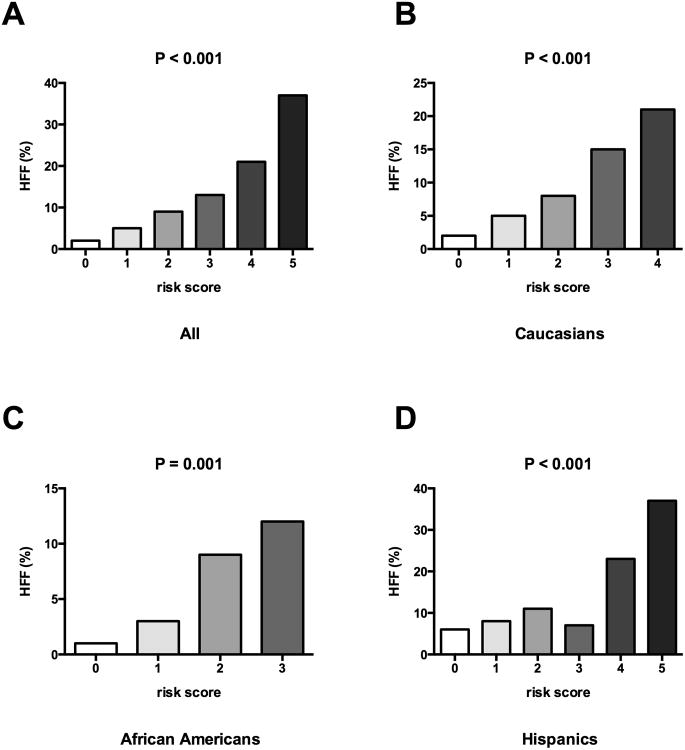
Because PNPLA3 rs738409 and GCKR rs1260326 were previously associated with pediatric NAFLD (14-16), we evaluated whether there was a joint effect between the rs58542926 in the TM6SF2 and the variants in the PNPLA3 and in the GCKR. In all ethnic groups we observed that HFF % increased as the risk score was higher independent of age, gender and BMI z-score (P<0.001) (Figure 3).
Figure 3. Joint effect of PNPLA3 and GCKR on increased HFF.
Association between the genetic risk score calculated adding the risk alleles of rs58542926 in the TM6SF2, the rs738409 in the PNPLA3 and the rs1260326 in the GCKR and the hepatic fat content (%HFF in the overall population (A), in Caucasians (B), in African Americans (C), in Hispanics (D). P values adjusted for age, sex, BMI. Data are shown using the mean % within each group.
Plasma Lipoproteins: effect of rs58542926 TM6SF2 gene variant
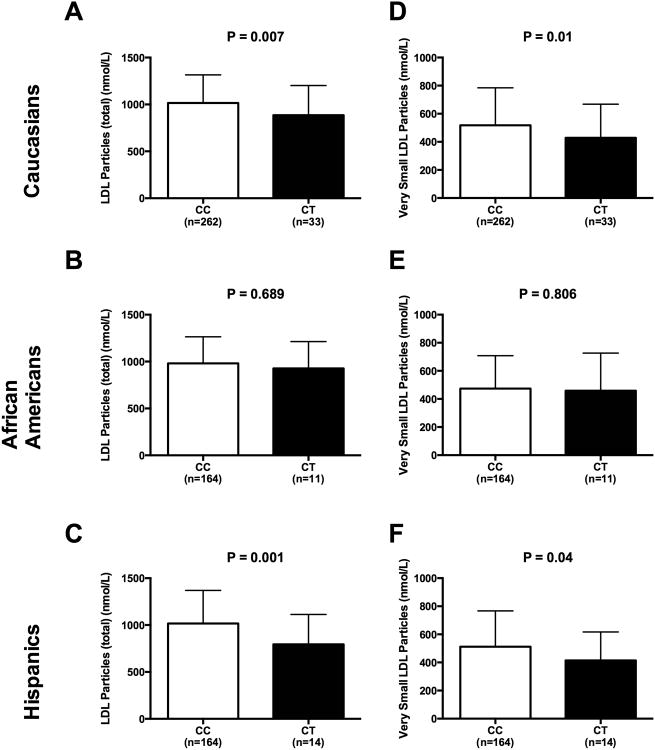
In the group of Caucasians, subjects carrying the T allele showed lower cholesterol levels (P=0.013), LDL particles (P=0.007), large LDL particles (P=0.819), small LDL particles (P=0.022), medium small LDL particles (P=0.057), and very small LDL particles (P=0.021), than C homozygous. Also in the Hispanic group, compared to the CC, the T allele carriers showed significantly lower levels of total cholesterol (P=0.002), VLDL (P<0.001), and LDL total particles (P=0.001), large LDL particles (P=0.017), small LDL particles (P=0.024), medium small LDL particles (P=0.005), very small LDL particles (P=0.044) and triglycerides (P=0.002) (Figure 4). We did not observe any differences in African American population for what concerns the lipoprotein subclasses.
Figure 4. Plasma Lipoproteins: effect of rs58542926 TM6SF2 gene variant.
Association in Caucasians, African Americans and Hispanics between TM6SF2 rs58542926 SNP and total LDL Particles (panel A-C) and Very Small LDL Particles (panel D-F). White bars indicate the CC and black bars indicate the T allele carriers. P values are adjusted for age, sex, and BMI. Data are shown as mean with SD.
Discussion
In this study, we found that obese children and adolescents carrying the minor allele for rs58542926 SNP have higher risk of showing hepatic steatosis and marked liver damage. We also calculated for the first time a genetic risk score for fatty liver by using three SNPs found associated with NAFLD in our population (PNPLA3 rs738409, GCKR rs1260326, TM6SF2 rs58542926) and observed that the higher the risk score, the higher the amount of intra-hepatic fat. Moreover, the carriers for the risk allele for the rs58542926 SNP showed a favorable lipid profile with significantly lower total cholesterol and LDL levels.
It is interesting to note that we did not observe an association between the TM6SF2 variant and the intra-hepatic fat in Hispanics, while the same variant in this group showed a strong association with ALT levels. In fact, although we observed a trend toward a higher hepatic fat content in subjects homozygous for the minor allele in Hispanics, this was far from being statistically significant (P=0.602). These data are consistent with what shown by Kozlitina et al. in the Dallas Heart Study (2) and can be explained by the fact that Hispanics show the highest prevalence of NAFLD among the studied population, meaning that to observe an association between the TM6SF2 SNP, which is a low frequency variant, and the phenotype a much larger sample size might be needed. Moreover, the fact that the association between this SNP and the ALT levels was strong in both studies independent from the degree of intra-hepatic fat content might suggests that in the Hispanic population the effect of the TM6SF2 variant could play a major role in liver injury more than in intra-hepatic fat accumulation.
Furthermore, we compared the results of our Caucasian children (n=402) with those shown by Grandone et al. (n=1010) (3). Our patients were slightly older and the prevalence of girls was higher (Supplemental table 5). For a better comparison of the data, we contacted the authors who kindly provided us with the r2 for the associations between the TM6SF2 rs58542926 and the ALT and lipids. As shown in the supplemental table 3, the variance of ALT and lipids explained by the rs58542926 SNP was similar in the two groups of children (American Caucasians and Italians) (Anna Grandone and Emanuele Miraglia del Giudice personal communication).
The TM6SF2 encodes a protein of 351 amino acids with 7–10 predicted transmembrane domains (17). The rs58542926 is characterized by a C-to-T substitution in the nucleotide 499 resulting in a Glutamic Acid to Lysine substitution at the codon 167 in the TM6SF2 gene (E167K). Protein subcellular localization studies demonstrated that the TM6SF2 protein is localized in the endoplasmic reticulum and in the ER-Golgi intermediate compartment of human liver cells (17). The rs58542926 variant is likely to cause a loss of function of the TM6SF2 encoded protein. In fact, in vitro and animal studies have shown that the TM6SF2 is a polytopic membrane protein and that the K167 variant form is misfolded and undergoes accelerated intracellular degradation (2). This results in the TM6SF2 loss of function, which in turn causes a reduced triglycerides secretion and an increased intra-hepatic triglycerides accumulation within multiple large lipid droplets in the liver tissue (2). The clinical consequence of the TM6SF2 loss of function is a higher risk of liver fat accumulation and liver damage (given the massive intra-hepatic accumulation of TGs) (4, 18), but a favorable lipid profile (given the low concentration of plasma TGs and LDL) (7, 19).
In fact, consistent with adult data (2, 5) we observed that obese children and adolescents also have a favorable lipid profile characterized by lower total cholesterol, VLDL, total LDL particles, large LDL particles, small LDL particles, medium small LDL particles and very small LDL particles, which are strong risk factors for myocardial infarction (20). Of note, we did not observe a statistically significant association between the subclasses of lipoproteins and the rs58542926 variant in African Americans. This might be due to the fact that African Americans tend to accumulate lower amounts of lipids in the liver probably owing a lower lipolysis from adipose tissue (21). Therefore, the effect of the rs58542926 SNP on lipoprotein accumulation in the hepatocytes is probably less marked in this ethnic group.
The rs58542926 variant confers protection against myocardial infarction and carotid plaques formation as it is linked to lower circulating lipoproteins (in particular small LDL and very small LDL) (22). Dongiovanni and colleagues extended these findings in a large, prospective cohort of morbidly obese individuals at high risk of cardiovascular diseases (CVD) with long-term follow-up and showed that carriers of the rs58542926 minor allele are protected against long term CVD by the reduced circulating levels of atherogenic lipoproteins (5). These findings might have therapeutic implications; in fact, recently, in a large cross-sectional cohort of at risk adults, it has been demonstrated that statin use is associated with protection against the liver damage related to NAFLD by down-regulation of the sterol regulatory element-binding protein 1c (SREBP1c), a key regulator of triglycerides synthesis (23), but the presence of the I148M PNPLA3 risk variant reduced statin beneficial effect, while rs58542926 TM6SF2 did not influence the beneficial effect of statins on NASH (23), which makes wonder about the possibility of treating T allele carriers with statin to inhibit liver damage, even if the lipid profile of these patients does not justify this treatment.
Strengths and limitations
This study has several strengths, such as 1) the extensive characterization of the metabolic phenotype of our cohort, 2) the young age of the patients, and therefore the absence of risk factors linked to alcohol consumption, aging and other comorbidities of obesity, and 3) the use of magnetic resonance imaging (MRI) measurement to assess HFF%. The major limitation of this study is the sample size: the rs58542926, in fact, is a rare variant, which is why we studied only a small number of patients carrying the risk allele within each ethnicity. Moreover, the follow-up time was not long enough to assess whether or not obese children carrying the rs58542926 variant might be protected in the long term from the development of CVD or whether the long exposition to obesity and its complications might negatively affect the beneficial effect of the genotype.
Conclusions and future directions
The rs58542926 variant is associated with early onset fatty liver disease and liver injury, but with a favorable cardiovascular profile. Further studies would help us to better understand the primary mechanism by which this mutation changes the structure of the protein and possibly to use it as a target to prevent liver damage and to ameliorate cardiovascular comorbidity in obese children and adolescents.
Supplementary Material
Acknowledgments
The authors are grateful to the patients and their families as well as to the Yale Center for Genome Analyses (YCGA) and Yale Center for Clinical Investigation (YCCI) and Hospital research Unit (HRU) personnel.
Financial support: This work has been made possible by the American Heart Association (AHA) through the 13SDG14640038, the 11CRP5620013 awards to NS and by the Yale Center for Clinical Investigation (2012 YCCI scholar award and YCCI just in time grant) to NS. NS is also funded by the Allen foundation. MG is funded by the EXTRA Program (University of Milano-Bicocca in collaboration with Assolombarda). SC is funded by NIH (grants R01-HD-40787, R01-HD-28016) and ADA (Distinguished Clinical Scientist Awards from the American Diabetes Association, DK-49230). This work was also made possible by DK045735 to the Yale Diabetes Research Center and Clinical and Translational Science Awards Grant UL1-RR- 024139 from the National Center for Advancing Translational Sciences, a component of the NIH, and NIH Roadmap for Medical Research. This paper's contents are solely the responsibility of the authors and do not necessarily represent the official view of NIH. NS is the guarantor of this work and as such had full access to all the data in the study and takes responsibility for the integrity of the data and the accuracy of the analyses.
List of abbreviations
- TM6SF2
transmembrane 6 superfamily member 2
- NAFLD
non-alcoholic fatty liver disease
- SNP
single-nucleotide polymorphism
- MRI
magnetic resonance imaging
- HFF
Hepatic Fat Fraction
- ALT
alanine aminotransferase
- LDL
Low Density Lipoprotein
- PCR
polymerase chain reaction
- OGTT
oral glucose tolerance test
- WBISI
whole-body insulin sensitivity index
- CK-18
cytokeratin 18
- BMI
body mass index
- HbA1c
hemoglobin A1c
- VLDL
very low density lipoprotein
- TG
Trygliceride
- CVD
cardiovascular diseases
- SREBP1c
sterol regulatory element-binding protein 1c
Contributor Information
Martina Goffredo, Email: martina.goffredo@yale.edu.
Sonia Caprio, Email: sonia.caprio@yale.edu.
Ariel E Feldstein, Email: afeldstein@ucsd.edu.
Ebe D'Adamo, Email: ebe.dadamo@yahoo.com.
Melissa M Shaw, Email: melissa.m.shaw@yale.edu.
Bridget Pierpont, Email: bridget.pierpont@yale.edu.
Mary Savoye, Email: mary.savoye@yale.edu.
Hongyu Zhao, Email: hongyu.zhao@yale.edu.
Allen E. Bale, Email: allen.bale@yale.edu.
References
- 1.Angulo P. Nonalcoholic fatty liver disease. N Engl J Med. 2002;346:1221–1231. doi: 10.1056/NEJMra011775. [DOI] [PubMed] [Google Scholar]
- 2.Kozlitina J, Smagris E, Stender S, Nordestgaard BG, Zhou HH, Tybjaerg-Hansen A, Vogt TF, et al. Exome-wide association study identifies a TM6SF2 variant that confers susceptibility to nonalcoholic fatty liver disease. Nat Genet. 2014;46:352–356. doi: 10.1038/ng.2901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Grandone A, Cozzolino D, Marzuillo P, Cirillo G, Di Sessa A, Ruggiero L, Di Palma MR, et al. TM6SF2 Glu167Lys polymorphism is associated with low levels of LDL-cholesterol and increased liver injury in obese children. Pediatr Obes. 2015 doi: 10.1111/ijpo.12032. [DOI] [PubMed] [Google Scholar]
- 4.Liu YL, Reeves HL, Burt AD, Tiniakos D, McPherson S, Leathart JB, Allison ME, et al. TM6SF2 rs58542926 influences hepatic fibrosis progression in patients with non-alcoholic fatty liver disease. Nat Commun. 2014;5:4309. doi: 10.1038/ncomms5309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dongiovanni P, Petta S, Maglio C, Fracanzani AL, Pipitone R, Mozzi E, Motta BM, et al. Transmembrane 6 superfamily member 2 gene variant disentangles nonalcoholic steatohepatitis from cardiovascular disease. Hepatology. 2015;61:506–514. doi: 10.1002/hep.27490. [DOI] [PubMed] [Google Scholar]
- 6.Romeo S, Kozlitina J, Xing C, Pertsemlidis A, Cox D, Pennacchio LA, Boerwinkle E, et al. Genetic variation in PNPLA3 confers susceptibility to nonalcoholic fatty liver disease. Nat Genet. 2008;40:1461–1465. doi: 10.1038/ng.257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Speliotes EK, Yerges-Armstrong LM, Wu J, Hernaez R, Kim LJ, Palmer CD, Gudnason V, et al. Genome-wide association analysis identifies variants associated with nonalcoholic fatty liver disease that have distinct effects on metabolic traits. PLoS Genet. 2011;7:e1001324. doi: 10.1371/journal.pgen.1001324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Expert Committee on the D, Classification of Diabetes M. Report of the expert committee on the diagnosis and classification of diabetes mellitus. Diabetes Care. 2003;26(Suppl 1):S5–20. doi: 10.2337/diacare.26.2007.s5. [DOI] [PubMed] [Google Scholar]
- 9.Matsuda M, DeFronzo RA. Insulin sensitivity indices obtained from oral glucose tolerance testing: comparison with the euglycemic insulin clamp. Diabetes Care. 1999;22:1462–1470. doi: 10.2337/diacare.22.9.1462. [DOI] [PubMed] [Google Scholar]
- 10.Cali AM, De Oliveira AM, Kim H, Chen S, Reyes-Mugica M, Escalera S, Dziura J, et al. Glucose dysregulation and hepatic steatosis in obese adolescents: is there a link? Hepatology. 2009;49:1896–1903. doi: 10.1002/hep.22858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fishbein MH, Gardner KG, Potter CJ, Schmalbrock P, Smith MA. Introduction of fast MR imaging in the assessment of hepatic steatosis. Magn Reson Imaging. 1997;15:287–293. doi: 10.1016/s0730-725x(96)00224-x. [DOI] [PubMed] [Google Scholar]
- 12.Kleiner DE, Brunt EM, Van Natta M, Behling C, Contos MJ, Cummings OW, Ferrell LD, et al. Design and validation of a histological scoring system for nonalcoholic fatty liver disease. Hepatology. 2005;41:1313–1321. doi: 10.1002/hep.20701. [DOI] [PubMed] [Google Scholar]
- 13.Brunt EM, Janney CG, Di Bisceglie AM, Neuschwander-Tetri BA, Bacon BR. Nonalcoholic steatohepatitis: a proposal for grading and staging the histological lesions. Am J Gastroenterol. 1999;94:2467–2474. doi: 10.1111/j.1572-0241.1999.01377.x. [DOI] [PubMed] [Google Scholar]
- 14.Santoro N, Kursawe R, D'Adamo E, Dykas DJ, Zhang CK, Bale AE, Cali AM, et al. A common variant in the patatin-like phospholipase 3 gene (PNPLA3) is associated with fatty liver disease in obese children and adolescents. Hepatology. 2010;52:1281–1290. doi: 10.1002/hep.23832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Santoro N, Zhang CK, Zhao H, Pakstis AJ, Kim G, Kursawe R, Dykas DJ, et al. Variant in the glucokinase regulatory protein (GCKR) gene is associated with fatty liver in obese children and adolescents. Hepatology. 2012;55:781–789. doi: 10.1002/hep.24806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Valenti L, Alisi A, Galmozzi E, Bartuli A, Del Menico B, Alterio A, Dongiovanni P, et al. I148M patatin-like phospholipase domain-containing 3 gene variant and severity of pediatric nonalcoholic fatty liver disease. Hepatology. 2010;52:1274–1280. doi: 10.1002/hep.23823. [DOI] [PubMed] [Google Scholar]
- 17.Mahdessian H, Taxiarchis A, Popov S, Silveira A, Franco-Cereceda A, Hamsten A, Eriksson P, et al. TM6SF2 is a regulator of liver fat metabolism influencing triglyceride secretion and hepatic lipid droplet content. Proc Natl Acad Sci U S A. 2014;111:8913–8918. doi: 10.1073/pnas.1323785111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Roh YS, Loomba R, Seki E. The TM6SF2 variants, novel genetic predictors for nonalcoholic steatohepatitis. Gastroenterology. 2015;148:252–254. doi: 10.1053/j.gastro.2014.11.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Surakka I, Horikoshi M, Magi R, Sarin AP, Mahajan A, Lagou V, Marullo L, et al. The impact of low-frequency and rare variants on lipid levels. Nat Genet. 2015;47:589–597. doi: 10.1038/ng.3300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Rizzo M, Berneis K. Small, dense low-density-lipoproteins and the metabolic syndrome. Diabetes Metab Res Rev. 2007;23:14–20. doi: 10.1002/dmrr.694. [DOI] [PubMed] [Google Scholar]
- 21.Nielsen SR, Sumner AE, Miller BV, 3rd, Turkova H, Klein S, Jensen MD. Free fatty acid flux in African-American and Caucasian adults--effect of sex and race. Obesity (Silver Spring) 2013;21:1836–1842. doi: 10.1002/oby.20322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Holmen OL, Zhang H, Fan Y, Hovelson DH, Schmidt EM, Zhou W, Guo Y, et al. Systematic evaluation of coding variation identifies a candidate causal variant in TM6SF2 influencing total cholesterol and myocardial infarction risk. Nat Genet. 2014;46:345–351. doi: 10.1038/ng.2926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Dongiovanni P, Petta S, Mannisto V, Margherita Mancina R, Pipitone R, Karja V, Maggioni M, et al. Statin use and nonalcoholic steatohepatitis in at risk individuals. J Hepatol. 2015 doi: 10.1016/j.jhep.2015.05.006. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.