Abstract
Purpose
Macromolecular proton fraction (MPF) mapping is a quantitative MRI method that reconstructs parametric maps of a relative amount of macromolecular protons causing the magnetization transfer (MT) effect and provides a biomarker of myelination in neural tissues. This study aimed to develop a high-resolution whole-brain MPF mapping technique utilizing a minimal possible number of source images for scan time reduction.
Methods
The described technique is based on replacement of an actually acquired reference image without MT saturation by a synthetic one reconstructed from R1 and proton density maps, thus requiring only three source images. This approach enabled whole-brain three-dimensional MPF mapping with isotropic 1.25×1.25×1.25 mm3 voxel size and scan time of 20 minutes. The synthetic reference method was validated against standard MPF mapping with acquired reference images based on data from 8 healthy subjects.
Results
Mean MPF values in segmented white and gray matter appeared in close agreement with no significant bias and small within-subject coefficients of variation (<2%). High-resolution MPF maps demonstrated sharp white-gray matter contrast and clear visualization of anatomical details including gray matter structures with high iron content.
Conclusions
Synthetic reference method improves resolution of MPF mapping and combines accurate MPF measurements with unique neuroanatomical contrast features.
Keywords: magnetic resonance imaging, macromolecular proton fraction, magnetization transfer, white matter, gray matter, myelin
INTRODUCTION
Macromolecular proton fraction (MPF) is a key biophysical parameter determining the magnetization transfer (MT) effect within the two-pool model (1) and describing a relative amount of protons characterized by semi-solid molecular motion and involved into magnetic cross-relaxation with free water protons. Based on a number of animal studies (2–8), MPF has been recognized as a quantitative MRI parameter closely associated with the myelin content in neural tissues. While MPF maps can be obtained in vivo using a variety of methods (a detailed overview can be found in (9)), they have a common limitation of either prohibitive scan time or limited spatial coverage and resolution. A fast method allowing whole-brain MPF mapping in humans based on a single MT-weighted image was recently proposed (9). This technique has been clinically tested in multiple sclerosis (10) and mild traumatic brain injury (mTBI) (11) studies and demonstrated a promise as a robust clinically-targeted myelin imaging approach. Particularly, the mTBI study (11) reported microscopic post-traumatic demyelination in normal-appearing brain tissues after blast injury with anatomical patterns consistent with the bio-mechanical mechanism of injury and the largest effect at the white-gray matter junction. In the clinical multiple-sclerosis study (10), MPF mapping revealed significant loss of myelin in both normal-appearing white matter (WM) and gray matter (GM) and demonstrated the primary clinical relevance of cortical GM demyelination. However, in its current implementation (9), MPF mapping has a limitation of relatively low spatial resolution available within clinically acceptable scan time. The goal of this study was to develop and test a high-resolution whole-brain MPF mapping method based on the single-point approach (9) and utilizing a minimal possible number of source images for scan time reduction. Specifically, a briefly presented earlier (12) procedure for reconstruction of MPF maps where only three source images are needed is described in detail.
METHODS
Algorithm for reconstruction of MPF maps from three source images
In the single-point MPF mapping method (9,12), the pulsed magnetization transfer matrix equation is iteratively solved by the Gauss-Newton method with standardized constraints for non-adjustable two-pool model parameters and corrections for B1 and B0 field inhomogeneities to yield an MPF map. As input data, a single-point MPF measurement requires a spoiled gradient echo (GRE) MT-weighted image with off-resonance saturation, a reference GRE image without off-resonance saturation for data normalization, and an independently acquired R1 (R1=1/T1) map. The widely used time efficient approach to measure R1 is the two-point variable flip angle (FA) method (13) employing a T1-weighted and a proton density (PD)-weighted spoiled GRE images. The key idea of the proposed algorithm is to synthesize a reference GRE image from R1 and PD maps with the repetition time (TR) and FA corresponding to those for the MT-weighted image according to the Ernst equation. If variable FA data are processed with B1 correction, a B1 map can also be used to calculate the synthetic reference image with actual flip angles in each voxel. As such, if a synthetic reference image is used, only three source images (MT-, T1-, and PD-weighted) are needed for MPF mapping. In summary, the main steps of the proposed algorithm are as follows:
1) Reconstruct R1 and PD maps from two variable FA images with B1 correction by fitting Ernst equation;
2) Compute a synthetic reference image from R1, PD, and B1 maps;
3) Reconstruct an MPF map from an MT-weighted image normalized to the synthetic reference image and an R1 map while applying B0 and B1 corrections according to the originally described procedure (9).
In this study, the above algorithm was used to design a time-efficient protocol for high-resolution three-dimensional (3D) whole-brain MPF mapping and validated by comparing it with the standard technique (9) based on the actually acquired reference image.
Participants
Eight healthy volunteers (mean age ± standard deviation (SD): 44.6 ± 12.2 years, age range: 29-66 years, 4 females/4 males) participated in this study. The study was approved by the Institutional Review Board, and written informed consent was obtained from all participants.
MRI Protocol
MRI data acquisition was performed on a 3.0 Tesla whole-body scanner (Achieva; Philips Medical Systems, Best, Netherlands) with an 8-channel phased-array head coil. In the GRE pulse sequence used for source image acquisition and the actual flip-angle imaging (AFI) sequence for B1 field mapping (14), the optimized spoiling regimen (15) were implemented with the excitation pulse phase increments of 169° for GRE and 39° for AFI while applying spoiling gradients with the maximal areas achievable within the prescribed TR. PD- and T1-weighted GRE images were acquired with TR = 21 ms and FA = 4° and 25°, respectively. MT-weighted images were acquired with TR = 28 ms and FA = 10°. Off-resonance saturation was achieved by applying the single-lobe sinc pulse with Gaussian apodization and following parameters: offset frequency 4 kHz, effective saturation FA = 560°, and duration 12 ms. Reference GRE images were obtained with the same TR = 28 ms and FA = 10° and without a saturation pulse. All images were acquired in the 3D mode with non-selective excitation, FOV = 240×240×180 mm3, and isotropic voxel size of 1.25×1.25×1.25 mm3. To increase signal-to-noise ratio, dual-echo acquisition (first/second echo time (TE1/TE2) = 2.3 ms/6.9 ms) was used for all the above scans with subsequent averaging of individual echo images during reconstruction (16). 3D dual-echo B0 maps (TR/TE1/TE2 = 20/2.3/3.3 ms, FA = 10°) (17) and AFI B1 maps (first/second TR (TR1/TR2) = 40/160 ms, TE = 2.3 ms, FA = 60°) (14) were obtained in the same geometry with voxel sizes of 2.5×2.5×2.5 mm3 and 2.5×2.75×5.0 mm3, respectively. Parallel imaging (SENSE) was used for all scans in two phase encoding directions with acceleration factors 1.5 (anterior-posterior) and 1.2 (left-right). Scan times were 4 min 21 s for the T1- and PD-weighted images, 5 min 48 s for the MT-weighted and reference images, 2 min 8 s for the B0 map, and 3 min 26 s for the B1 map, thus resulting in about 20 min acquisition time for the entire protocol without the reference image. Additionally, standard anatomical high-resolution 3D Magnetization Prepared Rapid Acquisition Gradient Echo (MPRAGE) T1-weighted images (TR/TE = 5.9/2.8 ms, FA = 8°, inversion time (TI) = 900 ms, shot interval = 2500 ms, voxel size = 1×1×1 mm3, scan time 4 min 44 s) were acquired with same FOV for comparison purposes.
Image processing and analysis
Reconstruction of MPF maps was performed using custom-written C-language software. FMRIB Software Library (FSL; http://www.fmrib.ox.ac.uk/fsl) (18) was used for segmentation of MPF maps and tissue masking. MPF maps were reconstructed with the standard method based on the acquired reference image and the synthetic reference method described above. Previously determined (9) constraints for non-adjustable two-pool model parameters and their combinations were used in both algorithms as follows: cross-relaxation rate constant R = 19.0 s-1, product of R1 and T2 of free water protons R1T2F = 0.022, and T2 of bound macromolecular protons T2B = 10 μs. Before reconstruction, non-brain tissues were removed from source images by applying a brain mask created from the PD-weighted image using the brain extraction tool (BET) (19) in FSL software. Additionally, cerebrospinal fluid (CSF) was segmented out from R1 maps based on a threshold value of R1 = 0.33 s-1 (equivalent to T1 = 3000 ms). MPF maps reconstructed with both methods were segmented into WM and GM. Segmentation was performed by an automated segmentation tool (FAST) (20) using FSL software in the native image space with the Markov random field weighting parameter 0.25. To account for potentially incomplete CSF segmentation by R1 threshold and exclude voxels containing partial volume of CSF (PVCSF), the third mixed tissue class was also prescribed. Tissue classes were defined by specifying initial tissue-type priors with the following MPF values: 12% for WM, 6% for GM, and 1% for PVCSF. Resulting binary segmentation masks were used to calculate mean MPF in WM and GM.
Statistical analysis
Statistical analysis was carried out in SPSS software (SPSS Inc, Chicago, Ill). Normality of data distribution was checked using the Shapiro-Wilk test to justify parametric analyses. An agreement between MPF map reconstruction techniques with acquired and synthetic reference images was assessed using Bland-Altman plots. The bias between MPF values in segmented WM and GM measured from MPF maps reconstructed by the two methods was examined using the one sample t-test for the mean difference between paired measurements. The limits of agreement were calculated as the mean difference ± 1.96SD of the mean difference. To estimate variability between measurements from the two reconstruction options, the within-subject coefficients of variation (CV) was calculated as the percentage ratio of SD to the mean of paired measurements. The Levene’s test was used to assess equality of variances. P values less than 0.05 were considered statistically significant. Data are presented as mean ± SD.
RESULTS
Agreement between MPF map reconstruction methods
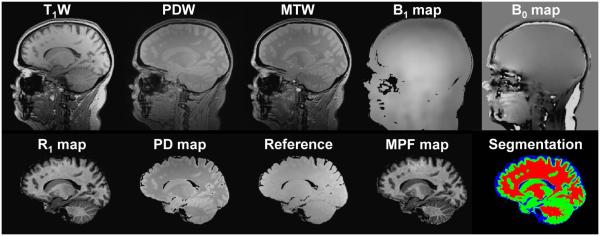
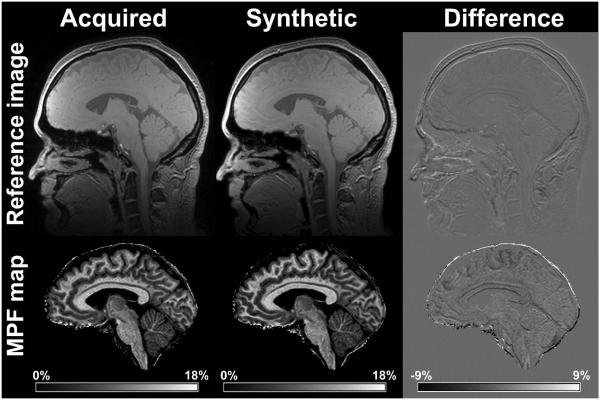
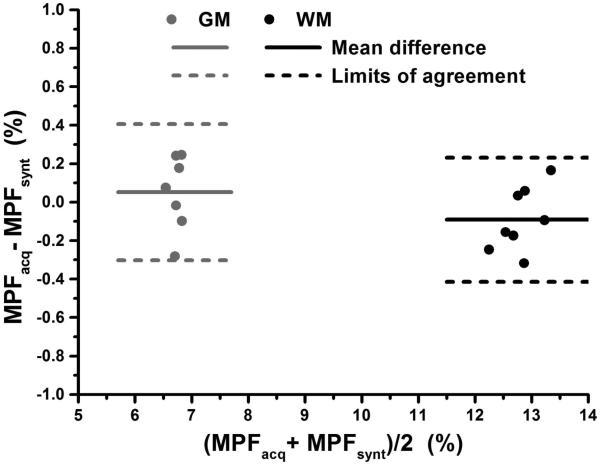
The minimal set of three source images, B0 and B1 field maps, and corresponding parametric maps generated by the algorithm based on these input data are exemplified in Fig. 1. Visual similarity between acquired and synthetic reference images and MPF maps obtained with both reconstruction algorithms is illustrated in Fig. 2. Note that only minor discrepancies associated with sub-voxel misregistration are seen on the tissue borders. Results of quantitative MPF measurements in segmented WM and GM (example tissue masks are shown in Fig. 1) based on the maps reconstructed with acquired and synthetic reference images are summarized in Table 1, and Bland-Altman plots are presented in Fig. 3. MPF in WM and GM demonstrated close agreement with no significant bias between reconstruction methods, equal variances, and small within-subject coefficients of variation. MPF measurements in brain tissues are also close to the previously reported values for healthy controls based on a lower-resolution MPF mapping technique (10,11).
Fig. 1.
Example source images, parametric maps, and results of an MPF map segmentation. Top row: source T1-weighted (T1W), PD-weighted (PDW), and MT-weighted (MTW) GRE images and B1 and B0 field maps. Bottom row: R1 and PD maps reconstructed from T1W and PDW images, synthetic reference image, resulting MPF map, and color-coded binary segmentation masks, where red, green, and blue colors correspond to WM, GM, and PVCSF tissue classes, respectively All processed images (bottom row) are presented with removed extra-cranial tissues and CSF.
Fig. 2.
Comparison between acquired and synthetic reference images (top row) and MPF maps reconstructed with them (bottom row). Left column: acquired reference image and corresponding MPF map; middle column: synthetic reference image and corresponding MPF map; right column: subtraction images. The synthetic reference image was calculated without brain masking for comparison purposes. Grayscale ranges correspond to MPF of 0-18% for the maps and −9-+9% for their difference.
Table 1.
Comparison between MPF measurements with acquired and synthetic reference images in segmented brain tissues.
| Value | WM | GM |
|---|---|---|
| Mean MPF with acquired reference (%) | 12.77 ± 0.41 | 6.73 ± 0.15 |
| Mean MPF with synthetic reference (%) | 12.86 ± 0.31 | 6.68 ± 0.14 |
| Mean difference (bias) (%) | −0.09 ± 0.16 | 0.05 ± 0.18 |
| Limits of agreement (%) | −0.41; 0.23 | −0.30; 0.41 |
| Significance for the bias (P) | 0.16 | 0.44 |
| Significance for inequality of variances (P) | 0.61 | 0.64 |
| Within-subject coefficient of variation (%) | 0.99 | 1.86 |
Fig. 3.
Bland-Altman plots comparing mean MPF values in segmented WM (black symbols) and GM (gray symbols) between reconstructions with acquired and synthetic reference images. Solid and dashed lines correspond to the mean difference and limits of agreement, respectively.
Neuroanatomical features of the MPF contrast
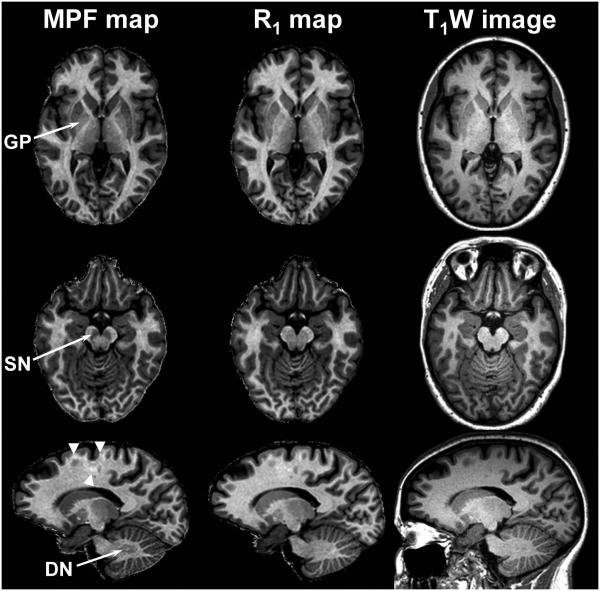
High-resolution whole-brain MPF maps demonstrate sharp contrast between WM and GM and clear visualization of anatomical details (Figs. 1 and 2). While MPF contrast appears generally similar to heavily T1-weighted contrast, two key distinctions were observed. The most notable feature of MPF maps is their capability to produce pure GM contrast for subcortical structures with high iron content, such as globus pallidus, substantia nigra, and dentate nucleus, as illustrated in Fig. 4. These structures appear clearly hypointense on MPF maps compared to WM, similar to the rest of GM. In contrast, iron-rich GM structures are undistinguishable or poorly distinguishable from surrounding WM on both T1-weighted anatomical images and R1 maps. Another feature of MPF maps is the contrast variations within WM due to variable degree of myelination (Fig. 4). Such an inherent WM signal variability is much less clear or invisible on R1 maps and T1-weighted images.
Fig. 4.
Reformatted sections of a 3D MPF map (left column), R1 map (middle column), and T1-weighted MPRAGE image (right column) comparing tissue contrast for iron-rich GM anatomical structures. Arrows indicate the globus pallidus (GP), substantia nigra (SN), and dentate nucleus (DN). Arrowheads indicate intrinsic variations of MPF within WM.
DISCUSSION
This study introduces the most time-efficient to date MPF mapping technique based on the reconstruction algorithm that utilizes the minimal possible number of source images. The task of reconstructing an MPF map within the previously described single-point method (9) can be conceptualized as the solution of the set of equations (matrix pulsed steady-state MT equation and Ernst equation) with respect to the three variables (MPF, R1, and PD). In the proposed approach, the reconstruction is performed as the non-linear least-squares solution with the three types of input images characterized by the contrast weightings highlighting the effects of the above variables (MT-, T1-, and, PD-weighted). Based on the proposed algorithm, the feasibility of high-resolution whole-brain 3D MPF mapping with clinically acceptable scan time has been demonstrated. In the described implementation, 3D MPF maps were obtained with isotropic voxel size of 1.25×1.25×1.25 mm3, which approaches the current standards of structural 3D imaging in various neuroscience applications and was unachievable for MPF mapping previously.
While the total scan time for the described protocol is still relatively long (20 minutes), further improvements in the time efficiency are possible. Particularly, the used a coil with a relatively modest performance (8 channels) did not allow implementing a parallel imaging protocol with high acceleration factors. It is expected that modern coils with 32 or more channels would allow considerable reduction of the scan time or further improvement in spatial resolution. A reduction of the TR in the component sequences is also possible. While the current protocol design relies on the optimal radiofrequency and gradient spoiling requirements (15), limitations of the gradient system (28 mT/m per channel) resulted in a relatively long TR, particularly for T1- and PD-weighted scans. Shorter TR can be achieved with stronger gradient systems, which are currently available on many 3 Tesla MRI platforms. Alternatively, more relaxed spoiling conditions can be implemented (21), though the potential effect of spoiling errors on the accuracy of MPF measurements needs to be investigated. For serial clinical applications, the proposed approach can be used with a lower spatial resolution, similar to the earlier described protocol (10). In such an implementation, whole-brain MPF mapping based on the three source images including field mapping sequences can be accomplished in 6-8 minutes.
An emerging area of application of the described MPF mapping method is the studies of GM myelination. This topic has recently attracted significant interest in both neuroanatomical and clinical contexts. A considerable heterogeneity of myelin content across different cortical areas (22) and age-related changes (23) have been demonstrated. Clinical relevance of GM demyelination in multiple sclerosis has been recently shown (10), suggesting the principal role of cortical demyelination for the development of disability and conversion from the relapsing-remitting to the secondary progressive disease phenotype. GM demyelination may also be an important pathological factor in various neurological disorders, such as Alzheimer disease (24,25), Huntington disease (26), and traumatic brain injury (11). A unique advantage of MPF for assessment of GM demyelination is its insensitivity to the variable iron content in neural tissues. This study demonstrated that MPF maps allow adequate representation of GM contrast in iron-rich subcortical structures. An increase of iron content in such structures is known for many neurological conditions including multiple sclerosis, Alzheimer, Parkinson, and Huntington diseases (26–29) and associated with aging (30,31). Age- and disease-related variations of the iron content also occur in cortical GM (28,31–33). Furthermore, demyelination and iron accumulation may be pathogenically linked in neurodegenerative diseases (24,26). Alternative quantitative (34–36) or semi-quantitative (22) magnetic resonance methods for myelin imaging are based on relaxation properties of tissues and, therefore, are inherently sensitive to a decrease of relaxation times T1, T2, and T2* caused by iron. As such, these methods are difficult or impossible to use in studies targeted at the precise assessment of GM demyelination in the presence of the confounding effect of iron deposition, especially in the anatomic structures with intrinsically high iron content. In combination with high spatial resolution achievable with the described approach, insensitivity of MPF contrast to iron makes MPF mapping an ultimate tool for studies of both cortical and subcortical GM demyelination in a variety of neurological conditions.
Besides quantitative tissue characterization, high-resolution MPF mapping has a potential to become a useful technique for various neuroscience applications including segmentation of cortical, subcortical, cerebellar, and brain stem structures. The use of MPF maps as source images in such applications may be advantageous due to a very high tissue contrast and insensitivity to iron. Another important advantage of MPF maps as source images is the inherent correction of field and coil reception non-uniformities. Specifically, spatial variations of B0 and B1 fields are corrected based on the rigorous physical model by directly accommodating corresponding field maps into the image reconstruction workflow. The effect of the coil reception profile is eliminated by normalization to the synthetic reference image and does not propagate into resulting MPF maps. As such, the use of MPF maps eliminates the need in computationally-intense bias field correction algorithms and allows utilization of native quantitative voxel values in various post-processing tasks. On the other hand, neuroanatomical applications of MPF mapping may require further improvements in spatial resolution, precise registration of component images, and development of specialized MPF-based atlases for particular anatomical structures.
This study has two limitations. First, it did not consider the effect of bi-exponential relaxation on R1 measurements. This effect has been shown to bias both R1 and the two-pool model parameters in quantitative MT imaging (37,38). However, it is unlikely that the analysis based on bi-exponential relaxation would change the conclusions of this study about an agreement between reconstruction methods, because the same R1 maps have been used in both reconstruction algorithms. Additionally, both R1 and MPF values can be corrected for errors caused by bi-exponential relaxation with a high degree of approximation using recalculation equations from Ref. (38). Second, the sample size was relatively small, and therefore, a minor bias between acquired and synthetic reference reconstructions cannot be excluded. Particularly, somewhat larger distinctions between MPF maps can be appreciated in certain WM regions on difference images (Fig. 2). At the same time, such effects can be caused by sub-voxel misregistration, particularly due to brain pulsation, which is characterized by displacements of about 0.5 mm (39).
CONCLUSIONS
This study demonstrates the feasibility of reconstructing MPF maps from three source images, which constitute the minimal possible input dataset for the MPF determination. The described method enables high-resolution whole-brain quantitatively accurate 3D MPF mapping in clinically acceptable time. High-resolution MPF maps combine unique neuroanatomical contrast features and quantitative information about myelination of both white and gray matter. The described methodology can be applied in a variety of fundamental and clinical neuroscience studies where non-invasive quantitative assessment of brain myelination is of interest.
ACKNOWLEDGEMENTS
The author acknowledges financial support from the National Institutes of Health (grant R21EB016135) for software development, National Multiple Sclerosis Society (grant RG 4864A1/1) for data acquisition, and Russian Science Foundation (project №14-45-00040) for data analysis and manuscript preparation.
REFERENCES
- 1.Henkelman RM, Huang X, Xiang QS, Stanisz GJ, Swanson SD, Bronskill MJ. Quantitative interpretation of magnetization transfer. Magn Reson Med. 1993;29:759–766. doi: 10.1002/mrm.1910290607. [DOI] [PubMed] [Google Scholar]
- 2.Janve VA, Zu Z, Yao SY, Li K, Zhang FL, Wilson KJ, Ou X, Does MD, Subramaniam S, Gochberg DF. The radial diffusivity and magnetization transfer pool size ratio are sensitive markers for demyelination in a rat model of type III multiple sclerosis (MS) lesions. Neuroimage. 2013;74:298–305. doi: 10.1016/j.neuroimage.2013.02.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Odrobina EE, Lam TY, Pun T, Midha R, Stanisz GJ. MR properties of excised neural tissue following experimentally induced demyelination. NMR Biomed. 2005;18:277–284. doi: 10.1002/nbm.951. [DOI] [PubMed] [Google Scholar]
- 4.Ou X, Sun SW, Liang HF, Song SK, Gochberg DF. The MT pool size ratio and the DTI radial diffusivity may reflect the myelination in shiverer and control mice. NMR Biomed. 2009;22:480–487. doi: 10.1002/nbm.1358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Rausch M, Tofts P, Lervik P, Walmsley A, Mir A, Schubart A, Seabrook T. Characterization of white matter damage in animal models of multiple sclerosis by magnetization transfer ratio and quantitative mapping of the apparent bound proton fraction f. Mult Scler. 2009;15:16–27. doi: 10.1177/1352458508096006. [DOI] [PubMed] [Google Scholar]
- 6.Samsonov A, Alexander AL, Mossahebi P, Wu YC, Duncan ID, Field AS. Quantitative MR imaging of two-pool magnetization transfer model parameters in myelin mutant shaking pup. Neuroimage. 2012;62:1390–1398. doi: 10.1016/j.neuroimage.2012.05.077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Thiessen JD, Zhang Y, Zhang H, Wang L, Buist R, Del Bigio MR, Kong J, Li XM, Martin M. Quantitative MRI and ultrastructural examination of the cuprizone mouse model of demyelination. NMR Biomed. 2013;26:1562–1581. doi: 10.1002/nbm.2992. [DOI] [PubMed] [Google Scholar]
- 8.Underhill HR, Rostomily RC, Mikheev AM, Yuan C, Yarnykh VL. Fast bound pool fraction imaging of the in vivo rat brain: Association with myelin content and validation in the C6 glioma model. Neuroimage. 2011;54:2052–2065. doi: 10.1016/j.neuroimage.2010.10.065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yarnykh VL. Fast macromolecular proton fraction mapping from a single off-resonance magnetization transfer measurement. Magn Reson Med. 2012;68:166–178. doi: 10.1002/mrm.23224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yarnykh VL, Bowen JD, Samsonov A, Repovic P, Mayadev A, Qian P, Gangadharan B, Keogh BP, Maravilla KR, Jung Henson LK. Fast Whole-Brain Three-dimensional Macromolecular Proton Fraction Mapping in Multiple Sclerosis. Radiology. 2015;274:210–220. doi: 10.1148/radiol.14140528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Petrie EC, Cross DJ, Yarnykh VL, et al. Neuroimaging, behavioral, and psychological sequelae of repetitive combined blast/impact mild traumatic brain injury in Iraq and Afghanistan war veterans. J Neurotrauma. 2014;31:425–436. doi: 10.1089/neu.2013.2952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yarnykh VL. In Proceedings of the 22th Annual Meeting of ISMRM-ESMRMB. Milan; Italy: 2014. Fast high-resolution whole-brain macromolecular proton fraction mapping using a minimal number of source images; p. 3335. [Google Scholar]
- 13.Deoni SC, Rutt BK, Peters TM. Rapid combined T1 and T2 mapping using gradient recalled acquisition in the steady state. Magn Reson Med. 2003;49:515–526. doi: 10.1002/mrm.10407. [DOI] [PubMed] [Google Scholar]
- 14.Yarnykh VL. Actual flip-angle imaging in the pulsed steady state: a method for rapid three-dimensional mapping of the transmitted radiofrequency field. Magn Reson Med. 2007;57:192–200. doi: 10.1002/mrm.21120. [DOI] [PubMed] [Google Scholar]
- 15.Yarnykh VL. Optimal radiofrequency and gradient spoiling for improved accuracy of T1 and B1 measurements using fast steady-state techniques. Magn Reson Med. 2010;63:1610–1626. doi: 10.1002/mrm.22394. [DOI] [PubMed] [Google Scholar]
- 16.Helms G, Dechent P. Increased SNR and reduced distortions by averaging multiple gradient echo signals in 3D FLASH imaging of the human brain at 3T. J Magn Reson Imaging. 2009;29:198–204. doi: 10.1002/jmri.21629. [DOI] [PubMed] [Google Scholar]
- 17.Skinner TE, Glover GH. An extended two-point Dixon algorithm for calculating separate water, fat, and B0 images. Magn Reson Med. 1997;37:628–630. doi: 10.1002/mrm.1910370426. [DOI] [PubMed] [Google Scholar]
- 18.Smith SM, Jenkinson M, Woolrich MW, et al. Advances in functional and structural MR image analysis and implementation as FSL. Neuroimage. 2003;23(Suppl.1):S208, 219. doi: 10.1016/j.neuroimage.2004.07.051. [DOI] [PubMed] [Google Scholar]
- 19.Smith SM. Fast robust automated brain extraction. Hum Brain Mapp. 2002;17:143–155. doi: 10.1002/hbm.10062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhang Y, Brady M, Smith S. Segmentation of brain MR images through a hidden Markov random field model and the expectation-maximization algorithm. IEEE Trans Med Imaging. 2001;20:45–57. doi: 10.1109/42.906424. [DOI] [PubMed] [Google Scholar]
- 21.Preibisch C, Deichmann R. Influence of RF spoiling on the stability and accuracy of T1 mapping based on spoiled FLASH with varying flip angles. Magn Reson Med. 2009;61:125–135. doi: 10.1002/mrm.21776. [DOI] [PubMed] [Google Scholar]
- 22.Glasser MF, Van Essen DC. Mapping human cortical areas in vivo based on myelin content as revealed by T1- and T2-weighted MRI. J Neurosci. 2011;31:11597–11616. doi: 10.1523/JNEUROSCI.2180-11.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Grydeland H, Walhovd KB, Tamnes CK, Westlye LT, Fjell AM. Intracortical myelin links with performance variability across the human lifespan: results from T1- and T2-weighted MRI myelin mapping and diffusion tensor imaging. J Neurosci. 2013;33:18618–18630. doi: 10.1523/JNEUROSCI.2811-13.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bartzokis G. Alzheimer’s disease as homeostatic responses to age-related myelin breakdown. Neurobiol Aging. 2011;32:1341–1371. doi: 10.1016/j.neurobiolaging.2009.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Carmeli C, Donati A, Antille V, Viceic D, Ghika J, von Gunten A, Clarke S, Meuli R, Frackowiak RS, Knyazeva MG. Demyelination in mild cognitive impairment suggests progression path to Alzheimer’s disease. PLoS One. 2013;8:e72759. doi: 10.1371/journal.pone.0072759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bartzokis G, Lu PH, Tishler TA, Fong SM, Oluwadara B, Finn JP, Huang D, Bordelon Y, Mintz J, Perlman, S. Myelin breakdown and iron changes in Huntington’s disease: pathogenesis and treatment implications. Neurochem Res. 2007;32:1655–1664. doi: 10.1007/s11064-007-9352-7. [DOI] [PubMed] [Google Scholar]
- 27.Bakshi R, Benedict RH, Bermel RA, Caruthers SD, Puli SR, Tjoa CW, Fabiano AJ, Jacobs L. T2 hypointensity in the deep gray matter of patients with multiple sclerosis: a quantitative magnetic resonance imaging study. Arch Neurol. 2002;59:62–68. doi: 10.1001/archneur.59.1.62. [DOI] [PubMed] [Google Scholar]
- 28.Brass SD, Chen NK, Mulkern RV, Bakshi R. Magnetic resonance imaging of iron deposition in neurological disorders. Top Magn Reson Imaging. 2006;17:31–40. doi: 10.1097/01.rmr.0000245459.82782.e4. [DOI] [PubMed] [Google Scholar]
- 29.Michaeli S, Oz G, Sorce DJ, Garwood M, Ugurbil K, Majestic S, Tuite P. Assessment of brain iron and neuronal integrity in patients with Parkinson’s disease using novel MRI contrasts. Mov Disord. 2007;22:334–340. doi: 10.1002/mds.21227. [DOI] [PubMed] [Google Scholar]
- 30.Bilgic B, Pfefferbaum A, Rohlfing T, Sullivan EV, Adalsteinsson E. MRI estimates of brain iron concentration in normal aging using quantitative susceptibility mapping. Neuroimage. 2012;59:2625–2635. doi: 10.1016/j.neuroimage.2011.08.077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rodrigue KM, Haacke EM, Raz N. Differential effects of age and history of hypertension on regional brain volumes and iron. Neuroimage. 2011;54:750–759. doi: 10.1016/j.neuroimage.2010.09.068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hirai T, Korogi Y, Sakamoto Y, Hamatake S, Ikushima I, Takahashi M. T2 shortening in the motor cortex: effect of aging and cerebrovascular diseases. Radiology. 1996;199:799–803. doi: 10.1148/radiology.199.3.8638008. [DOI] [PubMed] [Google Scholar]
- 33.Zhu WZ, Zhong WD, Wang W, Zhan CJ, Wang CY, Qi JP, Wang JZ, Lei T, Quantitative MR. phase-corrected imaging to investigate increased brain iron deposition of patients with Alzheimer disease. Radiology. 2009;253:497–504. doi: 10.1148/radiol.2532082324. [DOI] [PubMed] [Google Scholar]
- 34.Deoni SCL, Rutt BK, Arun T, Pierpaoli C, Jones DK. Gleaning multicomponent T1 and T2 information from steady-state imaging data. Magn Reson Med. 2008;60:1372–1387. doi: 10.1002/mrm.21704. [DOI] [PubMed] [Google Scholar]
- 35.Hwang D, Kim DH, Du YP. In vivo multi-slice mapping of myelin water content using T2* decay. Neuroimage. 2010;52:198–204. doi: 10.1016/j.neuroimage.2010.04.023. [DOI] [PubMed] [Google Scholar]
- 36.MacKay A, Whittall K, Mädler J, Li D, Paty D, Graeb D. In vivo visualization of myelin water in brain by magnetic resonance. Magn Reson Med. 1994;31:673–677. doi: 10.1002/mrm.1910310614. [DOI] [PubMed] [Google Scholar]
- 37.Ou X, Gochberg DF. MT effects and T1 quantification in single-slice spoiled gradient echo imaging. Magn Reson Med. 2008;59:835–845. doi: 10.1002/mrm.21550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Mossahebi P, Yarnykh VL, Samsonov A. Analysis and correction of biases in cross-relaxation MRI due to biexponential longitudinal relaxation. Magn Reson Med. 2013;71:830–838. doi: 10.1002/mrm.24677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Poncelet BP, Wedeen VJ, Weisskoff RM, Cohen MS. Brain parenchyma motion: measurement with cine echo-planar MR imaging. Radiology. 1992;185:645–651. doi: 10.1148/radiology.185.3.1438740. [DOI] [PubMed] [Google Scholar]