Abstract
Autism spectrum disorder (ASD) is one of the most complex behavioral disorders with a strong genetic influence. The objectives of this article are to review the current status of genetic research in ASD, and to provide information regarding the potential candidate genes, mutations, and genetic loci possibly related to pathogenesis in ASD. Investigations on monogenic causes of ASD, candidate genes among common variants, rare de novo mutations, and copy number variations are reviewed. The current possible clinical applications of the genetic knowledge and their future possibilities are highlighted.
Keywords: Autism spectrum disorder, Syndromic autism, de novo mutations, Genetic diagnosis
INTRODUCTION
Autism spectrum disorder (ASD) is a neurodevelopmental disorder characterized by very early onset of dysfunction in social communication and interaction, repetitive behavior, and limited interest. It is now believed that ASD is a result of complex gene-environment interactions, with strong and clear genetic influences. Studies of twin pairs, high-risk infant siblings, families, and populations have estimated concordance rates and segregation of the disorder within families. The concordance rate was reported as 60-70% in monozygous twins and as 5-30% in siblings; this is in agreement with a recent large prospective study revealing a recurrence rate of 18% in infant siblings and of 33% in multiplex families [1,2]. However, it is currently believed that over 50% of the risk of developing ASD is attributed to genetic variation [3,4].
Advances in genetic technologies, large cohort studies, and widespread database sharing have contributed to the discovery and validation of causative genes in ASD [5]. Knowledge from genetic studies of ASD also provides insight into other neurodevelopmental disorders, as ASD shares both behavioral characteristics and endophenotypes. However, ASD is one of the most heterogeneous neurodevelopmental disorders, with great variation observed in behavioral manifestations and cognitive profiles, which makes determination of the single most important genetic risk factor extremely difficult.
Identifying biomarkers has been one of the primary goals of biological research of ASD, and current research efforts are directed predominantly toward the identification of markers for risk and early diagnosis [6]. There have been intense research efforts to identify the genetic basis of ASD, with an assumption that the genetic markers can be utilized as essential biomarkers in the diagnosis of, and the development of pharmacological treatments for, ASD. The objective of this paper is to review the current knowledge of genetic variations in ASD, and its role in the identification of genetic biomarkers of ASD for diagnostic and therapeutic purposes.
GENETIC VARIANTS
Common variants
The genetic architecture of ASD is diverse in frequency (common vs. rare variation), mode of inheritance (inherited vs. de novo variation), type of variation (single nucleotide, indel, or copy number variation [CNV]) and mode of action (dominant, recessive, or additive) [3,7]. Common variation refers to genetic variation from the reference genome, which is present in >1% of the population. Common variants with small effects are thought to act additively in the development of complex traits in ASD [8]. One recent investigation reported that the liability of ASD is mostly attributed to common variation in the genetic architecture, and that rare de novo mutations contribute to individual liability (49% of common inherited variants, 3% of de novo, 3% of rare inherited variants, and 41% of unaccounted) [7].
Confirmation of the most specific, consistently replicated, and highly effective common variants involved in the pathogenesis of ASD is another issue. The first molecular genetic studies of autism were candidate gene association studies that aimed to discover common genetic variants in the form of single-nucleotide polymorphisms (SNPs). However, a large disadvantage of this is that it requires existing physiological, biochemical, or functional knowledge, which is either finite or unavailable [9]. These investigations have been hindered by inadequate sample sizes and sparse genotyping, resulting in a lack of reproducible markers except for a few plausible genes [9,10].
The most consistently reported genes among the common variants include the gamma-aminobutyric acid (GABA) A receptor, beta 3 (GABRB3); oxytocin receptor (OXTR); reelin (RELN); serotonin transporter (SLC6A4); N-methyl-D-aspartate receptor (NMDA; GRIN2B); arginine vasopressin receptor 1A (AVPR1A); engrailed homeobox 2 (EN2); integrin, beta 3 (platelet glycoprotein IIIa, antigen CD61; ITGB3); met proto-oncogene (hepatocyte growth factor receptor; MET); and contactin-associated protein-like 2 (CNTCAP2) genes [11,12,13,14,15,16,17,18,19,20,21,22]. GABRB3, which is localized to chromosome 15q11-q13 and is involved in genome instability, gene expression, imprinting, and recombination, was investigated in the first era of ASD genetic research [13,23]. This region became a major subject of attention because deletion of this locus is related to monogenic causes of ASD, Prader-Willi syndrome, and Algelman's syndrome, and because GABA may be a pharmacological therapeutic target [13,24,25]. Oxytocin acts as a neuromodulator in the central nervous system, and induces social/affective bonding in animal models. The OXTR is a promising biomarker candidate, due to its genetic variants, functions on behavior, and positive results in human clinical trials [17,26,27].
However, these candidate gene studies also revealed that common variation has a weak effect when individual SNPs are investigated. A genome-wide association study (GWAS) avoids the need for a priori hypotheses for the primary cause of illness, and is a more appropriate approach for genetic studies of complex disorders such as autism [10]. GWASs have been applied to psychiatric disorders with complex phenotypes, and several variants have been reported [28]. Several GWASs have been conducted for ASD, and a few well-designed studies reported that common genetic variants on 5p14.1 and 5p15 were highlighted and replicated in two independent samples, each carrying a small increased risk (OR 1.2) or protective effect (OR 0.6) [29,30,31]. A significant association was observed with the CDH9 and CDH10 genes, but replications of this association was inconsistent [29,30,31,32,33]. Several studies identified significant SNP markers that were replicated in two or more independent samples and were associated with specific phenotypes of ASD, but the effect sizes were relatively small [29,30,31,32,33,34,35]. However, significant genome-wide results were not consistently reproduced across studies and ethnicities [33,35].
Those inconsistencies can be attributed to the phenotypic heterogeneity of ASD and to relatively small sample sizes. Researchers attempted to decrease phenotypic heterogeneity by subphenotyping or using quantitative phenotypes, but this was unsuccessful in enhancing the substantial power of GWAS, resulting in the necessity of very large sample sizes, such as 50,000 individuals [5,33,34].
Rare variants and monogenic autism
Rare variation is genetic variation that is present in the population at a frequency of ≤1%. ASD can be expressed as the behavioral manifestation of known genetic syndromes, called syndromic autism, as opposed to idiopathic autism, which does not have known genetic causes. Syndromic autism often has dysmorphic features characterized by the genetic syndrome it belongs to and equal male:female ratios, unlike idiopathic autism, which occurs 4-5 times more frequently in males than in females [36]. Single-gene disorders, including fragile X (mutations in FMR1), tuberous sclerosis complex (mutations in TSC1 and TSC2), Dup15q syndrome, deletions in the 16p11.2 region, Rett syndrome (mutation in MeCP2), and neurofibromatosis (mutations in NF1), are detected in 3-5% of subjects with ASD, and are well-known as having an ASD phenotype as well as comorbid intellectual disabilities and epilepsy [37].
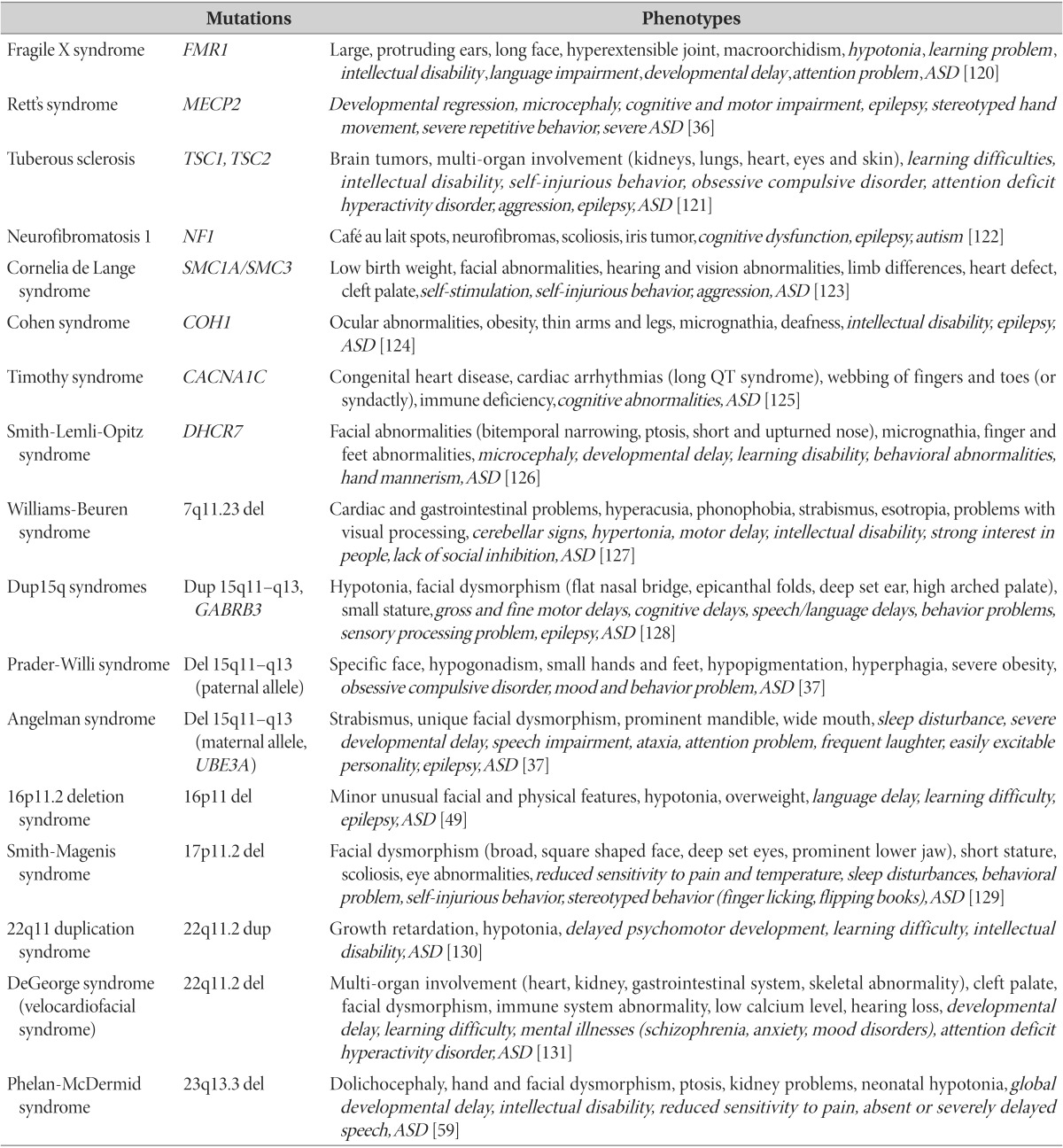
Recent development of whole-exome sequencing (WES) techniques has revealed that more than 25% of individuals with ASD have identifiable, causative, and protein-disrupting rare genetic mutations [5]. However, single mutations account for no more than 1% of cases, mainly due to phenotypic heterogeneity and variable penetrance. Though the prevalence is not strikingly high, syndromic autism helps to understand core deficits of ASD as one of the phenotypes that specific genetic mutations carry, and acts as a gateway to explore the genetic etiology of ASD. The representative examples of monogenic autism and their clinical implications are summarized in Table 1.
Table 1. Examples of monogenic "syndromic" autism and related phenotypes.
ASD, autism spectrum disorder.
Copy number variation
Copy number variants (CNVs) are variations (duplication or deletion) in chromosomal structure of greater than 1,000 nucleotides, usually a section of DNA with a length from 1 kb to several Mb. CNVs can be either common or rare, transmitted or de novo, and are widely distributed in human genome, accounting for a substantial proportion of genetic variation [5,38]. Studies have revealed an increased frequency of CNVs in individuals with ASD compared to normal controls and several de novo CNVs in children with autism, suggesting excessive genomic instability. The frequency of de novo CNVs in ASD has been reported as 3-19% in ASD from simplex and multiplex families, compared to approximately 1% in healthy controls [39,40,41].
Genomic imbalances associated with ASD are classified as recurrent and nonrecurrent events. Recurrent events are non-allelic homologous recombination, with reciprocal dosage imbalances (deletion and duplication) in different individuals [42]. Notable examples of recurrent CNVs are microdeletions and duplications in chromosome 1q21, 15q13, and 16p11.2, and microdeletion syndrome in chromosomes 2p15-2p16.1, 17p11.2, and 17q12 [43,44,45,46,47,48,49]. CNVs are associated with a wide range of phenotypic heterogeneity, including dysmorphic features, intellectual disabilities, language impairments, attention problems, hyperactivity, aggression and other behavioral problems, mood disorders, and schizophrenia, which implicates those variants might not be a specific cause of social disability in ASD [18].
Synaptic genes
Of the genetic variations studied regarding ASD, the most consistently reported genetic abnormalities are mutations in synaptic genes, including neuroligins (NLGN), SH3 and multiple ankyrin repeat domains (SHANK), neurexin (NRXN) families, and contactin associated protein-like 2 (CNTNAP2) [50,51,52,53,54,55,56,57,58,59,60,61,62]. Mutations in synaptic genes are not specific to ASD, and are also found in other neuropsychiatric disorders, such as schizophrenia and Alzheimer's disease [63,64]. However, as these neuropsychiatric conditions share common features with ASD, such as cognitive dysfunction, limited emotional expression, and lack of social reciprocity, synaptic dysfunction is still considered a common pathway of these major, chronic neuropsychiatric illnesses [5,18].
NLGNs are known to act as splice site-specific ligands for beta-neurexins and be involved in the formation and remodeling of central nervous system synapses (http://www.ncbi.nlm.nih.gov/gene/54413). The identification of a de novo, loss of function mutation in neuroligin 4, X-linked (NLGN4X) in an affected mother that was transmitted to two affected boys first suggested the possibility of synaptic dysfunction involvement in ASD [65]. This was followed by the identification of a single-base missense mutation of NLGN3 in another family [53]. These findings have been replicated in other studies, and NLGN3, NLGN4, and NLGN4Y were found to be possibly associated with ASD. However, mutation of those genes in ASD is relatively low (0.6-3.3%), and the clinical phenotypes and neurobiological characteristics of these mutations are also quite diverse, including ASD, intellectual disabilities, and Tourette syndrome, and inconsistent across ethnicities [51,54,55,61,62].
A second family of genes possibly associated with ASD is the SHANK genes (SHANK1, SHANK2, and SHANK3), encoding synaptic proteins that may function as molecular scaffolds in the postsynaptic density of excitatory synapses (http://www.ncbi.nlm. nih.gov/gene/22941). SHANK3 is the most widely studied, but SHANK1 and SHANK2 are also implicated by de novo deletions observed in subjects with ASD [50,57,58,59]. Durant et al. (2007) reported eight non-synonymous mutations in ASD patients that were not present in healthy controls; rare de novo mutations in SHANK3 located in chromosome 22q13.3 were identified in probands and families with ASD in many studies [50]. Rare mutations and genomic deletions have been reported in different SHANK3 loci, with a frequency of 0.2-0.8% of probands in ASD [50,57,66,67,68,69]. Mutations in SHANK3 are gaining attention, as they are related to Phelan-McDermid syndrome (PMS) and 22q13 deletion syndrome, and are one of the known genetic causes of ASD. PMS is characterized by autism or autistic-like behavior in more than 50% of subjects, and is accompanied by neurological deficits, including global developmental delay, moderate to severe intellectual impairment, absent or severely delayed speech, and neonatal hypotonia [59].
The transmission pattern of SHANK3 mutations is variable; inheritance from healthy parents and existence in unaffected siblings were reported [50,57,67]. Recently, Nemirovsky et al. (2015) reported germline mosaicism for a heterozygous cytosine deletion in exon 21 of SHANK3 by whole-genome sequencing in three male siblings from a segregated family exhibiting phenotypes of severe intellectual disability, absence of language, autism spectrum symptoms, and epilepsy [58]. As with other potential candidates, the associated phenotypes of SHANK3 mutations are not specific for ASD, but SHANK3 is regarded as one of the potential causative genes and therapeutic targets of ASD, based on animal and cellular model studies.
Other important synaptic genes are NRXN1, NRXN2 and NRXN3, encoding neuroligins. This trans-synaptic complex is required for efficient neurotransmission, and they are involved in the formation of synaptic contacts by interaction with neurexins [60]. Neuroligin aggregation is synaptogenic, but exhibits specificity: NLGN1, NLGN3 and NLGN4 link only to glutamatergic postsynaptic proteins, but NLGN2 links to both glutamatergic and GABAergic postsynaptic proteins [52]. In the earlier era of ASD genetic studies, CNVs were found to disrupt the locus containing NRXN1, but this was inconsistent, with a high unaffected carrier frequency of deletions [70,71,72,73]. More recently, there have been relatively large cohort studies that describe a higher rate of deletions in the NRXN1 region located in the probands of chromosome 2p16.3 associated with ASD, compared to healthy controls, with an overrepresentation of small-sized inverted repeats [72,74]. Shared psychopathologies related to the deletions were developmental delays, speech delays, abnormal behaviors, including ASD, and some degree of dysmorphism [72].
CNTNAP2 is another candidate gene suggested to be associated with ASD by human and animal model studies. Family-based association studies identified a common variant (rs7794745) that was associated with increased risk of autism, and another study revealed an amino acid substitution in the CNTNAP2 protein in children with autism [75,76,77]. Variation of the CNTNAP2 gene and age at first word, a language development phenotype, are both related to autism rather than to the diagnosis itself, and raises the implication that genetic variants of a quantitative phenotype of ASD interact with FOXP2 [75]. The functional relevance of CNTNAP2 genetic variants has been validated in animal models; CNTNAP2 (-/-) mice exhibited deficits in all three core behavioral deficits of ASD, as well as hyperactivity and epileptic seizures, and improved repetitive behavior of mutant mice [78]. A large-sized GWAS study failed to demonstrate a significant association of the marker noted in the previous studies, and no associations between rare heterogeneous mutations of CNTNAP2 and ASD were observed [29,79]. However, a recent investigation revealed the possible involvement of novel functional variants of the 5'-promotor region, mediated by alterations in transcription-binding sites in subjects with ASD. CNTNAP2 is still regarded as one of the potential causative genes of ASD that warrants further research [80].
These findings indicate the possibility that ASD might be caused by abnormalities in synaptic plasticity, as indicated by proteins that play essential roles in synaptic development and modification. There is evidence that NRXN (presynaptic) and NLGN (postsynaptic) interact as synaptic adhesion molecules, providing mechanistic support in synaptic formation and maintenance [81]. Alternate splicing of NLGN controls selective binding with α- and β-NRXN, provides synaptic diversity, and regulates function. Alternate splicing also controls the variable interaction of NRXN with other postsynaptic ligands, such as in glutamatergic, GABA-ergic, and cholinergic synapses [81,82]. The SHANK family includes postsynaptic scaffolding proteins, which link multiple receptor signaling complexes and the actin cytoskeleton that are essential for maintaining synaptic function [83]. SHANK3 directly and indirectly binds to CNTN and NLGNs and interacts with presynaptic glutamatergic receptors [83,84]. The molecular function of CNTNAP2 is relatively unclear, but it is known to encode a neuronal transmembrane protein member of the NRXN superfamily that is involved in neural-glia interactions and clustering of potassium channels in myelinated axons (http://www.omim.org/entry/604569).
Overall, despite of low frequency of de novo mutations in affected individuals, inconsistencies in genetic analysis results, and phenotypic heterogeneity, synapse-related genes are crucial candidates of ASD, and provide baseline evidence for testing compounds that can enhance synaptogenesis for the treatment of the core symptoms of ASD [25].
Genetic networks
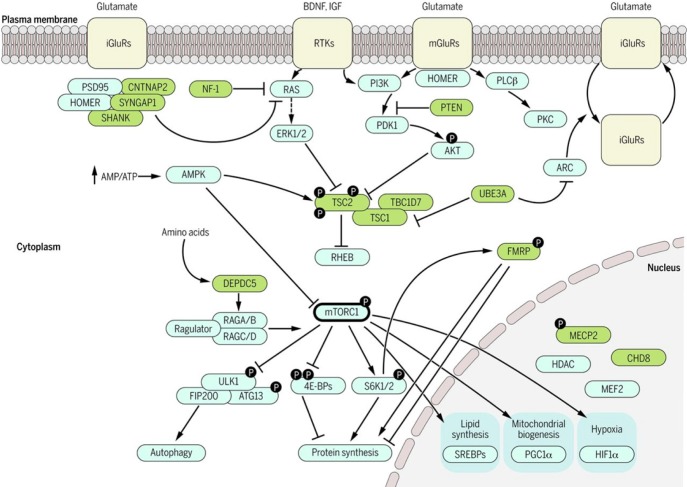
For some of the identified genes, genetic pathways were identified at the cellular and molecular level based on genetic network analyses and genetic functional studies. Besides synaptic development and function mentioned in the previous section, protein synthesis and metabolism, modulation of transcription process, chromatin remodeling, calcium signaling, and mTOR and oxytocin pathways have also been implicated [85,86,87]. This suggests that genes involved in ASD might be related each other in several convergent functional units, especially in neuronal development, modulation, and intracellular transcriptional mechanisms. As more and more causative genes of ASD are identified, their interaction within the context of functional significance would be clarified. Molecular pathways possibly involved in pathogenesis of ASD are summarized in Fig. 1 [86].
Fig. 1. Molecular pathways implicated in neurodevelopmental disorders. RTKs, receptor tyrosine kinases; iGluRs, metabotropic glutamate receptors; PGC-1α, peroxisome proliferator-activated receptor gamma coactivator 1-alpha; SREBP, sterol regulatory element-binding proteins; HIF1α, hypoxia-inducible factor 1 alpha; ULK1, unc-51-like kinase 1; ARC, activity-regulated cytoskeleton-associated protein; UBE3A, ubiquitin protein ligase E3A; MeCP2, methyl CpG binding protein 2; FMRP, fragile X mental retardation protein; PI3K/mTOR , phosphatidylinositol 3-kinase/mammalian target of rapamycin; PSD-95, postsynaptic density protein 95; CNTNAP2, Contactin-associated protein-like 2; NF1, neurofibromin 1; PLCβ, Abstract Phospholipase C β; SYNGAP1, Synaptic Ras GTPase-activating protein 1; ERK1/2, extracellular signal-regulated kinase; PTEN, Phosphatidylinositol-3,4,5-trisphosphate 3-phosphatase; PDK1, Pyruvate dehydrogenase lipoamide kinase isozyme 1; PKC, Paroxysmal kinesogenic choreoathetosis, neurological disorder Protein kinase C; AKT, Protein kinase B; AMPK, AMP-activated protein kinase; TSC2, Tuberous Sclerosis Complex 2; TSC1, tuberous sclerosis 1; RHEB, GTP-binding protein Rheb; DEPDC5, DEP domain-containing 5; mTORC1, mammalian target of rapamycin complex 1; mGluR, metabotropic glutamate receptor; SHANK, Shank protein; ATG13, Autophagy-related protein 13; HDAC, Histone deacetylases; CHD8, Chromodomain-helicase-DNA-binding protein 8; MEF2, myocyte enhancer factor-2; RAS, Ras protein; TBC1D7, TBC1 domain family, Member 7; 4E-BPs, eIF4E-binding proteins; FIP200, a ULK-interacting protein; S6K1/2, Anti-RIBOSOMAL S6 KINASE 1/2; HOMER, homer scaffolding protein; RAGA/B, Ras-related GTP binding A/B ortholog; RAGC/D, Ras-related GTP binding C/D ortholog. From M. Sahin and M. Sur, Genes, circuits, and precision therapies for autism and related neurodevelopmental disorders, Science 350, aab3897 (2015). DOI: 10.1126/science.aab3897. Reprinted with permission from The American Association for the Advancement of Science (AAAS).
Genes and brain circuits
Theoretically, ASD is an end-product of gene-environment interaction, mediated by changes in brain physiology, function, and morphology causing cognitive and behavioral dysfunction. Multiple brain areas involving facial recognition, emotion evaluation, empathy, mentalization, social cognition and behavior are known to be associated with ASD as part of the "social brain network" [88]. Alterations in brain connectivity and morphology might be a potential endophenotypes of ASD. However, not many human studies have explored associations between specific genetic variants and brain circuits or morphological phenotypes. One recent study examined phenotypic characteristics of subjects with ASD carrying germline heterogeneous PTEN mutations (PTEN-ASD); compared to subjects with idiopathic (non-PTEN) ASD and healthy controls, subjects with PTEN-ASD showed prominent cognitive dysfunction and white-matter abnormalities, mediated by reductions in the PTEN protein [89]. Moreover, significant differences in fMRI activation and deactivation patterns to social stimuli as well as functional and structural connectivity in the temporo-parietal region based on the existence of the rs1858830 MET risk allele (C/C) highlighted alterations in gene-brain pathway in ASD [14]. More data are necessary to understand the complex pathways connecting specific genetic mutation/genotype and brain changes, which have a direct impact on specific behavioral phenotypes of ASD.
GENETICS USED FOR DIAGNOSIS AND THERAPEUTIC TARGETS: CLINICAL REALM
The ultimate goal of the evaluation of genetic etiology in complex disorders is the discovery of biomarkers for risk assessment, diagnosis, and prediction of therapeutic responses and prognoses, and the development of therapeutic components. The current best estimate diagnosis of ASD is based on behavioral observation and developmental history, assisted by standardized diagnostic instruments. While it is strongly suggested that early intervention in ASD can promote better prognoses, reduce secondary behavioral complications, and even induce normalization of brain activity [90], diagnostic confirmation before 2 years of age based only on the behavioral observations has limitations. Thus, the identification of genetic markers will provide useful information to aid in the early diagnosis of ASD in infants and begin early intervention.
However, the current status of genetic diagnoses of ASD is insufficient in clinical utility and precision for general applications. While known genetic causes are identified in 20-25% of ASD, each of those mutations/variants are rare and account for only 1-2% of the probands [6,91,92], which means there are no single predictable genetic markers for the development of ASD. As described previously, scientists believe that ASD is the product of the interplay between multiple common and rare genetic variants, and that genetic diagnosis should involve a combination of multiple genetic markers as a form of targeted gene panels. There have been attempts to make a gene set to diagnose ASD: for example, Skafidas et al. (2012) identified a group of SNPs selected by pathway analysis, and applied machine learning to the identified SNPs to generate a predictive classifier for ASD diagnosis. By applying 237 highly significant SNPs to three independent cohorts, there was a high level of diagnostic accuracy observed in genetically homogenous populations (84.3-85.6%), but not in an ethnically distinct cohort of Han Chinese (56%) [93]. Despite small sample sizes and the preliminary nature of the methodology, other researchers applied gene panels on subgroups of ASD and observed 75-90% accuracy in classification [94]. These studies may imply that development of a diagnosis based on genetic markers needs consider ethnic diversity and phenotypic heterogeneity. There are several gene panels offered by clinical molecular laboratories, but their clinical validity has not yet been fully evaluated [95]. In the future, next generation sequencing (NGS) is expected to produce a level of resolution down to the single base pair level and to enhance the assay precision level, but diagnostic validity should proceed so that the technology works in a clinically valid way [96].
In its current status, clinical value is primarily focused on the identification of known genetic causes of ASD. It is recommended that once a clinical diagnosis of ASD is made, genetic testing should be initiated [96]. This includes single-gene tests for monogenic causes of ASD, including fragile X syndrome, tuberous sclerosis complex, Rett syndrome, Angelman syndrome, Prader-Willi syndrome, phosphatase and tensin homolog (PTEN)-associated disorders, Noonan syndrome, cortical dysplasia-focal epilepsy syndrome (associated with CNTNAP2), and Phelan-McDermid syndrome, by detecting point mutations, microdeletions, duplications, and large repeat expansions using sequencing, fluorescence in situ hybridization (FISH), and Southern blotting technologies [95]. An assay for CNVs with array comparative genomic hybridization (aCGH) is also available for known variations of ASD [96]. At the clinical level, practice guidelines of the American College of Medical Genetics (ACMG) recommends a three generation family history with pedigree analyses; an initial evaluation for known syndromes associated with ASD, especially if the subject has dysmorphic features or specific clinical indicators; a chromosomal microarray; and DNA testing for fragile X for all male children suspected of an ASD as the first tier genetic evaluation. ACMG recommends sequencing and duplication testing for the MECP2 gene in female patients, and PTEN testing for those with macrocephaly as second tier genetic testing [97,98].
There are ethical considerations of the clinical use of genetic testing and counseling for ASD. First, it is important that biomarker discovery, especially commercialization of biomarker data in autism, does not result in children being given a biological label that fixes and defines their potential and treatment [99,100]. Second, genetic biomarker results have a huge impact on parental decision-making toward reproduction; therefore, more research and communication is needed for a better understanding of parental needs and attitudes [100].
Pharmacological treatment of ASD is primarily focused on improving comorbid behavioral emotional symptoms, such as irritability, aggression, anxiety, tics, self-injurious behaviors, and epilepsy [101]. One of the ultimate goals of the discovery and validation of biomarkers is developing molecular therapeutics to treat the core symptoms of ASD, based on knowledge about disease modifying mechanisms. Identification of causative genes, especially from high-throughput methods such as NGS, is paving the way for developing pharmacological agents to treat the core symptoms of ASD, including abnormal reciprocal social interaction and communication. Several recent well-designed studies have modeled human genetic variants associated with ASD in mice, using SHANK3, CNTNAP2, MECP2, UBE3A, and FMR1 genes, and are refining potential therapeutic mechanisms [78,102,103,104,105,106]. These studies refine the function of genetic biomarkers of autism and relate them to behavioral phenotypes, and suggest potential target compounds for recovery of function. However, it is only the beginning of the discovery of therapeutic molecules for human subjects, covering the heterogeneity and complexity of the molecular pathways of autism.
A few empirical trials have begun for syndromic autism with well-known monogenic etiology, with hopes to expand the trial for idiopathic ASD. Tuberous sclerosis is one of the examples; the TSC1 and TSC2 genes encode proteins regulating the mTORC1 protein complex. mTOR plays an important role in protein synthesis, cell growth, and axon formation. mTOR inhibitors (including rapamycin and everolimus) are currently being tested for their effect on neurocognitive outcomes in children with tuberous sclerosis complex, with outcome measurements that include autism symptoms, cognition, language skills, and behavior [79,107]. Also clinical trials using mechanism-based targeted treatment for Dup15q syndrome (duplication of 15q11.2-q13) and fragile X syndrome are also under way [37,98,107].
Oxytocin is neuromodulatory hormone involved in various social behaviors in humans and animals, including parents-offspring bonding, affection, social recognition, and social antagonism [17]. Although inconsistencies in genetic studies of OXTR in ASD, positive associations have been reported in multiple studies and across multiple ethnicities. Significant hypermethylation of critical sites of OXTR was observed in subjects with ASD and in the brother of a proband with OXTR deletion implicated epigenetic modification of the OXTR gene in ASD [26,108,109]. OXTR knock-out animal models show various characteristics of social behavior including parental nurturing, pair bonding, and social memory as well as brain changes in the limbic and paralimbic regions [110,111]. Based on strong coherent evidence from human and animal studies across genetic, neural, and behavioral levels, oxytocin has been chosen as a pharmacological agent targeting core symptoms of ASD. A few randomized controlled trials have showed the efficacy of intravenous or nasal administration of oxytocin in core domains of ASD, such as social cognition and behavior. A few well-designed studies have reported enhanced performance in empathy tasks accompanied by increased anterior insula activity and coordination in the medical prefrontal cortex [112,113,114]. Although there are unmet limitations in the generalizability of subjects, establishment of dosage and route of administration, and maintenance of effect and safety in children, oxytocin might be a potential therapeutic agent for core symptoms of ASD with the most achievable clinical utility.
NGS IN ASD GENETICS
High-throughput technologies have facilitated gene discovery in ASD. The most recent development, NGS, which include whole-genome sequencing (WGS), whole-exome sequencing (WES), and targeted sequencing, promotes the precise identification of genetic variants at base-by-base levels. NGS promotes the identification of rare alleles to a degree not possible using genotyping platforms, and allows identification of single gene defects and partial variations of gene function [37,115]. NGS in ASD is still emerging; it is useful to identify novel de novo variants not observed by conventional methods. Recent studies have confirmed de novo CNV loci in large-sized cohorts of idiopathic ASD families, observed new candidate loci, and confirmed de novo mutations in subjects with ASD and other neuropsychological abnormalities [58,116,117,118]. Regarding diagnostic utility, one recent study reported complicated results that the diagnostic yield of WES is comparable to chromosomal microarray, while combined diagnostic yield is higher among children with more complex morphological phenotypes, emphasizing the importance of phenotypic classification [119].
Although NGS is not yet widespread due to its relatively high cost and the demand for techniques involving large-scale data and bioinformatics, it is predicted that large-scale investigations combining NGS technology and accumulated clinical data will facilitate the genetic study of ASD from diagnoses to targeted treatments in the future [5].
CONCLUSION
A wide range of genetic variation is involved in ASD, with interplays of gene-gene and gene-environment interactions. Both genotypic and phenotypic heterogeneity contribute to the difficulty in the thorough exploration and confirmation of causative genetic factors. However, recent technological developments, including NGS, and the accumulation of clinical information are bridging the gap in the application of genetic knowledge towards clinical practice.
ACKNOWLEDGMENTS
This work was supported by the Korea Healthcare Technology R&D Project (A120029) from the Ministry of Health and Welfare, Republic of Korea. The author has no conflict of interests to declare.
References
- 1.Bailey A, Le Couteur A, Gottesman I, Bolton P, Simonoff E, Yuzda E, Rutter M. Autism as a strongly genetic disorder: evidence from a British twin study. Psychol Med. 1995;25:63–77. doi: 10.1017/s0033291700028099. [DOI] [PubMed] [Google Scholar]
- 2.Ozonoff S, Young GS, Carter A, Messinger D, Yirmiya N, Zwaigenbaum L, Bryson S, Carver LJ, Constantino JN, Dobkins K, Hutman T, Iverson JM, Landa R, Rogers SJ, Sigman M, Stone WL. Recurrence risk for autism spectrum disorders: a Baby Siblings Research Consortium study. Pediatrics. 2011;128:e488–e495. doi: 10.1542/peds.2010-2825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.De Rubeis S, Buxbaum JD. Genetics and genomics of autism spectrum disorder: embracing complexity. Hum Mol Genet. 2015;24(R1):R24–R31. doi: 10.1093/hmg/ddv273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hallmayer J, Cleveland S, Torres A, Phillips J, Cohen B, Torigoe T, Miller J, Fedele A, Collins J, Smith K, Lotspeich L, Croen LA, Ozonoff S, Lajonchere C, Grether JK, Risch N. Genetic heritability and shared environmental factors among twin pairs with autism. Arch Gen Psychiatry. 2011;68:1095–1102. doi: 10.1001/archgenpsychiatry.2011.76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Geschwind DH, State MW. Gene hunting in autism spectrum disorder: on the path to precision medicine. Lancet Neurol. 2015;14:1109–1120. doi: 10.1016/S1474-4422(15)00044-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Voineagu I, Yoo HJ. Current progress and challenges in the search for autism biomarkers. Dis Markers. 2013;35:55–65. doi: 10.1155/2013/476276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gaugler T, Klei L, Sanders SJ, Bodea CA, Goldberg AP, Lee AB, Mahajan M, Manaa D, Pawitan Y, Reichert J, Ripke S, Sandin S, Sklar P, Svantesson O, Reichenberg A, Hultman CM, Devlin B, Roeder K, Buxbaum JD. Most genetic risk for autism resides with common variation. Nat Genet. 2014;46:881–885. doi: 10.1038/ng.3039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Klei L, Sanders SJ, Murtha MT, Hus V, Lowe JK, Willsey AJ, Moreno-De-Luca D, Yu TW, Fombonne E, Geschwind D, Grice DE, Ledbetter DH, Lord C, Mane SM, Martin CL, Martin DM, Morrow EM, Walsh CA, Melhem NM, Chaste P, Sutcliffe JS, State MW, Cook EH, Jr, Roeder K, Devlin B. Common genetic variants, acting additively, are a major source of risk for autism. Mol Autism. 2012;3:9. doi: 10.1186/2040-2392-3-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zhu M, Zhao S. Candidate gene identification approach: progress and challenges. Int J Biol Sci. 2007;3:420–427. doi: 10.7150/ijbs.3.420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Losh M, Sullivan PF, Trembath D, Piven J. Current developments in the genetics of autism: from phenome to genome. J Neuropathol Exp Neurol. 2008;67:829–837. doi: 10.1097/NEN.0b013e318184482d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sutcliffe JS, Delahanty RJ, Prasad HC, McCauley JL, Han Q, Jiang L, Li C, Folstein SE, Blakely RD. Allelic heterogeneity at the serotonin transporter locus (SLC6A4) confers susceptibility to autism and rigid-compulsive behaviors. Am J Hum Genet. 2005;77:265–279. doi: 10.1086/432648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ma DQ, Rabionet R, Konidari I, Jaworski J, Cukier HN, Wright HH, Abramson RK, Gilbert JR, Cuccaro ML, Pericak-Vance MA, Martin ER. Association and gene-gene interaction of SLC6A4 and ITGB3 in autism. Am J Med Genet B Neuropsychiatr Genet. 2010;153B:477–483. doi: 10.1002/ajmg.b.31003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kim SA, Kim JH, Park M, Cho IH, Yoo HJ. Association of GABRB3 polymorphisms with autism spectrum disorders in Korean trios. Neuropsychobiology. 2006;54:160–165. doi: 10.1159/000098651. [DOI] [PubMed] [Google Scholar]
- 14.Rudie JD, Hernandez LM, Brown JA, Beck-Pancer D, Colich NL, Gorrindo P, Thompson PM, Geschwind DH, Bookheimer SY, Levitt P, Dapretto M. Autism-associated promoter variant in MET impacts functional and structural brain networks. Neuron. 2012;75:904–915. doi: 10.1016/j.neuron.2012.07.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Penagarikano O, Geschwind D. CNTNAP2 and autism spectrum disorders. In: Powel CM, editor. The autisms. New York, NY: Oxford University Press; 2013. pp. 274–285. [Google Scholar]
- 16.Yang SY, Cho SC, Yoo HJ, Cho IH, Park M, Yoe J, Kim SA. Family-based association study of microsatellites in the 5flanking region of AVPR1A with autism spectrum disorder in the Korean population. Psychiatry Res. 2010;178:199–201. doi: 10.1016/j.psychres.2009.11.007. [DOI] [PubMed] [Google Scholar]
- 17.Yamasue H. Promising evidence and remaining issues regarding the clinical application of oxytocin in autism spectrum disorders. Psychiatry Clin Neurosci. 2015 doi: 10.1111/pcn.12364. in press. [DOI] [PubMed] [Google Scholar]
- 18.Persico AM, Napolioni V. Autism genetics. Behav Brain Res. 2013;251:95–112. doi: 10.1016/j.bbr.2013.06.012. [DOI] [PubMed] [Google Scholar]
- 19.Wang L, Jia M, Yue W, Tang F, Qu M, Ruan Y, Lu T, Zhang H, Yan H, Liu J, Guo Y, Zhang J, Yang X, Zhang D. Association of the ENGRAILED 2 (EN2) gene with autism in Chinese Han population. Am J Med Genet B Neuropsychiatr Genet. 2008;147B:434–438. doi: 10.1002/ajmg.b.30623. [DOI] [PubMed] [Google Scholar]
- 20.Lee EJ, Choi SY, Kim E. NMDA receptor dysfunction in autism spectrum disorders. Curr Opin Pharmacol. 2015;20:8–13. doi: 10.1016/j.coph.2014.10.007. [DOI] [PubMed] [Google Scholar]
- 21.Yoo HJ, Cho IH, Park M, Yang SY, Kim SA. Family based association of GRIN2A and GRIN2B with Korean autism spectrum disorders. Neurosci Lett. 2012;512:89–93. doi: 10.1016/j.neulet.2012.01.061. [DOI] [PubMed] [Google Scholar]
- 22.Skaar DA, Shao Y, Haines JL, Stenger JE, Jaworski J, Martin ER, DeLong GR, Moore JH, McCauley JL, Sutcliffe JS, Ashley-Koch AE, Cuccaro ML, Folstein SE, Gilbert JR, Pericak-Vance MA. Analysis of the RELN gene as a genetic risk factor for autism. Mol Psychiatry. 2005;10:563–571. doi: 10.1038/sj.mp.4001614. [DOI] [PubMed] [Google Scholar]
- 23.Buxbaum JD, Silverman JM, Smith CJ, Greenberg DA, Kilifarski M, Reichert J, Cook EH, Jr, Fang Y, Song CY, Vitale R. Association between a GABRB3 polymorphism and autism. Mol Psychiatry. 2002;7:311–316. doi: 10.1038/sj.mp.4001011. [DOI] [PubMed] [Google Scholar]
- 24.Martin ER, Menold MM, Wolpert CM, Bass MP, Donnelly SL, Ravan SA, Zimmerman A, Gilbert JR, Vance JM, Maddox LO, Wright HH, Abramson RK, DeLong GR, Cuccaro ML, Pericak-Vance MA. Analysis of linkage disequilibrium in gamma-aminobutyric acid receptor subunit genes in autistic disorder. Am J Med Genet. 2000;96:43–48. doi: 10.1002/(sici)1096-8628(20000207)96:1<43::aid-ajmg9>3.0.co;2-3. [DOI] [PubMed] [Google Scholar]
- 25.Vorstman JA, Spooren W, Persico AM, Collier DA, Aigner S, Jagasia R, Glennon JC, Buitelaar JK. Using genetic findings in autism for the development of new pharmaceutical compounds. Psychopharmacology(Berl) 2014;231:1063–1078. doi: 10.1007/s00213-013-3334-z. [DOI] [PubMed] [Google Scholar]
- 26.Lerer E, Levi S, Salomon S, Darvasi A, Yirmiya N, Ebstein RP. Association between the oxytocin receptor (OXTR) gene and autism: relationship to Vineland Adaptive Behavior Scales and cognition. Mol Psychiatry. 2008;13:980–988. doi: 10.1038/sj.mp.4002087. [DOI] [PubMed] [Google Scholar]
- 27.Preti A, Melis M, Siddi S, Vellante M, Doneddu G, Fadda R. Oxytocin and autism: a systematic review of randomized controlled trials. J Child Adolesc Psychopharmacol. 2014;24:54–68. doi: 10.1089/cap.2013.0040. [DOI] [PubMed] [Google Scholar]
- 28.Collins AL, Sullivan PF. Genome-wide association studies in psychiatry: what have we learned? Br J Psychiatry. 2013;202:1–4. doi: 10.1192/bjp.bp.112.117002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ma D, Salyakina D, Jaworski JM, Konidari I, Whitehead PL, Andersen AN, Hoffman JD, Slifer SH, Hedges DJ, Cukier HN, Griswold AJ, McCauley JL, Beecham GW, Wright HH, Abramson RK, Martin ER, Hussman JP, Gilbert JR, Cuccaro ML, Haines JL, Pericak-Vance MA. A genome-wide association study of autism reveals a common novel risk locus at 5p14.1. Ann Hum Genet. 2009;73:263–273. doi: 10.1111/j.1469-1809.2009.00523.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wang K, Zhang H, Ma D, Bucan M, Glessner JT, Abrahams BS, Salyakina D, Imielinski M, Bradfield JP, Sleiman PM, Kim CE, Hou C, Frackelton E, Chiavacci R, Takahashi N, Sakurai T, Rappaport E, Lajonchere CM, Munson J, Estes A, Korvatska O, Piven J, Sonnenblick LI, Alvarez Retuerto AI, Herman EI, Dong H, Hutman T, Sigman M, Ozonoff S, Klin A, Owley T, Sweeney JA, Brune CW, Cantor RM, Bernier R, Gilbert JR, Cuccaro ML, McMahon WM, Miller J, State MW, Wassink TH, Coon H, Levy SE, Schultz RT, Nurnberger JI, Haines JL, Sutcliffe JS, Cook EH, Minshew NJ, Buxbaum JD, Dawson G, Grant SF, Geschwind DH, Pericak-Vance MA, Schellenberg GD, Hakonarson H. Common genetic variants on 5p14.1 associate with autism spectrum disorders. Nature. 2009;459:528–533. doi: 10.1038/nature07999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Weiss LA, Arking DE, Daly MJ, Chakravarti A, Arking DE, Brune CW, West K, O'Connor A, Hilton G, Tomlinson RL, West AB, Cook EH, Jr, Chakravarti A, Weiss LA, Green T, Chang SC, Gabriel S, Gates C, Hanson EM, Kirby A, Korn J, Kuruvilla F, McCarroll S, Morrow EM, Neale B, Purcell S, Sasanfar R, Sougnez C, Stevens C, Altshuler D, Gusella J, Santangelo SL, Sklar P, Tanzi R, Daly MJ, Anney R, Bailey AJ, Baird G, Battaglia A, Berney T, Betancur C, Bölte S, Bolton PF, Brian J, Bryson SE, Buxbaum JD, Cabrito I, Cai G, Cantor RM, Cook EH, Jr, Coon H, Conroy J, Correia C, Corsello C, Crawford EL, Cuccaro ML, Dawson G, de Jonge M, Devlin B, Duketis E, Ennis S, Estes A, Farrar P, Fombonne E, Freitag CM, Gallagher L, Geschwind DH, Gilbert J, Gill M, Gillberg C, Goldberg J, Green A, Green J, Guter SJ, Haines JL, Hallmayer JF, Hus V, Klauck SM, Korvatska O, Lamb JA, Laskawiec M, Leboyer M, Le Couteur A, Leventhal BL, Liu XQ, Lord C, Lotspeich LJ, Maestrini E, Magalhaes T, Mahoney W, Mantoulan C, McConachie H, McDougle CJ, McMahon WM, Marshall CR, Miller J, Minshew NJ, Monaco AP, Munson J, Nurnberger JI, Jr, Oliveira G, Pagnamenta A, Papanikolaou K, Parr JR, Paterson AD, Pericak-Vance MA, Pickles A, Pinto D, Piven J, Posey DJ, Poustka A, Poustka F, Regan R, Reichert J, Renshaw K, Roberts W, Roge B, Rutter ML, Salt J, Schellenberg GD, Scherer SW, Sheffield V, Sutcliffe JS, Szatmari P, Tansey K, Thompson AP, Tsiantis J, Van Engeland H, Vicente AM, Vieland VJ, Volkmar F, Wallace S, Wassink TH, Wijsman EM, Wing K, Wittemeyer K, Yaspan BL, Zwaigenbaum L, Morrow EM, Yoo SY, Sean Hill R, Mukaddes NM, Balkhy S, Gascon G, Al-Saad S, Hashmi A, Ware J, Joseph RM, LeClair E, Partlow JN, Barry B, Walsh CA, Pauls D, Moilanen I, Ebeling H, Mattila ML, Kuusikko S, Jussila K, Ignatius J, Sasanfar R, Tolouei A, Ghadami M, Rostami M, Hosseinipour A, Valujerdi M, Santangelo SL, Andresen K, Winkloski B, Haddad S, Kunkel L, Kohane Z, Tran T, Won Kong S, O'Neil SB, Hanson EM, Hundley R, Holm I, Peters H, Baroni E, Cangialose A, Jackson L, Albers L, Becker R, Bridgemohan C, Friedman S, Munir K, Nazir R, Palfrey J, Schonwald A, Simmons E, Rappaport LA, Gauthier J, Mottron L, Joober R, Fombonne E, Rouleau G, Rehnstrom K, von Wendt L, Peltonen L Gene Discovery Project of Johns Hopkins & the Autism Consortium. A genome-wide linkage and association scan reveals novel loci for autism. Nature. 2009;461:802–808. doi: 10.1038/nature08490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Anney R, Klei L, Pinto D, Regan R, Conroy J, Magalhaes TR, Correia C, Abrahams BS, Sykes N, Pagnamenta AT, Almeida J, Bacchelli E, Bailey AJ, Baird G, Battaglia A, Berney T, Bolshakova N, Bölte S, Bolton PF, Bourgeron T, Brennan S, Brian J, Carson AR, Casallo G, Casey J, Chu SH, Cochrane L, Corsello C, Crawford EL, Crossett A, Dawson G, de Jonge M, Delorme R, Drmic I, Duketis E, Duque F, Estes A, Farrar P, Fernandez BA, Folstein SE, Fombonne E, Freitag CM, Gilbert J, Gillberg C, Glessner JT, Goldberg J, Green J, Guter SJ, Hakonarson H, Heron EA, Hill M, Holt R, Howe JL, Hughes G, Hus V, Igliozzi R, Kim C, Klauck SM, Kolevzon A, Korvatska O, Kustanovich V, Lajonchere CM, Lamb JA, Laskawiec M, Leboyer M, Le Couteur A, Leventhal BL, Lionel AC, Liu XQ, Lord C, Lotspeich L, Lund SC, Maestrini E, Mahoney W, Mantoulan C, Marshall CR, McConachie H, McDougle CJ, McGrath J, McMahon WM, Melhem NM, Merikangas A, Migita O, Minshew NJ, Mirza GK, Munson J, Nelson SF, Noakes C, Noor A, Nygren G, Oliveira G, Papanikolaou K, Parr JR, Parrini B, Paton T, Pickles A, Piven J, Posey DJ, Poustka A, Poustka F, Prasad A, Ragoussis J, Renshaw K, Rickaby J, Roberts W, Roeder K, Roge B, Rutter ML, Bierut LJ, Rice JP, Salt J, Sansom K, Sato D, Segurado R, Senman L, Shah N, Sheffield VC, Soorya L, Sousa I, Stoppioni V, Strawbridge C, Tancredi R, Tansey K, Thiruvahindrapduram B, Thompson AP, Thomson S, Tryfon A, Tsiantis J, Van Engeland H, Vincent JB, Volkmar F, Wallace S, Wang K, Wang Z, Wassink TH, Wing K, Wittemeyer K, Wood S, Yaspan BL, Zurawiecki D, Zwaigenbaum L, Betancur C, Buxbaum JD, Cantor RM, Cook EH, Coon H, Cuccaro ML, Gallagher L, Geschwind DH, Gill M, Haines JL, Miller J, Monaco AP, Nurnberger JI, Jr, Paterson AD, Pericak-Vance MA, Schellenberg GD, Scherer SW, Sutcliffe JS, Szatmari P, Vicente AM, Vieland VJ, Wijsman EM, Devlin B, Ennis S, Hallmayer J. A genome-wide scan for common alleles affecting risk for autism. Hum Mol Genet. 2010;19:4072–4082. doi: 10.1093/hmg/ddq307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Connolly JJ, Glessner JT, Hakonarson H. A genome-wide association study of autism incorporating autism diagnostic interview-revised, autism diagnostic observation schedule, and social responsiveness scale. Child Dev. 2013;84:17–33. doi: 10.1111/j.1467-8624.2012.01838.x. [DOI] [PubMed] [Google Scholar]
- 34.Chaste P, Klei L, Sanders SJ, Hus V, Murtha MT, Lowe JK, Willsey AJ, Moreno-De-Luca D, Yu TW, Fombonne E, Geschwind D, Grice DE, Ledbetter DH, Mane SM, Martin DM, Morrow EM, Walsh CA, Sutcliffe JS, Lese Martin C, Beaudet AL, Lord C, State MW, Cook EH, Jr, Devlin B. A genome-wide association study of autism using the Simons Simplex Collection: Does reducing phenotypic heterogeneity in autism increase genetic homogeneity? Biol Psychiatry. 2015;77:775–784. doi: 10.1016/j.biopsych.2014.09.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Liu X, Shimada T, Otowa T, Wu YY, Kawamura Y, Tochigi M, Iwata Y, Umekage T, Toyota T, Maekawa M, Iwayama Y, Suzuki K, Kakiuchi C, Kuwabara H, Kano Y, Nishida H, Sugiyama T, Kato N, Chen CH, Mori N, Yamada K, Yoshikawa T, Kasai K, Tokunaga K, Sasaki T, Gau SS. Genome-wide Association Study of Autism Spectrum Disorder in the East Asian Populations. Autism Res. doi: 10.1002/aur.1536. in press. [DOI] [PubMed] [Google Scholar]
- 36.Hagberg B. Rett syndrome: clinical peculiarities and biological mysteries. Acta Paediatr. 1995;84:971–976. doi: 10.1111/j.1651-2227.1995.tb13809.x. [DOI] [PubMed] [Google Scholar]
- 37.Veltman MW, Craig EE, Bolton PF. Autism spectrum disorders in Prader-Willi and Angelman syndromes: a systematic review. Psychiatr Genet. 2005;15:243–254. doi: 10.1097/00041444-200512000-00006. [DOI] [PubMed] [Google Scholar]
- 38.Liu X, Takumi T. Genomic and genetic aspects of autism spectrum disorder. Biochem Biophys Res Commun. 2014;452:244–253. doi: 10.1016/j.bbrc.2014.08.108. [DOI] [PubMed] [Google Scholar]
- 39.Glessner JT, Wang K, Cai G, Korvatska O, Kim CE, Wood S, Zhang H, Estes A, Brune CW, Bradfield JP, Imielinski M, Frackelton EC, Reichert J, Crawford EL, Munson J, Sleiman PM, Chiavacci R, Annaiah K, Thomas K, Hou C, Glaberson W, Flory J, Otieno F, Garris M, Soorya L, Klei L, Piven J, Meyer KJ, Anagnostou E, Sakurai T, Game RM, Rudd DS, Zurawiecki D, McDougle CJ, Davis LK, Miller J, Posey DJ, Michaels S, Kolevzon A, Silverman JM, Bernier R, Levy SE, Schultz RT, Dawson G, Owley T, McMahon WM, Wassink TH, Sweeney JA, Nurnberger JI, Coon H, Sutcliffe JS, Minshew NJ, Grant SF, Bucan M, Cook EH, Buxbaum JD, Devlin B, Schellenberg GD, Hakonarson H. Autism genome-wide copy number variation reveals ubiquitin and neuronal genes. Nature. 2009;459:569–573. doi: 10.1038/nature07953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Marshall CR, Noor A, Vincent JB, Lionel AC, Feuk L, Skaug J, Shago M, Moessner R, Pinto D, Ren Y, Thiruvahindrapduram B, Fiebig A, Schreiber S, Friedman J, Ketelaars CE, Vos YJ, Ficicioglu C, Kirkpatrick S, Nicolson R, Sloman L, Summers A, Gibbons CA, Teebi A, Chitayat D, Weksberg R, Thompson A, Vardy C, Crosbie V, Luscombe S, Baatjes R, Zwaigenbaum L, Roberts W, Fernandez B, Szatmari P, Scherer SW. Structural variation of chromosomes in autism spectrum disorder. Am J Hum Genet. 2008;82:477–488. doi: 10.1016/j.ajhg.2007.12.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sebat J, Lakshmi B, Malhotra D, Troge J, Lese-Martin C, Walsh T, Yamrom B, Yoon S, Krasnitz A, Kendall J, Leotta A, Pai D, Zhang R, Lee YH, Hicks J, Spence SJ, Lee AT, Puura K, Lehtimäki T, Ledbetter D, Gregersen PK, Bregman J, Sutcliffe JS, Jobanputra V, Chung W, Warburton D, King MC, Skuse D, Geschwind DH, Gilliam TC, Ye K, Wigler M. Strong association of de novo copy number mutations with autism. Science. 2007;316:445–449. doi: 10.1126/science.1138659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Talkowski ME, Minikel EV, Gusella JF. Autism spectrum disorder genetics: diverse genes with diverse clinical outcomes. Harv Rev Psychiatry. 2014;22:65–75. doi: 10.1097/HRP.0000000000000002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kumar RA, Sudi J, Babatz TD, Brune CW, Oswald D, Yen M, Nowak NJ, Cook EH, Christian SL, Dobyns WB. A de novo 1p34.2 microdeletion identifies the synaptic vesicle gene RIMS3 as a novel candidate for autism. J Med Genet. 2010;47:81–90. doi: 10.1136/jmg.2008.065821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Liang JS, Shimojima K, Ohno K, Sugiura C, Une Y, Ohno K, Yamamoto T. A newly recognised microdeletion syndrome of 2p15-16.1 manifesting moderate developmental delay, autistic behaviour, short stature, microcephaly, and dysmorphic features: a new patient with 3.2 Mb deletion. J Med Genet. 2009;46:645–647. doi: 10.1136/jmg.2008.059220. [DOI] [PubMed] [Google Scholar]
- 45.Liu X, Malenfant P, Reesor C, Lee A, Hudson ML, Harvard C, Qiao Y, Persico AM, Cohen IL, Chudley AE, Forster-Gibson C, Rajcan-Separovic E, Lewis ME, Holden JJ. 2p15-p16.1 microdeletion syndrome: molecular characterization and association of the OTX1 and XPO1 genes with autism spectrum disorders. Eur J Hum Genet. 2011;19:1264–1270. doi: 10.1038/ejhg.2011.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Miller DT, Shen Y, Weiss LA, Korn J, Anselm I, Bridgemohan C, Cox GF, Dickinson H, Gentile J, Harris DJ, Hegde V, Hundley R, Khwaja O, Kothare S, Luedke C, Nasir R, Poduri A, Prasad K, Raffalli P, Reinhard A, Smith SE, Sobeih MM, Soul JS, Stoler J, Takeoka M, Tan WH, Thakuria J, Wolff R, Yusupov R, Gusella JF, Daly MJ, Wu BL. Microdeletion/duplication at 15q13.2q13.3 among individuals with features of autism and other neuropsychiatric disorders. J Med Genet. 2009;46:242–248. doi: 10.1136/jmg.2008.059907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Pagnamenta AT, Wing K, Sadighi Akha E, Knight SJ, Bölte S, Schmötzer G, Duketis E, Poustka F, Klauck SM, Poustka A, Ragoussis J, Bailey AJ, Monaco AP International Molecular Genetic Study of Autism Consortium. A 15q13.3 microdeletion segregating with autism. Eur J Hum Genet. 2009;17:687–692. doi: 10.1038/ejhg.2008.228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Rajcan-Separovic E, Harvard C, Liu X, McGillivray B, Hall JG, Qiao Y, Hurlburt J, Hildebrand J, Mickelson EC, Holden JJ, Lewis ME. Clinical and molecular cytogenetic characterisation of a newly recognised microdeletion syndrome involving 2p15-16.1. J Med Genet. 2007;44:269–276. doi: 10.1136/jmg.2006.045013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Weiss LA, Shen Y, Korn JM, Arking DE, Miller DT, Fossdal R, Saemundsen E, Stefansson H, Ferreira MA, Green T, Platt OS, Ruderfer DM, Walsh CA, Altshuler D, Chakravarti A, Tanzi RE, Stefansson K, Santangelo SL, Gusella JF, Sklar P, Wu BL, Daly MJ Autism Consortium. Association between microdeletion and microduplication at 16p11.2 and autism. N Engl J Med. 2008;358:667–675. doi: 10.1056/NEJMoa075974. [DOI] [PubMed] [Google Scholar]
- 50.Durand CM, Betancur C, Boeckers TM, Bockmann J, Chaste P, Fauchereau F, Nygren G, Rastam M, Gillberg IC, Anckarsäter H, Sponheim E, Goubran-Botros H, Delorme R, Chabane N, Mouren-Simeoni MC, de Mas P, Bieth E, Rogé B, Héron D, Burglen L, Gillberg C, Leboyer M, Bourgeron T. Mutations in the gene encoding the synaptic scaffolding protein SHANK3 are associated with autism spectrum disorders. Nat Genet. 2007;39:25–27. doi: 10.1038/ng1933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Gauthier J, Bonnel A, St-Onge J, Karemera L, Laurent S, Mottron L, Fombonne E, Joober R, Rouleau GA. NLGN3/NLGN4 gene mutations are not responsible for autism in the Quebec population. Am J Med Genet B Neuropsychiatr Genet. 2005;132B:74–75. doi: 10.1002/ajmg.b.30066. [DOI] [PubMed] [Google Scholar]
- 52.Graf ER, Zhang X, Jin SX, Linhoff MW, Craig AM. Neurexins induce differentiation of GABA and glutamate postsynaptic specializations via neuroligins. Cell. 2004;119:1013–1026. doi: 10.1016/j.cell.2004.11.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Jamain S, Quach H, Betancur C, Råstam M, Colineaux C, Gillberg IC, Soderstrom H, Giros B, Leboyer M, Gillberg C, Bourgeron T, Gillberg C, Råstam M, Gillberg C, Nydén A, Söderström H, Leboyer M, Betancur C, Philippe A, Giros B, Colineaux C, Cohen D, Chabane N, Mouren-Siméoni MC, Brice A, Sponheim E, Spurkland I, Skjeldal OH, Coleman M, Pearl PL, Cohen IL, Tsiouris J, Zappella M, Menchetti G, Pompella A, Aschauer H, Van Maldergem L Paris Autism Research International Sibpair Study. Mutations of the X-linked genes encoding neuroligins NLGN3 and NLGN4 are associated with autism. Nat Genet. 2003;34:27–29. doi: 10.1038/ng1136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Laumonnier F, Bonnet-Brilhault F, Gomot M, Blanc R, David A, Moizard MP, Raynaud M, Ronce N, Lemonnier E, Calvas P, Laudier B, Chelly J, Fryns JP, Ropers HH, Hamel BC, Andres C, Barthélémy C, Moraine C, Briault S. X-linked mental retardation and autism are associated with a mutation in the NLGN4 gene, a member of the neuroligin family. Am J Hum Genet. 2004;74:552–557. doi: 10.1086/382137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Lawson-Yuen A, Saldivar JS, Sommer S, Picker J. Familial deletion within NLGN4 associated with autism and Tourette syndrome. Eur J Hum Genet. 2008;16:614–618. doi: 10.1038/sj.ejhg.5202006. [DOI] [PubMed] [Google Scholar]
- 56.Maćkowiak M, Mordalska P, Wędzony K. Neuroligins, synapse balance and neuropsychiatric disorders. Pharmacol Rep. 2014;66:830–835. doi: 10.1016/j.pharep.2014.04.011. [DOI] [PubMed] [Google Scholar]
- 57.Moessner R, Marshall CR, Sutcliffe JS, Skaug J, Pinto D, Vincent J, Zwaigenbaum L, Fernandez B, Roberts W, Szatmari P, Scherer SW. Contribution of SHANK3 mutations to autism spectrum disorder. Am J Hum Genet. 2007;81:1289–1297. doi: 10.1086/522590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Nemirovsky SI, Córdoba M, Zaiat JJ, Completa SP, Vega PA, González-Morón D, Medina NM, Fabbro M, Romero S, Brun B, Revale S, Ogara MF, Pecci A, Marti M, Vazquez M, Turjanski A, Kauffman MA. Whole genome sequencing reveals a de novo SHANK3 mutation in familial autism spectrum disorder. PLoS One. 2015;10:e0116358. doi: 10.1371/journal.pone.0116358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Phelan K, McDermid HE. The 22q13.3 Deletion Syndrome (Phelan-McDermid Syndrome) Mol Syndromol. 2012;2:186–201. doi: 10.1159/000334260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Reissner C, Klose M, Fairless R, Missler M. Mutational analysis of the neurexin/neuroligin complex reveals essential and regulatory components. Proc Natl Acad Sci USA. 2008;105:15124–15129. doi: 10.1073/pnas.0801639105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Talebizadeh Z, Lam DY, Theodoro MF, Bittel DC, Lushington GH, Butler MG. Novel splice isoforms for NLGN3 and NLGN4 with possible implications in autism. J Med Genet. 2006;43:e21. doi: 10.1136/jmg.2005.036897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Xu X, Xiong Z, Zhang L, Liu Y, Lu L, Peng Y, Guo H, Zhao J, Xia K, Hu Z. Variations analysis of NLGN3 and NLGN4X gene in Chinese autism patients. Mol Biol Rep. 2014;41:4133–4140. doi: 10.1007/s11033-014-3284-5. [DOI] [PubMed] [Google Scholar]
- 63.Shankar GM, Li S, Mehta TH, Garcia-Munoz A, Shepardson NE, Smith I, Brett FM, Farrell MA, Rowan MJ, Lemere CA, Regan CM, Walsh DM, Sabatini BL, Selkoe DJ. Amyloid-beta protein dimers isolated directly from Alzheimer's brains impair synaptic plasticity and memory. Nat Med. 2008;14:837–842. doi: 10.1038/nm1782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Stephan KE, Friston KJ, Frith CD. Dysconnection in schizophrenia: from abnormal synaptic plasticity to failures of self-monitoring. Schizophr Bull. 2009;35:509–527. doi: 10.1093/schbul/sbn176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Zhang C, Milunsky JM, Newton S, Ko J, Zhao G, Maher TA, Tager-Flusberg H, Bolliger MF, Carter AS, Boucard AA, Powell CM, Südhof TC. A neuroligin-4 missense mutation associated with autism impairs neuroligin-4 folding and endoplasmic reticulum export. J Neurosci. 2009;29:10843–10854. doi: 10.1523/JNEUROSCI.1248-09.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Boccuto L, Lauri M, Sarasua SM, Skinner CD, Buccella D, Dwivedi A, Orteschi D, Collins JS, Zollino M, Visconti P, Dupont B, Tiziano D, Schroer RJ, Neri G, Stevenson RE, Gurrieri F, Schwartz CE. Prevalence of SHANK3 variants in patients with different subtypes of autism spectrum disorders. Eur J Hum Genet. 2013;21:310–316. doi: 10.1038/ejhg.2012.175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Durand CM, Perroy J, Loll F, Perrais D, Fagni L, Bourgeron T, Montcouquiol M, Sans N. SHANK3 mutations identified in autism lead to modification of dendritic spine morphology via an actin-dependent mechanism. Mol Psychiatry. 2012;17:71–84. doi: 10.1038/mp.2011.57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Mameza MG, Dvoretskova E, Bamann M, Hönck HH, Güler T, Boeckers TM, Schoen M, Verpelli C, Sala C, Barsukov I, Dityatev A, Kreienkamp HJ. SHANK3 gene mutations associated with autism facilitate ligand binding to the Shank3 ankyrin repeat region. J Biol Chem. 2013;288:26697–26708. doi: 10.1074/jbc.M112.424747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Uchino S, Waga C. SHANK3 as an autism spectrum disorder-associated gene. Brain Dev. 2013;35:106–110. doi: 10.1016/j.braindev.2012.05.013. [DOI] [PubMed] [Google Scholar]
- 70.Szatmari P, Paterson AD, Zwaigenbaum L, Roberts W, Brian J, Liu XQ, Vincent JB, Skaug JL, Thompson AP, Senman L, Feuk L, Qian C, Bryson SE, Jones MB, Marshall CR, Scherer SW, Vieland VJ, Bartlett C, Mangin LV, Goedken R, Segre A, Pericak-Vance MA, Cuccaro ML, Gilbert JR, Wright HH, Abramson RK, Betancur C, Bourgeron T, Gillberg C, Leboyer M, Buxbaum JD, Davis KL, Hollander E, Silverman JM, Hallmayer J, Lotspeich L, Sutcliffe JS, Haines JL, Folstein SE, Piven J, Wassink TH, Sheffield V, Geschwind DH, Bucan M, Brown WT, Cantor RM, Constantino JN, Gilliam TC, Herbert M, Lajonchere C, Ledbetter DH, Lese-Martin C, Miller J, Nelson S, Samango-Sprouse CA, Spence S, State M, Tanzi RE, Coon H, Dawson G, Devlin B, Estes A, Flodman P, Klei L, McMahon WM, Minshew N, Munson J, Korvatska E, Rodier PM, Schellenberg GD, Smith M, Spence MA, Stodgell C, Tepper PG, Wijsman EM, Yu CE, Rogé B, Mantoulan C, Wittemeyer K, Poustka A, Felder B, Klauck SM, Schuster C, Poustka F, Bölte S, Feineis-Matthews S, Herbrecht E, Schmötzer G, Tsiantis J, Papanikolaou K, Maestrini E, Bacchelli E, Blasi F, Carone S, Toma C, Van Engeland H, de Jonge M, Kemner C, Koop F, Langemeijer M, Hijmans C, Staal WG, Baird G, Bolton PF, Rutter ML, Weisblatt E, Green J, Aldred C, Wilkinson JA, Pickles A, Le Couteur A, Berney T, McConachie H, Bailey AJ, Francis K, Honeyman G, Hutchinson A, Parr JR, Wallace S, Monaco AP, Barnby G, Kobayashi K, Lamb JA, Sousa I, Sykes N, Cook EH, Guter SJ, Leventhal BL, Salt J, Lord C, Corsello C, Hus V, Weeks DE, Volkmar F, Tauber M, Fombonne E, Shih A, Meyer KJ Autism Genome Project Consortium. Mapping autism risk loci using genetic linkage and chromosomal rearrangements. Nat Genet. 2007;39:319–328. doi: 10.1038/ng1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Bucan M, Abrahams BS, Wang K, Glessner JT, Herman EI, Sonnenblick LI, Alvarez Retuerto AI, Imielinski M, Hadley D, Bradfield JP, Kim C, Gidaya NB, Lindquist I, Hutman T, Sigman M, Kustanovich V, Lajonchere CM, Singleton A, Kim J, Wassink TH, McMahon WM, Owley T, Sweeney JA, Coon H, Nurnberger JI, Li M, Cantor RM, Minshew NJ, Sutcliffe JS, Cook EH, Dawson G, Buxbaum JD, Grant SF, Schellenberg GD, Geschwind DH, Hakonarson H. Genome-wide analyses of exonic copy number variants in a family-based study point to novel autism susceptibility genes. PLoS Genet. 2009;5:e1000536. doi: 10.1371/journal.pgen.1000536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Dabell MP, Rosenfeld JA, Bader P, Escobar LF, El-Khechen D, Vallee SE, Dinulos MB, Curry C, Fisher J, Tervo R, Hannibal MC, Siefkas K, Wyatt PR, Hughes L, Smith R, Ellingwood S, Lacassie Y, Stroud T, Farrell SA, Sanchez-Lara PA, Randolph LM, Niyazov D, Stevens CA, Schoonveld C, Skidmore D, MacKay S, Miles JH, Moodley M, Huillet A, Neill NJ, Ellison JW, Ballif BC, Shaffer LG. Investigation of NRXN1 deletions clinical and molecular characterization. Am J Med Genet A. 2013;161A:717–731. doi: 10.1002/ajmg.a.35780. [DOI] [PubMed] [Google Scholar]
- 73.Glahn DC, Williams JT, McKay DR, Knowles EE, Sprooten E, Mathias SR, Curran JE, Kent JW, Jr, Carless MA, Göring HH, Dyer TD, Woolsey MD, Winkler AM, Olvera RL, Kochunov P, Fox PT, Duggirala R, Almasy L, Blangero J. Discovering schizophrenia endophenotypes in randomly ascertained pedigrees. Biol Psychiatry. 2015;77:75–83. doi: 10.1016/j.biopsych.2014.06.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Chen X, Shen Y, Zhang F, Chiang C, Pillalamarri V, Blumenthal I, Talkowski M, Wu BL, Gusella JF. Molecular analysis of a deletion hotspot in the NRXN1 region reveals the involvement of short inverted repeats in deletion CNVs. Am J Hum Genet. 2013;92:375–386. doi: 10.1016/j.ajhg.2013.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Alarcón M, Abrahams BS, Stone JL, Duvall JA, Perederiy JV, Bomar JM, Sebat J, Wigler M, Martin CL, Ledbetter DH, Nelson SF, Cantor RM, Geschwind DH. Linkage, association, and gene-expression analyses identify CNTNAP2 as an autism-susceptibility gene. Am J Hum Genet. 2008;82:150–159. doi: 10.1016/j.ajhg.2007.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Arking DE, Cutler DJ, Brune CW, Teslovich TM, West K, Ikeda M, Rea A, Guy M, Lin S, Cook EH, Jr, Chakravarti A. A common genetic variant in the neurexin superfamily member CNTNAP2 increases familial risk of autism. Am J Hum Genet. 2008;82:160–164. doi: 10.1016/j.ajhg.2007.09.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Bakkaloglu B, O'Roak BJ, Louvi A, Gupta AR, Abelson JF, Morgan TM, Chawarska K, Klin A, Ercan-Sencicek AG, Stillman AA, Tanriover G, Abrahams BS, Duvall JA, Robbins EM, Geschwind DH, Biederer T, Gunel M, Lifton RP, State MW. Molecular cytogenetic analysis and resequencing of contactin associated protein-like 2 in autism spectrum disorders. Am J Hum Genet. 2008;82:165–173. doi: 10.1016/j.ajhg.2007.09.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Peñagarikano O, Abrahams BS, Herman EI, Winden KD, Gdalyahu A, Dong H, Sonnenblick LI, Gruver R, Almajano J, Bragin A, Golshani P, Trachtenberg JT, Peles E, Geschwind DH. Absence of CNTNAP2 leads to epilepsy, neuronal migration abnormalities, and core autism-related deficits. Cell. 2011;147:235–246. doi: 10.1016/j.cell.2011.08.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Murdoch JD, Gupta AR, Sanders SJ, Walker MF, Keaney J, Fernandez TV, Murtha MT, Anyanwu S, Ober GT, Raubeson MJ, DiLullo NM, Villa N, Waqar Z, Sullivan C, Gonzalez L, Willsey AJ, Choe SY, Neale BM, Daly MJ, State MW. No evidence for association of autism with rare heterozygous point mutations in Contactin-Associated Protein-Like 2 (CNTNAP2), or in Other Contactin-Associated Proteins or Contactins. PLoS Genet. 2015;11:e1004852. doi: 10.1371/journal.pgen.1004852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Chiocchetti AG, Kopp M, Waltes R, Haslinger D, Duketis E, Jarczok TA, Poustka F, Voran A, Graab U, Meyer J, Klauck SM, Fulda S, Freitag CM. Variants of the CNTNAP2 5 promoter as risk factors for autism spectrum disorders: a genetic and functional approach. Mol Psychiatry. 2015;20:839–849. doi: 10.1038/mp.2014.103. [DOI] [PubMed] [Google Scholar]
- 81.Powel CM, Boucard AA. Neuroligins and neurexins: bridging the synaptic cleft in autism. In: Powel CM, Monteggia LM, editors. The autisms. New York, NY: Oxford University Press; 2013. pp. 214–239. [Google Scholar]
- 82.Boucard AA, Chubykin AA, Comoletti D, Taylor P, Südhof TC. A splice code for trans-synaptic cell adhesion mediated by binding of neuroligin 1 to alpha- and beta-neurexins. Neuron. 2005;48:229–236. doi: 10.1016/j.neuron.2005.08.026. [DOI] [PubMed] [Google Scholar]
- 83.Powel CM. Shank gene family and autism. In: Powel CM, Monteggia LM, editors. The autisms. Oxford University Press: New York, NY; 2013. pp. 176–188. [Google Scholar]
- 84.Böckers TM, Mameza MG, Kreutz MR, Bockmann J, Weise C, Buck F, Richter D, Gundelfinger ED, Kreienkamp HJ. Synaptic scaffolding proteins in rat brain. Ankyrin repeats of the multidomain Shank protein family interact with the cytoskeletal protein alpha-fodrin. J Biol Chem. 2001;276:40104–40112. doi: 10.1074/jbc.M102454200. [DOI] [PubMed] [Google Scholar]
- 85.Ebrahimi-Fakhari D, Sahin M. Autism and the synapse: emerging mechanisms and mechanism-based therapies. Curr Opin Neurol. 2015;28:91–102. doi: 10.1097/WCO.0000000000000186. [DOI] [PubMed] [Google Scholar]
- 86.Sahin M, Sur M. Genes, circuits, and precision therapies for autism and related neurodevelopmental disorders. Science. 2015;350:aab3897. doi: 10.1126/science.aab3897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Feldman R, Monakhov M, Pratt M, Ebstein RP. Oxytocin Pathway Genes: Evolut ionar y Ancient System Impacting on Human Affiliation, Sociality, and Psychopathology. Biol Psychiatry. :pii: S0006-3223(15)00656-3. doi: 10.1016/j.biopsych.2015.08.008. [DOI] [PubMed] [Google Scholar]
- 88.Kennedy DP, Adolphs R. Perception of emotions from facial expressions in high-functioning adults with autism. Neuropsychologia. 2012;50:3313–3319. doi: 10.1016/j.neuropsychologia.2012.09.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Frazier TW, Embacher R, Tilot AK, Koenig K, Mester J, Eng C. Molecular and phenotypic abnormalities in individuals with germline heterozygous PTEN mutations and autism. Mol Psychiatry. 2015;20:1132–1138. doi: 10.1038/mp.2014.125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Dawson G, Jones EJ, Merkle K, Venema K, Lowy R, Faja S, Kamara D, Murias M, Greenson J, Winter J, Smith M, Rogers SJ, Webb SJ. Early behavioral intervention is associated with normalized brain activity in young children with autism. J Am Acad Child Adolesc Psychiatry. 2012;51:1150–1159. doi: 10.1016/j.jaac.2012.08.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Abrahams BS, Geschwind DH. Advances in autism genetics: on the threshold of a new neurobiology. Nat Rev Genet. 2008;9:341–355. doi: 10.1038/nrg2346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Jeste SS, Geschwind DH. Disentangling the heterogeneity of autism spectrum disorder through genetic findings. Nat Rev Neurol. 2014;10:74–81. doi: 10.1038/nrneurol.2013.278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Skafidas E, Testa R, Zantomio D, Chana G, Everall IP, Pantelis C. Predicting the diagnosis of autism spectrum disorder using gene pathway analysis. Mol Psychiatry. 2014;19:504–510. doi: 10.1038/mp.2012.126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Hu VW, Lai Y. Developing a predictive gene classifier for autism spectrum disorders based upon differential gene expression profiles of phenotypic subgroups. N Am J Med Sci(Boston) 2013;6 doi: 10.7156/najms.2013.0603107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Jiang YH, Wang Y, Xiu X, Choy KW, Pursley AN, Cheung SW. Genetic diagnosis of autism spectrum disorders: the opportunity and challenge in the genomics era. Crit Rev Clin Lab Sci. 2014;51:249–262. doi: 10.3109/10408363.2014.910747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Heil KM, Schaaf CP. The genetics of Autism Spectrum Disorders--a guide for clinicians. Curr Psychiatry Rep. 2013;15:334. doi: 10.1007/s11920-012-0334-3. [DOI] [PubMed] [Google Scholar]
- 97.Schaefer GB, Mendelsohn NJ Professional Practice and Guidelines Committee. Clinical genetics evaluation in identifying the etiology of autism spectrum disorders: 2013 guideline revisions. Genet Med. 2013;15:399–407. doi: 10.1038/gim.2013.32. [DOI] [PubMed] [Google Scholar]
- 98.Baker E, Jeste SS. Diagnosis and management of autism spectrum disorder in the era of genomics: rare disorders can pave the way for targeted treatments. Pediatr Clin North Am. 2015;62:607–618. doi: 10.1016/j.pcl.2015.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Singh I, Rose N. Biomarkers in psychiatry. Nature. 2009;460:202–207. doi: 10.1038/460202a. [DOI] [PubMed] [Google Scholar]
- 100.Walsh P, Elsabbagh M, Bolton P, Singh I. In search of biomarkers for autism: scientific, social and ethical challenges. Nat Rev Neurosci. 2011;12:603–612. doi: 10.1038/nrn3113. [DOI] [PubMed] [Google Scholar]
- 101.Doyle CA, McDougle CJ. Pharmacotherapy to control behavioral symptoms in children with autism. Expert Opin Pharmacother. 2012;13:1615–1629. doi: 10.1517/14656566.2012.674110. [DOI] [PubMed] [Google Scholar]
- 102.Chao HT, Chen H, Samaco RC, Xue M, Chahrour M, Yoo J, Neul JL, Gong S, Lu HC, Heintz N, Ekker M, Rubenstein JL, Noebels JL, Rosenmund C, Zoghbi HY. Dysfunction in GABA signalling mediates autism-like stereotypies and Rett syndrome phenotypes. Nature. 2010;468:263–269. doi: 10.1038/nature09582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Dölen G, Osterweil E, Rao BS, Smith GB, Auerbach BD, Chattarji S, Bear MF. Correction of fragile X syndrome in mice. Neuron. 2007;56:955–962. doi: 10.1016/j.neuron.2007.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Huang HS, Allen JA, Mabb AM, King IF, Miriyala J, Taylor-Blake B, Sciaky N, Dutton JW, Jr, Lee HM, Chen X, Jin J, Bridges AS, Zylka MJ, Roth BL, Philpot BD. Topoisomerase inhibitors unsilence the dormant allele of Ube3a in neurons. Nature. 2012;481:185–189. doi: 10.1038/nature10726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Smith DG, Ehlers MD. Mining and modeling human genetics for autism therapeutics. Curr Opin Neurobiol. 2012;22:902–910. doi: 10.1016/j.conb.2012.03.004. [DOI] [PubMed] [Google Scholar]
- 106.Smith SE, Zhou YD, Zhang G, Jin Z, Stoppel DC, Anderson MP. Increased gene dosage of Ube3a results in autism traits and decreased glutamate synaptic transmission in mice. Sci Transl Med. 2011;3:103ra97. doi: 10.1126/scitranslmed.3002627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Sahin M. Targeted treatment trials for tuberous sclerosis and autism: no longer a dream. Curr Opin Neurobiol. 2012;22:895–901. doi: 10.1016/j.conb.2012.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Gregory SG, Connelly JJ, Towers AJ, Johnson J, Biscocho D, Markunas CA, Lintas C, Abramson RK, Wright HH, Ellis P, Langford CF, Worley G, Delong GR, Murphy SK, Cuccaro ML, Persico A, Pericak-Vance MA. Genomic and epigenetic evidence for oxytocin receptor deficiency in autism. BMC Med. 2009;7:62. doi: 10.1186/1741-7015-7-62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Wu S, Jia M, Ruan Y, Liu J, Guo Y, Shuang M, Gong X, Zhang Y, Yang X, Zhang D. Positive association of the oxytocin receptor gene (OXTR) with autism in the Chinese Han population. Biol Psychiatry. 2005;58:74–77. doi: 10.1016/j.biopsych.2005.03.013. [DOI] [PubMed] [Google Scholar]
- 110.Takayanagi Y, Yoshida M, Bielsky IF, Ross HE, Kawamata M, Onaka T, Yanagisawa T, Kimura T, Matzuk MM, Young LJ, Nishimori K. Pervasive social deficits, but normal parturition, in oxytocin receptor-deficient mice. Proc Natl Acad Sci U S A. 2005;102:16096–16101. doi: 10.1073/pnas.0505312102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Higashida H, Lopatina O, Yoshihara T, Pichugina YA, Soumarokov AA, Munesue T, Minabe Y, Kikuchi M, Ono Y, Korshunova N, Salmina AB. Oxytocin signal and social behaviour: comparison among adult and infant oxytocin, oxytocin receptor and CD38 gene knockout mice. J Neuroendocrinol. 2010;22:373–379. doi: 10.1111/j.1365-2826.2010.01976.x. [DOI] [PubMed] [Google Scholar]
- 112.Yatawara CJ, Einfeld SL, Hickie IB, Davenport TA, Guastella AJ. The effect of oxytocin nasal spray on social interaction deficits observed in young children with autism: a randomized clinical crossover trial. Mol Psychiatry. 2015 doi: 10.1038/mp.2015.162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Watanabe T, Kuroda M, Kuwabara H, Aoki Y, Iwashiro N, Tatsunobu N, Takao H, Nippashi Y, Kawakubo Y, Kunimatsu A, Kasai K, Yamasue H. Clinical and neural effects of six-week administration of oxytocin on core symptoms of autism. Brain. 2015;138:3400–3412. doi: 10.1093/brain/awv249. [DOI] [PubMed] [Google Scholar]
- 114.Guastella AJ, Hickie IB. Oxytocin treatment, circuitry and autism: a critical review of the literature placing oxytocin into the autism context. Biol Psychiatry. 2015:pii: S0006-3223(15)00543-0. doi: 10.1016/j.biopsych.2015.06.028. [DOI] [PubMed] [Google Scholar]
- 115.Buxbaum JD, Daly MJ, Devlin B, Lehner T, Roeder K, State MW Autism Sequencing Consortium. The autism sequencing consortium: large-scale, high-throughput sequencing in autism spectrum disorders. Neuron. 2012;76:1052–1056. doi: 10.1016/j.neuron.2012.12.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Butler MG, Rafi SK, Hossain W, Stephan DA, Manzardo AM. Whole exome sequencing in females with autism implicates novel and candidate genes. Int J Mol Sci. 2015;16:1312–1335. doi: 10.3390/ijms16011312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Chapman NH, Nato AQ, Jr, Bernier R, Ankenman K, Sohi H, Munson J, Patowary A, Archer M, Blue EM, Webb SJ, Coon H, Raskind WH, Brkanac Z, Wijsman EM. Whole exome sequencing in extended families with autism spectrum disorder implicates four candidate genes. Hum Genet. 2015;134:1055–1068. doi: 10.1007/s00439-015-1585-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Moreno-Ramos OA, Olivares AM, Haider NB, de Autismo LC, Lattig MC. Whole-Exome Sequencing in a South American Cohort Links ALDH1A3, FOXN1 and Retinoic Acid Regulation Pathways to Autism Spectrum Disorders. PLoS One. 2015;10:e0135927. doi: 10.1371/journal.pone.0135927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Tammimies K, Marshall CR, Walker S, Kaur G, Thiruvahindrapuram B, Lionel AC, Yuen RK, Uddin M, Roberts W, Weksberg R, Woodbury-Smith M, Zwaigenbaum L, Anagnostou E, Wang Z, Wei J, Howe JL, Gazzellone MJ, Lau L, Sung WW, Whitten K, Vardy C, Crosbie V, Tsang B, D'Abate L, Tong WW, Luscombe S, Doyle T, Carter MT, Szatmari P, Stuckless S, Merico D, Stavropoulos DJ, Scherer SW, Fernandez BA. Molecular diagnostic yield of chromosomal microarray analysis and whole-exome sequencing in children with autism spectrum disorder. JAMA. 2015;314:895–903. doi: 10.1001/jama.2015.10078. [DOI] [PubMed] [Google Scholar]
- 120.Koukoui SD, Chaudhuri A. Neuroanatomical, molecular genetic, and behavioral correlates of fragile X syndrome. Brain Res Rev. 2007;53:27–38. doi: 10.1016/j.brainresrev.2006.06.001. [DOI] [PubMed] [Google Scholar]
- 121.Curatolo P, Porfirio MC, Manzi B, Seri S. Autism in tuberous sclerosis. Eur J Paediatr Neurol. 2004;8:327–332. doi: 10.1016/j.ejpn.2004.08.005. [DOI] [PubMed] [Google Scholar]
- 122.Garg S, Lehtonen A, Huson SM, Emsley R, Trump D, Evans DG, Green J. Autism and other psychiatric comorbidity in neurofibromatosis type 1: evidence from a population-based study. Dev Med Child Neurol. 2013;55:139–145. doi: 10.1111/dmcn.12043. [DOI] [PubMed] [Google Scholar]
- 123.Moss J, Howlin P, Magiati I, Oliver C. Characteristics of autism spectrum disorder in Cornelia de Lange syndrome. J Child Psychol Psychiatry. 2012;53:883–891. doi: 10.1111/j.1469-7610.2012.02540.x. [DOI] [PubMed] [Google Scholar]
- 124.Howlin P, Karpf J, Turk J. Behavioural characteristics and autistic features in individuals with Cohen Syndrome. Eur Child Adolesc Psychiatry. 2005;14:57–64. doi: 10.1007/s00787-005-0416-4. [DOI] [PubMed] [Google Scholar]
- 125.Splawski I, Yoo DS, Stotz SC, Cherry A, Clapham DE, Keating MT. CACNA1H mutations in autism spectrum disorders. J Biol Chem. 2006;281:22085–22091. doi: 10.1074/jbc.M603316200. [DOI] [PubMed] [Google Scholar]
- 126.Bukelis I, Porter FD, Zimmerman AW, Tierney E. Smith-Lemli-Opitz syndrome and autism spectrum disorder. Am J Psychiatry. 2007;164:1655–1661. doi: 10.1176/appi.ajp.2007.07020315. [DOI] [PubMed] [Google Scholar]
- 127.Tordjman S, Anderson GM, Botbol M, Toutain A, Sarda P, Carlier M, Saugier-Veber P, Baumann C, Cohen D, Lagneaux C, Tabet AC, Verloes A. Autistic disorder in patients with Williams-Beuren syndrome: a reconsideration of the Williams-Beuren syndrome phenotype. PLoS One. 2012;7:e30778. doi: 10.1371/journal.pone.0030778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Koochek M, Harvard C, Hildebrand MJ, Van Allen M, Wingert H, Mickelson E, Holden JJ, Rajcan-Separovic E, Lewis ME. 15q duplication associated with autism in a multiplex family with a familial cryptic translocation t(14;15) (q11.2;q13.3) detected using array-CGH. Clin Genet. 2006;69:124–134. doi: 10.1111/j.1399-0004.2005.00560.x. [DOI] [PubMed] [Google Scholar]
- 129.Laje G, Morse R, Richter W, Ball J, Pao M, Smith AC. Autism spectrum features in Smith-Magenis syndrome. Am J Med Genet C Semin Med Genet. 2010;154C:456–462. doi: 10.1002/ajmg.c.30275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Lo-Castro A, Galasso C, Cerminara C, El-Malhany N, Benedetti S, Nardone AM, Curatolo P. Association of syndromic mental retardation and autism with 22q11.2 duplication. Neuropediatrics. 2009;40:137–140. doi: 10.1055/s-0029-1237724. [DOI] [PubMed] [Google Scholar]
- 131.Radoeva PD, Coman IL, Salazar CA, Gentile KL, Higgins AM, Middleton FA, Antshel KM, Fremont W, Shprintzen RJ, Morrow BE, Kates WR. Association between autism spectrum disorder in individuals with velocardiofacial (22q11.2 deletion) syndrome and PRODH and COMT genotypes. Psychiatr Genet. 2014;24:269–272. doi: 10.1097/YPG.0000000000000062. [DOI] [PMC free article] [PubMed] [Google Scholar]