Abstract
TaGS5 genes were cloned from bread wheat and were physically mapped on 3AS and 3DS. Sequencing results revealed that a SNP was found in the sixth exon of TaGS5-A1 gene. The SNP resulted in amino acid change from alanine to serine at the 303 bp position of TaGS5-A1. These two alleles were designated as TaGS5-A1a (alanine at the 303 bp position) and TaGS5-A1b genes (serine at the 303-bp position). Analysis of association of TaGS5-A1 alleles with agronomic traits indicated that cultivars with TaGS5-A1b possessed wider kernel width and higher thousand-kernel weight, as well as significantly lower plant height, spike length, and internode length below spike than those of cultivars with TaGS5-A1a over 3 years. These trait differences between TaGS5-A1a and TaGS5-A1b genotypes were larger in landraces than in modern cultivars. This finding suggested that TaGS5 gene played an important role in modulating yield-related traits in the landraces, which possibly resulted from numerous superior genes gathering in modern cultivars after strong artificial selection. The preferred TaGS5-A1b haplotype underwent very strong positive selection in Chinese modern wheat breeding, but not in Chinese landraces. Expression analysis of the TaGS5-A1 gene indicated that TaGS5-A1b allele possessed significantly higher expression level than TaGS5-A1b allele in differently developmental seeds. This study could provide relatively superior genotype in view of agronomic traits in wheat breeding programs. Likewise, this study could offer important information for the dissection of molecular and genetic basis of yield-related traits.
Keywords: bread wheat, TaGS5 gene, yield-related traits, kernel size, RT-PCR
Introduction
Wheat is an important food crop in the world, and improvement of wheat yield has always been an essential breeding target for wheat breeders over the past years. As one of the three wheat yield elements, thousand-kernel weight (TKW) is considered to have an important influence on wheat yield and could be determined by the kernel size. To date, some important yield-related genes (GS3, GS5, GW2, GW3, GW5, GW6, GW8, GIF1, and Ghd7) associated with kernel size were gradually discovered and cloned in rice (Li et al., 2004a,b, 2011; Fan et al., 2006; Xie et al., 2006; Song et al., 2007; Wang et al., 2008, 2012b; Weng et al., 2008; Xue et al., 2008; Guo et al., 2009; Takano-Kai et al., 2009, 2013). For example, the GS5 gene, encoding a putative serine carboxypeptidase, was cloned in rice as a positive regulator of grain size by regulating grain width, filling, and weight (Li et al., 2011). More recently, ZmGS5 gene also were cloned by in silico cloning and was found to be associated with kernel weight and cell number in maize (Liu et al., 2015). Meanwhile, some co-orthologs associated with kernel size and weight were successfully cloned in maize (Li et al., 2010). These factors facilitated the cloning of genes related to kernel size in polyploidy wheat.
Hexaploid wheat has a larger genome size (≈17.9 Gb) compared with rice (≈400 Mb) and maize (≈3 Gb; Varshney et al., 2006). Therefore, direct cloning of yield-related genes from hexaploid wheat without reference genes was difficult. However, a large number of yield-related quantitative trait loci (QTLs) were identified in polyploid wheat. These QTLs were mapped on almost all of the chromosomes, including group 3 in bread wheat (Varshney et al., 2000; Wang et al., 2009; Zhang et al., 2010, 2012; Wu et al., 2012; Cui et al., 2014; Liu et al., 2014). Moreover, a number of yield-related genes were cloned from polyploid wheat based on in silico cloning. Wheat sucrose synthase two orthologous gene (TaSus2) was isolated and found to be significantly associated with TKW (Jiang et al., 2011). Two putative cytokinin oxidase genes (TaCKX2.1 and TaCKX2.2) were cloned and showed intimate relationship to kernel number per spike of bread wheat (Zhang et al., 2011). A cell wall invertase gene TaCwi-A1 could explain 4.8% of phenotypic variance for kernel weight over 2 years (Ma et al., 2010). A TaGW2 gene obtained by in silico cloning was significantly associated with TKW, heading, and maturity in bread wheat (Su et al., 2010; Hong et al., 2014; Simmonds et al., 2014). TaGW2 also negatively regulated kernel weight by controlling gene expression level during seed development (Qin et al., 2014).
In this study, we successfully obtained TaGS5 genes from A and D genomes of bread wheat by in silico cloning and mapped them on the short arm of chromosomes 3A and 3D. Analysis of association with agronomic traits indicated that wheat cultivars with TaGS5-A1b allele possess relatively preferred agronomic traits. The purpose of this study is to illustrate association of allelic variation of TaGS5-A1 gene with agronomic traits and uncover the relatively superior TaGS5-A1 alleles in view of agronomic traits in order to provide useful information for the selection of relatively preferable genotype in view of kernel size and plant height in wheat breeding program.
Results
Cloning of TaGS5 gene in bread wheat
Based on the cDNA sequence of OsGS5 in rice, a number of expressed sequence tags (ESTs) were collected from transcriptomes of durum and bread wheat cultivars, as well as NCBI database by full alignment. A putative full-length TaGS5 cDNA sequence was generated by assembling above ESTs. Furthermore, a 1446 bp cDNA sequence was successfully amplified in the cDNA of Chinese Spring with primer set TaGS5−P1 (Table 1), which was designed according to the above-mentioned putative TaGS5 cDNA sequence.
Table 1.
Primers used for the identification of TaGS5 gene in this study.
| Primer | Primer sequence (5′–3′) | Annealing temperature (°C) | PCR fragment size (bp) |
|---|---|---|---|
| TaGS5-P1 | Forward: CAAGCCACTCACTCTCACAT Reverse: TCCTTGAACTCATTTTGGGTCA |
57 | 1533 |
| TaGS5-P2 | Forward: CAAGCCACTCACTCTCACAT Reverse: GATCAGCGCTATCCCTTCTG |
57 | 1436 |
| TaGS5-P3 | Forward: AGCCACTCACTCTCACATTTG Reverse: AGAAGGAATGTGTCGATCAGC |
58 | 1448 |
| TaGS5-P4 | Forward: AGCCAAGCCACTCACTCT Reverse: CTCTCCTTGAACTCATTTTGGG |
56 | 1504 |
| TaGS5-P5 | Forward: GCGAACCAAGACAAGCAG Reverse: CCTTGTACTGCGGAAACCTC |
56 | 930 |
| TaGS5-P6 | Forward: CTTCTGAGCTAGGACCTCTC Reverse: ACAAGGTCAGCTAGTTGTGG |
56 | 1226 |
| TaGS5-P7 | Forward: ACATCCTCTGACCTCACCAA Reverse: GATACAACTGCATGGCTCCA |
57 | 1427 |
| TaGS5-P8 | Forward: TCATTATGTGCCACAACTAGCT Reverse: AGTACCGAAAAGTTGTACGACT |
57 | 1225 |
| TaGS5-P9 | Forward: TGTCAATGGGATGTTGCCTG Reverse: TCATCGGTGTGTAGGAAGCTG |
58 | 1162 |
| TaGS5-P10 | Forward: TCATACACACATAATCCAGTCGA Reverse: GATCGTGGGTGTTGCATCTAT |
55 | 800 |
| TaGS5-P11 | Forward: GACTTAGAACCACGACAGCC Reverse: CGTAGCATCCATCGGCATG |
57 | 1086 |
| TaGS5-P12 | Forward: GAGCACAAGAGTGAAGCGAGATGG Reverse: CGTTGTTGGCGTATGCGTCTGA |
59 | 1400 |
| TaGS5-P13 | Forward: AAGGTCGGGCAAAGTCTATG Reverse: CGAGGAGAAAGAGAGCAAGGA |
56 | 1000 |
| TaGS5-P14 | Forward: GAAGGCCAGCACATACATCA Reverse: TGTGCCACCTGTCATTTCTT |
57 | 2127 |
| 18s | Forward: CCTGCGGCTTAATTGACTC Reverse: GTTAGCAGGCTGAGGTCTCG |
56 | 150 |
| TaGS5-P15 | Forward: GAAGGCCAGCACATACATCA Reverse: GCTGCTGATGTTTGTCCA |
56 | 276 |
| TaGS5-P16 | Forward: TAGAGCCTCAAACTGGACCG Reverse: AGATGCTGATGATGTTTGTCCA |
56 | 127 |
To further obtain full-length TaGS5 genomic DNA sequence, five primer sets (TaGS5−P5 to TaGS5−P9) were designed according to TaGS5cDNA sequence. Exon positions were deduced by full alignment of TaGS5 cDNA sequence and OsGS5 gDNA sequence. Five corresponding fragments were successfully amplified in gDNA of Chinese Spring with primer sets TaGS5−P5 to TaGS5−P9. A total of 70 subclones were sequenced from both directions after ligation with T-Easy vector. Genomic DNAs of T. urartu, Ae. speltoids, T. tauschii, and durum wheat were used to map the genomic location of each subclone. Based on the positioned sequences of subclones, two full-length TaGS5 gDNA sequences were successfully assembled on A and D genomes of Chinese Spring and were designated as TaGS5-A1 and TaGS5-D1 genes (Figure S1).
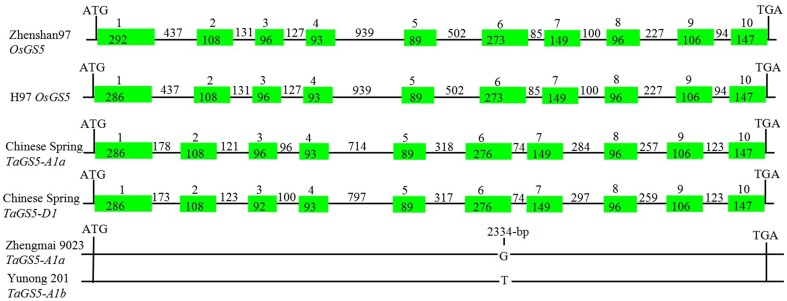
Physical mapping by a set of nullisomic–tetrasomic lines and ditelosomic lines of Chinese Spring indicated that TaGS5-A1 and TaGS5-D1 genes were located on the short arm of chromosomes 3A and 3D, respectively. Sequencing blast of TaGS5-A1 and TaGS5-D1 genes in URGI database of Chinese Spring (http://wheat-urgi.versailles.inra.fr/) indicated that TaGS5-A1 gene is identical to contig TA_454_ch3A|scaffold21314_length_7500 on the chromosome 3A, whereas TaGS5-D1 gene is identical to contig IWGSC_3DS|IWGSC_chr3DS_ab_k95_contigs_longerthan_200_2595062 on the chromosome 3DS. This finding suggested that the sequences and chromosome locations of TaGS5-A1 and TaGS5-D1 genes were very reliable. Further analysis of gDNA sequences of TaGS5-A1 with 3611 bp and TaGS5-D1 with 3705 bp indicated that both TaGS5-A1 and TaGS5-D1 genes were composed of 10 exons and 9 introns (Figure 1). The deduced amino acid sequence showed that both TaGS5-A1 and TaGS5-D1 genes could encode a 481 aa protein with six domains as predicted using software SMART (http://smart.embl-heidelberg.de/). These domains were alpha amylase inhibitor domain at 22–74 interval, putative carbohydrate binding domain at 102–163 interval, RIIalpha (regulatory subunit portion of type II PKA R-subunit) domain at 153–183 interval, fibrinogen-related (FBG) domain at 234–383 interval, Bowman–Birk type proteinase inhibitor domain at 260–296 interval, and phosphoinositide 3-kinase region postulated to contain C2 domain at 286–422 interval.
Figure 1.
Schematic representation of the structures of the TaGS5 and OsGS5 genes in bread wheat and rice. Filled boxes and lines represent exons and introns, respectively.
Molecular characterization of TaGS5 genes in chinese bread wheat
Four primer sets (TaGS5−P10 to TaGS5−P13, Table 1) were used to analyze nucleotide polymorphism of TaGS5-A1 promoter sequence in the 90 selected wheat cultivars. No fragment difference was found in the promoter of TaGS5-A1 gene. Furthermore, 20 wheat cultivars, which were composed of 10 cultivars with relatively large kernel size, and 10 cultivars with relatively small kernel size, were selected to sequence cDNA sequences of TaGS5-A1 and TaGS-D1 genes amplified with primer set TaGS5−P1 and TaGS5−P10–P13 (Table 1). Sequencing results revealed that a SNP of G/T was found at position 907 bp of cDNA sequences (Figure 2), which was in the predicted FBG domain of TaGS5-A1 gene. Furthermore, five primer sets (TaGS5−P5–P9, Table 1) were used to identify allelic variations in TaGS5-A1 and TaGS5-D1 genomic DNA sequences among all surveyed cultivars, but no other polymorphism was found. Further analysis of the SNP indicated that the G/T mutation occurred at position 2334 bp in the sixth exon of TaGS5-A1 genomic DNA sequence. Sequencing results confirmed the reliability of the SNP. This SNP resulted in an amino acid change from alanine to serine. The TaGS5-A1 allele with G at position 907 bp of TaGS5-A1 cDNA sequence was designated as TaGS5-A1a, whereas the TaGS5-A1 allele with T at position 907 bp of TaGS5-A1 cDNA sequence was designated as TaGS5-A1b according to the nomenclature of McIntosh et al. (2007).
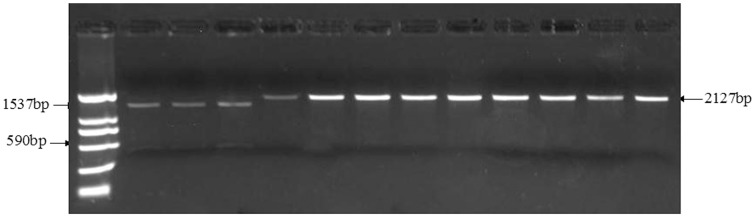
Figure 2.
Identification of TaGS5-A1a and TaGS5-A1b alleles by digestion of restriction enzyme Bbv1.
The alternation of G to T (GCAGC to TCAGC) in TaGS5-A1 gene broke the digestion site of restriction enzyme Bbv1 (GCAGC). Therefore, restriction enzyme Bbv1 was used to digest 2127 bp fragments amplified with primer set TaGS5_P13 (Table 1) in different wheat cultivars to distinguish TaGS5-A1a and TaGS5-A1b alleles. After digestion by BbvI, cultivars with both 590 and 1537 bp fragments possessed TaGS5-A1a allele, and cultivars with only 2127 bp fragment possessed TaGS5-A1b allele (Figure 2).
Association between TaGS5-A1 alleles and kernel size and other agronomic traits
Based on the results of PCR amplification with primer set TaGS5_P14 and digestion by enzyme BbvI, 15 and 26 out of 41 landraces, respectively, belonged to TaGS5-A1a and TaGS5-A1b alleles, and 70 and 252 out of 322 modern cultivars belonged to TaGS5-A1a and TaGS5-A1b alleles, respectively. Variance analysis in the surveyed landraces indicated that plant height, panicle length, and internode length below spike of cultivars with TaGS5-A1a alleles were relatively higher than those with TaGS5-A1b alleles. Moreover, TKW, kernel length, and kernel width of wheat landraces with TaGS5-A1b alleles were significantly higher than those of wheat cultivars with TaGS5-A1a alleles over 3 years. The grain length/grain width (GL/GW) ratio of landraces with TaGS5-A1a was also significantly higher than that of cultivars with TaGS5-A1b over 3 years (Table 2).
Table 2.
Comparison of agronomic traits of bread cultivars with TaGS5-A1a and TaGS5-A1b alleles.
| Trait | 2013 | 2014 | 2015 | ||||
|---|---|---|---|---|---|---|---|
| TaGS5-A1a | TaGS5-A1b | TaGS5-A1a | TaGS5-A1b | TaGS5-A1a | TaGS5-A1b | ||
| Sample number | 85 | 278 | 85 | 278 | 85 | 278 | |
| Plant height (cm) | 81.39a | 75.64a | 96.58A | 77.72B | 100.25A | 92.19B | |
| Spike length (cm) | 10.24a | 9.81b | 10.02A | 9.14B | 10.90a | 10.38b | |
| Internode length below spike (cm) | 27.23A | 25.04B | 31.62a | 29.75a | 32.77A | 30.30B | |
| Spikelet number per spike | 19.03a | 19.35a | 19.56a | 19.88a | 19.11a | 19.32a | |
| Total | Kernel number per spike | 44.82a | 43.00a | 51.56a | 53.07a | 48.26a | 48.09a |
| Kernel length (mm) | 6.89a | 6.85a | 7.00a | 6.93a | 6.93a | 6.81a | |
| Kernel width (mm) | 3.22B | 3.29A | 3.46a | 3.72a | 3.38a | 3.42a | |
| Kernel length/kernel width ratio | 2.15A | 2.09B | 2.03A | 1.93B | 2.06a | 2.00b | |
| Thousand-kernel weight (g) | 40.86b | 42.94a | 49.24a | 50.72a | 41.52a | 42.52a | |
| Sample number | 15 | 26 | 15 | 26 | 15 | 26 | |
| Plant height (cm) | 103.89a | 92.49a | 117.00A | 98.44B | 123.66A | 103.36B | |
| Spike length (cm) | 10.25a | 9.90a | 10.33a | 9.94a | 10.87a | 10.35a | |
| Internode length below spike (cm) | 31.98a | 30.19a | 40.10a | 34.35b | 37.14a | 34.81a | |
| Spikelet number per spike | 18.96a | 19.44a | 19.62a | 19.85a | 19.64a | 19.28a | |
| Landraces | Kernel number per spike | 46.76a | 41.94a | 50.12a | 47.37a | 50.33a | 47.32a |
| Kernel length (mm) | 6.48b | 6.81a | 6.53b | 6.96a | 6.43b | 6.73a | |
| Kernel width (mm) | 3.01b | 3.16a | 3.23B | 3.51A | 3.21b | 3.40a | |
| Kernel length/kernel width ratio | 2.16a | 2.16a | 2.05a | 1.99a | 2.01a | 1.99a | |
| Thousand-kernel weight (g) | 33.52b | 39.07a | 40.46B | 46.65A | 35.73b | 41.21a | |
| Sample number | 70 | 252 | 70 | 252 | 70 | 252 | |
| Plant height (cm) | 76.57a | 73.90a | 80.06a | 75.58b | 95.23a | 91.04b | |
| Spike length (cm) | 10.24a | 9.80b | 9.96A | 9.06B | 10.90a | 10.38b | |
| Internode length below spike (cm) | 26.21a | 24.51b | 29.80a | 29.28b | 31.83a | 29.84b | |
| Spikelet number per spike | 19.04a | 19.34a | 19.55a | 19.89a | 18.99a | 19.32a | |
| Modern cultivars | Kernel number per spike | 44.40a | 43.11a | 51.87a | 53.66a | 47.82a | 48.12a |
| Kernel length (mm) | 6.98a | 6.85b | 7.10A | 6.93B | 7.03A | 6.82B | |
| Kernel width (mm) | 3.26a | 3.31a | 3.53a | 3.75a | 3.416a | 3.418a | |
| Kernel length/kernel width ratio | 2.15A | 2.08B | 2.02A | 1.92B | 2.07A | 2.00B | |
| Thousand-kernel weight(g) | 42.44a | 43.34a | 51.12a | 51.14a | 42.76a | 42.66a | |
Uppercase and lowercase letters after numbers showed extreme (P < 0.01) and significant (P < 0.05) differences, respectively.
Variation analysis in surveyed modern cultivars showed that plant height, spike length, and internode length below spike of cultivars with TaGS5-A1a alleles were also relatively higher than those with TaGS5-A1b alleles (Table 2). However, the differences between the traits of cultivars with two TaGS5-A1 alleles sharply reduced in the modern cultivars when compared with the landraces (Table 2). Furthermore, the kernel length and GL/GW ratio of cultivars with TaGS5-A1a were significantly higher than that of cultivars with TaGS5-A1b over 3 years. Kernel width and TKW of cultivars with TaGS5-A1b were slightly higher than those with TaGS5-A1a in 2013 and 2014 (Table 2). This finding suggested that TaGS5-A1b is a relatively preferred allele in view of agronomic traits in bread wheat. This allele for high TKW probably underwent strong positive selection in Chinese modern wheat breeding because of its high percentage (78.2% in the modern cultivars and 63.4% in the landraces). Additionally, no significant difference existed between the spikelet number per spike and kernel number per spike of the landraces and modern cultivars with TaGS5-A1a and TaGS5-A1b over 3 years (Table 2).
To further determine the influence of TaGS5-A1 alleles on kernel size and other agronomic traits in modern cultivars, a F10 RIL population was examined using TaGS5-A1 CAPS marker. Results showed that 64 and 89 out of 153 wheat inbred lines belonged to TaGS5-A1a and TaGS5-A1b alleles, respectively. The average of each trait of the lines under two locations was used to analyze the association of TaGS5-A1 alleles with kernel size and other agronomic traits (Table 3). Variance analysis also indicated that plant height (92.2 cm), spike length (11.1 cm), and internode length below spike (39.2 cm) of the lines with TaGS5-A1a alleles were significantly higher than those (88.1, 10.07, and 36.6 cm, respectively) of the lines with TaGS5-A1b alleles (P < 0.05; Table 3). Further analysis indicated that kernel length of lines with TaGS5-A1b (7.08 mm) were slightly narrower than that of lines with TaGS5-A1a (7.09 mm), and kernel width of lines with TaGS5-A1b (3.52 mm) were slightly wider than that of lines with TaGS5-A1a (3.47 mm), but these differences were not significant. However, the average of TKW of the lines with TaGS5-A1b alleles (45.8 g) was significantly higher than that of the lines with TaGS5-A1a alleles (44.9 g; P < 0.05). The difference mainly resulted from relatively wider kernel width of the lines with TaGS5-A1b allele (Table 3). In addition, the GL/GW ratio of the lines with TaGS5-A1a alleles (2.04) was significantly higher than that of the lines with TaGS5-A1b alleles (2.01; P < 0.05). This result is consistent with the association between TaGS5-A1 alleles and agronomic traits in landraces and modern cultivars, as mentioned above.
Table 3.
Comparison of agronomic traits of lines with TaGS5-A1a and TaGS5-A1b alleles using the F10RIL population of UC 1110/ PI 610750.
| Trait | F10 RIL population | |
|---|---|---|
| TaGS5-A1a | TaGS5-A1b | |
| Sample number | 64 | 89 |
| Plant height (cm) | 92.2a | 88.1b |
| Spike length (cm) | 11.1a | 10.7b |
| Internode length below spike (cm) | 39.2a | 36.6a |
| Spikelet number per spike | 18.1a | 18.7a |
| Kernel number per spike | – | – |
| Kernel length (mm) | 7.09a | 7.08a |
| Kernel width (mm) | 3.47a | 3.52a |
| Kernel length/kernel width ratio | 2.04a | 2.01b |
| Thousand-kernel weight (g) | 44.9b | 45.8a |
Lowercase letters after numbers showed and significant (P < 0.05) differences, respectively.
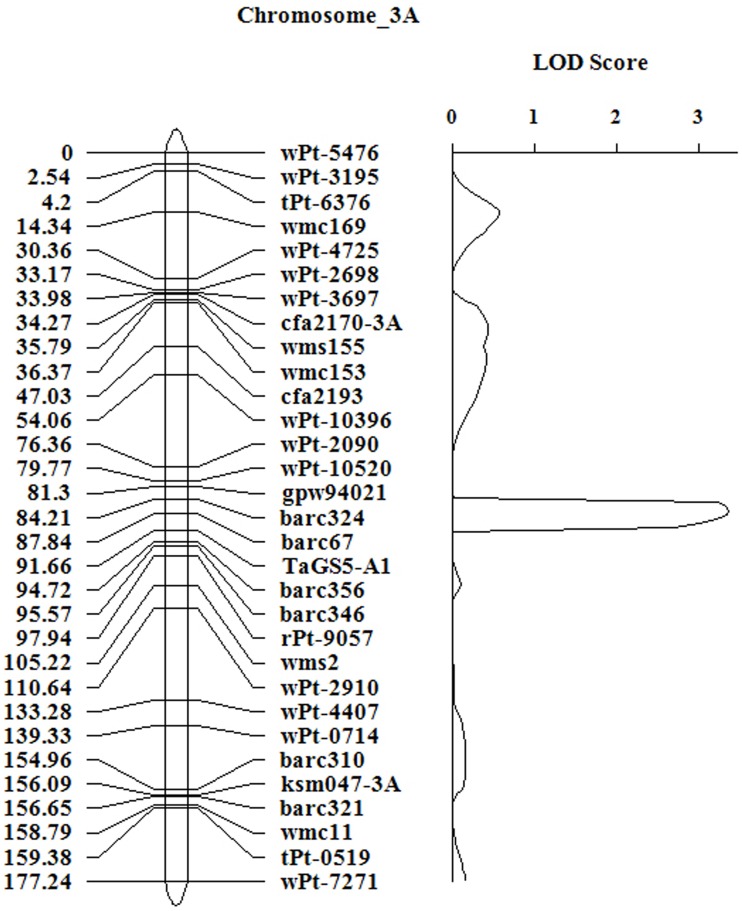
To confirm the chromosomal location and effect of TaGS5-A1 gene in bread wheat, TaGS5-A1 gene was further mapped using the F10 RIL population (Figure 4). Linkage analysis showed that a major QTL for TKW was mapped on chromosome 3AS, between simple sequence repeat markers Barc356 and Barc67 (C-3AS2-0.23). The 8.2% of the phenotypic variance in the F10 populations over two environments was explained with a logarithm of odds (LOD) score of 3.36 (Figure 3).
Figure 3.
Linkage map of TaGS5-A1 gene on 3AS and logarithm of odds contours obtained by composite interval mapping for quantitative trait loci on thousand-kernel weight in the F10 RIL population of UC 1110/ PI 610750. The data of thousand-kernel weight were averaged over two locations.
Expression analysis of TaGS5-A1a and TaGS5-A1b genotypes
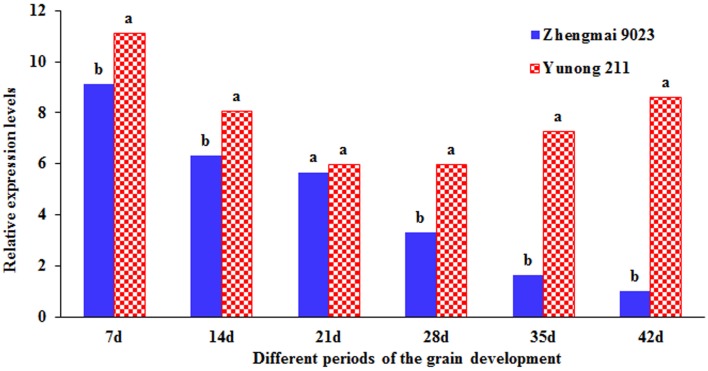
Two cultivars, namely, Zhengmai 9023 with TaGS5-A1a and Yunong 211 with TaGS5-A1b, were selected to identify expression levels at six different developmental stages of the seeds (Figure 4). Quantitative real-time PCR (qRT-PCR) indicated that the relative expression level of TaGS5-A1b was higher than that of TaGS5-A1a in all six stages.
Figure 4.
Relative expression level of TaGS5-A1 gene in seeds of different developmental stages of Yunong 211 and Zhengmai 9023. Different letters on the top of the bars indicated the significant difference at 5% probability level.
Discussion
Wheat yield is usually the most important trait for wheat breeders over the long-term improvement of bread wheat as it reflects the culmination of all the processes of vegetative and reproductive growth, as well as their interactions with the edaphic and aerial environments (Quarrie et al., 2006). Wheat yield greatly increased through continuous artificial selection; especially, improvement of lodge resistance sharply increased wheat production by taking advantage of dwarf genes (Rht-B1 and Rht-D1) in the 1960s and 1970s (Peng et al., 1999), which resulted in the worldwide “Green Revolution.” However, wheat yield has not significantly improved in the past decade because of the increasingly narrow genetic basis of wheat germplasm in current wheat breeding program. Therefore, discovering new gene germplasms with relatively superior agronomic traits is markedly beneficial in improving wheat yield. In this study, we cloned TaGS5 gene from bread wheat and identified two alleles at the TaGS5-A1 locus. Furthermore, allelic variation of TaGS5-A1 gene was intimately associated with agronomic traits. However, this association showed significant difference in landraces and modern cultivars. The larger differences of the agronomic traits between TaGS5-A1a and TaGS5-A1b genotypes in the landraces suggested that TaGS5 gene played an important role in modulating yield-related traits in the landraces. The differences may possibly be caused by more relatively superior genes related to yield traits gathered in the modern Chinese cultivars after strong artificial selection in view of agronomic traits (Wang et al., 2012a).
Chinese landraces usually show higher plant height, easy lodging, and lower yield compared with modern cultivars. However, landraces are precious germplasm for their aspects as almost unselected populations, including yield-related traits. The preferred TaGS5-A1b genotype for high TKW was more prevalent in modern cultivars than in landraces, indicating that TaGS5-A1b genotype underwent very strong positive selection in modern Chinese wheat breeding. Interestingly, a paradox seemed to occur on the association between TaGS5-A1 gene and kernel length, i.e., wheat landraces with TaGS5-A1b allele possessed longer kernel length than that of landraces with TaGS5-A1a allele, but the comparison result was the exact opposite in the modern Chinese cultivars. Kernel length increase in the landraces possibly resulted from improvement of lodging resistance because cultivars with TaGS5-A1b allele showed 10–20% lower plant height than cultivars with TaGS5-A1a allele.
Chinese wheat is mainly planted in 10 agro-ecological zones, which are further divided into 26 sub-zones. Winter, facultative, and spring wheats are sown both in autumn and spring. Of all agro-ecological zones, the Yellow and Huai wheat production regions, which cover all of Henan and parts of Shandong, Hebei, Shanxi, Anhui, and Jiangsu Provinces, are the most important and largest wheat production zone with 60–70% of the total harvested area and the total wheat production. Successful utilization of dwarfing genes (Rht1, Rht2, and Rht8) and the 1B/1R translocation made significant contributions to yield improvement in the wheat-producing region of the Yellow and Huai Valleys, as indicated by Zhou et al. (2007). However, yield increase has been slowing down in recent years because of homogenization of parents in wheat breeding program of this region. The germplasm in this study was from the Yellow and Huai wheat regions. Most of the germplasms were currently popular cultivars or advanced lines, and some used to play the important role as parents in wheat breeding in the Yellow and Huai wheat regions. Therefore, uncovering yield-related genes and understanding their association with agronomic traits could provide useful information for further improvement of yield in the wheat breeding program of the Yellow and Huai wheat regions of China.
In this study, TaGS5 genes were physically mapped on the chromosome of 3A and 3D and were closely associated with kernel size, TKW, plant height, spike length, and internode length below spike. To date, previous studies indicated that almost all chromosomes of bread wheat harbored yield-related QTLs, including group 3 (Wang et al., 2009; Wu et al., 2012; Zhang et al., 2012; Cui et al., 2014; Liu et al., 2014). In particular, a number of QTLs in 3A and 3D chromosome were identified to control plant height, spike length, spike per plant, kernel number per spike, kernel weight per spike, kernel length, kernel width, and TKW (Cui et al., 2014; Liu et al., 2014). These QTLs account for 5.26–15.82% of the phenotypic variation. A TKW-related QTL was previously mapped to be linked with Barc356 on chromosome 3A in bread wheat (Cui et al., 2014). Liu et al. (2014) indicated that a QTL in the interval of Barc324-Xwmc428 on chromosome 3A was detected to be associated with plant height in five environments. In this study, TaGS5-A1 gene was intimately linked to Barc356 and Barc324 in the F10 RIL population. However, more research is required to further evaluate the apparent association of the TaGS5 loci with agronomic traits.
The great influence of single-nucleotide change on genes has been studied for many years; a single-nucleotide change could result in a strong influence on genes and further cause apparent change of phenotype (Slade et al., 2005; Uauy et al., 2009; Jiao et al., 2010; Kurowska et al., 2011; Rawat et al., 2012; Chen et al., 2014; Dabhi and Mistry, 2014; Sharp and Dong, 2014). Jiao et al. (2010) indicated that a point mutation in the third exon of OsSPL14 gene generated an “ideal” rice plant with a reduced tiller number, increased lodging resistance, and enhanced kernel yield. In the present study, TaGS5-A1 gene exhibited a single-nucleotide change belonging to non-synonymous mutation in the sixth exon, resulting in the change of kernel size, and other agronomic traits. Evidence also suggested that non-synonymous mutations could directly or indirectly destabilize the amino acid interactions and hydrogen bond networks and finally affecting protein function (Dabhi and Mistry, 2014). Therefore, the discovery of the TaGS5-A1b allele will be useful for the study of interaction relationship of the yield-related genes because the non-synonymous mutation in the FBG domain of TaGS5-A1 gene might change the structure of the TaGS5-A1 protein.
Materials and methods
Plant materials
A total of 363 wheat cultivars were planted in 2012–2013, 2013–2014, and 2014–2015 cropping seasons at the experimental field of Zhengzhou Scientific Research and Education Center of Henan Agricultural University (N34.9°, E113.6°). Accessions used in this study were composed of 41 landraces and 322 modern cultivars or advanced lines from the Yellow and Huai wheat regions of China. The field experimental design was performed with random complete block design with two replicates. Each plot contained four 200 cm long rows with 23 cm between neighboring rows and 10 cm between neighboring plants (Chen et al., 2013a,b). Test plots were managed according to local practices. The plant height, spike length, internode length below spike, and spikelet number of 10 spikes of each cultivar surveyed were investigated in the fields before the plants were harvested. After harvest, the average kernel number of 10 spikes of each cultivar surveyed, as well as the kernel length, kernel width, GL/GW ratio, and TKW of two plants of each cultivar surveyed, was investigated using naturally dried materials.
An entire set of Chinese Spring nullisomic–tetrasomic lines and ditelosomic lines were used to map the TaGS5 genes on chromosomes of Chinese Spring. Professor Jorge Dubcovsky from the University of California, Davis, kindly provided F10RIL population (UC 1110 × PI 610750) composed of 153 lines, which were used for further physical mapping of TaGS5-A1 and analysis of association between TaGS5-A1 alleles and kernel size. The RIL population was planted in October 2013 and was harvested in the June 2014 cropping season at Anyang (N36.1°, E114.5°) and Zhengzhou (N34.9°, E113.6°); the population grew well at both locations. No lodge occurred in the entire field experiment. Agronomic traits (e.g., plant height, spike length, spikelet number per spike, kernel length, kernel width, and TKW) were investigated as mentioned above.
DNA extraction and polymerase chain reaction amplification parameters
The total genomic DNA of all wheat cultivars surveyed in this study was extracted using CTAB method (Chen et al., 2010). PCR amplifications were performed in a Bio-Rad S1000 or ABI 9700 thermal cyclers. The PCR reaction system with total volumes of 25 μL was composed of 0.5 μL of each primer (10 pmol/μL), 100 ng genomic DNA, 2.5 μL 10 × Taq buffer (Mg2+ plus), 0.5 μL dNTP (2.5 mM), and 1.25 U Taq DNA polymerases (Tiangen Biotech, Beijing Co., Ltd.). The PCR reaction procedure consisted of three stages. The first stage involved 94°C for 5 min. The second stage included 94°C for 30 s, annealing (55–59°C) for 30 s, and 72°C for 1–2 min, performing 35 cycles. The last stage was 72°C for 7 min. PCR products were analyzed and separated on 1.0–1.5% (w/v) agarose gels, stained with ethidium bromide, and visualized using UV light. The expected PCR products were purified using the SanPrep Column DNA Gel Extraction Kit (Shanghai Biological Technology Co., Ltd.). All purified PCR products were ligated into pMD18-T vector (TaKaRa Biotechnology Co., Ltd., Dalian) and were transformed into cells of the Escherichia coli DH-5α strain. Plasmids containing targeted fragments were extracted using Plasmid Rapid Isolation Kit (Shanghai Biological Technology Co., Ltd.) and were sequenced for 12 clones for each sample using Shanghai Sangon Biotech Co., Ltd. The sequencing reliability of each subclone was checked using Chromas 2.4.3 (http://technelysium.com.au/?page_id=13) and FinchTV 1.5.0 (http://www.geospiza.com/Products/finchtv.shtml).
Primer designing for cloning TaGS5 genes in bread wheat
Based on the cDNA sequence of OsGS5 gene (JN256056 and JN256055) in rice, several new contigs were obtained by alignment in a durum wheat transcriptome (kindly provided by Professor Jorge Dubcovsky) and a hexaploid wheat transcriptome of Yunong 201 (unpublished data in our lab). In addition, several wheat ESTs were gained by blasting cDNA sequence of OsGS5 in NCBI database. The collected wheat sequences and the cDNA sequence of OsGS5 were aligned and assembled using DNAMAN Version 6.0 software (http://www.softlandsl.com/free/dnaman+6+full.html). Based on the assembled sequence, four pairs of primer sets (TaGS5−P1, TaGS5−P2, TaGS5−P3, and TaGS5−P4; Table 1) were designed to amplify the full length of the cDNA sequence of GS5 gene in Chinese Spring. Finally, a primer set (TaGS5−P1) successfully amplified full-length TaGS5 cDNA sequence in Chinese Spring. Based on the obtained cDNA sequence of TaGS5 gene, five new primer sets (TaGS5−P5, TaGS5−P6, TaGS5−P7, TaGS5−P8, and TaGS5−P9; Table 1) were designed to amplify TaGS5 genomic DNA sequence. All primers (Table 1) in this study were designed using Premier Primer 3.0 (http://primer3.ut.ee/) and Premier Primer 5.0 and were synthesized by Shanghai Biological Technology Co., Ltd. (http://www.sangon.com/).
Development of CAPS marker for identification of TaGS5-A1 alleles
Genomic-specific primer set TaGS5−P14 (Table 1) was designed for the development of CAPS marker to distinguish T/G at the six exons of the TaGS5-A1 gDNA sequence. Amplification fragments were purified and were digested by Bbv1 restriction enzyme (New England Biolabs Co., Ltd). The reaction system of Bbv1 restriction enzyme with total volumes of 50 μL were composed of 5 μL 10 × NEB buffer II, 1 μL Bbv1 restriction enzyme, 2 μL PCR production, and 42 μL dd H2O. The mixed solutions were stably kept at 37°C for 40 min in water bath tank (HH-S4A Double-row4-hole). Finally, the digested products were separated on 1.5% (w/v) agarose gels.
Quantitative real-time PCR analysis of TaGS5-A1a and TaGS5-A1b genotypes
The total RNA of three-leaf seedlings was extracted using Trizol reagent and was reverse-transcripted to cDNA through PrimeScript RT reagent kit with gDNA Eraser (TaKaRa Biotechnology Co., Ltd, Dalian), following the kit instruction. A pair of specific primers (TaGS5−P1, Table 1) was used by reverse transcriptional PCR. The PCR products were linked to pGEM18-T vector by pGEM18-T simple vector kit (TaKaRa: D103A). E. coli (TaGS5-cDNA-T) were cultured for 6–8 h at 37°C and sequenced by Shanghai Biological Technology Co., Ltd.
Gene-specific primers TaGS5-P15 and TaGS5-P16 were designed to examine the expression levels of TaGS5-A1a and TaGS5-A1b genotypes. 18s gene was selected as internal control with the primer 18s (Table 1). The expression profiles of TaGS5-A1a and TaGS5-A1b genotypes were measured using the cDNA samples from seeds of different developmental stages (i.e., 7d, 14d, 21d, 28d, 35d, and 42d after anthesis) of cultivars Zhengmai 9023 with TaGS5-A1a and Yunong 211 with TaGS5-A1b. Bio-Rad iQ5 Sequence Detection System (Applied Biosynthesis, CA, USA) with SYBR Premix Ex TaqII (TaKaRa Biotechnology Co., Ltd, Dalian) was used to perform qRT-PCR. The PCR conditions consisted of an initial denaturation step for 2 min at 94°C, followed by 40 cycles of 10 s at 95°C, 10 s at 56°C, 20 s at 72°C, and a final extension of 10 min at 72°C. The 2−ΔΔCT method was used to normalize and calibrate transcript values relative to the endogenous 18s control. Six independent samples with triplicate repeats were analyzed.
QTL mapping and statistical analyses
Physical mapping and QTL analysis of TaGS5-A1 genes in the F10 population were performed using IciMapping 4.0 software (http://www.isbreeding.net/). Detailed genetic map of the F10 population was in Lowe et al. (2011) and we only used markers from 3A chromosome to map traits surveyed. QTL scanning was performed using inclusive composite interval mapping through stepwise regression by considering all marker information on chromosome 3 using 2.5 as the threshold LOD score (Meng et al., 2015).
Analysis of variance was conducted using PROC MIXED in the Statistical Analysis System (SAS Institute Inc., 2000) with genotype classes as categorical variables to derive the means of agronomic traits for each class and to test the significant level for the two classes. The differences of agronomic traits among different genotypes were tested by least significant range multiple comparisons.
Author contributions
FC designed the project. SW performed experiment. XZ and DC investigated agronomic traits. FC and SW wrote the paper.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
This project was funded by the 973 projects (2014CB160303 and 2014CB138105), Program for New Century Excellent Talents in University (NCET-13-0776) of China and Program for Innovative Research Team (in Science and Technology) in University of Henan Province (14IRTSTHN010).
Supplementary material
The Supplementary Material for this article can be found online at: http://journal.frontiersin.org/article/10.3389/fpls.2015.01166
References
- Chen F., Gao M. X., Zhang J. H., Zuo A. H., Shang X. L., Cui D. Q. (2013a). Molecular characterization of vernalization and response genes in bread wheat from the Yellow and Huai Valley of China. BMC Plant Biol. 13:199. 10.1186/1471-2229-13-199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen F., Li H., Cui D. Q. (2013b). Discovery, distribution and diversity of Puroindoline-D1 genes in bread wheat from five countries (Triticum aestivum L.). BMC Plant Biol. 13:125. 10.1186/1471-2229-13-125 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen F., Xu H. X., Zhang F. Y., Xia X. C., He Z. H., Wang D. W., et al. (2010). Physical mapping of puroindoline b-2 genes and molecular characterization of a novel variant in durum wheat (Triticum turgidum L.). Mol. Breed. 28, 153–161. 10.1007/s11032-010-9469-2 [DOI] [Google Scholar]
- Chen L., Hao L., Parry M. A. J., Phillips A. L., Hu Y. G. (2014). Progress in TILLING as a tool for functional genomics and improvement of crops. J. Integr. Plant Biol. 56, 425–443. 10.1111/jipb.12192 [DOI] [PubMed] [Google Scholar]
- Cui F., Zhao C., Ding A., Li J., Wang L., Li X., et al. (2014). Construction of an integrative linkage map and QTL mapping of grain yield-related traits using three related wheat RIL populations. Theor. Appl. Genet. 127, 659–675. 10.1007/s00122-013-2249-8 [DOI] [PubMed] [Google Scholar]
- Dabhi B., Mistry K. N. (2014). In silico analysis of single nucleotide polymorphism (SNP) in human TNF-alpha gene. Meta Gene. 2, 586–595. 10.1016/j.mgene.2014.07.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan C., Xing Y., Mao H., Lu T., Han B., Xu C., et al. (2006). GS3, a major QTL for grain length and weight and minor QTL for grain width and thickness in rice, encodes a putative transmembrane protein. Theor. Appl. Genet. 112, 1164–1171. 10.1007/s00122-006-0218-1 [DOI] [PubMed] [Google Scholar]
- Guo L., Ma L., Jiang H., Zeng D., Hu J., Wu L., et al. (2009). Genetic analysis and fine mapping of two genes for grain shape and weight in rice. J. Integr. Plant Biol. 51, 45–51. 10.1111/j.1744-7909.2008.00793.x [DOI] [PubMed] [Google Scholar]
- Hong Y., Chen L., Du L. P., Su Z., Wang J., Ye X., et al. (2014). Transcript suppression of TaGW2 increased grain width and weight in bread wheat. Funct. Integr. Genomics 14, 341–349. 10.1007/s10142-014-0380-5 [DOI] [PubMed] [Google Scholar]
- Jiang Q., Hou J., Hao C., Wang L., Ge H., Dong Y., et al. (2011). The wheat (T. aestivum) sucrose synthase 2 gene (TaSus2) active in endosperm development is associated with yield traits. Funct. Integr. Genomics 11, 49–61. 10.1007/s10142-010-0188-x [DOI] [PubMed] [Google Scholar]
- Jiao Y., Wang Y., Xue D., Wang J., Yan M., Liu G., et al. (2010). Regulation of OsSPL14 by OsmiR156 defines ideal plant architecture in rice. Nat. Genet. 42, 541–544. 10.1038/ng.591 [DOI] [PubMed] [Google Scholar]
- Kurowska M., Daszkowska-Golec A., Gruszka D., Marzec M., Szurman M., Szarejko I., et al. (2011). TILLING - a shortcut in functional genomics. J. Appl. Genet. 52, 371–390. 10.1007/s13353-011-0061-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J., Thomson M., McCouch S. R. (2004a). Fine mapping of a grain-weight quantitative trait locus in the pericentromeric region of rice chromosome 3. Genetics 168, 2187–2195. 10.1534/genetics.104.034165 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J., Xiao J., Grandillo S., Jiang L., Wan Y., Deng Q., et al. (2004b). QTL detection for rice grain quality traits using an interspecific backcross population derived from cultivated Asian (O. sativa L.) and African (O. glaberrima S.) rice. Genome 47, 697–704. 10.1139/g04-029 [DOI] [PubMed] [Google Scholar]
- Li Q., Li L., Yang X., Warburton M. L., Bai G., Dai J., et al. (2010). Relationship, evolutionary fate and function of two maize co-orthologs of rice GW2 associated with kernel size and weight. BMC Plant Biol. 10:143. 10.1186/1471-2229-10-143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y., Fan C., Xing Y., Jiang Y., Luo L., Sun L., et al. (2011). Natural variation in GS5 plays an important role in regulating grain size and yield in rice. Nat. Genet. 43, 1266–1269. 10.1038/ng.977 [DOI] [PubMed] [Google Scholar]
- Liu G., Jia L. J., Lu L. H., Qin D. D., Zhang J. P., Guan P. F., et al. (2014). Mapping QTLs of yield-related traits using RIL population derived from common wheat and Tibetan semi-wild wheat. Theor. Appl. Genet. 127, 2415–2432. 10.1007/s00122-014-2387-7 [DOI] [PubMed] [Google Scholar]
- Liu J., Deng M., Guo H., Raihan S., Luo J., Xu Y., et al. (2015). Maize orthologs of rice GS5 and their trans-regulator are associated with kernel development. J. Integr. Plant Biol. 57, 943–953. 10.1111/jipb.12421 [DOI] [PubMed] [Google Scholar]
- Lowe L., Jankuloski L., Chao S., Chen X., See D., Dubcovsky J. (2011). Mapping and validation of QTL which confer partial resistance to broadly virulent post-2000 North American races of stripe rust in hexaploid wheat. Theor. Appl. Genet. 123, 143–157. 10.1007/s00122-011-1573-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma D., Yan J., He Z., Wu L., Xia X. (2010). Characterization of a cell wall invertase gene TaCwi-A1 on common wheat chromosome 2A and development of functional markers. Mol. Breed. 29, 43–52. 10.1007/s11032-010-9524-z [DOI] [Google Scholar]
- McIntosh R. A., Devos K. M., Dubcovsky J., Rogers W. J., Morris C. F., Appels R., et al. (2007). Catalogue of gene symbols for wheat: 2007 supplement. Available online at: http://wheat.pw.usda.gov/ggpages/wgc/2007upd.html
- Meng L., Li H., Zhang L., Wang J. (2015). QTL IciMapping: integrated software for genetic linkage map construction and quantitative trait locus mapping in bi-parental populations. Crop J. 3, 265–279. 10.1016/j.cj.2015.01.001 [DOI] [Google Scholar]
- Peng J., Richards D. E., Hartley N. M., Murphy G. P., Devos K. M., Flintham J. E., et al. (1999). 'Green revolution' genes encode mutant gibberellin response modulators. Nature 400, 256–261. 10.1038/22307 [DOI] [PubMed] [Google Scholar]
- Qin L., Hao C. Y., Hou J., Wang Y. Q., Li T., Wang L. F., et al. (2014). Homologous haplotypes, expression, genetic effects and geographic distribution of the wheat yield gene TaGW2. BMC Plant Biol. 14:107. 10.1186/1471-2229-14-107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quarrie S. A., Pekic Quarrie S., Radosevic R., Rancic D., Kaminska A., Barnes J. D., et al. (2006). Dissecting a wheat QTL for yield present in a range of environments: from the QTL to candidate genes. J. Exp. Bot. 57, 2627–2637. 10.1093/jxb/erl026 [DOI] [PubMed] [Google Scholar]
- Rawat N., Sehgal S. K., Joshi A., Rothe N., Wilson D. L., McGraw N., et al. (2012). A diploid wheat TILLING resource for wheat functional genomics. BMC Plant Biol. 12:205. 10.1186/1471-2229-12-205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- SAS Institute Inc (2000). SAS Procedures Guide, Version 8. Cary, NC: SAS Institute Inc. [Google Scholar]
- Sharp P., Dong C. (2014). TILLING for plant breeding. Methods Mol. Biol. 1145, 155–165. 10.1007/978-1-4939-0446-4_13 [DOI] [PubMed] [Google Scholar]
- Simmonds J., Scott P., Leverington-Waite M., Turner A. S., Brinton J., Korzun V., et al. (2014). Identification and independent validation of a stable yield and thousand grain weight QTL on chromosome 6A of hexaploid wheat (Triticum aestivum L.). BMC Plant Biol. 14:191. 10.1186/s12870-014-0191-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slade A. J., Fuerstenberg S. I., Loeffler D., Steine M. N., Facciotti D. (2005). A reverse genetic, nontransgenic approach to wheat crop improvement by TILLING. Nat. Biotechnol. 23, 75–81. 10.1038/nbt1043 [DOI] [PubMed] [Google Scholar]
- Song X. J., Huang W., Shi M., Zhu M. Z., Lin H. X. (2007). A QTL for rice grain width and weight encodes a previously unknown RING-type E3 ubiquitin ligase. Nat. Genet. 39, 623–630. 10.1038/ng2014 [DOI] [PubMed] [Google Scholar]
- Su Z., Hao C., Wang L., Dong Y., Zhang X. (2010). Identification and development of a functional marker of TaGW2 associated with grain weight in bread wheat (Triticum aestivum L.). Theor. Appl. Genet. 122, 211–223. 10.1007/s00122-010-1437-z [DOI] [PubMed] [Google Scholar]
- Takano-Kai N., Jiang H., Kubo T., Sweeney M., Matsumoto T., Kanamori H., et al. (2009). Evolutionary history of GS3, a gene conferring grain length in rice. Genetics 182, 1323–1334. 10.1534/genetics.109.103002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takano-Kai N., Jiang H., Powell A., McCouch S., Takamure I., Furuya N., et al. (2013). Multiple and independent origins of short seeded alleles of GS3 in rice. Breed. Sci. 63, 77–85. 10.1270/jsbbs.63.77 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uauy C., Paraiso F., Colasuonno P., Tran R. K., Tsai H., Berardi S., et al. (2009). A modified TILLING approach to detect induced mutations in tetraploid and hexaploid wheat. BMC Plant Biol. 9:115. 10.1186/1471-2229-9-115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varshney R. K., Hoisington D. A., Tyagi A. K. (2006). Advances in cereal genomics and applications in crop breeding. Trends Biotechnol. 24, 490–499. 10.1016/j.tibtech.2006.08.006 [DOI] [PubMed] [Google Scholar]
- Varshney R. K., Prasad M., Roy J. K., Kumar Harjit-Singh N., Dhaliwal H. S., Balyan H. S., et al. (2000). Identification of eight chromosomes and a microsatellite marker on 1AS associated with QTL for grain weight in bread wheat. Theor. Appl. Genet. 100, 1290–1294. 10.1007/s001220051437 [DOI] [Google Scholar]
- Wang E., Wang J., Zhu X., Hao W., Wang L., Li Q., et al. (2008). Control of rice grain-filling and yield by a gene with a potential signature of domestication. Nat. Genet. 40, 1370–1374. 10.1038/ng.220 [DOI] [PubMed] [Google Scholar]
- Wang L., Ge H., Hao C., Dong Y., Zhang X. (2012a). Identifying loci influencing 1000-kernel weight in wheat by microsatellite screening for evidence of selection during breeding. PLoS ONE 7:e29432. 10.1371/journal.pone.0029432 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang R. X., Hai L., Zhang X. Y., You G. X., Yan C. S., Xiao S. H. (2009). QTL mapping for grain filling rate and yield-related traits in RILs of the Chinese winter wheat population Heshangmai × Yu8679. Theor. Appl. Genet. 118, 313–325 10.1007/s00122-008-0901-5 [DOI] [PubMed] [Google Scholar]
- Wang S., Wu K., Yuan Q., Liu X., Liu Z., Lin X., et al. (2012b). Control of grain size, shape and quality by OsSPL16 in rice. Nat. Genet. 44, 950–954. 10.1038/ng.2327 [DOI] [PubMed] [Google Scholar]
- Weng J., Gu S., Wan X., Gao H., Guo T., Su N., et al. (2008). Isolation and initial characterization of GW5, a major QTL associated with rice grain width and weight. Cell Res. 18, 1199–1209. 10.1038/cr.2008.307 [DOI] [PubMed] [Google Scholar]
- Wu X. S., Chang X. P., Jing R. L. (2012). Genetic insight into yield-associated traits of wheat grown in multiple rain-fed environments. PLoS ONE 7:e31249. 10.1371/journal.pone.0031249 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie X., Song M. H., Jin F., Ahn S. N., Suh J. P., Hwang H. G., et al. (2006). Fine mapping of a grain weight quantitative trait locus on rice chromosome 8 using near-isogenic lines derived from a cross between Oryza sativa and Oryza rufipogon. Theor. Appl. Genet. 113, 885–894. 10.1007/s00122-006-0348-5 [DOI] [PubMed] [Google Scholar]
- Xue W., Xing Y., Weng X., Zhao Y., Tang W., Wang L., et al. (2008). Natural variation in Ghd7 is an important regulator of heading date and yield potential in rice. Nat. Genet. 40, 761–767. 10.1038/ng.143 [DOI] [PubMed] [Google Scholar]
- Zhang J., Liu W., Yang X., Gao A., Li X., Wu X., et al. (2011). Isolation and characterization of two putative cytokinin oxidase genes related to grain number per spike phenotype in wheat. Mol. Biol. Rep. 38, 2337–2347. 10.1007/s11033-010-0367-9 [DOI] [PubMed] [Google Scholar]
- Zhang L., Zhao Y. L., Gao L. F., Zhao G. Y., Zhou R. H., Zhang B. S., et al. (2012). TaCKX6-D1, the ortholog of rice OsCKX2, is associated with grain weight in hexaploid wheat. New Phytol. 195, 574–584. 10.1111/j.1469-8137.2012.04194.x [DOI] [PubMed] [Google Scholar]
- Zhang L. Y., Liu D. C., Guo X. L., Yang W. L., Sun J. Z., Wang D. W., et al. (2010). Genomic distribution of quantitative trait loci for yield and yield-related traits in common wheat. J. Integr. Plant Biol. 52, 996–1007. 10.1111/j.1744-7909.2010.00967.x [DOI] [PubMed] [Google Scholar]
- Zhou Y., He Z., Sui X., Xia X., Zhang X., Zhang G. (2007). Genetic improvement of grain yield and associated traits in the northern China winter wheat region from 1960 to 2000. Crop Sci. 47, 245–253. 10.2135/cropsci2006.03.0175 [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.