Abstract
Here we report that promoter mutations in telomerase (TERT), the most common noncoding mutations in cancer, give rise to monoallelic expression of TERT. Through deep RNA sequencing, we find that TERT activation in human cancer cell lines can occur in either mono- or biallelic manner. Without exception, hotspot TERT promoter mutations lead to the re-expression of only one allele, accounting for approximately half of the observed cases of monoallelic TERT expression. Furthermore, we show that monoallelic TERT expression is highly prevalent in certain tumor types and widespread across a broad spectrum of cancers. Taken together, these observations provide insights into the mechanisms of TERT activation and the ramifications of noncoding mutations in cancer.
Introduction
In healthy tissues, expression of the telomerase gene (TERT) is generally restricted to the germline and stem cells. However, TERT activation, necessary for the maintenance of unlimited replicative potential, occurs commonly in many cancers. Recurrent mutations in the TERT promoter region were first discovered in melanoma and have since been found to be the most common somatic mutations in many cancers.1, 2, 3, 4 Two mutually exclusive cytidine-to-thymidine mutations, C228T and C250T (chr5:1 295 228 C>T and 1 295 250 C>T; hg19), positioned at 124 and 146 base pairs upstream of the ATG translational start site of TERT, respectively, are found in ~70% of melanomas, 80–90% of glioblastoma multiforme, 60% of hepatocellular carcinoma, 60% of bladder cancer, 70% of basal cell carcinoma, 50% of cutaneous squamous cell carcinoma and up to 30% of thyroid cancers. Other recurrent but much less frequent mutations in the TERT promoter (occurring at positions chr5: 1 295 228 C>A, 1 295 242–1 295 243 CC>TT and 1 295 161A>C) have also been observed.5 All of these mutations create putative consensus ETS transcription factor binding sites (GGAA/T) and are hypothesized as a mechanism of TERT activation.1, 2 Subsequent studies in glioblastoma and bladder cancer demonstrated that TERT promoter mutations correlate with higher TERT mRNA and protein expression and elevated telomerase activity.6 Furthermore, recent data demonstrate that the mutant TERT promoter can be bound and activated by GABP, an ETS transcription factor.7 The recurrent TERT promoter mutations also prevent silencing of TERT upon differentiation of stem cells.8 For this study, we used whole-genome DNA and deep RNA sequencing data from 329 cancer cell lines representing a wide range of tumor types from the Cancer Cell Line Encyclopedia (CCLE)9, 10 to interrogate the TERT locus and determine the modes of TERT activation.
Results and discussion
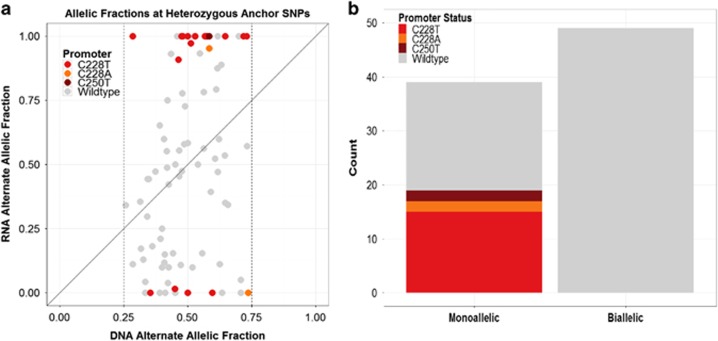
To analyze TERT expression, we first identified cell lines with heterozygous ‘anchor' single-nucleotide polymorphisms (SNPs) (defined here as having at least three reference and alternate reads with a DNA alternate allelic fraction between 0.25 and 0.75) in the DNA sequences of expressed exonic and untranslated regions of the TERT gene. Cell lines with insufficient RNA sequencing coverage of anchor SNPs (i.e., fewer than eight reads) and cell lines with high-level copy-number amplification or deletion of the TERT locus were excluded from downstream analyses. In total, 88 CCLE lines met these criteria and had at least one heterozygous SNP that would allow evaluation of an allelic bias in RNA expression (Supplementary Table 1). The number of sequencing reads that supported the reference or alternative alleles were counted both in whole-genome sequencing and RNA-Seq data and used to quantify allelic imbalance (Figure 1a). In total, 39 out of 88 cell lines (44.3%) showed a monoallelic pattern of TERT expression with a cutoff of one allele being expressed at least 10-fold higher than the other allele in RNA versus DNA.
Figure 1.
Identification of cancer cell lines with monoallelic TERT expression. (a) Fraction of sequencing reads harboring nucleotide that differs from reference human genome for heterozygous anchor SNPs. DNA and RNA alternate allelic fractions are plotted along the x axis and y axis, respectively. Analysis was performed on 88 cell lines with a DNA alternate allelic fraction between 0.25 and 0.75 in the DNA sequences of expressed exonic and untranslated regions of the TERT gene. Strong deviation from the diagonal indicates allelic bias in expression. Individual points represent samples, and those harboring hotspot promoter mutations are highlighted. Nineteen cell lines harbored one of the hotspot promoter mutations C228T, C228A or C250T and were found within the 88 cell lines that met the criteria for assessment of allelic bias. (b) Bar plot of promoter mutation status in cell lines with monoallelic and biallelic expression of the TERT gene. TERT expression was classified as monoallelic in samples with heterozygous anchor SNPs in the TERT gene for which expression of a major allele was >10-fold higher than expression of the minor allele in RNA versus DNA.
We then investigated the relationship between the monoallelic expression pattern and known mechanisms of TERT activation. To identify the recurrent TERT promoter mutations, we used whole-genome sequencing data from the CCLE to interrogate the TERT promoter region in 329 cell lines, of which 316 lines had sufficient coverage at all previously defined hotspots (Supplementary Table 2). In total, 60 lines were found to harbor known TERT promoter mutations (Supplementary Table 2 and Supplementary Figure 1). Promoter mutation status was determined by computing allele counts at genomic loci for five known recurrent somatic mutations in the TERT gene promoter region (positions 1 295 191, 1 295 228, 1 295 242, 1 295 243 and 1 295 250 in chromosome 5; Supplementary Table 2). Samples in which mutations were detected at any of these positions were classified as promoter-mutant. Consistent with previous reports, all detected hotspot mutations were mutually exclusive.
We hypothesized that the TERT promoter mutations would lead to activation of only one allele. Indeed, we found that all of the cell lines harboring TERT promoter mutations showed monoallelic expression (Figure 1b). The correlation of monoallelic expression with the known hotspot TERT promoter mutations was highly statistically significant (Fisher's exact test, one-tailed; P<10−8) and overall, nearly half (48.7% 19/39) of the cell lines that showed monoallelic expression could be accounted for by the presence of the recurrent TERT promoter mutations (Figure 1b). We did not observe a significant difference in TERT mRNA expression levels in TERT promoter-mutant-positive cell lines compared with TERT promoter wild-type cell lines (Supplementary Figure 2).
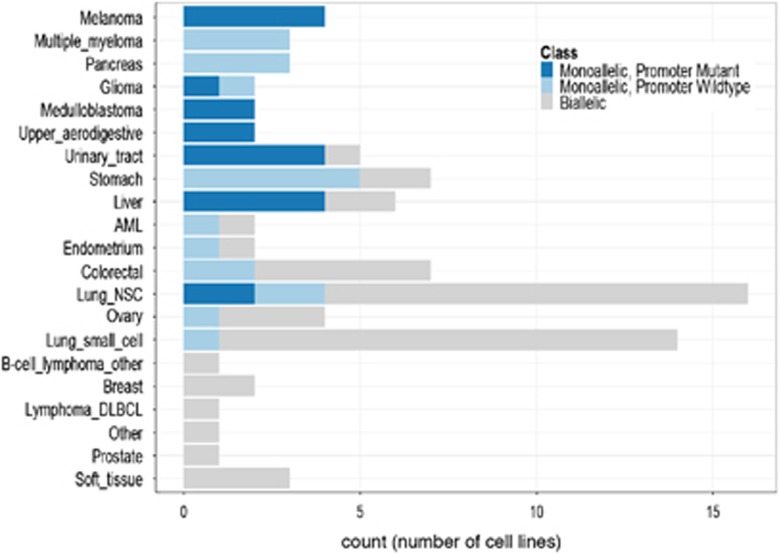
We also found that the prevalence of monoallelic expression of TERT varied markedly among different cell lineages (Figure 2). Cancer types such as melanoma, multiple myeloma, pancreas, upper aerodigestive tract, urinary tract and stomach almost exclusively showed monoallelic expression of TERT. Overall, in over 70% of lineages, at least one cell line showed evidence of monoallelic activation of TERT. Among cell lines with monoallelic TERT expression, we found a highly significant difference in the distribution across lineages when compared with cell lines with biallelic expression of TERT for both promoter-mutant and promoter-wild-type cell lines (P=1.59 × 10−6 and P=0.006, respectively; Fisher–Freeman–Halton test; Figure 2). We did not observe significant differences in mRNA expression levels between cell lines that displayed monoallelic expression or biallelic expression of TERT. These data suggest that somatic monoallelic activation of TERT may show evidence of a transcriptional compensation phenomenon that has been observed in monoallelic expression in other models (Supplementary Figure 3).11
Figure 2.
Distribution of cell lines with monoallelic TERT expression across tissue lineages. We found a highly significant difference in the distribution of samples with monoallelic TERT expression across lineages when compared with cell lines with biallelic TERT expression for both promoter-mutant and promoter-wild-type cell lines (P=1.59 × 10−6 and P=0.006, respectively; Fisher–Freeman–Halton test).
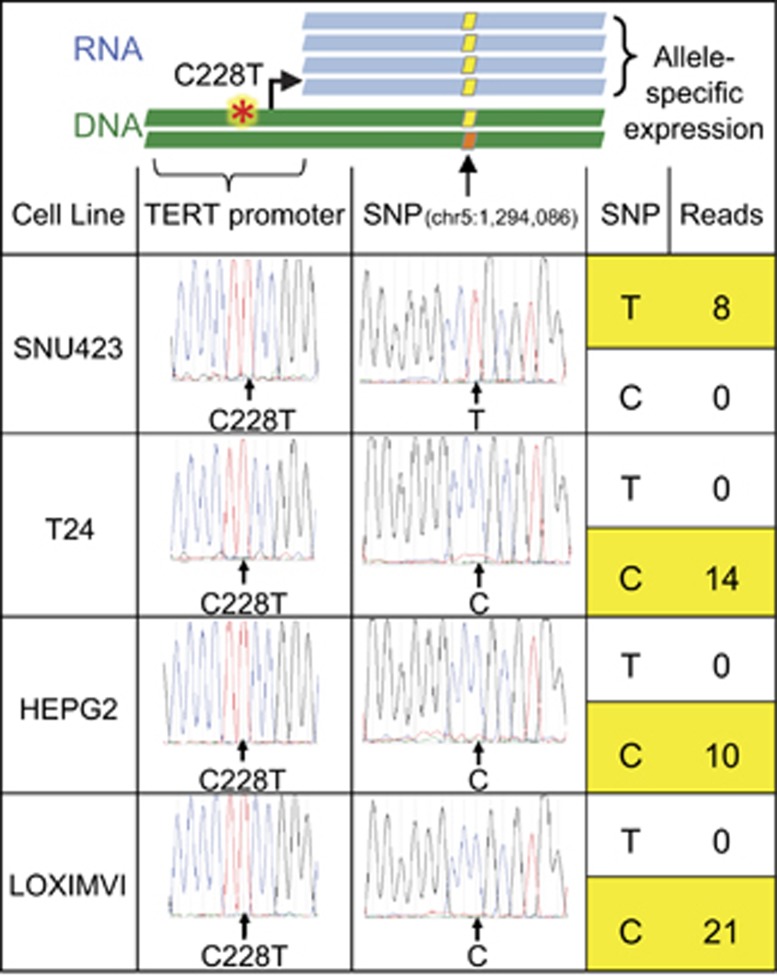
To test whether the promoter mutations occurred in cis with the alleles, which showed monoallelic expression, we subcloned the region of the TERT gene encompassing the heterozygous ‘anchor' SNP (rs2736098) at position chr5:1 294 086 and the promoter mutation C228T (chr5:1 295 228 C>T) in four unrelated cell lines. Using Sanger sequencing, we found that the allele that showed monoallelic expression at this position was in cis with the mutant promoter allele and not the wild-type promoter allele in all lines tested (P=0.05; Student's T-test; Figure 3).
Figure 3.
Relationship between TERT promoter mutations and allele-specific expression. DNA was isolated from cell lines with a TERT promoter mutation and nearby heterozygous exon 2 anchor SNP (first column) using the DNeasy Blood and Tissue Kit (Qiagen, Hilden, Germany). The genomic region containing the TERT promoter and anchor SNP was PCR amplified, gel purified and cloned using the Zero Blunt PCR Cloning Kit (Life Technologies, Carlsbad, CA, USA). Sanger sequencing chromatograms of individual clones demonstrate the TERT promoter mutation (second column) in cis with one of the alleles at the anchor SNP (third column). RNA sequencing confirmed allele-specific expression by identifying exclusive expression of the SNP that was in cis with the TERT promoter mutation (fourth column).
Although all cell lines with TERT promoter mutations showed a monoallelic pattern of TERT expression, these cell lines accounted for only half of the samples that exhibited strong allelic bias. Monoallelic expression in the remaining samples could be potentially explained by two major mechanisms—unidentified cis-acting genetic alterations that affect only one chromosome, or mitotically stable epigenetic activation of one allele. To investigate the first possibility, we analyzed the 10 kb region upstream of the TERT gene. We failed to detect a significant enrichment in proximal cis events (point mutations or rearrangements) in lines with an unexplained monoallelic pattern of TERT activation as compared with lines with biallelic expression of TERT (Supplementary Figure 4 and Supplementary Data). These data suggest that other proximal cis-acting genetic alterations do not appear to explain monoallelic expression of TERT in the cancer cell lines we analyzed that do not harbor a hotspot mutation in the TERT promoter. However, we cannot rule out the possibility that some of the substitutions found within the promoter or that genomic rearrangements beyond the upstream 10 kb could potentially contribute to monoallelic expression.
Activation of TERT expression is a crucial step in the progression of many cancers and understanding the molecular mechanisms of such activation is important for the understanding of tumorigenesis. Recent genome-wide analyses have highlighted that noncoding mutations in the TERT promoter are among the most common mutations in human cancer.12, 13, 14 We observed that the monoallelic mode of TERT activation is widespread, with half of cases explained by the known promoter mutations and certain lineages showing almost exclusive monoallelic expression of TERT. These observations highlight the heterogeneity of molecular mechanisms to activate TERT, some of which are still to be discovered. Here we show that the integration of DNA and RNA sequencing to determine monoallelic expression can be used to identify and characterize cis-acting noncoding mutations in cancer. While our results establish TERT monoallelic expression in cell lines across many tumor types, confirmation of these results in primary tumors will be needed. Whether other mechanisms such as random monoallelic expression or epigenetic mechanisms account for the non-promoter-mutant-associated TERT monoallelic expression remains an active area of investigation.15, 16, 17 Aside from TERT, recent studies demonstrate that other promoter regions may be recurrently mutated in cancer.18, 19, 20 Our findings suggest that somatic monoallelic activation of TERT is a common mechanism of TERT expression and that TERT promoter mutations drive monoallelic expression of TERT.
Acknowledgments
We thank Drs JL Stern and TR Cech for helpful discussions. This work was supported by a research grant from Novartis Institutes for Biomedical Research. FWH is supported by the Prostate Cancer Foundation Young Investigator Award, ASCO Young Investigator Award and the Department of Defense. The BAM files for the CCLE whole genome sequencing data corresponding to the TERT locus can be accessed at ftp://tertguest:broad@ftp.broadinstitute.org.1
Drs Sellers and Stegmeier are employees of Novartis. Dr Garraway has received research funding form Novartis and holds equity in Foundation Medicine. The other authors declare no conflict of interest.
Footnotes
Supplementary Information accompanies this paper on the Oncogenesis website (http://www.nature.com/oncsis).
Supplementary Material
References
- 1Huang FW, Hodis E, Xu MJ, Kryukov GV, Chin L, Garraway LA. Highly recurrent TERT promoter mutations in human melanoma. Science 2013; 6122: 957–959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2Horn S, Figl A, Rachakonda PS, Fischer C, Sucker A, Gast A et al. TERT promoter mutations in familial and sporadic melanoma. Science 2013; 6122: 959–961. [DOI] [PubMed] [Google Scholar]
- 3Killela PJ, Reitman ZJ, Jiao Y, Bettegowda C, Agrawal N, Diaz LA Jr et al. TERT promoter mutations occur frequently in gliomas and a subset of tumors derived from cells with low rates of self-renewal. Proc Natl Acad Sci USA 2013; 15: 6021–6026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4Vinagre J, Almeida A, Pópulo H, Batista R, Lyra J, Pinto V et al. Frequency of TERT promoter mutations in human cancers. Nat Commun 2013; 4: 2185. [DOI] [PubMed] [Google Scholar]
- 5Heidenreich B, Rachakonda PS, Hemmink K, Kumar R. TERT promoter mutations in cancer development. Curr Opin Genet Dev 2014; 24: 30–37. [DOI] [PubMed] [Google Scholar]
- 6Borah S, Xi L, Zaug AJ, Powell NM, Dancik GM, Cohen SB et al. TERT promoter mutations and telomerase reactivation in urothelial cancer. Science 2015; 6225: 1006–1010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7Bell RJ, Rube HT, Kreig A, Mancini A, Fouse SD, Nagarajan RP et al. The transcription factor GABP selectively binds and activates the mutant TERT promoter in cancer. Science 2015; 6238: 1036–1039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8Chiba K, Johnson JZ, Vogan JM, Wagner T, Boyle JM, Hockemeyer D. Cancer-associated TERT promoter mutations abrogate telomerase silencing. eLife 2015; 4: e07918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9Barretina J, Caponigro G, Stransky N, Venkatesan K, Margolin AA, Kim S et al. The Cancer Cell Line Encyclopedia enables predictive modelling of anticancer drug sensitivity. Nature 2012; 7391: 603–607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10CGHub data repository. Available at: https://cghub.ucsc.edu/datasets/ccle.html (last accessed on May 2015).
- 11Eckersley-Maslin MA, Thybert D, Bergmann JH, Marioni JC, Flicek P, Spector DL. Random monoallelic gene expression increases upon embryonic stem cell differentiation. Dev Cell 2014; 28: 351–365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12Weinhold N, Jacobsen A, Schultz N, Sander C, Lee W. Genome-wide analysis of noncoding regulatory mutations in cancer. Nat Genet 2014; 46: 1160–1165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13Fredriksson NJ, Ny L, Nilsson JA, Larsson E. Systematic analysis of noncoding somatic mutations and gene expression alterations across 14 tumor types. Nat Genet 2014; 46: 1258–1263. [DOI] [PubMed] [Google Scholar]
- 14Melton C, Reuter JA, Spacek DV, Snyder M. Recurrent somatic mutations in regulatory regions of human cancer genomes. Nat Genet 2015; 47: 710–716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15Gimelbrant A, Hutchinson JN, Thompson BR, Chess A. Widespread monoallelic expression on human autosomes. Science 2007; 5853: 1136–1140. [DOI] [PubMed] [Google Scholar]
- 16Eckersly-Maslin MA, Spector DL. Random monoallelic expression: regulating gene expression one allele at a time. Trends Genet 2015; 6: 237–244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17Castelo-Branco P, Choufani S, Mack S, Gallagher D, Zhang C, Lipman T et al. Methylation of the TERT promoter and risk stratification of childhood brain tumours: an integrative genomic and molecular study. Lancet Oncol 2013; 6: 534–542. [DOI] [PubMed] [Google Scholar]
- 18Smith KS, Yadav VK, Pedersen BS, Shaknovich R, Geraci MW, Pollard KS et al. Signatures of accelerated somatic evolution in gene promoters in multiple cancer types. Nucleic Acids Res 2015; 11: 5307–5317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19Mathelier A, Lefebvre C, Zhang AW, Arenillas DJ, Ding J, Wasserman WW et al. Cis-regulatory somatic mutations and gene-expression alteration in B-cell lymphomas. Genome Biol 2015; 17: 1966–1967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20Katainen R, Dave K, Pitkänen E, Palin K, Kivioja T, Välimäki N et al. CTCF/cohesin-binding sites are frequently mutated in cancer. Nat Genet 2015; 47: 818–821. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.