Abstract
Protein–protein interactions (PPIs) and microRNA (miRNA)–target interactions are important for deciphering the mechanisms of tumorigenesis. However, current PPI databases do not support cancer-specific analysis. Also, no available databases can be used to retrieve cancer-associated miRNA–target interactions. As the pathogenesis of human cancers is affected by several miRNAs rather than a single miRNA, it is needed to uncover miRNA synergism in a systems level. Here for each cancer type, we constructed a miRNA–miRNA functionally synergistic network based on the functions of miRNA targets and their topological features in that cancer PPI network. And for the first time, we report the cancer-specific database CancerNet (http://bis.zju.edu.cn/CancerNet), which contains information about PPIs, miRNA–target interactions and functionally synergistic miRNA–miRNA pairs across 33 human cancer types. In addition, PPI information across 33 main normal tissues and cell types are included. Flexible query methods are allowed to retrieve cancer molecular interactions. Network viewer can be used to visualize interactions that users are interested in. Enrichment analysis tool was designed to detect significantly overrepresented Gene Ontology categories of miRNA targets. Thus, CancerNet serves as a comprehensive platform for assessing the roles of proteins and miRNAs, as well as their interactions across human cancers.
Introduction
Cancer is a complex disease characterized by a large number of molecular interaction alterations.1 Distinguishing the bona fide drivers of cancer phenotypes has proven to be a daunting task, which is further exacerbated by the complexity of elucidating how such drivers interact synergistically to elicit cancer phenotypes. Thus, a systems biology approach, the analysis of multilevel molecular interactions, is required to understand the pathogenesis of human cancers.2, 3
Protein–protein interaction (PPI) networks provide a global picture of cellular functions and biological processes,4 the dysfunction of some interactions causes many diseases, including cancer. Therefore, the use of PPI networks has become one of the major and powerful approaches to elucidate the molecular mechanisms underlying the complex diseases on the system level.5, 6 Recently, Barshir et al.7 developed the TissueNet including tissue-specific and tissue-wide PPIs across 16 human normal tissues. Veres et al.8 constructed a cellular compartment-specific database for PPI network analysis. However, these databases do not provide cancer-related PPIs. Thus, a disease analysis is not allowed. However, it is valuable to construct a more comprehensive database to store context-specific PPIs across varieties of cancer and normal tissues. This is especially the case because of the tremendous increase in human protein interaction data, as well as expression data in both cancer and normal tissues produced by large-scale projects, such as The Cancer Genome Atlas9 and NIH Roadmap Epigenomics Mapping Project.10
MicroRNAs (miRNAs), as master gene regulators, have crucial roles in disease-associated processes.11, 12 A single miRNA is capable of regulating >200 mRNAs, and a single mRNA may be regulated by multiple miRNAs,13 but a substantial fraction of these interactions may depend on the cell type and/or context14 and function in specific tissues.15 Therefore, identification of context-dependent miRNA–target interactions is quite important to study the pathogenesis of complex diseases.16, 17 Mammalian miRNAs can influence gene expression not only by inhibiting protein translation but also by decreasing target mRNA levels that causing a negative correlation between the expression levels of miRNAs and their target mRNAs.18, 19, 20 As a result, the inverse expression relationship between miRNAs and mRNAs are frequently used to predict miRNA targets.21, 22, 23 MiRNA expression broadly contributes to tissue specificity of mRNA expression in many human tissues.24 Therefore, by integrating gene expression data across a variety of cancers into the global miRNA–target regulatory network, cancer-specific and cancer-wide miRNA–target interactions can be inferred.
Multiple miRNAs can synergistically regulate one or more pathways by targeting common or functionally similar targets.25 In addition, the synergism may change in different diseases. Therefore, detecting the miRNA synergism across diverse cancer types is essential for uncovering pathogenesis of human cancers in a global sense. Several methods have been developed to discover miRNA synergism. Xu et al.25 constructed a miRNA–miRNA functionally synergistic network via co-regulating functional modules while Li et al.26 detected synergistic miRNA regulatory modules by overlapping neighborhood expansion. These studies have demonstrated the importance of miRNA synergism. However, the researchers only focused on several cancer types. Therefore, a pattern analysis across diverse cancer types is unavailable. Here we present a novel approach to identify miRNA synergism across human cancer types. For each cancer, functional similarity between miRNAs was estimated by measuring the similarity of their associated target genes, and the proximity of target genes in the PPI network was further detected. Our methods considered both the biological functions of miRNA targets and their topological structure in the PPI network. A systematic insight into the nature and scale of the potential synergistic interactions is essential for the study of complex diseases.
In this study, we developed a new database CancerNet, which is a cancer-specific database that provides multilevel molecular interactions across diverse cancer types. Users can retrieve cancer-wide or cancer-specific PPIs, miRNA–target interactions and functionally synergistic miRNAs using multiple query methods. CancerNet also provides enrichment analysis for miRNA targets and a network visualization tool. Above all, CancerNet serves as an important data resource that can help researchers to understand the regulatory mechanisms or interaction patterns between different molecules across diverse cancer types.
Results
Overview of CancerNet
CancerNet aims to provide cancer-specific molecular interaction networks across multiple cancer types. Currently, 33 human cancer types are included. The interactions contain PPIs, miRNA–target interactions and miRNA–miRNA synergistic interactions. Experimentally detected PPIs were assembled from five major PPI databases and miRNA–target interactions were considered as the combination of the predicted targets from six algorithms and two experimentally validated data sets, amounting to 185 589 PPIs and 3 249 385 miRNA–target interactions, respectively. Synergistic miRNA pairs were predicted according to the functions of target genes as well as their proximity in the PPI network. By integrating expression data in different cancer samples and information from Gene Ontology (GO) annotations, cancer-wide and cancer-specific molecular interactions were identified. CancerNet offers a unique platform for assessing the roles of human proteins and miRNAs, as well as their interactions across human cancers.
Query and result description
Query
A flexible and user-friendly query method is provided by CancerNet, users can query it using one molecule to retrieve its interaction partners per cancer, or using a pair of interacting molecules to retrieve the cancer types that the interaction appears in. For each molecule, two types of identifier can be used. In case of a miRNA, the name or accession number in miRBase can be used, while in case of a gene, the official symbol or Entrez ID can be used.
Result list
CancerNet output data provide lists of interactions and detail information about the interactions. Expression levels and functional similarity score are provided for the PPI and miRNA–miRNA result list, respectively. And for the miRNA–target result list, the Pearson correlation coefficient and P-value are offered. In addition, users can check the cancer specificity of each interaction from the last column of the result list. Specifically, for each functionally similar miRNA pair, users can further explore the enriched GO terms of their targets.
Visualization
One molecule may interact with multiple molecules in the same context. Therefore, these interactions form a complex regulatory network. To visualize these networks, a graphical network viewer based on the Cytoscape Web program27 was developed. Edges (interactions) are colored distinctly according to their associated cancer types. In addition, users can use a special toolkit of ‘My PPI/miRNA-Target/miRNA-miRNA Network', which contains previously stored interactions of interest. Consequently, this toolkit supports users in discovering specific biological network formed by molecular interactions that involved in regulation of the interrelated processes.
Enrichment analysis
Functional enrichment analysis of miRNA target genes are widely used to detect the miRNA functions. A ‘GO Enrichment Analysis' module has been developed for miRNA–target list based on GO annotation. The χ2-test and Fisher's exact test were used to evaluate the significance of enrichment for GO terms. This module facilitates users to explore the biological functions of target gene set that they are interested in.
Biological applications
Multilevel molecular interactions are involved in tumorigenesis. Therefore, deciphering these interactions is essential for cancer research. CancerNet enables researchers to comprehensively view the interactions formed by the molecules of interest across diverse cancer types. Combination treatment with these interactions has an important role in cancer research.28 In this section, we will show how to use CancerNet to find useful information.
MiR-1 and miR-133a have been frequently found to be co-downregulated in several types of cancer.29, 30, 31, 32, 33, 34 Furthermore, overexpression of both of them inhibited cancer cell proliferation and induced cancer cell apoptosis.32, 35, 36 Therefore, there must be a functional link between miR-1 and miR-133a. Next, this miRNA pair will be further discussed using our system.
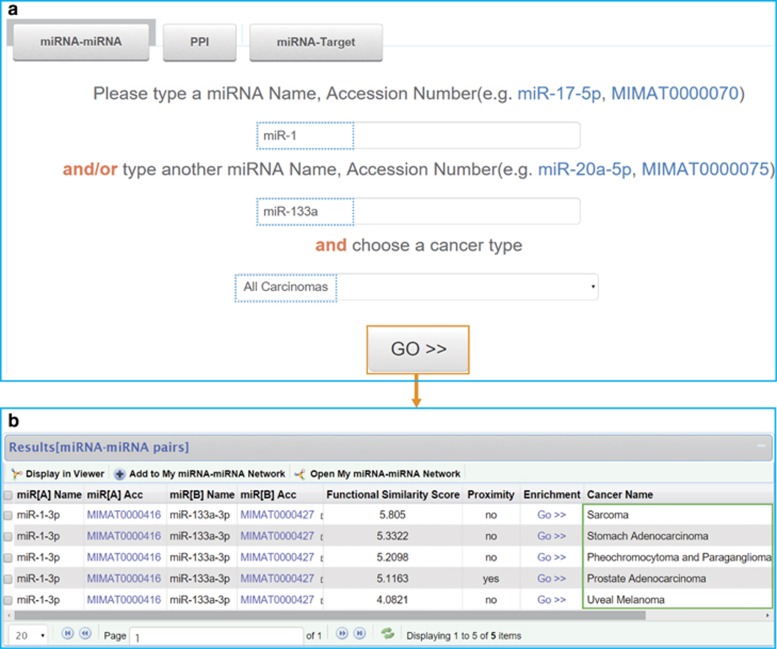
First, we are curious about in which cancer types this miRNA pair exists, so miR-1 and miR-133a were queried simultaneously in CancerNet (Figure 1). From the result page, we could find that miR-1 and miR-133a are functionally synergistic in five cancer types: sarcoma; stomach adenocarcinoma; pheochromocytoma–paraganglioma; prostate adenocarcinoma; and uveal melanoma. Among them, it has been revealed that both miR-1 and miR-133a are related to sarcoma,30 stomach adenocarcinoma37, 38 and prostate adenocarcinoma.39 MiR-133b, which is from the miR-133 family (miR-133a and miR-133b), potentially regulates pathways related to pheochromocytoma–paraganglioma.40 In addition, expression level of both miR-1 and miR-133a altered in uveal melanoma.41 Accordingly, the synergism between miR-1 and miR-133a revealed by CancerNet is consistent with previous studies.
Figure 1.
Synergism between miR-1 and miR-133a. (a) Screenshots of the search section. The search engine includes three parts: miRNA–miRNA; PPI; and miRNA–target, allowing users to query functionally synergistic miRNA pairs, PPIs and miRNA–target interactions, respectively, across diverse cancer types. As an example, the miR-1 and miR-133a were queried. (b) Result list of miR-1 and miR-133a. Synergism between miR-1 and miR-133a exists in five types of cancers, shown in green box.
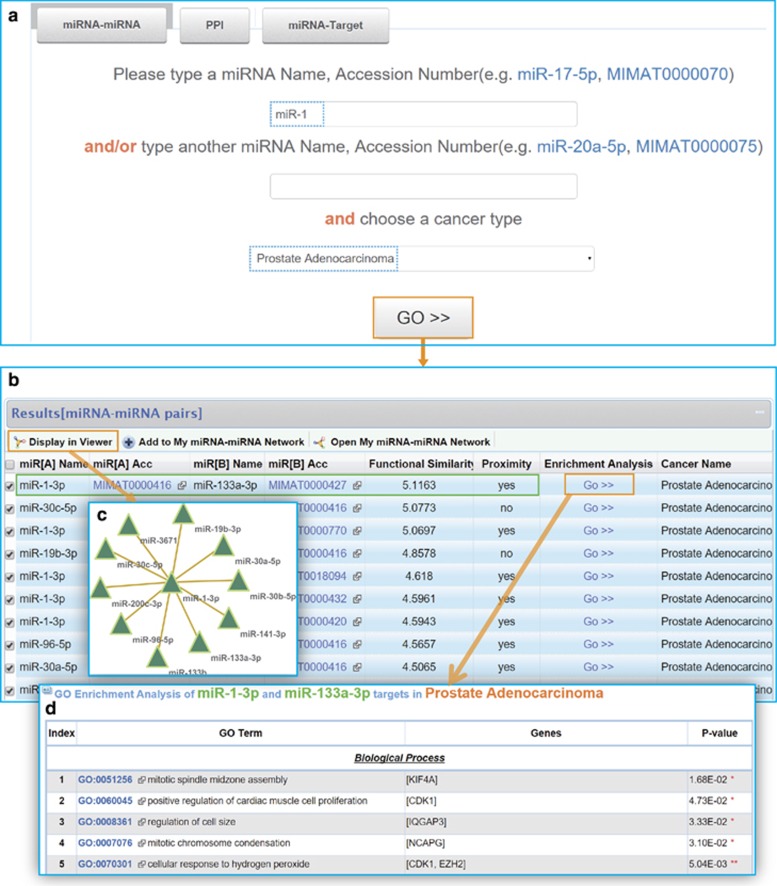
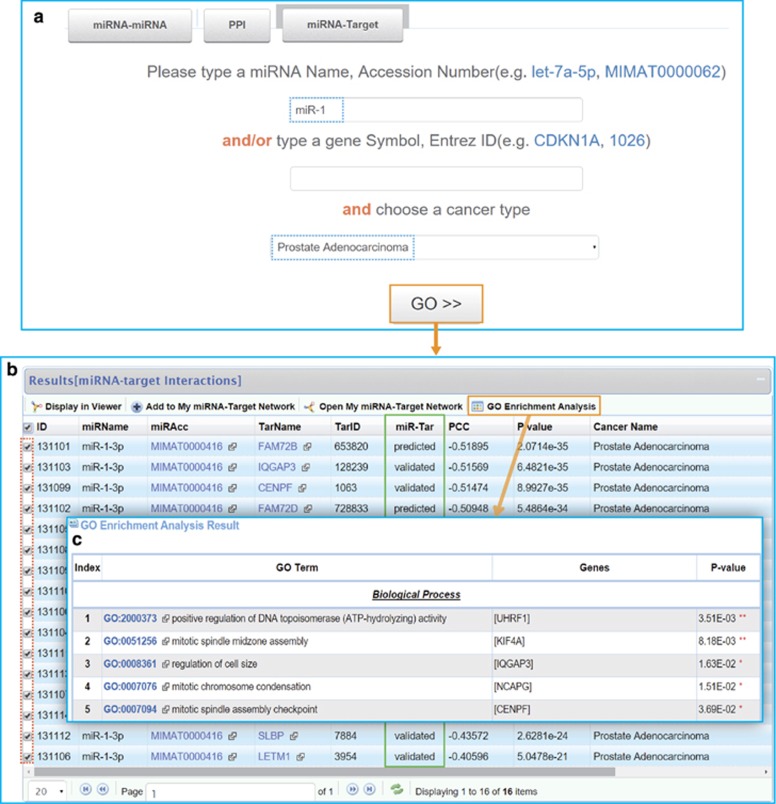
Second, we would like to further detect all the miRNAs that are functionally synergistic with miR-1 or miR-133a in a certain cancer type such as prostate adenocarcinoma. This can be easily achieved by our system. Here we only show the results of miR-1 (Figure 2). Of all the pairs formed by miR-1 in prostate adenocarcinoma, miR-133a and miR-1 share the highest functional similarity score that means their functions are closely linked. Besides, their targets are proximate in the PPI network. Then we want to find out the pathways that their targets are associated with. To realize this, a functional enrichment analysis tool has been embedded in the result page that allows users to discover the significant GO terms for the targets of each miRNA pair. The targets of miR-1 and miR-133a are enriched in cancer-related GO biological processes such as mitotic cell cycle (P=1.13E-08), cell division (P=2.54E-08) and mitotic nuclear division (P=9.74E-08). Our system also provides a graphic viewer to visualize interactions that users are interest in. In addition, the targets of miR-1 or miR-133a in prostate adenocarcinoma could be further queried (Figure 3). For miR-1, most miRNA–target interactions are experimentally validated (12/16). Similarly, we can select the targets of interest and perform a functional enrichment analysis.
Figure 2.
Functionally synergistic miRNA pairs formed by miR-1. (a) Screenshots of the search page. (b) Result list of miR-1. MiR-1 and miR-133a share the highest functional similarity score, shown in the green box. (c) Functionally synergistic miRNA–miRNA network (top 10 are displayed). (d) Results of functional enrichment analysis.
Figure 3.
Targets of miR-1 in prostate adenocarcinoma. (a) Screenshots of the search page. (b) Result list of miR-1 targets. Most miRNA–target interactions (12/16) have been experimentally validated, shown in green box. (c) Results of functional enrichment analysis.
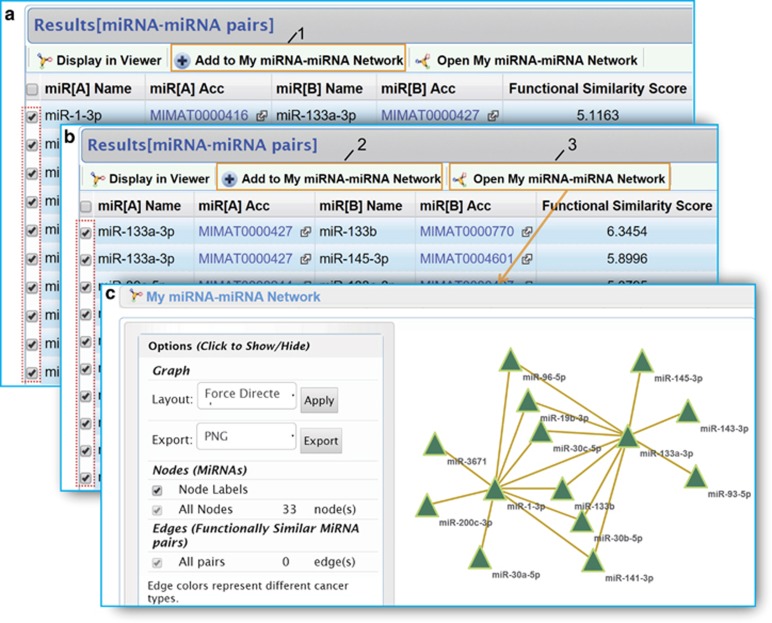
Finally, CancerNet provides a way to study functionally synergistic miRNAs on the system level. For example, the common partners that are synergistic with miR-1 and miR-133a in prostate adenocarcinoma can be visualized by using ‘My miRNA-miRNA Network' module (Figure 4). The six common partners (miR-19b, miR-30b, miR-30c, miR-96, miR-133b and miR-141) are all related with prostate cancer. MiR-19b promotes prostate cell proliferation with regulation of PTEN and its downstream signals.42 Overexpression of miR-30 in prostate cancer cells suppresses epithelial-mesenchymal transition (EMT) phenotypes and inhibits cell migration and invasion.43 Upregulation of miR-96 enhances cellular proliferation of prostate cancer cells through FOXO1.44 Overexpression of miR-133b in LNCaP cells boosts cell proliferation and cell cycle progression, but inhibits apoptosis.45 MiR-141 modulates androgen receptor transcriptional activity in human prostate cancer cells through targeting the small heterodimer partner protein.46 Together with miR-1 and miR-133a, these miRNAs form a functionally synergistic clique in prostate cancer and jointly function in tumorigenesis. Therefore, a combinatorial study of these synergistic miRNAs is important for understanding the mechanisms of tumorigenesis and finding bona fide biomarkers for cancer diagnosis and prognosis.
Figure 4.
Network visualization of the common partners that are synergistic with miR-1 and miR-133a in prostate adenocarcinoma. (a) Result list of miR-1 partners. Top 10 items are added to ‘My miRNA-miRNA Network'. (b) Result list of miR-133a partners. Top 10 items are added to ‘My miRNA-miRNA Network'. (c) ‘My miRNA-miRNA Network'.
From our system, we could find out that co-expression of miR-1 and miR-133a is meaningful. They synergistically regulate functionally related genes or pathways in several cancer types. Understanding the synergism between them is quite useful to elucidate the roles they have in tumorgenesis.
Discussion
Knowledge of molecular interactions is quite useful for discovering the functions of molecules and the processes they are involved in. However, there is little systematic insight into the nature and scale of the potential interactions in human cancers. Although human PPIs are accessible through several public databases, these databases do not specify the human cancer types in which these PPIs take place. In addition, there are no databases can be used to retrieve cancer-specific miRNA–target interactions as well as functionally synergistic miRNA pairs. However, by integrating high-throughput sequencing data, we can discover these interactions across diverse cancer types. To discover the miRNA synergism, we measured the functional similarity of targets in the miRNA–target network and the target proximity in the PPI network in each cancer type. These results could help researchers to elucidate the roles miRNAs have in tumorigenesis from a system-level perspective.
In summary, CancerNet provides a unique data resource for the analysis of PPI networks, miRNA–target regulatory networks and functionally synergistic miRNA–miRNA networks across diverse cancer types. Studying these cancer-specific interactions will help researchers to detect the interaction patterns across human cancers, discover the true cancer-related regulatory modules and understand the mechanisms of tumorigenesis. In addition, it allows researchers to quickly identify poorly annotated miRNAs or protein-coding genes interacted with molecules, which have already been well studied. Therefore, our system provides guidance on studying the functions of these molecules, especially their roles in human cancers, and has significant implications for miRNA combination therapy of human cancers.
Materials and Methods
Expression data sources and processing
Level 3 miRNA-Seq and RNA-Seq data were obtained from the The Cancer Genome Atlas data portal (https://tcga-data.nci.nih.gov/tcga/tcgaHome2.jsp), including 33 cancer types, 8296 tumor samples in all. Reads per million miRNA mapped values were used to represent miRNA expression levels. For RNA-Seq data, upper quartile normalized gene counts were calculated using the RSEM algorithm,47 RSEM's gene level expression estimates were multiplied by 1 000 000 to obtain transcript per million (TPM) estimates for each gene, and TPM estimates were transformed to log-space by taking log2(TPM+1). Normal tissue and cell line RNA-Seq data were downloaded from Illumina Body Map 2.048 and NIH Roadmap Epigenomics Mapping Project.10 Raw reads were mapped to hg19 using TopHat 2.0.1149 and gene expression levels were quantified by Cufflinks 2.2.0.50
Protein–protein interactions
We compiled experimentally identified PPIs from BioGRID,51 DIP,52 HPRD,53 IntAct54 and MINT.55 Only physical interactions were retained and these data are amounted to 185 589 PPIs between 16 278 human proteins. For each cancer type, genes were considered expressed if their transformed expression level was equal to or above 2 (in log2(TPM+1) scale) in at least 80% samples. Whereas, for each normal tissue or cell type, mean expression values were calculated and genes with at least two FPKM (fragments per kilobase of exon per million fragments mapped) were considered expressed. Then, a PPI was associated with a cancer or normal tissue if the two pair mates were both found to be expressed in that cancer or normal tissue.
MiRNA–target interactions
MiRNA sequences were extracted from the miRBase database (release 21).56 MiRNA target genes were acquired from six common miRNA target-predicting programs, including DIANA-microT,57 miRDB,58 PITA,59 RNAhybrid,60 TargetScan61 and miRanda.62 In order to improve the reliability of the predicted target genes, only targets predicted by at least two of these programs were retained and further incorporated with two experimentally validated data sets from miRecords63 and miRTarBase.64 Next, the miRNA–target interactions existing in a cancer type were produced according to the inverse expression relationships between miRNAs and their target mRNAs. Thus, the Pearson correlation coefficients were calculated for each miRNA–target pair, whose threshold was set to <−0.4 and corresponding P-value was set to <0.01.
MiRNA–miRNA synergism
To identify functionally synergistic miRNA pairs in a specific type of cancer, the functional similarity of targets was first calculated. Then, the proximity of targets in that cancer PPI network was measured.
The functional similarity scores were calculated for each miRNA pair using FastSemSim (http://sourceforge.net/p/fastsemsim) based on the GO biological process terms,65 and the threshold was set to >5. We used information content to evaluate semantic similarity of GO terms, as defined by Resnik.66 Here for each miRNA in a certain cancer type, only the targets existing in the miRNA–target interaction network from that cancer were used.
For each pair of miRNAs, if their targets tend to be proximate in the PPI network, we considered that these two miRNAs share common functions as closely linked nodes tend to form a function module in the PPI network.67 We presented a method to measure the proximity of targets in a network, which considered the proximity between two target sets as well as the local proximity of all the targets. Therefore, the proximity between one target and one target set has to be defined. Here we let ‘t' represents one target and let ‘TS' represents one target set. Then we define the proximity between t and TS, p(t,TS), as the sum of shortest path length from t to each member of TS, for example, TS={ts1, ts2, …tsk}. It is calculated as follows:
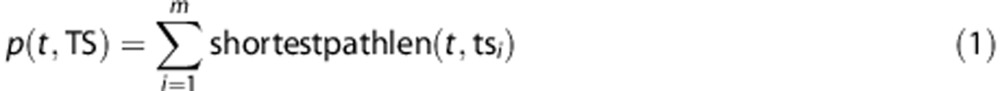 |
Next, if target set A contains m genes and target set B contains n genes, then the proximity between A and B is defined as
 |
And the local proximity formed by A and B is defined as
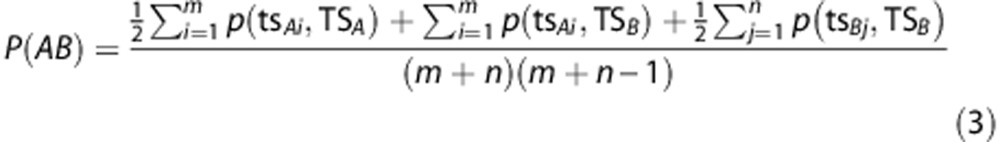 |
which measures the local connectivity among all the genes from A and B. Then we set the threshold for P(A,B) and P(AB) to <1.5 and <0.7, respectively, to determine whether two miRNAs have a synergistic relationship.
Acknowledgments
We thank Dr Björn Sommer for helpful discussions during his stay in Hangzhou. This work was in part supported by the National Natural Sciences Foundation of China (No. 31371328, 31450110068, 31571366), Science Technology Department of Zhejiang Province (No. 2015C32057), CSC and DAAD (PPP program 2015-2016), the Fundamental Research Funds for the Central Universities, 111 Project and the Program for Innovative Research Team in Zhejiang University.
The authors declare no conflict of interest.
References
- 1Hornberg JJ, Bruggeman FJ, Westerhoff HV, Lankelma J. Cancer: a Systems Biology disease. Biosystems 2006; 83: 81–90. [DOI] [PubMed] [Google Scholar]
- 2Kitano H. Systems biology: a brief overview. Science 2002; 295: 1662–1664. [DOI] [PubMed] [Google Scholar]
- 3Goh KI, Cusick ME, Valle D, Childs B, Vidal M, Barabasi AL. The human disease network. Proc Natl Acad Sci USA 2007; 104: 8685–8690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4Vidal M, Cusick ME, Barabasi AL. Interactome networks and human disease. Cell 2011; 144: 986–998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5Ideker T, Sharan R. Protein networks in disease. Genome Res 2008; 18: 644–652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6Ortutay C, Vihinen M. Identification of candidate disease genes by integrating Gene Ontologies and protein-interaction networks: case study of primary immunodeficiencies. Nucleic Acids Res 2009; 37: 622–628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7Barshir R, Basha O, Eluk A, Smoly IY, Lan A, Yeger-Lotem E. The TissueNet database of human tissue protein-protein interactions. Nucleic Acids Res 2013; 41: D841–D844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8Veres DV, Gyurko DM, Thaler B, Szalay KZ, Fazekas D, Korcsmaros T et al. ComPPI: a cellular compartment-specific database for protein-protein interaction network analysis. Nucleic Acids Res 2015; 43: D485–D493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9Cancer Genome Atlas Research N, Weinstein JN, Collisson EA, Mills GB, Shaw KR, Ozenberger BA et al. The Cancer Genome Atlas Pan-Cancer analysis project. Nat Genet 2013; 45: 1113–1120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10Bernstein BE, Stamatoyannopoulos JA, Costello JF, Ren B, Milosavljevic A, Meissner A et al. The NIH Roadmap Epigenomics Mapping Consortium. Nat Biotechnol 2010; 28: 1045–1048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11Calin GA, Dumitru CD, Shimizu M, Bichi R, Zupo S, Noch E et al. Frequent deletions and down-regulation of micro-RNA genes miR15 and miR16 at 13q14 in chronic lymphocytic leukemia. Proc Natl Acad Sci USA 2002; 99: 15524–15529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell 2004; 116: 281–297. [DOI] [PubMed] [Google Scholar]
- 13Jin H, Tuo W, Lian H, Liu Q, Zhu XQ, Gao H. Strategies to identify microRNA targets: new advances. Nat Biotechnol 2010; 27: 734–738. [DOI] [PubMed] [Google Scholar]
- 14Bartel DP. MicroRNAs: target recognition and regulatory functions. Cell 2009; 136: 215–233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15Hsieh WJ, Lin FM, Huang HD, Wang H. Investigating microRNA-target interaction-supported tissues in human cancer tissues based on miRNA and target gene expression profiling. PLoS ONE 2014; 9: e95697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16Genovese G, Ergun A, Shukla SA, Campos B, Hanna J, Ghosh P et al. microRNA regulatory network inference identifies miR-34a as a novel regulator of TGF-beta signaling in glioblastoma. Cancer Discov 2012; 2: 736–749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17Yang D, Sun Y, Hu L, Zheng H, Ji P, Pecot CV et al. Integrated analyses identify a master microRNA regulatory network for the mesenchymal subtype in serous ovarian cancer. Cancer Cell 2013; 23: 186–199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18Bagga S, Bracht J, Hunter S, Massirer K, Holtz J, Eachus R et al. Regulation by let-7 and lin-4 miRNAs results in target mRNA degradation. Cell 2005; 122: 553–563. [DOI] [PubMed] [Google Scholar]
- 19Lim LP, Lau NC, Garrett-Engele P, Grimson A, Schelter JM, Castle J et al. Microarray analysis shows that some microRNAs downregulate large numbers of target mRNAs. Nature 2005; 433: 769–773. [DOI] [PubMed] [Google Scholar]
- 20Guo H, Ingolia NT, Weissman JS, Bartel DP. Mammalian microRNAs predominantly act to decrease target mRNA levels. Nature 2010; 466: 835–840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21Huang JC, Babak T, Corson TW, Chua G, Khan S, Gallie BL et al. Using expression profiling data to identify human microRNA targets. Nat Methods 2007; 4: 1045–1049. [DOI] [PubMed] [Google Scholar]
- 22Gennarino VA, Sardiello M, Avellino R, Meola N, Maselli V, Anand S et al. MicroRNA target prediction by expression analysis of host genes. Genome Res 2009; 19: 481–490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23Jacobsen A, Silber J, Harinath G, Huse JT, Schultz N, Sander C. Analysis of microRNA-target interactions across diverse cancer types. Nat Struct Mol Biol 2013; 20: 1325–1332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24Sood P, Krek A, Zavolan M, Macino G, Rajewsky N. Cell-type-specific signatures of microRNAs on target mRNA expression. Proc Natl Acad Sci USA 2006; 103: 2746–2751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25Xu J, Li CX, Li YS, Lv JY, Ma Y, Shao TT et al. MiRNA-miRNA synergistic network: construction via co-regulating functional modules and disease miRNA topological features. Nucleic Acids Res 2011; 39: 825–836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26Li Y, Liang C, Wong KC, Luo J, Zhang Z. Mirsynergy: detecting synergistic miRNA regulatory modules by overlapping neighbourhood expansion. Bioinformatics 2014; 30: 2627–2635. [DOI] [PubMed] [Google Scholar]
- 27Lopes CT, Franz M, Kazi F, Donaldson SL, Morris Q, Bader GD. Cytoscape Web: an interactive web-based network browser. Bioinformatics 2010; 26: 2347–2348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28Noguchi S, Yasui Y, Iwasaki J, Kumazaki M, Yamada N, Naito S et al. Replacement treatment with microRNA-143 and -145 induces synergistic inhibition of the growth of human bladder cancer cells by regulating PI3K/Akt and MAPK signaling pathways. Cancer Lett 2013; 328: 353–361. [DOI] [PubMed] [Google Scholar]
- 29Ambs S, Prueitt RL, Yi M, Hudson RS, Howe TM, Petrocca F et al. Genomic profiling of microRNA and messenger RNA reveals deregulated microRNA expression in prostate cancer. Cancer Res 2008; 68: 6162–6170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30Rao PK, Missiaglia E, Shields L, Hyde G, Yuan B, Shepherd CJ et al. Distinct roles for miR-1 and miR-133a in the proliferation and differentiation of rhabdomyosarcoma cells. FASEB J 2010; 24: 3427–3437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31Nohata N, Hanazawa T, Kikkawa N, Sakurai D, Sasaki K, Chiyomaru T et al. Identification of novel molecular targets regulated by tumor suppressive miR-1/miR-133a in maxillary sinus squamous cell carcinoma. Int J Oncol 2011; 39: 1099–1107. [DOI] [PubMed] [Google Scholar]
- 32Yoshino H, Chiyomaru T, Enokida H, Kawakami K, Tatarano S, Nishiyama K et al. The tumour-suppressive function of miR-1 and miR-133a targeting TAGLN2 in bladder cancer. Br J Cancer 2011; 104: 808–818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33Chen WS, Leung CM, Pan HW, Hu LY, Li SC, Ho MR et al. Silencing of miR-1-1 and miR-133a-2 cluster expression by DNA hypermethylation in colorectal cancer. Oncol Rep 2012; 28: 1069–1076. [DOI] [PubMed] [Google Scholar]
- 34Caruso S, Bazan V, Rolfo C, Insalaco L, Fanale D, Bronte G et al. MicroRNAs in colorectal cancer stem cells: new regulators of cancer stemness? Oncogenesis 2012; 1: e32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35Nasser MW, Datta J, Nuovo G, Kutay H, Motiwala T, Majumder S et al. Down-regulation of micro-RNA-1 (miR-1) in lung cancer. Suppression of tumorigenic property of lung cancer cells and their sensitization to doxorubicin-induced apoptosis by miR-1. J Biol Chem 2008; 283: 33394–33405. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 36Kawakami K, Enokida H, Chiyomaru T, Tatarano S, Yoshino H, Kagara I et al. The functional significance of miR-1 and miR-133a in renal cell carcinoma. Eur J Cancer 2012; 48: 827–836. [DOI] [PubMed] [Google Scholar]
- 37Han C, Zhou Y, An Q, Li F, Li D, Zhang X et al. MicroRNA-1 (miR-1) inhibits gastric cancer cell proliferation and migration by targeting MET. Tumour Biol 2015; 36: 6715–6723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38Cheng Z, Liu F, Wang G, Li Y, Zhang H, Li F. miR-133 is a key negative regulator of CDC42-PAK pathway in gastric cancer. Cell Signal 2014; 26: 2667–2673. [DOI] [PubMed] [Google Scholar]
- 39Kojima S, Chiyomaru T, Kawakami K, Yoshino H, Enokida H, Nohata N et al. Tumour suppressors miR-1 and miR-133a target the oncogenic function of purine nucleoside phosphorylase (PNP) in prostate cancer. Br J Cancer 2012; 106: 405–413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40de Cubas AA, Leandro-Garcia LJ, Schiavi F, Mancikova V, Comino-Mendez I, Inglada-Perez L et al. Integrative analysis of miRNA and mRNA expression profiles in pheochromocytoma and paraganglioma identifies genotype-specific markers and potentially regulated pathways. Endocr Relat Cancer 2013; 20: 477–493. [DOI] [PubMed] [Google Scholar]
- 41Venza M, Dell'Aversana C, Visalli M, Altucci L, Teti D, Venza I. Identification of microRNA expression patterns in cutaneous and uveal melanoma cell lines. Tumori 2014; 100: e4–e7. [DOI] [PubMed] [Google Scholar]
- 42Tian L, Fang YX, Xue JL, Chen JZ. Four microRNAs promote prostate cell proliferation with regulation of PTEN and its downstream signals in vitro. PLoS ONE 2013; 8: e75885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43Kao CJ, Martiniez A, Shi XB, Yang J, Evans CP, Dobi A et al. miR-30 as a tumor suppressor connects EGF/Src signal to ERG and EMT. Oncogene 2014; 33: 2495–2503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44Haflidadottir BS, Larne O, Martin M, Persson M, Edsjo A, Bjartell A et al. Upregulation of miR-96 enhances cellular proliferation of prostate cancer cells through FOXO1. PLoS ONE 2013; 8: e72400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45Li X, Wan X, Chen H, Yang S, Liu Y, Mo W et al. Identification of miR-133b and RB1CC1 as independent predictors for biochemical recurrence and potential therapeutic targets for prostate cancer. Clin Cancer Res 2014; 20: 2312–2325. [DOI] [PubMed] [Google Scholar]
- 46Xiao J, Gong AY, Eischeid AN, Chen D, Deng C, Young CY et al. miR-141 modulates androgen receptor transcriptional activity in human prostate cancer cells through targeting the small heterodimer partner protein. Prostate 2012; 72: 1514–1522. [DOI] [PubMed] [Google Scholar]
- 47Li B, Dewey CN. RSEM: accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinformatics 2011; 12: 323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48Bradley RK, Merkin J, Lambert NJ, Burge CB. Alternative splicing of RNA triplets is often regulated and accelerates proteome evolution. PLoS Biol 2012; 10: e1001229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49Kim D, Pertea G, Trapnell C, Pimentel H, Kelley R, Salzberg SL. TopHat2: accurate alignment of transcriptomes in the presence of insertions, deletions and gene fusions. Genome Biol 2013; 14: R36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50Trapnell C, Williams BA, Pertea G, Mortazavi A, Kwan G, van Baren MJ et al. Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat Biotechnol 2010; 28: 511–515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51Stark C, Breitkreutz BJ, Chatr-Aryamontri A, Boucher L, Oughtred R, Livstone MS et al. The BioGRID Interaction Database: 2011 update. Nucleic Acids Res 2011; 39: D698–D704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52Salwinski L, Miller CS, Smith AJ, Pettit FK, Bowie JU, Eisenberg D. The Database of Interacting Proteins: 2004 update. Nucleic Acids Res 2004; 32: D449–D451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53Keshava Prasad TS, Goel R, Kandasamy K, Keerthikumar S, Kumar S, Mathivanan S et al. Human Protein Reference Database—2009 update. Nucleic Acids Res 2009; 37: D767–D772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54Kerrien S, Aranda B, Breuza L, Bridge A, Broackes-Carter F, Chen C et al. The IntAct molecular interaction database in 2012. Nucleic Acids Res 2012; 40: D841–D846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55Licata L, Briganti L, Peluso D, Perfetto L, Iannuccelli M, Galeota E et al. MINT, the molecular interaction database: 2012 update. Nucleic Acids Res 2012; 40: D857–D861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56Kozomara A, Griffiths-Jones S. miRBase: annotating high confidence microRNAs using deep sequencing data. Nucleic Acids Res 2014; 42: D68–D73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57Maragkakis M, Reczko M, Simossis VA, Alexiou P, Papadopoulos GL, Dalamagas T et al. DIANA-microT web server: elucidating microRNA functions through target prediction. Nucleic Acids Res 2009; 37: W273–W276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58Wong N, Wang X. miRDB: an online resource for microRNA target prediction and functional annotations. Nucleic Acids Res 2015; 43: D146–D152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59Kertesz M, Iovino N, Unnerstall U, Gaul U, Segal E. The role of site accessibility in microRNA target recognition. Nat Genet 2007; 39: 1278–1284. [DOI] [PubMed] [Google Scholar]
- 60Kruger J, Rehmsmeier M. RNAhybrid: microRNA target prediction easy, fast and flexible. Nucleic Acids Res 2006; 34: W451–W454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61Lewis BP, Burge CB, Bartel DP. Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell 2005; 120: 15–20. [DOI] [PubMed] [Google Scholar]
- 62Enright AJ, John B, Gaul U, Tuschl T, Sander C, Marks DS. MicroRNA targets in Drosophila. Genome Biol 2003; 5: R1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63Xiao F, Zuo Z, Cai G, Kang S, Gao X, Li T. miRecords: an integrated resource for microRNA-target interactions. Nucleic Acids Res 2009; 37: D105–D110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64Hsu SD, Tseng YT, Shrestha S, Lin YL, Khaleel A, Chou CH et al. miRTarBase update 2014: an information resource for experimentally validated miRNA-target interactions. Nucleic Acids Res 2014; 42: D78–D85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65Gene Ontology C. The Gene Ontology in 2010: extensions and refinements. Nucleic Acids Res 2010; 38: D331–D335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66Resnik P. Using information content to evaluate semantic similarity in a taxonomy. Proceedings of the 14th international joint conference on Artificial intelligence 1995; 1: 448–453. [Google Scholar]
- 67Girvan M, Newman ME. Community structure in social and biological networks. Proc Natl Acad Sci USA 2002; 99: 7821–7826. [DOI] [PMC free article] [PubMed] [Google Scholar]