Graphical abstract
Abbreviations: MRSA, methicillin-resistant Staphylococcus aureus; MDR, multi-drug resistant; VipTx-I and VipTx-II, viperatoxins I and II; PLA2, phospholipase A2; MTXs, myotoxins; MALDI-TOF/MS, matrix-assisted laser desorption ionization-time of flight/mass spectrometer; MH, Mueller Hinton; TS, Tryptic Soya; MICs, minimum inhibitory concentrations; SEM, scanning electron microscopy; TEM, transmission electron microscopy
Keywords: Bactericidal, Daboia russelli russelli, Phospholipase A2, Viperatoxin-I, Viperatoxin-II
Highlights
-
•
Two novel viperatoxins (VipTx-I and VipTx-II) from Indian Russell’s viper snake venom were purified and characterized.
-
•
VipTx-II but not VipTx-I showed strong antimicrobial effects against S. aureus and Burkholderia pseudomallei (strains KHW/TES), Proteus vulgaris and P. mirabilis.
-
•
In broth dilution assays, VipTx-II had a potent bactericidal effect at the lowest dilutions against B. pseudomallei (strains KHW/TES), S. aureus and P. mirabilis.
-
•
Protein-induced bactericidal potency was closely associated with pore formation and membrane damage.
-
•
These proteins showed a low level of cytotoxic effects on human cells.
Abstract
Infections caused by methicillin-resistant Staphylococcus aureus (MRSA) have become a rising threat to public health. There is an urgent need for development of promising new therapeutic agents against drug resistant bacteria like S. aureus. This report discusses purification and characterization of proteins from Indian Russell’s viper snake venom. Novel 15-kDa proteins called “Viperatoxin” (VipTx-I and VipTx-II) were extracted from the whole venom and evaluated using in vitro antimicrobial experiments. The N-terminal amino acid sequence of “Viperatoxin” showed high sequence homology to daboiatoxin isolated from the same venom and also matched phospholipase A2 (PLA2) enzymes isolated from other snake venoms. In an in vitro plate assay, VipTx-II but not VipTx-I showed strong antimicrobial effects against S. aureus and Burkholderia pseudomallei (KHW & TES), Proteus vulgaris and P. mirabilis. The VipTx-II was further tested by a broth-dilution assay at 100–3.1 μg/ml concentrations. The most potent bactericidal effect was found at the lowest dilutions (MICs of 6.25 μg/ml) against B. pseudomallei, S. aureus and P. vulgaris (MICs of 12.25 μg/ml). Electron microscopic investigation revealed that the protein-induced bactericidal potency was closely associated with pore formation and membrane damage, even at the lowest concentrations (<20 μg/ml). The toxin caused a low level of cytotoxic effects as observed in human (THP-1) cells at higher concentrations. Molecular weight determinations of VipTx-II by sodium dodecyl sulfate-polyacrylamide gel electrophoresis showed one major, along with a few minor bands. The results indicate that VipTx-II plays a significant role in bactericidal and membrane damaging effects in vitro. Non-cytotoxic properties on human cells highlight it as a promising candidate for further evaluation of antimicrobial potential in vivo.
1. Introduction
Infections caused by methicillin-resistant Staphylococcus aureus (MRSA) have become a very important threat to public health [1]. It can cause severe disease, including necrotizing fasciitis, sepsis, endocarditis and pneumonia [2]. The Gram-negative bacterium Burkholderia pseudomallei causes not only human melioidosis [3] but also community-acquired bacteraemic pneumonia [4], septicemias and also high mortality due to septic shock [5]. The presence of septicaemia (44%) and major organ failure (48%) results in death as well as relapse in patients with inappropriate treatment [6]. B. pseudomallei are intrinsically resistant to many antimicrobial agents including first and second generations of cephalosporins, penicillin, macrolides, colistin, rifamycins and aminoglycosides [7]. The above drugs cause serious side effects such as nephrotoxicity and neurotoxicity. Therefore, there is an urgent need for the development of promising new therapeutic agents against drug-resistant bacteria.
Antimicrobial proteins and peptides are produced by all forms of living organisms and represent a novel class of antibiotics to treat infectious diseases [8]. Snake venoms are an extremely rich source of pharmacologically-active proteins with considerable clinical potential [9], [10]. Snake venoms from Viperidae species possess significant bactericidal inhibition [11]. Previous studies show that various venom proteins possess significant antimicrobial activity [12], [13].
Several types of secreted phospholipase A2 (sPLA2) reportedly exert potent bactericidal actions dependent upon their enzymatic activities [14]. sPLA2s have been implicated in lipid digestion to enhance host defence mechanisms that include antibacterial properties [15]. Many studies have demonstrated that the type-IIA sPLA2 is an endogenous antibiotic-like protein that kills bacteria [16]. The acidic PLA2 from Porthidium nasutum snake venom has antibacterial activity [17]. PLA2 homologues present in snake venoms, known as Lys49 PLA2s [18], also have bactericidal activity. Myotoxic PLA2 enzymes are also known to induce bactericidal activity against Escherichia coli and S. aureus [19], [20]. The bactericidal effect of PLA2 isolated from Bothrops snake venoms is reportedly due to its catalytic activity [21] but according to Lomonte et al. [19], the catalytically inactive myotoxic Lys49-PLA2 can also induce a bactericidal effect. PLA2 myotoxins purified from crotalidae snake venoms, including both Lys49 and Asp49-type isoforms, are bactericidal and thus indicate a common mechanism of action for the IIA PLA2 protein family. There are not only bactericidal properties of short cationic peptides derived from a snake venom Lys49-PLA2 [20], [22], but also anti-HIV [23] and anti-fungal activity of a PLA2-derived synthetic peptide variant against Candida albicans [24]. A group of antimicrobial peptides derived from the C-terminal sequence 115–129 of myotoxin II and its triple Tyr–Trp substituted peptide p115-W3, have been reported previously [22]. More tryptophan substitutions increased microbicidal potency against Gram-negative and Gram-positive bacteria [25]. Another study reported that the myotoxins (MTXs) of B. brazili and cationic synthetic peptides derived from the C-terminal region (115–129) can display antimicrobial effects against E. coli and C. albicans [26]. Thus, an enzymatically-independent bactericidal effect of PLA2 protein has also been demonstrated for a specific membrane-damaging protein site [27]. Another study shows that this peptide interacts with lipopolysaccharide (LPS) and lipid A from different Gram-negative bacteria, or with lipoteichoic acid from S. aureus, and relies on a membrane-permeabilizing mechanism to exert its bactericidal effects [27].
Indian Russell’s viper snake venom (Daboia russelli russelli) contains complex mixtures of many distinct proteins [28], ions, biogenic amines, polyamines, polypeptides, neurotoxins, cytolytic peptides, enzymes, thrombin-like proteinase [29], l-amino acid oxidase [30], procoagulant enzymes (factor X) [31], V activators [32], haemorrhagins [33], basic PLA2 and acidic PLA2 [34]. This viper venom is an enormous source of proteins/peptides that have not been fully explored for antimicrobial properties. In the present study, we purified two novel proteins (VipTx-I and VipTx-II) and determined their homogeneity by sodium dodecylsulphate polyacrylamide gel electrophoresis (SDS–PAGE), Matrix Assisted Laser Desorption/ionization-Time of Flight mass spectrometry (MALDI TOF/MS), N-terminal amino acid sequence, and antimicrobial activity with the latter mechanism of action determined by electron microscopy.
2. Experimental procedures
2.1. Chemicals
Chemicals and solvents were obtained from Fluka Chemie GmbH (Deisenhofen, Germany) and Merck (Darmstadt, Germany). Electrophoresis materials such as bromophenol blue, 2-mercaptoethanol, glycerol, sodium dodecylsulphate (SDS), Coomassie Brilliant Blue R-250, 30% acrylamide/bisacrylamide, ammonium persulphate and N,N,N,N-tetramethylethylenediamine (TEMED) were from Bio-Rad (Hercules, CA, USA). Dye reagent for protein assay (Bio-Rad) and all other reagents were from Sigma (St Louis, MO, USA).
2.2. Extraction of venom
Lyophilized venom of D. russelli russelli (Indian Russell’s viper) was purchased from commercial sources (Venom Supplies Pte Ltd, Tanunda, South Australia). The venom samples were collected in a sterile manner under strict laboratory conditions, and were transferred to microcentrifuge tubes, immediately frozen and lyophilized. The dried venom was normally packed and stored dark at −20 °C.
2.3. Purification of protein
Lyophilized whole crude venom (500 mg) of D. russelli russelli was dissolved with 10 ml of 50 mM (pH 7.4) Tris-hydrochloric acid (Tris–HCl) buffer. The suspension was centrifuged at 500 g at 4 °C for 15 min and filtered through a 0.22 μm syringe filter (Nalge Nunc International, Rochester, NY, USA) to remove any colloidal or particulate material. Aliquots of the yellowish clear supernatant were loaded on a Superdex G-75 column (1.6 × 40 cm; Amersham Pharmacia (GE Healthcare, Upsala, Sweden) previously equilibrated with the same buffer (50 mM Tris–HCl, pH 7.4). Fractions (2 ml) were collected at a flow rate of 15 ml/h. The absorbance of all fractions was monitored at 280 nm. Eight fractions (RV1-RV8) were collected from the single pool of venom fractionated by a G-75 gel-filtration column and aliquots taken for testing antibacterial and PLA2 activities, as well as protein measurement. The fraction (RV5) with highest antibacterial and PLA2 activities was further separated by using a C18 sepharose column (250 × 4.6 mm, 300 Å, 5 μm) with reverse-phase (RP)-high performance liquid chromatography (RP-HPLC) and resolved into four protein fractions (RV-F1, RV-F2, RV-F3, RV-F4) monitored at 254 nm. Of which, the active fraction RV-F4 was further purified by C8 sepharose column (Jupitor Phenomenex, Torrance, CA, USA) in 0.1% trifluoroacetic acid (TFA) in water eluted with a linear gradient of 80% acetonitrile (ACN) in 0.1% TFA at 215 nm. Pure protein fractions (VipTx-I and VipTx-II) were collected separately with a FC905 B fraction collector (0.5 ml per min) and designated as “viperatoxin”.
2.4. Protein assay
Protein concentrations of samples were determined by the method of Bradford [35], as modified by BioRad Laboratories (San Diego, CA, USA). Purified PLA2 samples were prepared at 4.0 mg/ml concentrations, using bovine gamma-globulin for the standard curve.
2.5. Protein analysis by SDS–PAGE
The purity of isolated VipTx-I and VipTx-II was verified by SDS–PAGE (4.5 stacking gel/14% separating gels – Tris-glycine running buffer) according to Laemmli, [36]. The fractions were diluted 1:1 with sample buffer (0.12 M Tris–HCl, pH 6.8 containing 2% SDS, 5% 2-mercapethanol, 10% glycerol, 0.02% bromophenol blue) and heated for 5 min in a boiling water bath. Electrophoresis was carried out at a constant current 20 mA for 2.5 h. The gel was fixed with 5% acetic acid overnight and stained for 2 h in 0.1% Coomassie Brilliant Blue R-250 in 5% acetic acid. Destaining was carried out in a solution containing 35% methanol and 7% acetic acid until the background became clear. The molecular weights of protein bands were determined using Bio-Rad SDS molecular weight markers.
2.6. Determination of molecular mass
Molecular weight analyses were performed primarily using a Perspective Biosystem matrix-assisted laser desorption ionization-time of flight (MALDI-TOF/MS) voyager-DE mass spectrometer (Framingham, MA), operated in delayed extraction mode. The enzymes (0.1 μl applied on a clean matrix plate) were analyzed using a saturated solution of α-cyano-4-hydroxycinnamic acid in acetone containing 1% TFA (Sigma, St. Louis, MO, USA). The proteins were selected in the mass range of 10,000–50,000 Da. Spectra were calibrated using calibration mixture 2 of the sequazyme peptide mass standards kit (AB SCIEX). MS-Fit was used for searches in the National Center for Biotechnological Information (NCBI) database. MALDI-TOF mass spectrometry was used for molecular weight determination.
2.7. Analysis of sequencing
Suitable enzymes were subject to N-terminal sequencing by Edman degradation using an Applied Biosystems 494 pulsed liquid-phase sequencer, equipped with an on-line 120 A PTH-amino acid analyzer at the National University of Singapore (NUS), Singapore. The resulting amino acid (AA) sequences were submitted to Basic Local Alignment Search Tool (BLAST) for sequence similarity search (http://web.expasy.org/cgi-bin/blast/blast.pl) by using the ExPASy World Wide Web (WWW) molecular biology server of the Swiss Institute of Bioinformatics (SIB). When the N-terminal sequences of VipTx-I and VipTx-II were blasted for sequence similarity. The VipTx-I and VipTx-II masses were different from that reported for D. russelli russelli (Indian Russell’s viper) in an earlier report [34].
2.8. PLA2 enzyme activity (PLA2)
The Cayman chemical secretory PLA2 (sPLA2) assay kit was used for measuring PLA2. The PLA2 enzyme activity is also converted to μmoles of fatty acid released per min per mg phospholipase by decreased absorbance produced by a known amount of acid. A decrease in absorbance of 0.1 was obtained with 0.025 μmoles of HCl in the reaction mixture [37].
2.9. Antimicrobial assay
Clinical isolates of Gram-negative bacteria E. coli (ATCC 25922), Enterobacter aerogenes, Proteus vulgaris (ATCC27968), Proteus mirabilis (ATCC35491), Pseudomonas aeruginosa (ATCC27853), B. pseudomallei (TES21), B. pseudomallei (KHW22) and the Gram-positive bacterium S. aureus (ATCC 29213) were obtained from the Department of Microbiology, Yong Loo Lin School of Medicine, NUS, Singapore. The following antimicrobial agents: Streptomycin (30 μg/disc), Chloramphenicol (30 μg/disc), Ceftazidime (30 μg/disc), Penicillin (10 units) and Vancomycin (10 units) (Becton Dickinson Labware, USA) were included as positive controls. Blank discs with sterile double-distilled water served as a negative control [38]. Mueller Hinton (MH) and Tryptic Soya (TS) agar medium was purchased from Oxoids, UK. The bacterial cultures were spread and allowed to grow overnight at 37 °C on 20 ml MH or TS agar (pH 7.4) plates (100 mm diameter) prior to storage at 4 °C. Antimicrobial susceptibility was tested according to the method of Bauer et al [39]. Gram-negative bacteria (E. coli, E. aerogenes, P. vulgaris, P. mirabilis, P. aeruginosa) and Gram-positive bacteria (S. aureus) were grown in MH broth, while B. pseudomallei (TES and KHW) were grown in TS broth (OD600 1.0) which corresponds to 1.5 × 105–3.2 × 106 colony forming units (CFU/ml). Bacteria were incubated with VipTx-I and VipTx-II at 100 μg/ml concentrations on MH and TS solid agar plates incubated for 24 h at 37 °C. Bacterial inhibition zones were measured as millimeters in diameter (inhibitory zones).
2.9.1. Minimum inhibitory concentrations (MICs)
Preparation of bacterial inoculums from frozen suspensions were sub-cultured onto MH and TS agar plates and passaged twice prior to susceptibility testing. The bacteria were grown in MH broth for 5–7 h (exponential phase) before adjusting concentration to a 0.5 McFarland turbidity standard. The adjusted bacterial cultures were diluted to approximately 3.2 × 106 CFU/ml [17]. MICs were determined by the broth micro-dilution techniques [40], for which serial dilutions of VipTx-I and VipTx-II were prepared at 100, 50, 25, 12.5, 6.125, 3.078 μg/ml in 96-well microtiter trays with appropriate broths (MH & TS), whereas multi-drug resistant B. pseudomallei (TES & KHW) was tested at 3.078–100 μg/ml concentrations in TS broth. Three replicates were used for each dilution series that included control wells containing bacteria without VipTx-I or VipTx-II. A 200 μl aliquot of the 106 CFU/ml was added to each well (96-well plates) with 50 μl of VipTx-II. The culture trays were incubated at 37 °C for 24 h, the inhibition of bacterial growth was determined by measuring the absorbance at 600 nm (Sunrise Precision Microplate reader, Tecan Group Ltd, Mannedorf, Switzerland). The MICs were taken as the lowest concentration of VipTx-I or VipTx-II that inhibited visible growth. The results given are mean values of three independent determinations. After MIC measurement, each dilution of proteins treated with bacterial samples (20 μl) were spread on to MH and TS agar plates and incubated at 37 °C for 24 h. Minimum bactericidal concentrations (MBCs) were assayed at 100–3.078 μg/ml concentrations.
2.9.2. Scanning electron microscopy (SEM)
The structural changes induced by VipTx-II (100-3.078 μg/ml) on S. aureus, B. pseudomallei (KHW & TES), E. aerogenes, P. vulgaris, and P. mirabilis were studied using SEM as described earlier [41]. Each protein sample (50 μl) contained (bacteria) pre-incubated together for 30 min at 37 °C. The control received equivalent volumes of MH broth containing bacteria. After removing a small portion of these samples for CFU/ml measurements, the remainder was centrifuged for 10 min at 2800g. Pellets were resuspended and fixed with an equal volume of 2.5% glutaraldehyde in 1 mM phosphate buffer (pH 7.4) for 1 h. Immediately following addition of fixative solution, the sample tubes were mixed by gently inverting them up and down for several minutes to prevent clumping of cells. The cells were post-fixed for an additional hour with 1% osmium tetroxide (OsO4) and washed thrice in PBS. Samples (1 μl) were pipetted onto a sterile cover glass coated with poly-l lysine and left for 20–30 min. The section was dehydrated by a series of alcohol baths (25%, 50%, 75%, 90% 100%). The samples were transferred from 100% ethanol to a critical point dryer (Balzers CPD-030, Bal-Tec AG, Vaduz, Liechtenstein), and dried using liquid carbon dioxide. The samples were mounted on aluminum specimen supports with carbon adhesive tabs, and coated with a 10–15 nm thickness of gold using a sputter coater SC D005 (Bal-Tec, EM Technology and Application, Liechtenstein). Samples were examined with a Philips XL 30 FEG SEM (Electron Microscopy, Japan) using an accelerating voltage of 5–10 kV.
2.9.3. Transmission electron microscopy (TEM)
The structural changes induced by VipTx-II on S. aureus and B. pseudomallei (KHW) were studied using TEM as described earlier [42]. Bacterial cells suspended in 10 mM phosphate buffer (pH 7.4) treated with VipTx-II (6.25 μg/ml) were fixed with an equal volume of 2.5% glutaraldehyde in 10 mM phosphate buffer, pH 7.4. The fixed samples were stored overnight at 4 °C in fixative solution. The suspended cells were rinsed with 10 mM phosphate buffer, and dehydrated through a graded series of ethanol (25–100%). During the entire filtration, rinsing, and dehydration process, cells were covered with fluid to prevent air drying. The samples were transferred from 100% ethanol in to a critical point dryer, and dried using carbon dioxide. The samples were mounted on aluminum specimen supports with carbon adhesive tabs, and coated with gold-palladium metal (60:40 alloy and 15 nm thickness) using a Hummer X sputter coater (Bal-Tec, EM Technology and Application, Liechtenstein). Samples were examined with a (JEF 2220) TEM using an accelerating voltage of 5–10 kV.
2.9.4. Cell proliferation and cytotoxicity (MTT based) assay
The human macrophage cells (THP-1) were obtained from ATCC, (Virginia, USA). Sterile Dulbecco’s Modified Eagle’s Medium (DMEM), Fetal Bovine Serum (FBS), and 10 mM HEPES were purchased from the National University Medical Institute (NUMI), Singapore. All chemicals were of analytical and cell culture grade. THP-1 cells were cultured in 72 cm2 flasks at a density of 107 cells/ml in DMEM culture medium supplemented with 10% FBS, and 1 ml of HEPES. The cells adhered to the flask bottom overnight at 37 °C in a humidified atmosphere of 5% CO2 and 95% air. The culture medium was changed four times a week. To analyze the initial events of VipTx-I and VipTx-II upon cell viability, the proteins were applied to THP-1 cells at different concentrations (10,000–39 μg/ml) with varied time intervals (24 and 48 h), tetrazolium dye added and incubated for 30 min. Cell proliferation was assessed by measuring optical density (OD) using an ELISA plate reader at 490 nm. All assays were performed in triplicates and repeated thrice.
2.9.5. Cytolytic assay by lactate dehydrogenase (LDH)
Cytolytic effects of proteins on human acute monocytic leukemia cells were evaluated by measuring the release of LDH enzyme using a cytotoxicity detection kit (Roche Mannheim, Germany). Proteins (VipTx-I and VipTx-II) were added to THP-1 cells (106 cells/well) cultured on 96-well plates in DMEM medium (NUMI, Singapore) supplemented with 10% (vol/vol) FBS. The proteins (10,000–39 μg/ml) were added and further incubated with cells for 24 and 48 h. A 200 μl aliquot of the centrifuged supernatant obtained from each well was used for the quantification of cell death and lysis, based on the measurement of LDH activity released from the cytosol of damaged cells into the supernatant. The assay was performed in triplicate.
2.9.6. Statistical analysis
The results (mean ± S.D., n = 5) were statistically analyzed by one way ANOVA with repeated measures used to analyze factors influencing the size of the growth inhibition zones. The level of statistical significance was at ∗P < 0.01 and ∗∗P < 0.05 etc.
3. Results
3.1. Purification and characterization of protein
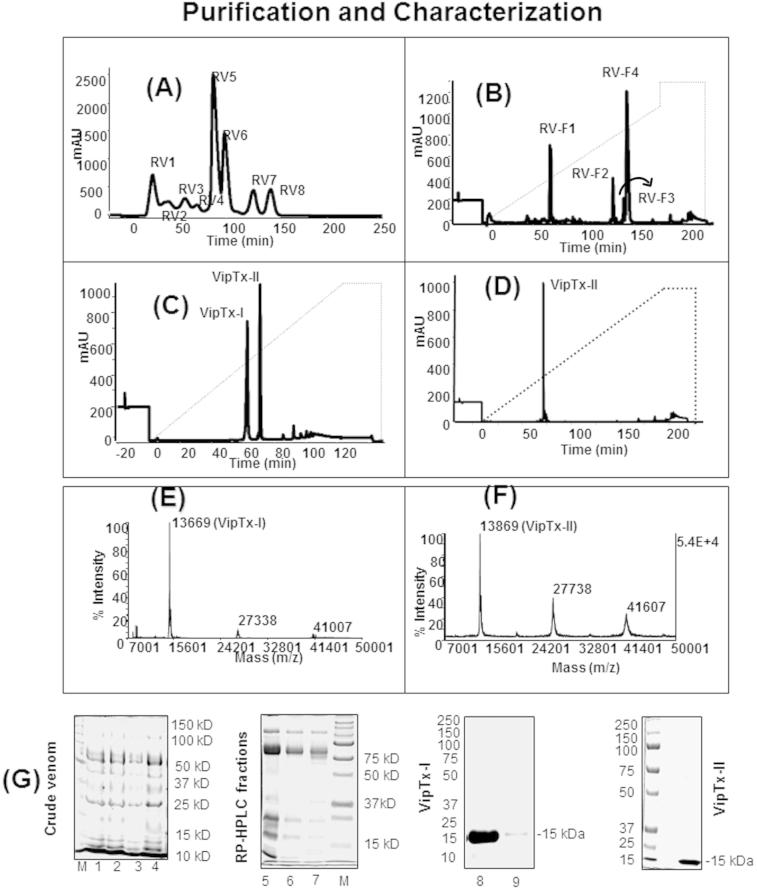
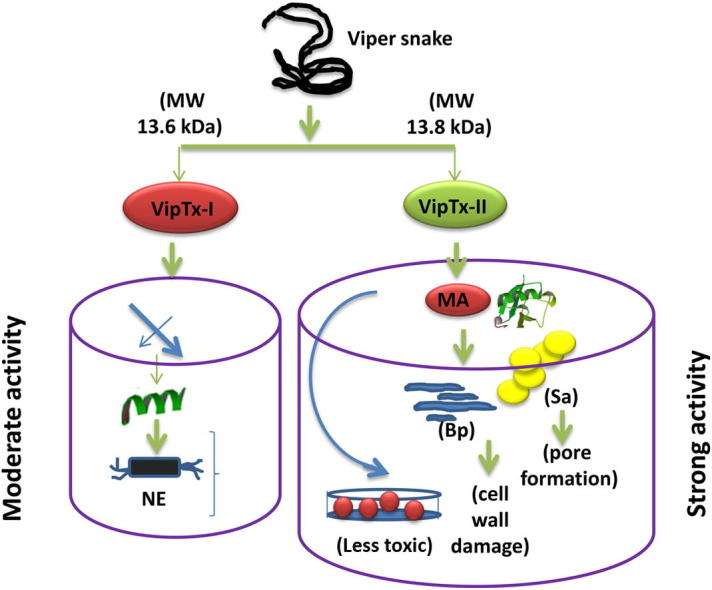
Viperatoxin was purified from the venom of Russell’s viper (D. russelli russelli) by gel-filtration chromatography on a Superdex G-75 column, yielding eight major protein peaks (Fig. 1A). All of the fractions (RV1 to RV8) were assayed for antibacterial activities, of which RV5 showed significant antibacterial and PLA2 activity versus RV4. The active fraction RV5 was further fractionated by reverse phase (RP) chromatography on Sepharose (C18 column), and resolved into four further fractions, namely RV-F1 to RV-F4 (Fig. 1B). The most active antibacterial fraction (RV-F4) was applied to Sepharose C18 and C8 reverse phase columns and resolved into two major purified proteins (Fig. 1C and D), subsequently designated as “Viperatoxins” VipTx-I and VipTx-II. The VipTx-II showed more phospholipase A2 enzymatic activity than the VipTx-I. However, the protein purity was assessed by mass spec MALDI-TOF/MS analysis showing the actual mass of VipTx-I (13669.93 daltons) and VipTx-II (13869.05 daltons) (Fig. 1E and F). Protein purity was assessed by SDS–PAGE, and molecular weight was estimated to be approximately 15 kDa (Fig. 1G).
Fig. 1.
(A) High performance liquid chromatography (HPLC) profiles of D. russellii russellii crude venom from a Superdex G-75 column, (B) fraction RV5 further separated by reverse-phase (RP)-HPLC spectrum of Sepharose C18 (RV-F1 to RV-F3) and (C and D) the most active fraction RV-F4 was further purified by C8 column and produced into two pure proteins namely Viperatoxin (VipTx-I and VipTx-II), (E and F) molecular weight of proteins were analyzed by MALDI-TOF/MS, (G) protein profile determined by sodium dodecyl sulphate–polyacrylamide gel electrophoresis (SDS–PAGE), lanes indicates: RV-CV Russell’s viper crude venom (1-4), lane (5-7) RP-HPLC fractions from C18 column, the homogeneity or purity of lane (8) VipTx-II, lane (9) VipTx-I (20 μg of protein loaded per lane) was performed by SDS–PAGE respectively.
3.2. Phospholipase A2 enzyme activity
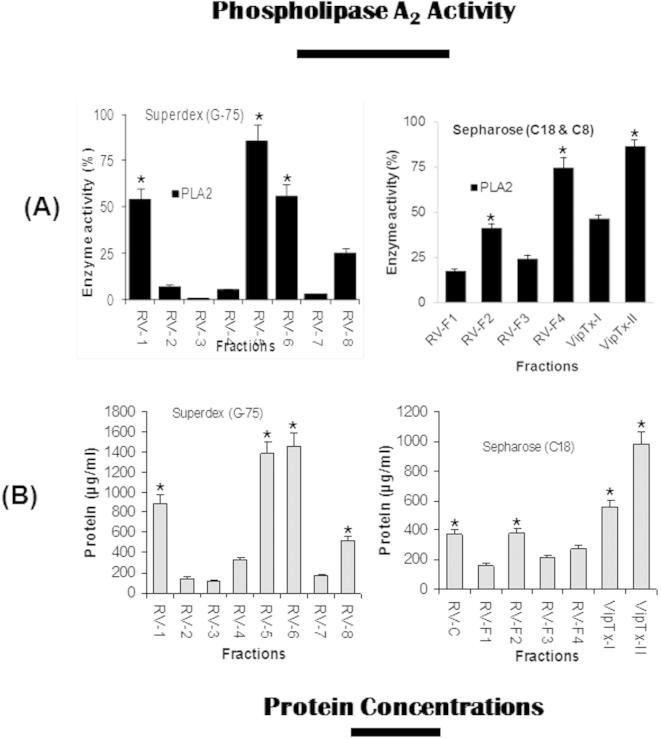
Phospholipase A2 (PLA2) enzyme was known to be a major component of snake venoms showed important toxic and pharmacological effects. In this study, eight PLA2 enzyme fractions were isolated from the crude venom such as RV1–RV8, of which fraction RV5 was displayed higher enzyme activity/bactericidal potency further purified by revere-phase chromatography (C18 column) resolved into four fractions (RV-F1–RV-F4). Similarly, enzymatically the most active fraction (RV-F4) was separated by C8 columns and yielded VipTx-I/VipTx-II proteins. Fascinatingly, there was not only a higher level of PLA2 enzymatic activity but also high proteins levels of VipTx-II determined in this assay system (Fig. S1 A and B).
3.3. Analysis of sequencing
The N-terminal amino acid (AA) residues of VipTx-I and VipTx-II were sequenced, and compared with those found in the expert protein analysis system (ExPASY) proteomics database using a basic local alignment search tool (BLAST) search alignment with several types of snake venom PLA2s (Table. 1). The AA sequences were matched exactly with the available sequences and its protein masses varied from the existing snake venom PLA2s. The sequence comparison shows that VipTx-II shares greatest sequence identity (60–86%) with a PLA2 from other vipers, and a high degree of sequence homology exists with the group RV-VIIIA PLA2s. In particular, the N-terminal residues of VipTx-II matched with existing PLA2s, but slight modification of one or two new AA residues found in the sequences are most likely due to post translational modifications. The VipTx-II also shared much sequence homology with the Asp-49 enzymes from several species. The BLAST search was matched with previously reported basic svPLA2s of the Viperidae. The N-terminal sequences (VipTx-II) were 91% identical to sp|P86368|PA23_DABRR (showing 5th in the alignment). These basic amino acids and hydrophobicity are essential for enhanced antimicrobial activity. Also, to the best of our knowledge, this is the first detailed report on the antimicrobial activity of Indian viper venom proteins along with their unique mechanisms of action.
Table 1.
Multiple sequence alignments for the N-terminal sequences of VipTx-I and VipTx-II phospholipase A2 (PLA2) from the Daboia russelli russelli was compared with existing snake venom basic phospholipase A2 sp|P59071|PA28_DABRR-VRV-PL-VIIIA of Daboia russelii (Russel’s viper), tr|D0VX11|D0VX11_DABRP-PLA2 VRV-PL-VIIIA (Daboia russellii pulchella), tr|B3RFI8|B3RFI8_9SAUR-PLA2 (Daboia russellii limitis), tr|A8CG84|A8CG84_DABRU-basic PLA2 Drk-b2 (svPLA2) of Daboia russelii (Russel’s viper or Vipera russelii), sp|P86368|PA23_DABRR-basic PLA2 of Daboia russelii (Russel’s viper), sp|P84674|PA25_DABRR-basic PLA2 VRV-PL-V of Daboia russelii (Russel’s viper), sp|P14424|PA2B_VIPAA- neutral PLA2 agkistrodotoxin ATX) of Agkistrodon halys or Gloydius halys (Chinese water mocassin), sp|P00626|PA2A_VIPAA-PLA2, ammodytoxin A [ATXA]of Vipera ammodytes ammodytes (Western sand viper) PLA2, sp|P11407|PA2C_VIPAA-ammodytoxin C [ATXC] of Vipera ammodytes ammodytes (Western sand viper), sp|P59171|PA25_ECHOC-acidic PLA2 of Echis ocellatus (Ocellated saw-scaled viper), basic PLA2 myotoxin III (Bothrops asper), sp|P20474|PA21_BOTAS-basic myotoxic PLA2 bothropstoxin-2 (BthTX-II) of Bothrops jararacussu (Jararacussu), sp|P45881|PA2B2_BOTJR-and sp|P86974|PA2D_BOTLC-basic PLA2 blD-PLA2) of Bothrops leucurus (White-tailed lancehead).
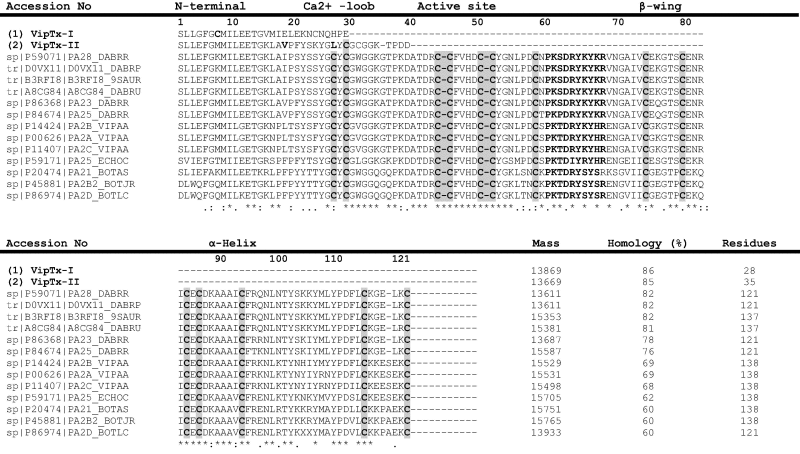 |
The ExPasy tools BLAST used for the CLUSTAL W (1.82) multiple sequence alignment of viper PLA2s (phosphatidylcholine 2-acylhydrolase).
3.4. In vitro antimicrobial activity
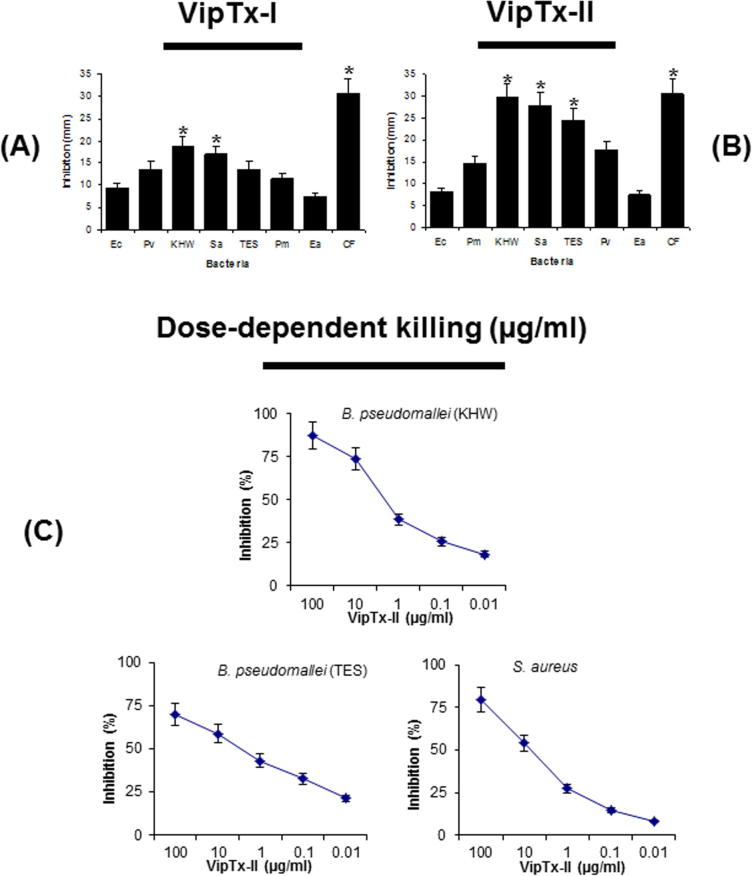
Purified proteins (VipTx-I and VipTx-II) were tested for their antibacterial properties against Gram-positive and Gram-negative bacteria at a 100 μg/ml concentration. The enzyme exhibited broad spectrum activity against a wide range of pathogenic organisms. The VipTx-II enzyme showed very strong antimicrobial action against S. aureus, B. pseudomallei (KHW), P. vulgaris, E. aerogenes, and P. mirabilis (Fig. 2A and B). The most promising activity of VipTx-II was compared with standard antibiotics (i.e. Ceftazidime, Chloramphenicol, Penicillin, Streptomycin, and Vancomycin). The inhibitory potential of VipTx-II was equal to that of standard antibiotics like Streptomycin, Chloramphenicol and Ceftazidime. However, VipTx-I exerted a very weak antimicrobial effect against all the tested bacteria. Particularly it’s devoid of activity against P. aeruginosa. However, this VipTx-II protein displayed the most potent antibacterial activity compared to that of the VipTx-I protein. Similarly, the antimicrobial activity of VipTx-II prompted us to conduct a further testing of MIC determinations by a broth dilution method.
Fig. 2.
In vitro antimicrobial activity against Gram-positive and Gram-negative bacterial cells were determined by standard procedure. (A) The VipTx-I protein displayed less activity against tested bacteria. (B) The VipTx-II protein showed most potent action against S. aureus and B. pseudomallei among the tested bacteria, the activity or inhibition zones were compared with a antibiotic. The activity of VipTx-II protein more or less equal when compared to that of commercial antibiotic. (C) The VipTx-II protein showed most potent action against S. aureus and B. pseudomallei (KHW & TES) and S. aureus among the tested bacteria, the activity or inhibition zones were compared with a variety of antibiotics. (D) The active protein was further tested in a dose-dependent (100–0.01 μg/ml) manner against the most sensitive organisms. Results exhibited that the protein exerted very strong bactericidal activity against B. pseudomallei (KHW) and S. aureus even at very low doses (10 μg/ml). Symbol denotes: Ec-Escherichia coli, Pv-Proteus vulgaris, KHW-Burkholderia pseudomallei, Sa-Staphylococcus aureus, TES-Burkholderia pseudomallei, Pm-Proteus mirabilis, Ea-Enterobacter aerogenes, STR-Streptomycin, CHL-Chloramphenicol, CF-Ceftazidime, PEN-Penicillin, VAN-Vancomycin.
3.4.1. Dose-dependent antimicrobial activity
Antibacterial susceptibility of the most effective protein (VipTx-II) was further assayed against multi-drug resistant (MDR) B. pseudomallei (strain of KHW) and S. aureus. The inhibitory potential of VipTx-II was equal against both types of bacteria; however, this protein was slightly more effective in controlling the growth of B. pseudomallei at 10 μg/ml (Fig. 2C).
3.4.2. Minimum inhibitory concentrations (MICs)
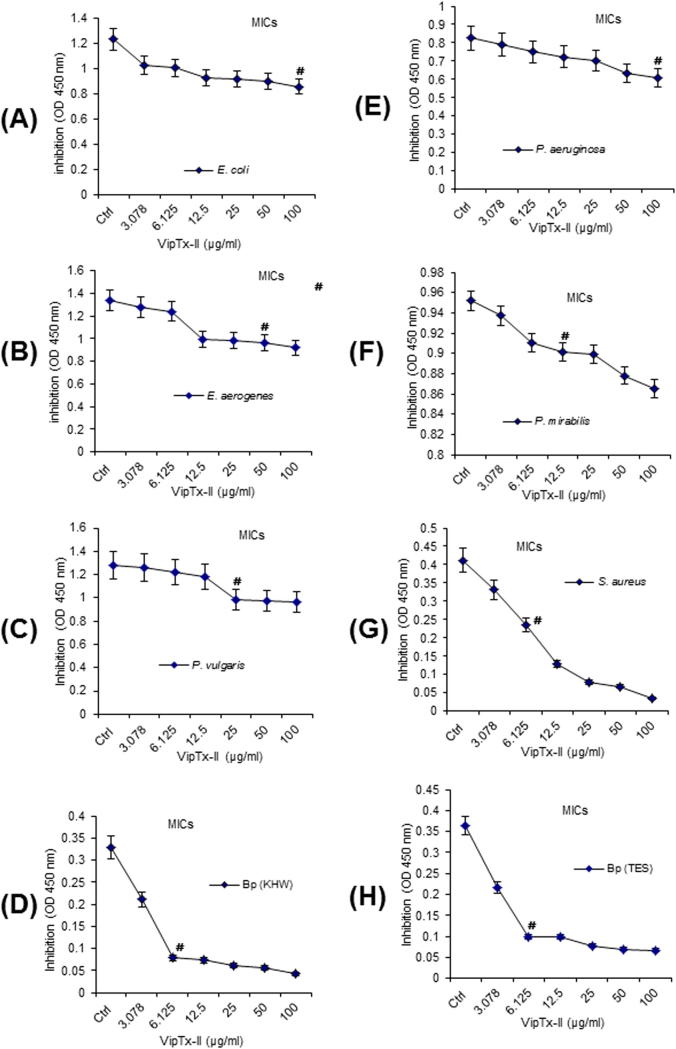
The MICs of proteins (VipTx-I and VipTx-II) were determined by broth-dilution assay with initial sample concentrations of 100, 50, 25, 12.5, 6.25, 3.125 and 1.56 μg/ml tested against Gram-positive and Gram-negative strains, including B. pseudomallei (TES & KHW). The MIC values are expressed as the lowest concentration that caused 100% bacterial growth inhibition. The VipTx-II exhibited marked activity against B. pseudomallei KHW & TES (MICs of 6.25 μg/ml), S. aureus, (MICs 6.25 μg/ml), P. mirabilis (MICs of 12.5 μg/ml), P. vulgaris (MICs of 25 μg/ml), whereas E. coli, E. aerogenes and P. aeruginosa showed weaker inhibitory MIC effects at all dilutions (100–1.56 μg/ml) shown in Fig. 3A–H. Interestingly, VipTx-II exhibited significant inhibition at the lowest dilution against B. pseudomallei and S. aureus (MIC of 6.125 μg/ml). The bactericidal (killing) potential of proteins was further quantified by a TS and MH broth dilution method (100–1.56 μg/ml) as shown in Table 2. The MBCs result revealed that VipTx-II exerted most significant inhibition against KHW strains of B. pseudomallei and S. aureus at the lowest dilutions (MBC 6.125 μg/ml) versus bacterial control. The VipTx-I only weakly killed multi-drug resistant strains of B. pseudomallei at all tested concentrations.
Fig. 3.
The MICs values were determined by a modified micro-broth dilution assay using VipTx-II against Gram-negative and Gram-positive bacteria. (a) VipTx-II inhibits S. aureus and B. pseudomallei in a dose-dependent fashion and less number of colonies was observed. The activity of VipTx-II was 10-fold greater against S. aureus and B. pseudomallei, each breaking point represents the mean of triplicate samples. VipTx-II inhibits S. aureus (MICs 6.125 μg/ml), B. pseudomallei KHW & TES (MICs 6.125 μg/ml), Proteus mirabilis (MICs 12.5 μg/ml) and Proteus vulgaris (MICs 25 μg/ml) more effectively versus P. aeruginosa, Escherichia coli and Enterobacter aerogenes.
Table 2.
Minimum bactericidal concentrations (MBCs) of viperatoxins isolated from the Indian Russell’s viper snake venom (Daboia russellii russellii).a
| Bacteria | MBCsa |
|
|---|---|---|
| VipTx-I (μg/ml) | VipTx-II (μg/ml) | |
| Escherichia coli | 100 | 100 |
| Enterobacter aerogenes | 100 | 100 |
| Proteus vulgaris | 50 | 12.5 |
| Proteus mirabilis | 50 | 12.5 |
| Pseudomonas aeruginosa | 100 | 100 |
| Staphylococcus aureus | 50 | 6.25 |
| Burkholderia pseudomallei (strain KHW) | 100 | 6.25 |
| Burkholderia pseudomallei (strain TES) | 100 | 6.25 |
The MBC values were determined by a modified micro-broth dilution assay using snake venom proteins (100, 50, 25, 12.5, 6.25, 3.125 μg/ml). Antimicrobial activity of viperatoxin (VipTx-I and VipTx-II) was determined by MBCs in a solution killing assay against Gram-negative and Gram-positive bacteria.
3.4.3. Mechanisms of action for VipTx-II
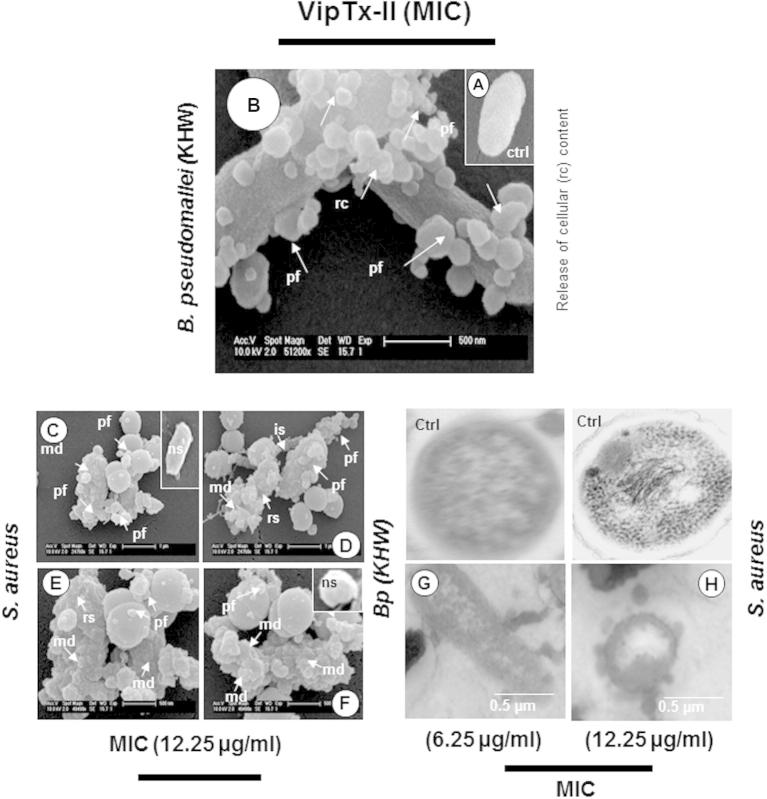
Smooth membrane structure was clearly evident in control bacteria without treatment, B. pseudomallei (KHW) treated with VipTx-II resulted in numerous mushroom-shaped blebs, thickening irregular shapes and retraction of cytoplasm, while bacteria appeared to be losing cell contents particularly at the division septa after 24 h (Fig. 4A and B). Particularly after the treatment of VipTx-II with Gram-positive (S. aureus), while fibrous and presumably cell contents (granular material) appeared to exude from the damaged membranes (Fig. 4C–F) and the inner membrane was difficult to discern. Bacteria treated with VipTx-II also lost of the membrane structure and the formed of blebs in B. pseudomallei strain KHW, clearly shown in transmission electron microscopy (Fig. 4G and H) than the control (ctrl). The ultra-structural studies have proven the pore-forming action of VipTx-II enzyme on bacteria. Generally, viper proteins induced specific structural and morphological changes versus untreated bacterial controls.
Fig. 4.
The mechanisms of action of antimicrobial protein was postulated by SEM examination. (A) Control – clear architecture, (B–H) The protein (VipTx-II) exerted excellent pore formation (blebs), disintegration of bacterial membranes heavily and release of cellular contents after exposure at 6.25 μg/ml (B. pseudomallei) and 12.25 μg/ml (S. aureus). (G and H) Transmission electron microscopic studies additionally damaged of cell wall and loss of cellular content after 24 h treatment of VipTx-II. Symbol denotes: pf-pore formation, rs-rough surface, md- membrane damage, is-irregular shapes, ns-normal surface, ctrl-control.
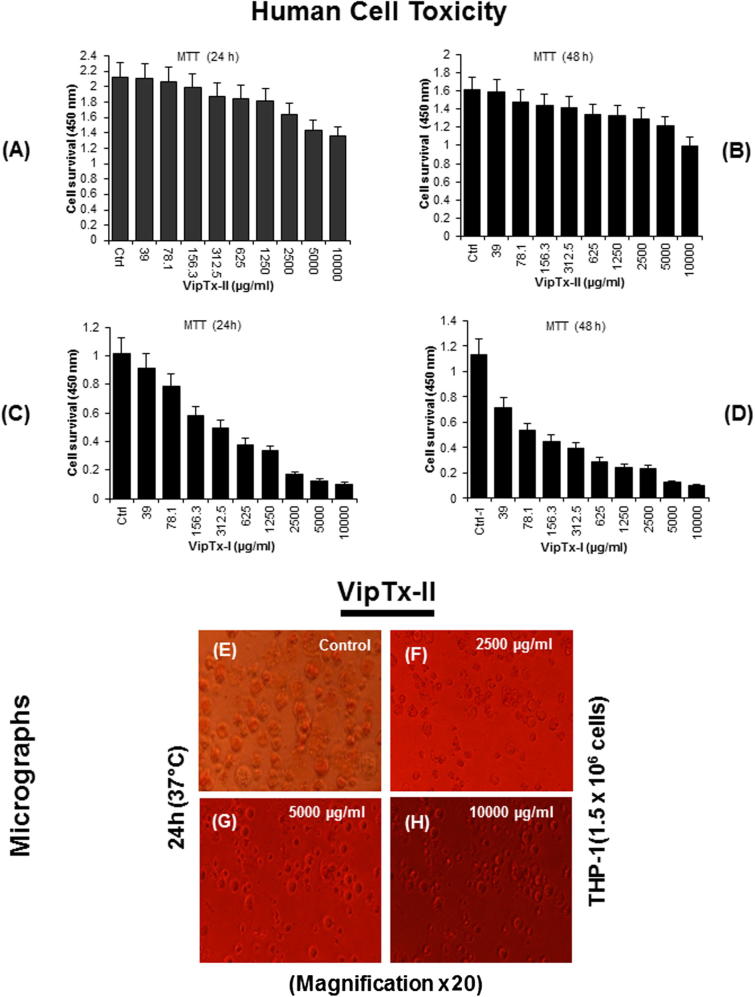
3.5. Morphological and cellular toxicity
Cytotoxicity results indicated that VipTx-II did not affect the cell viability up to a concentration of 1000 μg/ml. Human macrophages (THP-1) were not affected particularly at 625 μg/ml in a cytotoxicity assay. Exposure to VipTx-II revealed minimal cytotoxicity up to 625 μg/ml concentrations. The optimum dose of VipTx-II gradually decreased THP-1 cell proliferation after treatment with enzymes (Fig. 5A and B). However, cell survival decreased with increasing concentrations of VipTx-I from 39 to 10,000 μg/ml and EC50 1250 μg/ml (Fig. 5C and D). The morphological changes of the cells revealed that membrane disruption, lysis and significant cell death was evident at a 2500 μg/ml concentration of VipTx-I in a time and dose dependent (24 and 48 h) manner. Sixty percent of the THP-1 cells were inhibited by the exposure of VipTx-I after 48 h than in the control. However, the cytolytic levels were at higher concentrations up to 2500 μg/ml (Fig. 5E–H).
Fig. 5.
(A and B) Evaluation of MTT-based cytotoxicity of proteins incubated with human cells (THP-1) and various concentrations of VipTx-II after 24 h and 48 h incubation. Control (Ctrl) cells without treatment used as a control. (C and D) Cytotoxicity of protein of VipTx-I on THP-1cells were incubated with the different concentrations (10,000–39 μg/ml), VipTx-I showed severe reduction of cell proliferation and more toxicity up to 1250 μg/ml than the VipTx-II up to only 5000 μg/ml. Light micrograph showing the normal architecture of THP-1 cells, (E) THP-1 cells without treatment served as a control, (F–H) cells treated with 2500, 5000 and 10,000 μg/ml concentrations of protein VipTx-II caused morphological changes after exposure (Magnification ×20).
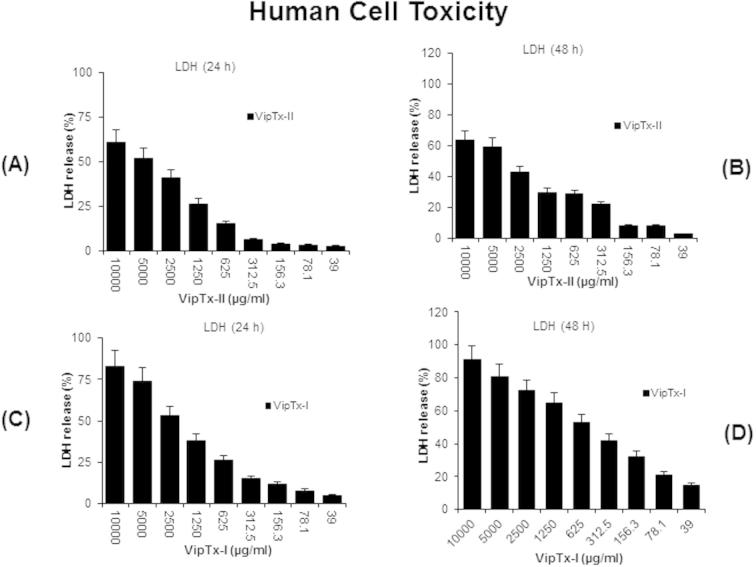
3.6. LDH assay
The LDH results revealed that THP-1 cells exposed to VipTx-II were not affected up to 1250 μg/ml concentrations (Fig. 6A and B). Significant cell death is evident at higher concentration (2500 μg/ml) in a dose and time-dependent manner (24 and 48 h), as a result there is more LDH release into the media. However, the cell proliferation was not affected especially at the optimal dose of VipTx-II (EC501250 μg/m1) than the VipTx-II (Fig. 6C and D). The optimum dose that inhibited bacterial proliferation did not affect the THP-1cells.
Fig. 6.
Cell death and cell lysis were determined by LDH activity released from the cytosol of damaged cells into the supernatant after exposure of proteins. (A–D) The VipTx-II protein did not lyse THP-1 cells exposed up to 1250 μg/ml doses, whereas the VipTx-I protein was induced higher percentage of cell death and more release of LDH in the culture supernatant.
4. Discussion
Bacterial resistance is a significant problem for treating infections, and thus there is a keen interest in research directed towards identifying novel agents to treat infections [43]. Most venomous animals contain a variety of venom proteins which participate in both digestion of prey and venom toxicity. Viperidae snake venoms represent a source of important bioactive molecules that have led to the development of diverse new drugs in clinical scenario. In this study, novel snake venom proteins were purified and designated as “Viperatoxin” (VipTx-I and VipTx-II) from the Indian Russell’s viper (D. russelli russelli). The N-terminal amino acid residues of VipTx-I and VipTx-II were sequenced, and compared with existing sequences in the ExPASY proteomics database using BLAST.
The sequence comparison shows that VipTx-II matched 60–86% homology with existing snake venom phospholipase A2s (svPLA2s). The molecular weight of VipTx-I and VipTX-II slightly differs with the previously reported protein masses from known PLA2s. Our results corroborate with the N-terminal sequences of B. neuwiedi pauloensis showed important homology with Asp49 basic myotoxic PLA2s from other snake venoms [44]. Whereas, Lys49 PLA2 (myotoxin I) elucidated from B. atrox venom displays very high level of homology with other Lys49 PLA2s, although its primary and three-dimensional structure show some difference in the C-terminal region [45]. Generally, characterized svPLA2s have a conserved fold with seven disulfide bridges and a histidine at the catalytic site, with calcium (Ca2+) bound at the active site [11], [28], [46]. The Russell’s viper svPLA2 structure also contains a Trp-31-containing loop (residues 25-34), β-wing consisting of double-stranded anti-parallel β-strands (residues 74-85) and C-terminal region 9 (residues 119-133). The crystal structures of complexes with transition analogs provide insights into the potential catalytic mechanisms with the presence of fatty acid in the hydrophobic channel [47].
Several recent studies have shown that antimicrobial proteins and peptides are produced by many living organisms and represent a novel class of antibiotics to treat infectious diseases [8]. Examples include crotamine, a myotoxin from venom of the South American rattlesnake, which is structurally related to beta-defensins. The later are antimicrobial peptides found in vertebrate animals [13], peptides from NajA naja venom [48], and a venom protein from the inland taipan which all exhibit antimicrobial activity. Previous studies have shown that the naturally occurring proteins display antimicrobial activity [12]. Remarkably, a group of Lys-49 PLA2s homologues present in snake venoms [19], are bactericidal even though they lack enzymatic activity. Another study shows that myotoxic PLA2s are bactericidal against E. coli and S. aureus [20].
In this study, VipTx-II exerted the most potent action against a multi-drug resistant strain (KHW) of B. pseudomallei as compared to the less resistant TES strains. The inhibitory potential of VipTx-II was quite equivalent to that of standard antibiotics such as streptomycin, chloramphenicol, ceftazidime and vancomycin versus VipTx-I. Further, we tested the antibacterial properties of VipTx-II and found that it killed several strains of Gram-positive and Gram-negative bacteria, with doses ranging from (100–0.01 μg/ml). The dose-dependant assay revealed bacterial killing by viper protein within 24 h and reached the maximum (75%) activity at a 10 μg/ml concentration. Our earlier study clearly demonstrated that viper proteins exert the most significant bactericidal effects against B. pseudomallei [49]. Our study, in contrast with the EcTx-I protein from the venoms of Saw-scaled viper species, demonstrated significant bactericidal inhibition of multi-drug resistance (MDR) B. pseudomallei (KHW) and E. aerogenes previously [38]. VipTx-II exerted the most significant inhibition against B. pseudomallei KHW strains even at the lowest dilutions (MICs 6.25 μg/ml). Interestingly, the VipTx-II protein showed very significant inhibitory effects against S. aureus, P. vulgaris and P. mirabilis even at 12.25 μg/ml doses (in a dose-dependent manner). Whereas, recently reported studies show that a basic protein of VRV-PL-V from Daboia russelli pulchella (venom PLA2 fraction V) effectively inhibits Gram-positive bacteria such as S. aureus and Bacillus subtilis at MICs 13-24 μg/ml versus Gram-negative E. coli, Vibrio cholerae, Klebsiella pneumoniae and Salmonella paratyphi [50]. Similarly the PLA2 fraction VIIIA of D. russelli pulchella (VRV-PL-VIIIa) also controls the growth of bacteria at 11–19 μg/ml doses [51], BnpTX-1/II is a basic myotoxic PLA2s obtained from B. neuwiedi pauloensis snake venom, BnpTX-1 showed the neurotoxic as well as antibacterial activity on E. coli and S. aureus [44]. The Lys49 phospholipase A2 (PLA2) of Bothrops atrox (myotoxin I) displays only a weak antibacterial activity on bacteria [45], whereas synthetic peptide derived from the Lys49 PLA2 of C-terminal segment of B. asper (myotoxin II) and tryptophan (Tyr–Trp) substitution enhances antimicrobial potency on Gram-negative (Salmonella typhimurium) and Gram-positive bacteria (S. aureus), with low cytotoxicity on skeletal muscle cells, C2C12 myoblasts [22]. Cardiotoxin 3 (CTX3) isolated from Naja naja atra (Taiwan cobra) exerts the most potent inhibitory effects against S. aureus than E. coli [52]. However, the VipTx-II displays the most potent inhibitory effects against B. pseudomallei KHW (MICs 6.25 μg/ml) as well as S. aureus, P. vulgaris and P. mirabilis (MICs 12.25 μg/ml) at very low concentrations compared to the existing PLA2s.
We found novel bactericidal mechanisms attributed to viper proteins (VipTx-I and VipTx-II) that induced pore formation on clinical isolates such as MDR B. pseudomallei (KHW strain). There were cellular changes that include membrane disintegration. Our study corroborate with membrane damage elicited by the binding of protein to the lipid membrane [53]. Recently, there have been suggestions that transmembrane pore formation is not the only mechanism of microbial killing [54]. Several earlier observations indicate that translocated protein or peptides can alter cytoplasmic membrane septum formation, inhibit cell-wall, nucleic-acid and protein synthesis or inhibit enzymatic activity [54]. Previously study of the mechanisms was clearly evidenced that the antimicrobial protein as well as peptides induced killing of microorganisms by severely damaging membrane and formation of pores on invading pathogens [38]. Therefore, sPLA2 is implicated in the lipid digestion as a host defence mechanism including the observed antibacterial activities [55].
Enzymatically-independent bactericidal effects of PLA2 has been demonstrated and mapped to a specific membrane-damaging protein site [20]. A number of Trp substituted peptides derived from svPLA2 can increase microbicidal potency against both Gram-negative and Gram-positive bacteria [25]. Previous studies also show that the interaction of this peptide with the lipopolysaccharide (LPS) and lipid A or lipoteichoic acid (LA) relies on a membrane-permeabilizing mechanism to exert its bactericidal effects [20]. In addition to Trp, Pro and Arg residues are also important in membrane disruption and/or cell entry. Thus these later Pro and Arg residues are an attractive template for designing novel antimicrobial peptides effective against a broad spectrum of microorganisms [56]. In addition, it has been reported that the insertion of positively-charged amino acids can result in big variations and high antibiotic activity. These peptides also significantly reduce hemolysis and cytotoxicity that correlate with decreased permeabilization of the zwitterionic phosphatidylcholine membrane, the major component of outer leaflets from red blood cells [56].
Several proteins and polypeptides of reptiles have common cytolytic properties, and these cytolysins provide an offensive armament for the animal defense. In this study, cell survival decreased with the increasing concentrations of viper protein (VipTx-I) at 39–10,000 μg/ml than VipTx-II. However, the enzyme (VipTx-II) has low level of toxicity for eukaryotic cells at higher concentrations. The morphological changes in THP-1 cells show membrane disruption, lysis and significant cell death apparent at 2500 μg/ml with VipTx-I in a time-dependent manner (24 and 48 h). Although, 60% of the cells were killed by the VipTx-I after 48 h, but the observed cytolytic effects were very high at higher protein concentrations (1250 μg/ml). This may be due to the mechanism for clearing free fatty acids that is toxic to the cell for retaining cellular energy reserve. Those fatty acids might be acylated by intracellular located enzymes to membrane-bound proteins. Acylation of these proteins can be potentially one of the mechanisms for disrupting membrane integrity and cellular signaling [57].
4.1. Mechanisms of actions
The antimicrobial potency of snake venom is largely unexplored. Naturally occurring snake venom proteins and peptides possess highly potent antimicrobial activity against B. pseudomallei [58]. A family of sPLA2 comprised of low molecular weight (13 kDa), disulfide-linked proteins [59] depend on Ca2+ for enzymatic activity and play an important role in innate immunity and killing of bacteria [60]. Antimicrobial proteins/peptides generally kill bacteria by permeabilizing and or disrupting their membranes [61]. The molecular basis for the activity and selectivity of these peptides has been studied in model membranes [61]. Cationic antimicrobial peptides interact preferentially with acidic lipids that are chiefly abundant and found in bacteria [38]. However, the basic proteins manifest strong amphiphilic properties on their molecular surface. For example, apolar tips of loops I-III form a hydrophobic zone flanked by a positively-charged Lys and Arg residues. Another membrane model study demonstrates that the hydrophobic bottom represents a principal membrane-binding motif for protein [62]. In addition, the structural defects in lipid bilayers induced by protein binding to the membrane can lead to the formation of pores [63]. The explanations above provide a basis for the major differences in cell specificity by various antimicrobial peptides/proteins. Interaction of peptides with bilayers alters the organization of the membranes and makes them more permeable to ions [64]. Overall, it is not only the nature of a protein and peptide, but also various characteristics of the cell membrane and metabolic state of the target cell, that ultimately determine the mechanism of antimicrobial activity.
In conclusion, we report for the first time a novel bactericidal protein, VipTx-II, from the crude venom of a Indian Russell’s viper. These protein possessed powerful antibacterial properties against Gram-positive and Gram-negative bacteria, in which VipTx-II was more effective against S. aureus and B. pseudomallei (KHW) than VipTx-I. Thus, the studies provide novel insights into the molecular mechanisms of action involving membrane damage and pore formation induced by viper VipTx-II. Furthermore, VipTx-II is not cytotoxic on human macrophages (THP-1) in comparison to VipTx-I. Further studies should help to completely solve the structure of these proteins that can be very useful to elucidate their detailed mechanisms of action in vivo. Thus, the observed antimicrobial activities of viper venom proteins/derived peptides could be further useful for developing potential antimicrobial candidates against drug-resistant Gram-negative and Gram positive bacteria.
Conflict of interest
The authors confirm that this article content has no conflicts of interest.
Author contributions
Conceived, designed, performed, analyzed data and wrote the manuscript: RPS. Provide facility and materials: AC, MZ, SAA, APK & GS. Corrected and fine-tuned of manuscript: BGS, HKLL, OLF, EGR.
Acknowledgements
This work was supported by Defence Science Technology Agency (DSTA) grant R-181-000-063-422 (DSO), Singapore. The Deanship of Scientific Research, College of Sciences Research Centre, King Saud University, Kingdom of Saudi also supported this work. This work was also supported by grants from the National Medical Research Council of Singapore [R-713-000-177-511], and by the NCIS Yong Siew Yoon Research Grant through donations from the Yong Loo Lin Trust to APK. APK was also supported by the National Research Foundation Singapore and the Singapore Ministry of Education under its Research Centres of Excellence initiative to Cancer Science Institute of Singapore, National University of Singapore. The author OLF is supported by CAPES, CNPq, FAPDF and FUNDECT.
Footnotes
Supplementary data associated with this article can be found, in the online version, at http://dx.doi.org/10.1016/j.fob.2015.10.004.
A. Supplementary data
Supplementary Fig. S1.
(A) Specific phospholipase A2 (PLA2) enzyme activity was determined for various fractions obtained from Superdex G-75 fractions (gel filtration) and RP-HPLC fractions (Sepharose C18 and C8). Chromatography of D. russellii russellii crude venom, fraction RV5 and RV6 showed more activity than the other fractions. The enzymatic activity of reverse-phase fractions (RV-F1 to RV-F3) were compared with activity of pure proteins (VipTx-I and VipTx-II), VipTx-II showed more activity than the fractions. (B) Proteins yields per fraction as well as pure proteins (VipTx-I and VipTx-II). Symbol denotes: RV-F-Russell’s viper fraction, VipTx-I – viperatoxin-I/II.
References
- 1.Moran G.J., Krishnadasan A., Gorwitz R.J., Fosheim G.E., McDougal L.K., Carey R.B. Methicillin-resistant S. aureus infections among patients in the emergency department. N. Engl. J. Med. 2006;355:666–674. doi: 10.1056/NEJMoa055356. [DOI] [PubMed] [Google Scholar]
- 2.Miller L.G., Perdreau-Remington F., Rieg G., Mehdi S., Perlroth J., Bayer A.S. Necrotizing fasciitis caused by community-associated methicillin-resistant Staphylococcus aureus in Los Angeles. N. Engl. J. Med. 2005;352:1445–1453. doi: 10.1056/NEJMoa042683. [DOI] [PubMed] [Google Scholar]
- 3.White N.J. Melioidosis. Lancet. 2003;361:1715–1722. doi: 10.1016/s0140-6736(03)13374-0. [DOI] [PubMed] [Google Scholar]
- 4.Subbalaxmi M.V., Chandra N., Rao M.N., Vemu L., Raju Y.S. Burkholderia pseudomallei: an uncommon cause of bacteraemic pneumoniA in A diabetic. Indian J. Chest Dis. Allied Sci. 2011;53:185–187. [PubMed] [Google Scholar]
- 5.Sawasdidoln C., Taweechaisupapong S., Sermswan R.W., Tattawasart U., Tungpradabkul S., Wongratanacheewin S. Growing BurholderiA pseudomallei in biofilm stimulating conditions significantly induces antimicrobial resistance. PLoS One. 2010;5:e9196. doi: 10.1371/journal.pone.0009196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Mukhopadhyay C., Chawla K., Krishna S., Nagalakshmi N., Rao S.P., Bairy I. Emergence of Burkholderia pseudomallei and pandrug-resistant non-fermenters from southern Karnataka. India. Trans. R. Soc. Trop. Med. Hyg. 2008;102:S12–S17. doi: 10.1016/S0035-9203(08)70005-1. [DOI] [PubMed] [Google Scholar]
- 7.Jenney A.W., Lum G., Fisher D.A., Currie B.J. Antibiotic susceptibility of Burkholderia pseudomallei from tropical northern AustraliA and implications for therapy of melioidosis. Int. J. Antimicrob. Agents. 2001;17:109–113. doi: 10.1016/s0924-8579(00)00334-4. [DOI] [PubMed] [Google Scholar]
- 8.Dawson R.M., Fox M.A., Atkins H.S., Liu C.Q. Potent antimicrobial peptides with selectivity for Bacillus anthracis over human erythrocytes. Int. J. Aantimicrob. Agents. 2011;38:237–242. doi: 10.1016/j.ijantimicag.2011.05.006. [DOI] [PubMed] [Google Scholar]
- 9.Koh D.C., Armugam A., Jeyaseelan K. Snake venom components and their applications in biomedicine. Cell. Mol. Life Sci. 2006;63:3030–3041. doi: 10.1007/s00018-006-6315-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sajevic T., Leonardi A., Križaj I. Haemostatically active proteins in snake venoms. Toxicon. 2011;57:627–645. doi: 10.1016/j.toxicon.2011.01.006. [DOI] [PubMed] [Google Scholar]
- 11.Ferreira B.L., Santos D.O., Andre Santos L., Rodrigues C.R., de Freitas C.C., Cabral L.M. Comparative analysis of viperidae venoms antibacterial profile: A short communication for proteomics. Evid. Based Complement. Alternat. Med. 2011:1–4. doi: 10.1093/ecam/nen052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.de Lima D., Abreu P.A., de Freitas C.C., Santos D.O., Borges R.O., dos Santos T.C. Snake venom: any clue for antibiotics and CAM? Evid. Based Complement. Alternat. Med. 2005;2:39–47. doi: 10.1093/ecam/neh063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Oguiura N., Boni-Mitake M., Affonso R., Zhang G. In vitro antibacterial and hemolytic activities of crotamine, A small basic myotoxin from rattlesnake Crotalus durissus. J. Antibiot. (Tokyo) 2011;64:327–331. doi: 10.1038/ja.2011.10. [DOI] [PubMed] [Google Scholar]
- 14.Zhao H., Kinnunen P.K. Modulation of the activity of secretory phospholipase A2 by antimicrobial peptides. Antimicrob. Agents Chemother. 2003;47:965–971. doi: 10.1128/AAC.47.3.965-971.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Valentin E., Lambeau G. What can venom phospholipase A2 tell us about the functional diversity of mammalian secreted phospholipases A2? Biochimie. 2000;82:815–831. doi: 10.1016/s0300-9084(00)01168-8. [DOI] [PubMed] [Google Scholar]
- 16.Wu Y., Raymond B., Goossens P.L., Njamkepo E., Guiso N., Paya M. Type-IIA secreted phospholipase A2 is an endogenous antibiotic-like protein of the host. Biochimie. 2010;92:583–587. doi: 10.1016/j.biochi.2010.01.024. [DOI] [PubMed] [Google Scholar]
- 17.Vargas L.J., Londoño M., Quintana J.C., Rua C., Segura C., Lomonte B. An acidic phospholipase A2 with antibacterial activity from Porthidium nasutum snake venom. Comp. Biochem. Physiol. B. Biochem. Mol. Biol. 2012;161:341–347. doi: 10.1016/j.cbpb.2011.12.010. [DOI] [PubMed] [Google Scholar]
- 18.Gutiérrez J.M., Lomonte B. Phospholipase A2 myotoxins from Bothrops snake venoms. In: Kini R.M., editor. Venom Phospholipase A2 Enzymes: Structure, Function, and Mechanism. John Wiley & Sons; Chichester, England: 1997. pp. 321–352. [Google Scholar]
- 19.Lomonte B., Angulo Y., Calderón L. An overview of lysine-49 phospholipase A2 myotoxins from crotalid snake venoms and their structural determinants of myotoxic action. Toxicon. 2003;42:885–901. doi: 10.1016/j.toxicon.2003.11.008. [DOI] [PubMed] [Google Scholar]
- 20.Páramo L., Lomonte B., Pizarro-Cerdá J., Bengoechea J.A., Gorvel J.P., Moreno E. Bactericidal activity of Lys49 and Asp49 myotoxic phospholipase A2 from Bothrops asper snake venom–synthetic Lys49 myotoxin II-(115–129)-peptide identifies its bactericidal region. Eur. J. Biochem. 1998;253:452–461. doi: 10.1046/j.1432-1327.1998.2530452.x. [DOI] [PubMed] [Google Scholar]
- 21.Soares A.M., Giglio J.R. Chemical modifications of phospholipases A2 from snake venoms: effects on catalytic and pharmacological properties. Toxicon. 2003;42:855–868. doi: 10.1016/j.toxicon.2003.11.004. [DOI] [PubMed] [Google Scholar]
- 22.Santamaría C., Larios S., Angulo Y., Pizarro-Cerda J., Gorvel J.P., Moreno E. Antimicrobial activity of myotoxin phospholipases A2 from crotalid snake venoms and synthetic peptide variants derived from their C-terminal region. Toxicon. 2005;45:807–815. doi: 10.1016/j.toxicon.2004.09.012. [DOI] [PubMed] [Google Scholar]
- 23.Villarrubia V.G., Costa L.A., Díez R.A. Secreted phospholipases A2 (sPLA2): friends or foes? Are they actors in antibacterial and anti HIV resistance? Med. Clin. (Barc) 2004;123:749–757. doi: 10.1016/s0025-7753(04)74656-4. [DOI] [PubMed] [Google Scholar]
- 24.Murillo L.A., Lan C.Y., Agabian N.M., Larios S., Lomonte B. Fungicidal activity of A phospholipaseA2 – derived synthetic peptide variant against CandidA albicans. Rev. Esp. Quimioter. 2007;20:330–333. [PubMed] [Google Scholar]
- 25.Araya C., Lomonte B. Antitumor effects of cationic synthetic peptides derived from Lys49 phospholipase A2 homologues of snake venoms. Cell Biol. 2007;3:263–268. doi: 10.1016/j.cellbi.2006.11.007. [DOI] [PubMed] [Google Scholar]
- 26.Costa T.R., Menaldo D.L., Oliveira C.Z., Santos-Filho N.A., Teixeira S.S., Nomizo A. Myotoxic phospholipases A2 isolated from Bothrops brazili snake venom and synthetic peptides derived from their C-terminal region: cytotoxic effect on microorganism and tumor cells. Peptides. 2008;29:1645–1656. doi: 10.1016/j.peptides.2008.05.021. [DOI] [PubMed] [Google Scholar]
- 27.Fonseca-Maldonado R., Ferreira T.L., Ward R.J. The bactericidal effect of human secreted group IID phospholipase A2 results from both hydrolytic and non-hydrolytic activities. Biochimie. 2012;94:1437–1440. doi: 10.1016/j.biochi.2012.03.020. [DOI] [PubMed] [Google Scholar]
- 28.Kang T.S., Georgieva D., Genov N., Murakami M.T., Sinha M., Kumar R.P. Enzymatic toxins from snake venom: structural characterization and mechanism of catalysis. FEBS J. 2011;278:4544–4576. doi: 10.1111/j.1742-4658.2011.08115.x. [DOI] [PubMed] [Google Scholar]
- 29.Nakayama D., Ben Ammar Y., Miyata T., Takeda S. Structural basis of coagulation factor V recognition for cleavage by RVV-V. FEBS Lett. 2011;585:3020–3025. doi: 10.1016/j.febslet.2011.08.022. [DOI] [PubMed] [Google Scholar]
- 30.Mandal S., Bhattacharyya D. Two l-amino acid oxidase isoenzymes from Russell’s viper (Daboia russelli russelli) venom with different mechanisms of inhibition by substrate analogs. FEBS J. 2008;275:2078–2095. doi: 10.1111/j.1742-4658.2008.06362.x. [DOI] [PubMed] [Google Scholar]
- 31.Suntravat M., Yusuksawad M., Sereemaspun A., Perez J.C., Nuchprayoon I. Effect of purified Russell’s viper venom-factor X activator (RVV-X) on renal hemodynamics, renal functions, and coagulopathy in rats. Toxicon. 2011;58:230–238. doi: 10.1016/j.toxicon.2011.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Segers K., Rosing J., Nicolaes G.A. Structural models of the snake venom factor V activators from Daboia russelli and Daboia lebetina. Proteins. 2006;64:968–984. doi: 10.1002/prot.21051. [DOI] [PubMed] [Google Scholar]
- 33.Maity G., Mandal S., Bhattacharjee P., Bhattacharyya D. Thermal detoxification of the venom from Daboia russelli russelli of Eastern IndiA with restoration of fibrinolytic activity. Toxicon. 2011;57:747–754. doi: 10.1016/j.toxicon.2011.02.008. [DOI] [PubMed] [Google Scholar]
- 34.Shevchenko A., Sunyaev S., Loboda A., Shevchenko A., Bork P., Ens W. Charting the proteomes of organisms with unsequenced genomes by MALDI-quadrupole time-of-flight mass spectrometry and BLAST homology searching. Anal. Chem. 2001;73:1917–1926. doi: 10.1021/ac0013709. [DOI] [PubMed] [Google Scholar]
- 35.Bradford M.M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 1976;72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- 36.Laemmli U.K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 37.Reynolds L.J., Hughes L.L., Dennis E.A. Analysis of human synovial fluid phospholipase A2 on short chain phosphatidylcholine-mixed micelles: development of A spectrophotometric assay suitable for A microtiterplate reader. Anal. Biochem. 1992;204:190–197. doi: 10.1016/0003-2697(92)90160-9. [DOI] [PubMed] [Google Scholar]
- 38.Perumal Samy R., Gopalakrishnakone P., Chow V.T.K., Ho B. Viper metalloproteinase (Agkistrodon halys pallas) with antimicrobial activity against multi-drug resistant human pathogens. J. Cell. Physiol. 2008;216:54–68. doi: 10.1002/jcp.21373. [DOI] [PubMed] [Google Scholar]
- 39.Bauer A.W., Kirby W.M., Sherris J.C. Antibiotic susceptibility testing by A standardized single disk method. Am. J. Clin. Pathol. 1966;45:493–496. [PubMed] [Google Scholar]
- 40.Lomonte B., Moreno E., Tarkowski A., Hanson L.A., Maccarana M. Neutralizing interaction between heparins and myotoxin II, A lysine 49 phospholipase A2 from Bothrops asper snake venom. Identification of A heparin-binding and cytolytic toxin region by the use of synthetic peptides and molecular modeling. J. Biol. Chem. 1994;269:29867–29873. [PubMed] [Google Scholar]
- 41.Motizuki M., Itoh T., Satoh T., Yokota S., Yamada M., Shimamura S. Lipid-binding and antimicrobial properties of synthetic peptides of bovine apolipoprotein A-II. Biochem. J. 1999;342:215–221. [PMC free article] [PubMed] [Google Scholar]
- 42.Matsuzaki K. Magainins as paradigm for the mode of action of pore forming polypeptides. Biochim. Biophys. Acta. 1998;1376:391–400. doi: 10.1016/s0304-4157(98)00014-8. [DOI] [PubMed] [Google Scholar]
- 43.Harris C.R., Thorarensen A. Advances in the discovery of novel antibacterial agents during the year 2002. Curr. Med. Chem. 2004;11:2213–2243. doi: 10.2174/0929867043364658. [DOI] [PubMed] [Google Scholar]
- 44.Rodrigues V.M., Marcussi S., Cambraia R.S., de Araujo A.L., Malta-Neto N.R., Hamaguchi A. Bactericidal and neurotoxic activities of two myotoxic phospholipases A2 from Bothrops neuwiedi pauloensis snake venom. Toxicon. 2004;44(3):305–314. doi: 10.1016/j.toxicon.2004.06.008. [DOI] [PubMed] [Google Scholar]
- 45.Nunez V., Arce V., Gutierrez J.M., Lomonte B. Structural and functional characterization of myotoxin I, A Lys49 phospholipase A2 homologue from the venom of the snake Bothrops atrox. Toxicon. 2004;44(1):91–101. doi: 10.1016/j.toxicon.2004.04.013. [DOI] [PubMed] [Google Scholar]
- 46.Singh G., Jasti J., Saravanan K., Sharma S., Kaur P., Srinivasan A. Crystal structure of the complex formed between A group I phospholipase A2 and A naturally occurring fatty acid at 2.7 Å resolution. Protein Sci. 2005;14:395–400. doi: 10.1110/ps.041115505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Chandra V., Jasti J., Kaur P., Dey S., Perbandt M., Srinivasan A. Crystal structure of A complex formed between a snake venom phospholipase A2 and A potent peptide inhibitor Phe–Leu–Ser–Tyr–Lys at 1.8 Å resolution. J. Biol. Chem. 2002;277:41079–41085. doi: 10.1074/jbc.M206130200. [DOI] [PubMed] [Google Scholar]
- 48.Sachidananda M.K., Murari S.K., Channe GowdA D. Characterization of an antibacterial peptide from Indian CobrA (NajA naja) J. Venom Anim. Toxins Trop. Dis. 2007;13:446–461. [Google Scholar]
- 49.Perumal Samy R., Pachiappan A., Gopalakrishnakone P., Thwin M.M., Hian Y.E., Chow V.T. In vitro antimicrobial activity of natural toxins and animal venoms tested against Burkholderia pseudomallei. BMC Infect. Dis. 2006;6:100. doi: 10.1186/1471-2334-6-100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Sudarshan S., Dhananjaya B.L. Antibacterial potential of A basic phospholipase A2 (VRV-PL-V) of Daboia russellii pulchella (Vipera russellii) venom. Biochemistry (Mosc) 2014;79(11):1237–1244. doi: 10.1134/S000629791411011X. [DOI] [PubMed] [Google Scholar]
- 51.Sudarshan S., Dhananjaya B.L. Antibacterial potential of A basic phospholipase A2 (VRV-PL-VIIIa) from Daboia russelii pulchella (Russell’s viper) venom. J. Venom Anim. Toxins Incl. Trop. Dis. 2015;21:17. doi: 10.1186/s40409-015-0014-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Chen L., Kao P., Fu Y., Lin S., Chang L. Membrane-damaging activity of Taiwan cobrA cardiotoxin 3 is responsible for its bactericidal activity. Toxicon. 2011;58(1):46–53. doi: 10.1016/j.toxicon.2011.04.021. [DOI] [PubMed] [Google Scholar]
- 53.Anderluh G., Lakey J.H. Disparate proteins use similar architectures to damage membranes. Trends Biochem. Sci. 2008;33:482–490. doi: 10.1016/j.tibs.2008.07.004. [DOI] [PubMed] [Google Scholar]
- 54.Brogden K.A. Antimicrobial peptides: pore formers or metabolic inhibitors in bacteria? Nat. Rev. Microbiol. 2005;238:239–250. doi: 10.1038/nrmicro1098. [DOI] [PubMed] [Google Scholar]
- 55.Yang S.T., Shin S.Y., Hahm K.S., Kim J.I. Design of perfectly symmetric Trp-rich peptides with potent and broad-spectrum antimicrobial activities. Int. J. Antimicrob. Agents. 2006;27:325–330. doi: 10.1016/j.ijantimicag.2005.11.014. [DOI] [PubMed] [Google Scholar]
- 56.Bechinger B. Structure and functions of channel-forming peptides: magainins, cecropins, melittin and alamethicin. J. Membr. Biol. 1997;156:197–211. doi: 10.1007/s002329900201. [DOI] [PubMed] [Google Scholar]
- 57.Suntravat M., Yusuksawad M., Sereemaspun A., Perez J.C., Nuchprayoon I. Effect of purified Russell’s viper venom-factor X activator (RVV-X) on renal hemodynamics, renal functions, and coagulopathy in rats. Toxicon. 2011;58:230–238. doi: 10.1016/j.toxicon.2011.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Oliveira D.G., Toyama M.H., Novello J.C., Beriam L.O., Marangoni S. Structural and functional characterization of basic PLA2 isolated from Crotalus durissus terrificus venom. J. Protein Chem. 2002;21:161–168. doi: 10.1023/a:1015320616206. [DOI] [PubMed] [Google Scholar]
- 59.Perumal Samy R., Gopalakrishnakone P., Thwin M.M., Chow T.K., Bow H., Yap E.H. Antibacterial activity of snake, scorpion and bee venoms: A comparison with purified venom phospholipase A2 enzymes. J. Appl. Microbiol. 2007;102:650–659. doi: 10.1111/j.1365-2672.2006.03161.x. [DOI] [PubMed] [Google Scholar]
- 60.Zhao H., Rinaldi A.C., Rufo A., Bozzi A., Kinnunen P.K.J., Di Giulio A. Structural and charge requirements for antimicrobial peptide insertion into biological and model membranes. In: Lazarovici P., Menestrina G., DallA Serra M., editors. Pore-forming peptides and protein toxins. Harwood Academic Publishers; Reading, UK: 2002. [Google Scholar]
- 61.Maget-Dana R., Lelievre D., Brack A. Surface active properties of amphiphilic sequential isopeptides: comparison between alpha-helical and beta-sheet conformations. Biopolymers. 1999;49:415–423. doi: 10.1002/(SICI)1097-0282(19990415)49:5<415::AID-BIP7>3.0.CO;2-J. [DOI] [PubMed] [Google Scholar]
- 62.Dubovskii P.V., Dementieva D.V., Bocharov E.V., Utkin Y.N., Arseniev A.S. Membrane binding motif of the P-type cardiotoxin. J. Mol. Biol. 2001;305:137–149. doi: 10.1006/jmbi.2000.4283. [DOI] [PubMed] [Google Scholar]
- 63.Konshina A.G., Boldyrev I.A., Utkin Y.N., Omelkov A.V., Efremov R.G. Snake cytotoxins bind to membranes viA interactions with phosphatidylserine head groups of lipids. PLoS One. 2011;6:e19064. doi: 10.1371/journal.pone.0019064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Kourie J.I., Shorthouse A.A. Properties of cytotoxic peptide-formed ion channels. Am. J. Physiol. Cell Physiol. 2000;278:C1063–C1087. doi: 10.1152/ajpcell.2000.278.6.C1063. [DOI] [PubMed] [Google Scholar]