Abstract
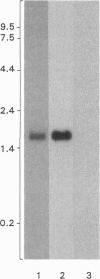
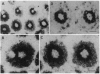
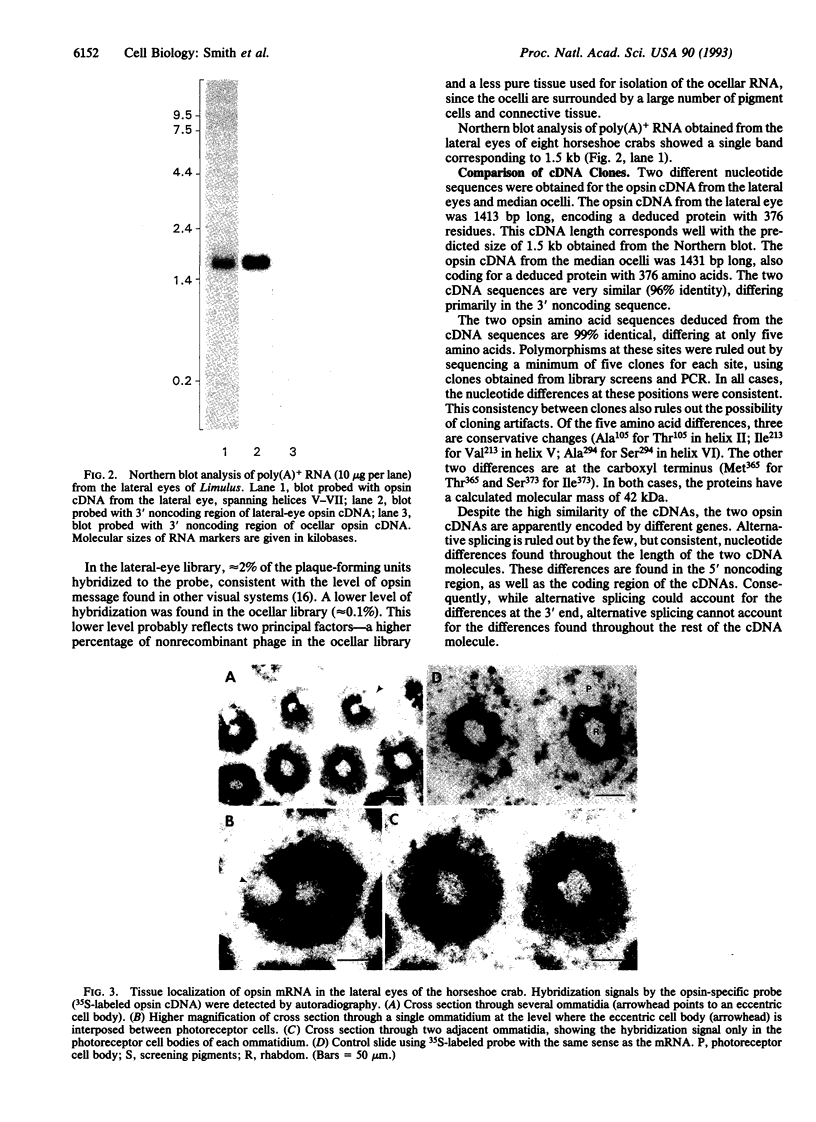
cDNA clones encoding opsins from the lateral eyes and median ocelli of the horseshoe crab, Limulus polyphemus, were isolated from cDNA libraries. The opsin cDNAs obtained from the lateral eye and ocellar libraries code for deduced proteins with 376 amino acids. The two cDNAs are 96% identical at the nucleic acid level, differing primarily at the 3' untranslated region, and are apparently the products of two separate genes. The deduced opsin proteins are 99% identical to each other, differing at only 5 amino acids. The opsins encoded by these cDNAs are most likely the protein moiety of the visible-wavelength rhodopsins in this animal. In the lateral eye, expression of the opsin gene is restricted to the photoreceptor cells of the ommatidia. Comparisons with opsins of other species show that the Limulus opsin proteins are most similar (53% identity) to the opsin from the R1-6 photoreceptors of flies.
Full text
PDF



Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Altschul S. F., Gish W., Miller W., Myers E. W., Lipman D. J. Basic local alignment search tool. J Mol Biol. 1990 Oct 5;215(3):403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- Archer S. N., Lythgoe J. N., Hall L. Rod opsin cDNA sequence from the sand goby (Pomatoschistus minutus) compared with those of other vertebrates. Proc Biol Sci. 1992 Apr 22;248(1321):19–25. doi: 10.1098/rspb.1992.0037. [DOI] [PubMed] [Google Scholar]
- Behrens M. E., Wulff V. J. Light-initiated responses of retinula and eccentric cells in the Limulus lateral eye. J Gen Physiol. 1965 Jul;48(6):1081–1093. doi: 10.1085/jgp.48.6.1081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bownds D. Site of attachment of retinal in rhodopsin. Nature. 1967 Dec 23;216(5121):1178–1181. doi: 10.1038/2161178a0. [DOI] [PubMed] [Google Scholar]
- Brown J. E., Rubin L. J., Ghalayini A. J., Tarver A. P., Irvine R. F., Berridge M. J., Anderson R. E. myo-Inositol polyphosphate may be a messenger for visual excitation in Limulus photoreceptors. Nature. 1984 Sep 13;311(5982):160–163. doi: 10.1038/311160a0. [DOI] [PubMed] [Google Scholar]
- Carulli J. P., Hartl D. L. Variable rates of evolution among Drosophila opsin genes. Genetics. 1992 Sep;132(1):193–204. doi: 10.1093/genetics/132.1.193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Church G. M., Gilbert W. Genomic sequencing. Proc Natl Acad Sci U S A. 1984 Apr;81(7):1991–1995. doi: 10.1073/pnas.81.7.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cowman A. F., Zuker C. S., Rubin G. M. An opsin gene expressed in only one photoreceptor cell type of the Drosophila eye. Cell. 1986 Mar 14;44(5):705–710. doi: 10.1016/0092-8674(86)90836-6. [DOI] [PubMed] [Google Scholar]
- Feinberg A. P., Vogelstein B. A technique for radiolabeling DNA restriction endonuclease fragments to high specific activity. Anal Biochem. 1983 Jul 1;132(1):6–13. doi: 10.1016/0003-2697(83)90418-9. [DOI] [PubMed] [Google Scholar]
- Franke R. R., König B., Sakmar T. P., Khorana H. G., Hofmann K. P. Rhodopsin mutants that bind but fail to activate transducin. Science. 1990 Oct 5;250(4977):123–125. doi: 10.1126/science.2218504. [DOI] [PubMed] [Google Scholar]
- Franke R. R., Sakmar T. P., Graham R. M., Khorana H. G. Structure and function in rhodopsin. Studies of the interaction between the rhodopsin cytoplasmic domain and transducin. J Biol Chem. 1992 Jul 25;267(21):14767–14774. [PubMed] [Google Scholar]
- HUBBARD R., WALD G. Cis-trans isomers of vitamin A and retinene in the rhodopsin system. J Gen Physiol. 1952 Nov;36(2):269–315. doi: 10.1085/jgp.36.2.269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- HUBBARD R., WALD G. Visual pigment of the horseshoe crab, Limulus polyphemus. Nature. 1960 Apr 16;186:212–215. doi: 10.1038/186212b0. [DOI] [PubMed] [Google Scholar]
- Hall M. D., Hoon M. A., Ryba N. J., Pottinger J. D., Keen J. N., Saibil H. R., Findlay J. B. Molecular cloning and primary structure of squid (Loligo forbesi) rhodopsin, a phospholipase C-directed G-protein-linked receptor. Biochem J. 1991 Feb 15;274(Pt 1):35–40. doi: 10.1042/bj2740035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Higgins D. G., Sharp P. M. CLUSTAL: a package for performing multiple sequence alignment on a microcomputer. Gene. 1988 Dec 15;73(1):237–244. doi: 10.1016/0378-1119(88)90330-7. [DOI] [PubMed] [Google Scholar]
- Higgins D. G., Sharp P. M. Fast and sensitive multiple sequence alignments on a microcomputer. Comput Appl Biosci. 1989 Apr;5(2):151–153. doi: 10.1093/bioinformatics/5.2.151. [DOI] [PubMed] [Google Scholar]
- Huber A., Smith D. P., Zuker C. S., Paulsen R. Opsin of Calliphora peripheral photoreceptors R1-6. Homology with Drosophila Rh1 and posttranslational processing. J Biol Chem. 1990 Oct 15;265(29):17906–17910. [PubMed] [Google Scholar]
- Karnik S. S., Sakmar T. P., Chen H. B., Khorana H. G. Cysteine residues 110 and 187 are essential for the formation of correct structure in bovine rhodopsin. Proc Natl Acad Sci U S A. 1988 Nov;85(22):8459–8463. doi: 10.1073/pnas.85.22.8459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirkwood A., Weiner D., Lisman J. E. An estimate of the number of G regulator proteins activated per excited rhodopsin in living Limulus ventral photoreceptors. Proc Natl Acad Sci U S A. 1989 May;86(10):3872–3876. doi: 10.1073/pnas.86.10.3872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levine E., Crain E., Robinson P., Lisman J. Nontransducing rhodopsin. J Gen Physiol. 1987 Oct;90(4):575–586. doi: 10.1085/jgp.90.4.575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacDonald R. J., Swift G. H., Przybyla A. E., Chirgwin J. M. Isolation of RNA using guanidinium salts. Methods Enzymol. 1987;152:219–227. doi: 10.1016/0076-6879(87)52023-7. [DOI] [PubMed] [Google Scholar]
- McMaster G. K., Carmichael G. G. Analysis of single- and double-stranded nucleic acids on polyacrylamide and agarose gels by using glyoxal and acridine orange. Proc Natl Acad Sci U S A. 1977 Nov;74(11):4835–4838. doi: 10.1073/pnas.74.11.4835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montell C., Jones K., Zuker C., Rubin G. A second opsin gene expressed in the ultraviolet-sensitive R7 photoreceptor cells of Drosophila melanogaster. J Neurosci. 1987 May;7(5):1558–1566. doi: 10.1523/JNEUROSCI.07-05-01558.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nathans J. Determinants of visual pigment absorbance: identification of the retinylidene Schiff's base counterion in bovine rhodopsin. Biochemistry. 1990 Oct 16;29(41):9746–9752. doi: 10.1021/bi00493a034. [DOI] [PubMed] [Google Scholar]
- Nathans J. The genes for color vision. Sci Am. 1989 Feb;260(2):42–49. doi: 10.1038/scientificamerican0289-42. [DOI] [PubMed] [Google Scholar]
- Neufeld T. P., Carthew R. W., Rubin G. M. Evolution of gene position: chromosomal arrangement and sequence comparison of the Drosophila melanogaster and Drosophila virilis sina and Rh4 genes. Proc Natl Acad Sci U S A. 1991 Nov 15;88(22):10203–10207. doi: 10.1073/pnas.88.22.10203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nolte J., Brown J. E. Ultraviolet-induced sensitivity to visible light in ultraviolet receptors of Limulus. J Gen Physiol. 1972 Feb;59(2):186–200. doi: 10.1085/jgp.59.2.186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Tousa J. E., Baehr W., Martin R. L., Hirsh J., Pak W. L., Applebury M. L. The Drosophila ninaE gene encodes an opsin. Cell. 1985 Apr;40(4):839–850. doi: 10.1016/0092-8674(85)90343-5. [DOI] [PubMed] [Google Scholar]
- Ovchinnikov YuA, Abdulaev N. G., Zolotarev A. S., Artamonov I. D., Bespalov I. A., Dergachev A. E., Tsuda M. Octopus rhodopsin. Amino acid sequence deduced from cDNA. FEBS Lett. 1988 May 9;232(1):69–72. doi: 10.1016/0014-5793(88)80388-0. [DOI] [PubMed] [Google Scholar]
- Richard E. A., Lisman J. E. Rhodopsin inactivation is a modulated process in Limulus photoreceptors. Nature. 1992 Mar 26;356(6367):336–338. doi: 10.1038/356336a0. [DOI] [PubMed] [Google Scholar]
- Sakmar T. P., Franke R. R., Khorana H. G. Glutamic acid-113 serves as the retinylidene Schiff base counterion in bovine rhodopsin. Proc Natl Acad Sci U S A. 1989 Nov;86(21):8309–8313. doi: 10.1073/pnas.86.21.8309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanger F., Nicklen S., Coulson A. R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci U S A. 1977 Dec;74(12):5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilden U., Kühn H. Light-dependent phosphorylation of rhodopsin: number of phosphorylation sites. Biochemistry. 1982 Jun 8;21(12):3014–3022. doi: 10.1021/bi00541a032. [DOI] [PubMed] [Google Scholar]
- Zuker C. S., Cowman A. F., Rubin G. M. Isolation and structure of a rhodopsin gene from D. melanogaster. Cell. 1985 Apr;40(4):851–858. doi: 10.1016/0092-8674(85)90344-7. [DOI] [PubMed] [Google Scholar]
- Zuker C. S., Montell C., Jones K., Laverty T., Rubin G. M. A rhodopsin gene expressed in photoreceptor cell R7 of the Drosophila eye: homologies with other signal-transducing molecules. J Neurosci. 1987 May;7(5):1550–1557. doi: 10.1523/JNEUROSCI.07-05-01550.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]