Abstract
Here we have developed a hypoxia response element driven imaging strategy that combined the hypoxia-driven expression of two optical reporters with different half-lives to detect temporal changes in hypoxia and hypoxia inducible factor (HIF) activity. For this purpose, human prostate cancer PC3 cells were transfected with the luciferase gene fused with an oxygen-dependent degradation domain (ODD-luc) and a variant of the enhanced green fluorescent protein (EGFP). Both ODD-luciferase and EGFP were under the promotion of a poly-hypoxia-response element sequence (5xHRE). The cells constitutively expressed tdTomato red fluorescent protein. For validating the imaging strategy, cells were incubated under hypoxia (1% O2) for 48 hours and then reoxygenated. The luciferase activity of PC3-HRE-EGFP/HRE-ODD-luc/tdtomato cells detected by bioluminescent imaging rapidly decreased after reoxygenation, whereas EGFP levels in these cells remained stable for several hours. After in vitro validation, PC3-HRE-EGFP/HRE-ODD-luc/tdtomato tumors were implanted subcutaneously and orthotopically in nude male mice and imaged in vivo and ex vivo using optical imaging in proof-of-principle studies to demonstrate differences in optical patterns between EGFP expression and bioluminescence. This novel "timer" imaging strategy of combining the short-lived ODD-luciferase and the long-lived EGFP can provide a time frame of HRE activation in PC3 prostate cancer cells and will be useful to understand the temporal changes in hypoxia and HIF activity during cancer progression and following treatments including HIF targeting strategies.
Abbreviations: BLI, bioluminescence imaging; CHX, cycloheximide; EGFP, enhanced green fluorescent protein; HIF, hypoxia-inducible factor; HRE, hypoxia-response element; Luc, luciferase; ODD, oxygen-dependent degradation domain; RFP, red fluorescent protein
Introduction
Hypoxia is frequently found in tumors and contributes to aggressiveness, resistance to treatment, and metastatic dissemination [1], [2], [3], [4]. The adaptive response of cancer cells to hypoxia is mediated through the stabilization of hypoxia inducible factors (HIFs) that increases the transcription of several genes mediating this response by binding to hypoxia response elements (HREs) in the promoter region of these genes [5]. HIF is a heterodimeric basic helix-loop-helix PAS (Per-ARNT-Sim) domain containing transcription factor that consists of one of three oxygen-regulated α-subunits, HIF-1α, HIF-2α, and HIF-3α, and a constitutively expressed β-subunit (HIF-β/ARNT) [6], [7]. The α-subunits are constitutively transcribed and translated, but are regulated at the protein level by oxygen-dependent hydroxylation of specific prolyl residues and degraded because of the presence of an oxygen-dependent degradation domain (ODD) [8]. The stabilization of HIF under hypoxia and its binding to the HRE provide a strategy to detect hypoxia by linking the HRE to the expression of optical reporter genes. Because increased HIF expression and stabilization can also occur under normoxia due to factors such as epi-/genetic HIF upregulation, PI3K-mTor pathway activation, and reactive oxygen species [9], [10], such a strategy can also be used to detect HIF activity [11]. Tumor hypoxia evolves because of poor vascularization [9], [10] but can resolve following increased angiogenesis and reoxygenation, making it important to track the dynamics of hypoxia and HIF activity in tumors.
Reporter genes that allow the visualization of molecular events in cells are widely used to track cells in vitro and in vivo when constitutively expressed [12], [13], [14]. Alternatively, these tools allow monitoring the activity of transcription factors such as HIF when specific promoters drive the expression of the reporter gene [15]. Among reporter genes, fluorescent proteins such as red and green fluorescent protein (RFP and GFP) are commonly used to study gene expression, protein trafficking, and cell localization in tissues [16]. In bioluminescence imaging (BLI), the firefly luciferase is the most commonly used reporter gene. In the presence of ATP and Mg++, luciferase catalyzes the oxidation of D-luciferin to oxyluciferin. This reaction also generates light (560 nm) [15], [17]. Although the administration of D-luciferin is required, BLI is a sensitive imaging modality that allows monitoring cell migration, proliferation, and/or activation of signaling pathways in vivo [15], [17].
The use of a destabilized variant of the enhanced green fluorescent protein (pd2EGFP) to characterize the relationship between hypoxia and vascularization in a PC3 prostate cancer xenograft model has been previously described [18], [19], [20]. The luciferase protein fused with ODD, under the promotion of five copies of HRE (5xHRE-ODD-luc), has also been previously used to monitor HIF-1 activity in xenograft tumors using BLI [11]. Addition of an ODD sequence strongly reduced the stability of the luciferase under normoxia, allowing dynamic monitoring of HIF-1 activity in vivo [11].
Here we have developed an imaging strategy based on the simultaneous HRE-driven expression of a long-lived EGFP and a short-lived luciferase fused to ODD in prostate cancer PC3 cells. Both reporter proteins were placed under the control of a 5xHRE sequence (schematic shown in Figure 1, A and B). PC3 cells also constitutively expressed tdTomato RFP to allow cell tracking (schematic shown in Figure 1B). This "timer" strategy can provide information on the temporal evolution of HIF activity and hypoxia in tumors. Because orthotopic tumor models better mimic clinical tumors compared with subcutaneous xenografts [21], [22], [23], we also performed in vivo and ex vivo imaging of the dual HIF reporter system (EGFP/ODD-luciferase) in PC3 tumors implanted subcutaneously and orthotopically in the prostate and observed patterns of EGFP and BLI colocalization as well as regions with low overlap in subcutaneous and orthotopic tumors and metastatic nodules.
Figure 1.
Description of the "timer" strategy for HIF imaging in PC3 prostate cancer cells. (A) Under normoxia, hydroxylation of the ODD of HIF-α triggers the degradation of this subunit. Under hypoxia, HIF-α stabilizes, and the dimer HIF-α/HIF-β binds to HRE sequences in the 5xHRE-ODD-luc and 5xHRE-EGFP constructs, resulting in the formation of long-lived EGFP and short-lived ODD-luciferase proteins. (B) Upon reoxygenation, the ODD sequence causes a rapid degradation of the ODD-luciferase fusion protein, whereas EGFP protein levels remain stable for longer periods. The RFP tdTomato is used as a constitutive reporter.
Material and Methods
Generation of the 5xHRE-ODD-luc Construct with a Puromycin Selection Marker
To construct the plasmid encoding the luciferase gene fused to ODD under the control of a poly-HRE sequence (5xHRE) with a puromycin resistance cassette, the 5xHRE-ODD-luc (firefly luciferase fused with ODD) sequence from a 5HREp-ODD-luc plasmid (kindly provided by Dr. H. Harada) was digested with restriction enzymes KpnI and XbaI (NEB, Ipswich, MA) and subcloned between KpnI and EcoRI of a pGL4.21 [luc2P/Puro] (Promega, Madison, WI) vector.
Generation of the PC3-HRE-EGFP/HRE-ODD-luc/tdTomato Cell Line
Human prostate cancer PC3 cells and PC3 cells expressing the enhanced green fluorescent protein variant pd2EGFP under the promotion of a 5xHRE sequence (PC3-HRE-EGFP) were transfected with the 5xHRE-ODD-luc construct (PC3-HRE-ODD-luc and PC3-HRE-EGFP/HRE-ODD-luc) using jetPRIME reagent (Polyplus transfection Inc., New York, NY) following the manufacturer’s protocol. Transfected cells were next selected using 1 μg/ml of puromycin.
PC3-HRE-EGFP/HRE-ODD-luc cells constitutively expressing the bright RFP tdTomato (PC3-HRE-EGFP/HRE-ODD-luc/tdTomato) were generated by lentiviral infection with the pRRL-Δluc-tdTomato plasmid. This plasmid was cotransfected in 293T cells with VSVG and ΔR8.2 plasmids to produce high-titer lentiviral particles.
Cell Culture and Hypoxia Treatment
PC3 cells were grown in RPMI medium with L-glutamine supplemented with 10% (v/v) fetal bovine serum. PC3-HRE-EGFP cells were supplemented with 400 μg/ml of G418, PC3-HRE-ODD-luc cells were supplemented with 1 μg/ml of puromycin, and PC3-HRE-EGFP/HRE-ODD-luc/tdTomato cells were supplemented with both 400 μg/ml of G418 and 1 μg/ml of puromycin.
Hypoxic treatment of cells was performed either by placing the plates in a modular incubator chamber (Billups-Rothenberg, Del Mar, CA), flushed at 2 psi for 3 minutes with a gas mixture of 1% O2/5% CO2, and N2 for the balance, or by treating cells with 200 μM of CoCl2 for 24 hours to chemically mimic hypoxia.
Fluorescence Imaging
For fluorescence quantification experiments, PC3-HRE-EGFP and PC3-HRE-EGFP/HRE-ODD-luc/tdTomato cells were seeded at low density in six-well plates and allowed to grow for 2 weeks. The resulting cell colonies were incubated at 1% O2 for 48 hours and imaged after reoxygenation using a Nikon inverted microscope equipped with a Nikon Coolpix digital camera (Nikon Instruments, Inc., Melville, NY) at 20 × magnification. Exposure times were kept constant during time-lapse imaging experiments. For EGFP imaging, background signal was subtracted using the rolling ball algorithm from ImageJ software (NIH), and brightness was adjusted. All image corrections were kept similar within experiments. For fluorescence quantification, regions of interest were manually drawn for each cell, and fluorescence quantification was performed using the following formula [24]:
Luciferase Activity Measurements
PC3-HRE-ODD-luc and PC3-HRE-EGFP/HRE-ODD-luc/tdTomato cells grown at 80% to 90% confluence in 6-well plates were washed with PBS and lysed in Cell Culture Lysis Reagent (Promega) at various time points after hypoxia treatments. The luciferase activity was measured in cell lysates using Luciferase Assay Systems (Promega) following manufacturer’s instructions. Luciferase activity measurements were performed using a Victor 3V plate reader (Perkin Elmer, Waltham, MA).
Western Blots
Whole cell extracts from 80% to 90% confluent 10-mm dishes were prepared in radioimmune precipitation buffer, fractionated by SDS-PAGE, transferred to a nitrocellulose membrane, and subjected to immunoblot assays using a monoclonal anti-EGFP, mouse antibody (dilution 1:2000, BD Biosciences, San Jose, CA), an anti-HIF-1α antibody (dilution 1:1000, Novus Biologicals, Denver, CO), and a monoclonal anti-actin mouse antibody (dilution 1:50000, Sigma, St. Louis, MO). Horseradish peroxidase–conjugated secondary antibody against either mouse or rabbit IgG was used at 1:2000 dilutions. The signal was visualized using ECL Plus reagents (Thermo Scientific, Rockford, IL). For experiments involving cycloheximide (CHX, Sigma), an inhibitor of protein synthesis, cell extracts from 105 cells were loaded per lane. Otherwise, protein quantification was performed before sample loading. CHX toxicity was assessed using the trypan blue exclusion test.
Animal Studies
All experimental animal protocols were approved by the Institutional Animal Care and Use Committee of the Johns Hopkins University School of Medicine. Animal studies were performed on adult nude male mice inoculated subcutaneously with 2 × 106 million cancer cells. For orthotopic implantation, a 1-mm3 piece of tumor tissue was microsurgically sutured into the prostate gland through a small incision as previously described [20]. Tumor pieces were obtained from a subcutaneous tumor and carefully dissected under a fluorescence microscope to remove hypoxic and necrotic tissue. Subcutaneous and orthotopic tumor volumes were approximately 400 to 500 mm3 at the time of imaging.
Optical Imaging
Optical imaging was performed using a Xenogen IVIS Spectrum system (Caliper Life Sciences, Hopkinton, MA). EGFP was detected using an excitation light of 500 nm and an emission filter set at 540 nm. tdTomato was detected using an excitation light of 570 nm and an emission filter set at 620 nm. For in vitro bioluminescence experiments, D-luciferin (150 μg/ml) (Promega) was added to samples before imaging. For in vivo BLI experiments, D-luciferin dissolved in PBS was injected intraperitoneally (3 mg/mouse) 15 minutes before imaging. Exposure times were kept constant within experiments. End point fluorescence and bioluminescence imaging of fresh 2-mm–thick tumor slices prepared with a tissue slicer was performed with the Xenogen system or using a 1 × objective on a fluorescence microscope (Nikon Ltd., Melville, NY). Fluorescence and bioluminescence quantification was performed on manually drawn regions of interest using the Living Image software (Perkin Elmer). The average radiant efficiency [(photons/s per cm2 per sr)/(μW/cm2)] was determined for fluorescence images, and the average radiance (photons/s per cm2 per sr) was measured for bioluminescence images.
Statistical Analyses
All results are represented as means ± SEM. Comparisons between groups were performed using Student’s t test when two groups were compared. One-way analysis of variance followed by Tukey's multiple comparison tests was used when more than two groups were compared. P < .05 was considered to be statistically significant. Statistical analyses, and EGFP and ODD-luciferase protein half-lives were determined using the GraphPad Prism software (La Jolla, CA).
Results
EGFP Is More Stable than ODD-luc in PC3-HRE-EGFP/HRE-ODD-luc/tdTomato Cells Following Reoxygenation
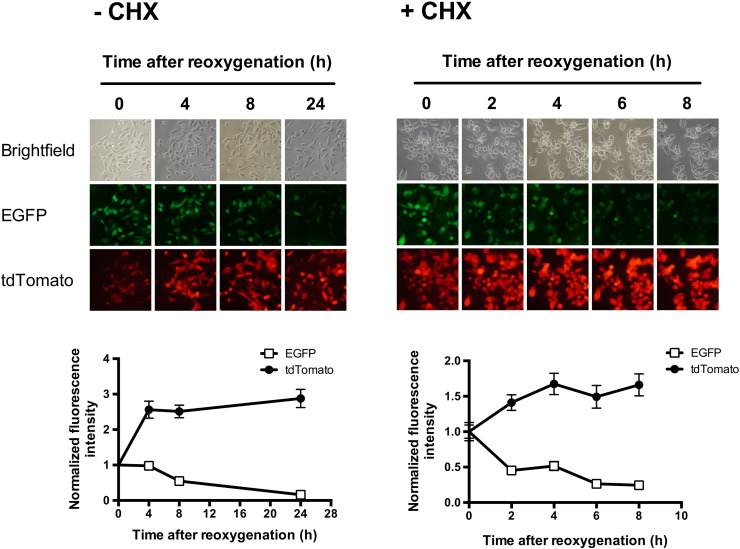
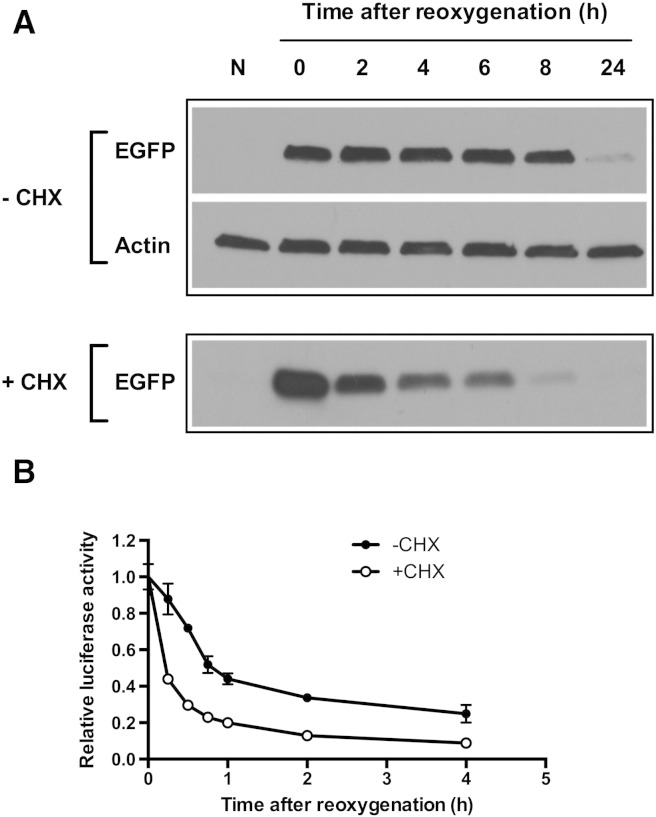
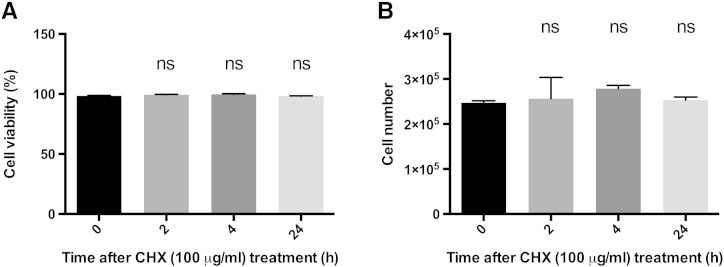
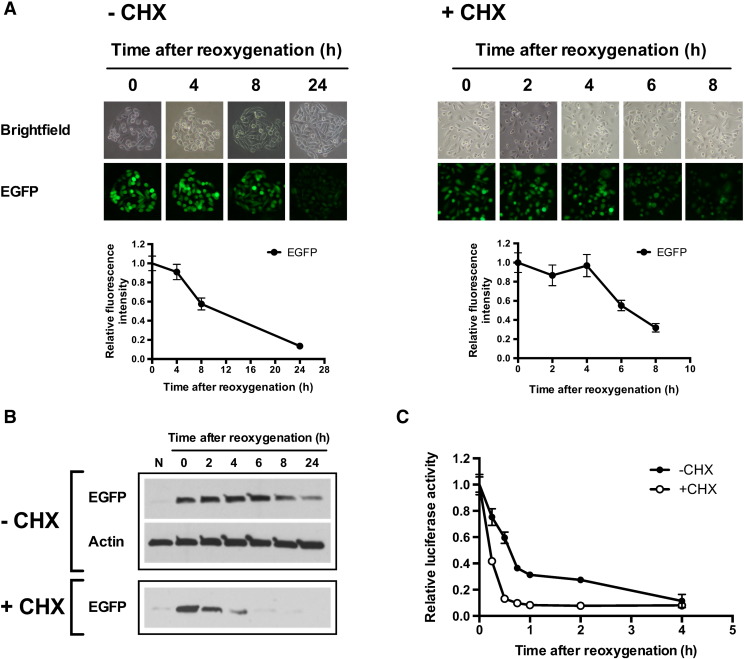
To characterize the stability of EGFP, PC3-HRE-EGFP/HRE-ODD-luc/tdTomato cells were incubated under hypoxia (48 hours at 1% O2) and then reoxygenated. As shown in Figure 2, bright green fluorescence was observed in cell colonies immediately after the hypoxia was released. The green fluorescence remained stable for 4 hours and then progressively started to decrease to undetectable levels within 24 hours after reoxygenation (Figure 2). The same experiment was repeated with cells treated with CHX after hypoxia to inhibit de novo protein synthesis. With CHX (100 μg/ml), EGFP fluorescence intensity was detected 4 hours after reoxygenation but was barely detectable 8 hours after reoxygenation (Figure 2). The EGFP degradation half-life was approximately 15 hours without CHX treatment and approximately 83 minutes in CHX-treated cells. In both control and CHX-treated cells, tdTomato fluorescence intensity was initially dim and progressively increased to become stable 4 hours after reoxygenation (Figure 2). The decrease in tdTomato fluorescence was observed when cells were incubated at 1% O2 but not when treated with 200 μM of CoCl2 for 24 hours (Figure S1). To confirm microscopy results, the stability of EGFP after reoxygenation was also characterized in immunoblots. As shown in Figure 3A, EGFP protein levels remained stable for 8 hours in cells without CHX after reoxygenation. On the other hand, EGFP protein levels decreased progressively in CHX-treated cells but remained detectable up to 6 hours after reoxygenation. CHX was found to be nontoxic in PC3 cells, and cell numbers remained stable after CHX addition (Figure S2).
Figure 2.
Time-lapse imaging of EGFP and tdTomato fluorescence in control (− CHX) and CHX-treated (+ CHX) PC3-HRE-GFP/HRE-ODD-luc/tdTomato cells after 48 hours at 1% O2. The upper panel shows representative images of the evolution of EGFP and tdTomato fluorescence in control and CHX-treated cell colonies after reoxygenation. The lower panel shows the quantification of EGFP (☐) and tdTomato (•) fluorescence in the corresponding control or CHX-treated cell colonies (100 μg/ml). EGFP and tdTomato intensities were normalized to values at time zero. Data are expressed as mean ± SEM.
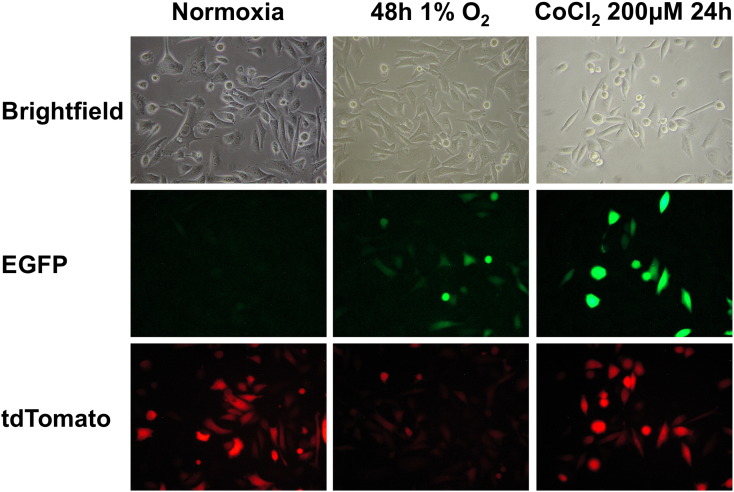
Figure S1.
Influence of oxygen on tdTomato fluorescence. Fluorescence images of PC3-HRE-GFP/HRE-ODD-luc/tdTomato cells incubated at 21% O2 (normoxia), at 1% O2 for 48 hours, or at 21% O2 in the presence of 200 μM CoCl2 for 24 hours (CoCl2 200 μM 24 hours) to chemically mimic hypoxia.
Figure 3.
Evolution of EGFP and ODD-luciferase expression in control (− CHX) and CHX-treated (+ CHX) PC3-HRE-GFP/HRE-ODD-luc/tdTomato cells after 48 hours at 1% O2. (A) Representative Western blot showing the decrease in EGFP proteins following reoxygenation in control and CHX-treated cells (100 μg/ml). (B) Evolution of luciferase activity in control (•) and CHX-treated cells (○, 100 μg/ml) after reoxygenation (n = 3). Data are expressed as mean ± SEM.
Figure S2.
Absence of toxicity of CHX on PC3 cells. (A) Viability and (B) proliferation of PC3 cells as a function of time in the presence of 100 μg/ml CHX in full medium (n = 3). Data are expressed as mean ± SEM. Not significant, P > .05.
We next examined the stability of the luciferase protein fused with ODD in PC3-HRE-EGFP/HRE-ODD-luc/tdTomato cells after a 48-hour incubation at 1% O2. Figure 3B shows that the luciferase activity quickly decreased after reoxygenation. After 4 hours of reoxygenation, the luciferase activity was 24.85 ± 4.81% of its original value in control cells and 8.80 ± 0.8% in CHX-treated cells (Figure 3B). The ODD-luciferase half-life was approximately 34 minutes in control cells without CHX treatment and approximately 11 minutes in CHX-treated cells.
The stability of EGFP and ODD-luc after reoxygenation was also characterized in PC3 cells carrying either the 5xHRE-EGFP construct (PC3-HRE-EGFP) or the 5xHRE-ODD-luc construct (PC3-HRE-ODD-luc). Briefly, the stability of both reporters was found to be similar in PC3-HRE-EGFP and PC3-HRE-ODD-luc cells compared with PC3-HRE-EGFP/HRE-ODD-luc/tdTomato cells (Figure S3).
Figure S3.
Stability of EGFP in PC3-HRE-EGFP and ODD-luciferase in PC3-HRE-ODD-luc cells following incubation at 1% O2 for 48 h. (A) Evolution of the EGFP fluorescence in representative cell colonies in control (CHX −) and CHX-treated (CHX +) PC3-HRE-EGFP cell colonies (100 μg/ml) after reoxygenation. (B) Representative Western blot showing the decrease in EGFP proteins following reoxygenation in control (CHX −) and CHX-treated (CHX +) PC3-HRE-EGFP cells (100 μg/ml). (C) Evolution of the luciferase activity in control (CHX −) (•) or CHX-treated (CHX +) PC3-HRE-ODD-luc cells (○, 100 μg/ml) after hypoxia (48 hours at 1% O2) (n = 3). Data are expressed as mean ± SEM.
High Expression of EGFP and Low Expression of ODD-luc in PC3-HRE-GFP/HRE-ODD-luc/tdTomato Cells Indicate a Reoxygenation Process
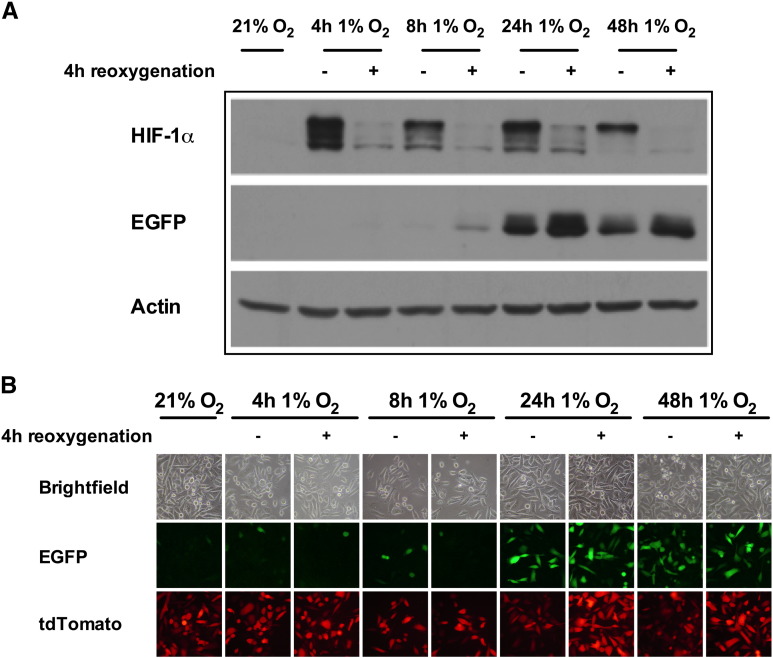
To validate the use of the long-lived EGFP and short-lived ODD-luciferase for discriminating hypoxia, normoxia, and reoxygenation in PC3 cells, PC3-HRE-EGFP/HRE-ODD-luc/tdTomato cells were incubated under hypoxia for 4, 8, 24, and 48 hours and then reoxygenated. Protein levels of HIF-1α, EGFP, and the luciferase activity of these cells were measured just after reoxygenation or 4 hours after hypoxia release. As shown in Figure 4A, 4 hours of hypoxia was sufficient to stabilize HIF-1α. However, EGFP expression could only be detected after 8 hours of hypoxia, and a strong EFGP induction was observed after a 24-hour incubation at 1% O2 (Figure 4, A and B). Whatever the duration of hypoxia, EGFP protein levels and EGFP fluorescence did not decrease 4 hours after reoxygenation (Figure 4, A and B). In agreement with previous results (Figure 2), the fluorescence of tdTomato was found to be reduced just after 24- and 48-hour incubation at 1% O2 but not 4 hours after reoxygenation (Figure 4B).
Figure 4.
Expression and stability of EGFP following different hypoxia-reoxygenation treatments in PC3-HRE-GFP/HRE-ODD-luc/tdTomato cells. (A) Representative Western blot showing HIF-1α and EGFP protein expression in PC3-HRE-GFP/HRE-ODD-luc/tdTomato cells incubated under normoxia, or under hypoxic conditions for 4, 8, 24, or 48 hours. For each hypoxic treatment, cells were immediately collected for Western blot analysis (−) or reoxygenated for 4 hours (+). (B) Representative images of EGFP and tdTomato fluorescence of PC3-HRE-GFP/HRE-ODD-luc/tdTomato cells following hypoxic treatments and reoxygenation for 4 hours.
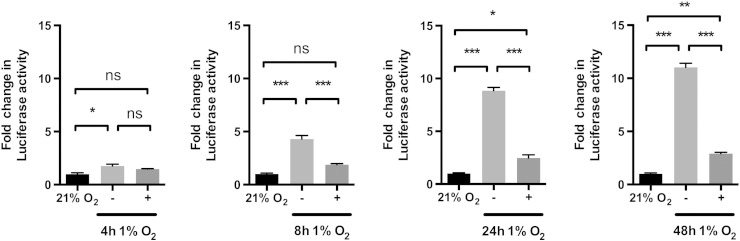
A 4-hour incubation of PC3-HRE-EGFP/HRE-ODD-luc/tdTomato cells at 1% O2 was sufficient to induce a 1.76 ± 0.17-fold increase in luciferase activity (Figure 5). However, the luciferase activity of PC3-HRE-EGFP/HRE-ODD-luc/tdTomato cells incubated at 1% O2 for 4 hours and reoxygenated for 4 hours was not statistically different from the hypoxia group (Figure 5). When cells were submitted to hypoxia for 8, 24, or 48 hours, a strong increase in the luciferase activity was measured in comparison with normoxic cells (Figure 5). A statistically significant drop in the luciferase activity was observed in cells reoxygenated for 4 hours after 8-, 24-, and 48-hour incubation at 1% O2 in comparison with hypoxic groups (Figure 5).
Figure 5.
Luciferase activity of PC3-HRE-GFP/HRE-ODD-luc/tdTomato cells following different hypoxia-reoxygenation treatments. PC3-HRE-GFP/HRE-ODD-luc/tdTomato cells were submitted to hypoxia treatments for 4, 8, 24, or 48 hours and immediately collected for luciferase assays (−) or reoxygenated for 4 hours (+) (n = 3). Data are expressed as mean ± SEM. *P < .05; **P < .01; ***P < .001; not significant (ns), P > .05.
Optical Imaging Allows HIF-1 Imaging in PC3-HRE-GFP/HRE-ODD-luc/tdTomato Cells
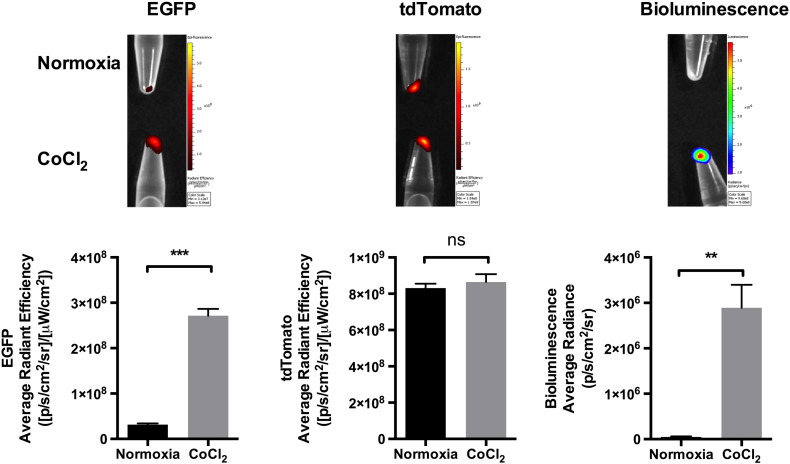
Using optical imaging, we next aimed to image simultaneously EGFP, tdTomato, and bioluminescence produced by PC3-HRE-EGFP/HRE-ODD-luc/tdTomato cells. For these experiments, microtubes containing CoCl2-treated (200 μM for 24 hours) or normoxic cell pellets (3×106 cells/tube) were imaged. As shown in Figure 6, a marked increase in the EGFP signal was observed in PC3-HRE-EGFP/HRE-ODD-luc/tdTomato cells treated with CoCl2, whereas a only a background autofluorescence signal was measured in normoxic cells. Similar levels of tdTomato signal intensity were detected in both normoxic and CoCl2-treated cells (Figure 6). A BLI signal was detected in CoCl2-treated but not in normoxic PC3-HRE-GFP/HRE-ODD-luc/tdTomato cells (Figure 6).
Figure 6.
In vitro fluorescence imaging and bioluminescence imaging of PC3-HRE-GFP/HRE-ODD-luc/tdTomato cells after CoCl2 addition. EGFP (false color), tdTomato, and bioluminescence imaging of control or CoCl2-treated (200 μM for 24 hours) PC3-HRE-GFP/HRE-ODD-luc/tdTomato cell pellets (3 × 106 cells/tube, n = 3). Exposure times were 1 second for EGFP, 0.1 second for tdTomato, and 1 minute for BLI. Data are expressed as mean ± SEM. *P < .05; **P < .01; ***P < .001; ns, P > .05.
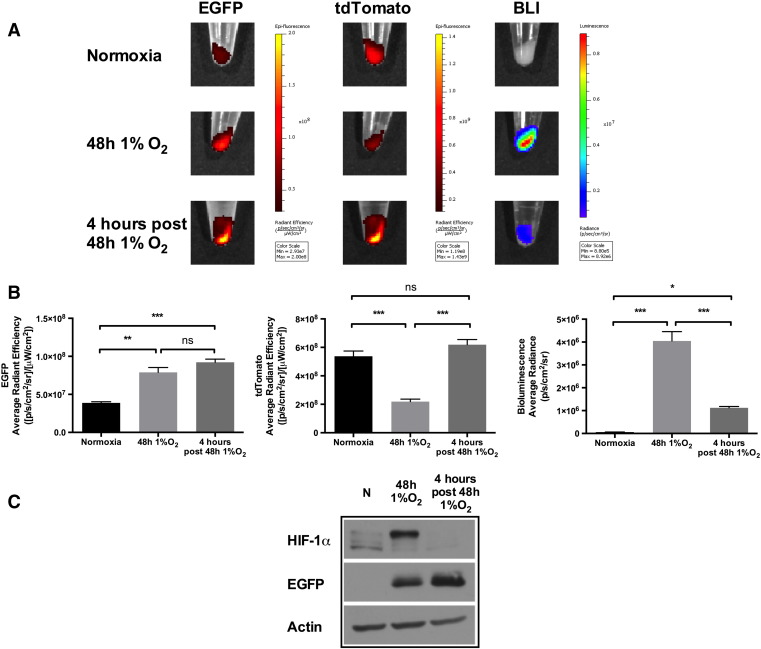
The Combination of Long-Lived EGFP and Short-Lived ODD-Luciferase Allows Differentiation between Normoxic, Hypoxic, and Reoxygenated PC3 Cells Using Optical Imaging
Taking advantage of the different stabilities of EGFP and ODD-luc proteins, we next aimed to visualize normoxic, hypoxic, and reoxygenated PC3-HRE-EGFP/HRE-ODD-luc/tdTomato cells using optical imaging. As shown in the representative images in Figure 7A and quantified intensities in Figure 7B, BLI activity was detected in hypoxic PC3-HRE-EGFP/HRE-ODD-luc/tdTomato cells but not in normoxic cells. After 4 hours of reoxygenation, the BLI signal significantly dropped in these cells (Figure 7, A and B). Representative EGFP images shown in Figure 7A and quantified intensities in Figure 7B demonstrate the significant increase of signal from hypoxic PC3-HRE-EGFP/HRE-ODD-luc/tdTomato cells compared with normoxic cells. Consistent with its long-lived feature and unlike the pattern from short-lived luciferase, the EGFP fluorescence was not statistically different in hypoxic PC3-HRE-EGFP/HRE-ODD-luc/tdTomato cells compared with reoxygenated cells (Figure 7, A and B). However, a decrease in tdTomato fluorescence was observed in hypoxic PC3-HRE-EGFP/HRE-ODD-luc/tdTomato cells in comparison with normoxic and reoxygenated cells (Figure 7, A and B). As shown in the representative immunoblot in Figure 7C, HIF-1α was stabilized in PC3-HRE-EGFP/HRE-ODD-luc/tdTomato cells after 48 hours at 1% O2 but not 4 hours after reoxygenation. EGFP protein levels were found to increase after a 48-hour hypoxia treatment and were sustained at 4 hours after reoxygenation (Figure 7C).
Figure 7.
Optical imaging of long-lived EGFP and short-lived ODD-luciferase in normoxic, hypoxic, and reoxygenated PC3-HRE-GFP/HRE-ODD-luc/tdTomato cells. (A) Representative optical images of PC3-HRE-GFP/HRE-ODD-luc/tdTomato pellets (3 × 106 cells/tube). EGFP is displayed in red. Cells were exposed to normoxia, 48 hours 1% O2, or 48 hours 1% O2 followed by 4-hour reoxygenation. (B) Quantification of EGFP, tdTomato, and BLI quantification of images acquired with the optical imaging scanner (n = 3 per group). (C) Representative immunoblots of PC3-HRE-GFP/HRE-ODD-luc/tdTomato cell lysates showing HIF-1α and EGFP protein levels under normoxia (N), 48 hours 1% O2, or 48 hours 1% O2 followed by 4-hour reoxygenation. Data are expressed as mean ± SEM. *P < .05; **P < .01; ***P < .001; ns, P > .05.
The "Timer" Imaging Method Based on the 5xHRE-EGFP and 5xHRE-ODD-luc Constructs Allows the Dynamic Visualization of HIF-1 Activation in PC3 Prostate Tumors
In proof-of-principle studies, PC3-HRE-EGFP/HRE-ODD-luc/tdTomato cells were inoculated subcutaneously in a group of mice to determine if differences in expression patterns of luciferase and EGFP were detected. As shown in the representative images, tumors where high intensity of BLI and EGFP regions colocalized were frequently observed (Figure 8A), which was confirmed in ex vivo tissue slices (Figure 8B). We also observed patterns in some tumors where high-intensity regions of BLI and EGFP did not colocalize, which was confirmed in the ex vivo tissue slices and indicated differences in the temporal evolution or resolution of HIF activity and hypoxia.
Figure 8.
Optical imaging patterns of two representative subcutaneous and orthotopic PC3-HRE-GFP/HRE-ODD-luc/tdTomato tumors together with a metastatic lesion. (A) Representative in vivo images of EGFP and tdTomato fluorescence and bioluminescence of mice with subcutaneous tumors. (B) Colocalization and mismatch between BLI and EGFP distributions in 2-mm–thick central tumor slice from each tumor in (A) imaged ex vivo using a Xenogen IVIS Spectrum imager and fluorescence microscopy. The scale bar represents 2 mm. (C) Representative in vivo images of EGFP and tdTomato fluorescence and bioluminescence of a mouse with a primary orthotopic tumor (P) and a peritoneal metastatic lesion (M). No EGFP or BLI is detected in the primary tumor unlike the metastatic lesion that shows both EGFP and BLI.
We additionally performed in vivo and ex vivo imaging of the dual HIF reporter system (EGFP/ODD-luciferase) in PC3 tumors implanted in the prostate. Representative in vivo images are presented in Figure 8C. Hypoxic tumor areas were not evident in the primary tumor, but both EGFP and BLI were observed in a peritoneal metastatic lesion.
Discussion
Here we have described an imaging strategy to determine the history of HIF activation in prostate cancer cells. The use of a "timer" reporter protein was first described in 2000 by Terskikh et al. [25]. The fluorescence of this timer, named E5, changed from green to red in a time-dependent manner after expression [25]. More recently, the simultaneous use of two fluorescent proteins with different maturation half-lives (fusion proteins or bicistronic expression) was described for monitoring the temporal changes of the activity of a given reporter [26], [27], [28]. Here, we have developed a timer strategy based on the combination of two reporter genes, regulated by an HIF-dependent promoter, with different degradation half-lives. Using optical imaging, we demonstrated that the combination of a long-lived EGFP and a short-lived luciferase fused to ODD allowed the identification of normoxic (EGFP −/luc −), hypoxic (EGFP +/luc +), and reoxygenated (EGFP +/luc −) prostate cancer cells. No cross talk between reporter proteins was observed during the multiplex imaging of EGFP, tdTomato, and bioluminescence in PC3 cells following a hypoxia challenge. The EGFP degradation half-life values of 15 hours without CHX treatment and approximately 83 minutes with CHX treatment are comparable to previously published values [29], as are the ODD-luciferase half-life values of approximately 34 minutes without CHX treatment and approximately 11 minutes in CHX-treated cells [11]. CHX treatment resulted in a steeper decline of luciferase activity and EGFP protein compared with untreated cells undergoing reoxygenation. This may be, in part, because the oxygen tensions may have taken longer to equilibrate and recover to normoxic levels during the reoxygenation process.
In the tumors, we observed colocalized regions of high BLI and high EGFP, as well as regions that did not colocalize. Regions with high BLI but low EGFP may represent early hypoxia, regions with high BLI and EGFP may represent long-term hypoxia, and regions with low BLI and high EGFP may represent reoxygenated regions that were previously hypoxic. Because the reporters are being expressed following HIF stabilization, it is also important to consider other mechanisms that may induce HIF stabilization in vivo such as reactive oxygen species that can be upregulated in tumors because of high oxidative stress [10], [30]. HIF stabilization due to oxidative stress may, however, result in a more uniform expression of the reporters unlike the distinct localized regions we observed in tumors in this study.
We previously observed that lymph node metastases arising from PC3-HRE-EGFP orthotopic tumors emitted green fluorescence, indicating the presence of hypoxic areas in metastases [20]. Here, following tumor implantation in the orthotopic site that is permissive to metastatic dissemination, we found no detectable hypoxia in the primary but both EGFP and BLI in a peritoneal metastasis. Based on the expression patterns of luciferase and EGFP, the "timer" strategy described here could be used to determine whether metastatic prostate cancer cells in the lymph nodes become hypoxic at the metastatic site or were already hypoxic in the primary tumor. Although serially imaging the luciferase reporter alone would detect the presence of hypoxia, this would not provide a longer-term history of whether hypoxia existed before reoxygenation in regions where there is no luciferase activity. This information is important to characterize the role of hypoxic environments in the primary tumor in the metastatic process.
The fluorescence of the constitutive tdTomato protein, used for cell tracking, was found to decrease at 1% O2 but remained constant after incubation with CoCl2. This can be explained by the fact that an oxidation step requiring O2 is needed in the maturation process of fluorescent proteins to produce light. Differences in maturation rates or oxygen requirements between fluorescent proteins may explain why such a phenomenon was more pronounced with tdTomato compared with EGFP [31], [32]. In another study, pd2EGFP fluorescence decreased but only at near-anoxic conditions (< 0.06% O2) [33]. The authors concluded that EGFP was a suitable reporter gene for monitoring tumor hypoxia [33]. Although oxygen and ATP are required for the generation of light by luciferase proteins, Harada et al. [11] have observed that the BLI signal coming from xenograft HeLa-HRE-ODD-luc tumors implanted in the mouse leg increased after ligation, consistent with the increased BLI signal of the HRE-ODD-luc construct in response to hypoxia observed in our study. In a separate study, the BLI signal from luciferase-positive 9L gliosarcoma cells was measured at various percentages of O2 [34]. Only small differences were found in cells exposed to 5% compared with 0.2% O2 [34], suggesting that ATP, rather than oxygen levels, affected the luciferase activity in these cells [34]. Sustained anoxia, on the other hand, will result in cell death and the absence of both EGFP and BLI signal.
Tumor hypoxia is a poor prognosticator that is associated with chemo- and radioresistance and recurrence [35], [36]. As a result, HIF targeting strategies are being evaluated in clinical trials (ClinicalTrials.gov identifier: NCT01111097). Briefly, chronic hypoxia results from the limited diffusion of oxygen to tumor cells distant from blood vessels [35], and acute hypoxia can result from vascular collapse due to increased tumor interstitial pressure [35]. The formation of hypoxia results in the upregulation of vascular endothelial growth factor [37] that can lead to increased angiogenesis and neovascularization and reoxygenation of hypoxic regions. Reoxygenation also occurs during therapy when cell death provides increased nutrients and oxygen to the remaining cells [38], [39]. Cycling hypoxia due to variations of red blood cell flux in tumor vessels has also been observed in tumors [40]. Endothelial cancer cells submitted to cycling hypoxia had increased levels of HIF-1 induction compared with cancer cells submitted to chronic hypoxia. This phenomenon was associated with an increased survival of cancer cells after radiotherapy in vivo [41].
Cycling hypoxia was found to result from both fast fluctuations of oxygenation (several cycles/hour) and slow fluctuations of oxygenation (over many hours and days). It was hypothesized that low-frequency variations in oxygen pressure in tumors could be due to vascular remodeling following angiogenesis [42], [43]. We observed that a 24-hour incubation of PC3-HRE-EGFP/HRE-ODD-luc/tdTomato cells at 1% O2 was required to detect EGFP using fluorescence microscopy. Our results also showed that the luciferase activity of PC3-HRE-EGFP/HRE-ODD-luc/tdTomato cells quickly dropped after reoxygenation, whereas EGFP levels remained stable for several hours. The dual EGFP/ODD-luciferase reporter system may be used to detect reoxygenation processes due to angiogenesis after long periods of hypoxia. Alternatively, the "timer" HIF imaging strategy can detect rapid variations of O2 after sustained hypoxia. The characterization of HIF inhibitors and radiosensitizing drugs that alleviate tumor hypoxia is another promising field of application for such an HIF imaging method [1], [43], [44], [45].
One limitation of the reporter system developed here arises from different sensing depths between EGFP and BLI. Unlike BLI, fluorescence from EGFP is affected by tissue attenuation during excitation and emission. As a result, in vivo variations in tissue thickness will play a role in the EGFP intensities that are detected. Unlike the in vivo data, because the ex vivo slices are of uniform thickness and only 2 mm thick, light attenuation may not be significant enough to pose a problem when comparing EGFP and BLI intensity distribution. Although the ex vivo slice data are ‘snapshots’ and do not allow longitudinal studies, they still provide temporal insights into HIF and hypoxia patterns in the tumor and in metastatic lesions. In addition, it was not possible to perform single cell imaging using our current in vivo optical imaging system, although fluorescence imaging of isolated cells tagged with fluorescent reporter proteins has been reported in vivo using intravital microscopy [46], [47], [48].
In conclusion, this novel "timer" imaging strategy of combining the short-lived ODD-luciferase and the long-lived EGFP has the potential to advance our understanding of temporal changes in hypoxia and HIF in the primary tumor and during the metastatic cascade, and for understanding therapy-based changes in tumor HIF expression, especially for treatments that target the HIF pathway.
The following are the supplementary data related to this article.
Acknowledgements
P. Danhier was supported by the Belgian American Educational Foundation (BAEF) and by the Fonds Spéciaux de la Recherche (Université catholique de Louvain). This work was supported by National Institutes of Health R01 CA73850, R01 CA82337, and P50 CA103175. We thank Dr. Marie-France Penet for assistance with the tumor studies and Dr. Venu Raman for originally developing the PC-3 HRE EGFP cells.
References
- 1.Gilkes DM, Semenza GL. Role of hypoxia-inducible factors in breast cancer metastasis. Future Oncol. 2013;9:1623–1636. doi: 10.2217/fon.13.92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Penet MF, Chen Z, Bhujwalla ZM. MRI of metastasis-permissive microenvironments. Future Oncol. 2011;7:1269–1284. doi: 10.2217/fon.11.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Semenza GL. Defining the role of hypoxia-inducible factor 1 in cancer biology and therapeutics. Oncogene. 2010;29:625–634. doi: 10.1038/onc.2009.441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gilkes DM, Semenza GL, Wirtz D. Hypoxia and the extracellular matrix: drivers of tumour metastasis. Nat Rev Cancer. 2014;14:430–439. doi: 10.1038/nrc3726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Fraga A, Ribeiro R, Principe P, Lopes C, Medeiros R. Hypoxia and Prostate Cancer Aggressiveness: A Tale With Many Endings. Clin Genitourin Cancer. 2015;13:295–301. doi: 10.1016/j.clgc.2015.03.006. [DOI] [PubMed] [Google Scholar]
- 6.Hu CJ, Sataur A, Wang L, Chen H, Simon MC. The N-terminal transactivation domain confers target gene specificity of hypoxia-inducible factors HIF-1alpha and HIF-2alpha. Mol Biol Cell. 2007;18:4528–4542. doi: 10.1091/mbc.E06-05-0419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Tian H, McKnight SL, Russell DW. Endothelial PAS domain protein 1 (EPAS1), a transcription factor selectively expressed in endothelial cells. Genes Dev. 1997;11:72–82. doi: 10.1101/gad.11.1.72. [DOI] [PubMed] [Google Scholar]
- 8.Berchner-Pfannschmidt U, Frede S, Wotzlaw C, Fandrey J. Imaging of the hypoxia-inducible factor pathway: insights into oxygen sensing. Eur Respir J. 2008;32:210–217. doi: 10.1183/09031936.00013408. [DOI] [PubMed] [Google Scholar]
- 9.Ranasinghe WK, Baldwin GS, Bolton D, Shulkes A, Ischia J, Patel O. HIF1alpha expression under normoxia in prostate cancer--which pathways to target? J Urol. 2015;193:763–770. doi: 10.1016/j.juro.2014.10.085. [DOI] [PubMed] [Google Scholar]
- 10.Krock BL, Skuli N, Simon MC. Hypoxia-induced angiogenesis: good and evil. Genes Cancer. 2011;2:1117–1133. doi: 10.1177/1947601911423654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Harada H, Kizaka-Kondoh S, Itasaka S, Shibuya K, Morinibu A, Shinomiya K, Hiraoka M. The combination of hypoxia-response enhancers and an oxygen-dependent proteolytic motif enables real-time imaging of absolute HIF-1 activity in tumor xenografts. Biochem Biophys Res Commun. 2007;360:791–796. doi: 10.1016/j.bbrc.2007.06.149. [DOI] [PubMed] [Google Scholar]
- 12.Winnard PT, Jr., Kluth JB, Raman V. Noninvasive optical tracking of red fluorescent protein-expressing cancer cells in a model of metastatic breast cancer. Neoplasia. 2006;8:796–806. doi: 10.1593/neo.06304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.O'Farrell AC, Shnyder SD, Marston G, Coletta PL, Gill JH. Non-invasive molecular imaging for preclinical cancer therapeutic development. Br J Pharmacol. 2013;169:719–735. doi: 10.1111/bph.12155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Danhier P, De Preter G, Magat J, Godechal Q, Porporato PE, Jordan BF, Feron O, Sonveaux P, Gallez B. Multimodal cell tracking of a spontaneous metastasis model: comparison between MRI, electron paramagnetic resonance and bioluminescence. Contrast Media Mol Imaging. 2014;9:143–153. doi: 10.1002/cmmi.1553. [DOI] [PubMed] [Google Scholar]
- 15.Brogan J, Li F, Li W, He Z, Huang Q, Li CY. Imaging molecular pathways: reporter genes. Radiat Res. 2012;177:508–513. doi: 10.1667/rr2918.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chudakov DM, Matz MV, Lukyanov S, Lukyanov KA. Fluorescent proteins and their applications in imaging living cells and tissues. Physiol Rev. 2010;90:1103–1163. doi: 10.1152/physrev.00038.2009. [DOI] [PubMed] [Google Scholar]
- 17.de Almeida PE, van Rappard JR, Wu JC. In vivo bioluminescence for tracking cell fate and function. Am J Physiol Heart Circ Physiol. 2011;301:H663–H671. doi: 10.1152/ajpheart.00337.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Raman V, Artemov D, Pathak AP, Winnard PT, Jr., McNutt S, Yudina A, Bogdanov A, Jr., Bhujwalla ZM. Characterizing vascular parameters in hypoxic regions: a combined magnetic resonance and optical imaging study of a human prostate cancer model. Cancer Res. 2006;66:9929–9936. doi: 10.1158/0008-5472.CAN-06-0886. [DOI] [PubMed] [Google Scholar]
- 19.Glunde K, Shah T, Winnard PT, Jr., Raman V, Takagi T, Vesuna F, Artemov D, Bhujwalla ZM. Hypoxia regulates choline kinase expression through hypoxia-inducible factor-1 alpha signaling in a human prostate cancer model. Cancer Res. 2008;68:172–180. doi: 10.1158/0008-5472.CAN-07-2678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Penet MF, Pathak AP, Raman V, Ballesteros P, Artemov D, Bhujwalla ZM. Noninvasive multiparametric imaging of metastasis-permissive microenvironments in a human prostate cancer xenograft. Cancer Res. 2009;69:8822–8829. doi: 10.1158/0008-5472.CAN-09-1782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yang M, Jiang P, Yamamoto N, Li L, Geller J, Moossa AR, Hoffman RM. Real-time whole-body imaging of an orthotopic metastatic prostate cancer model expressing red fluorescent protein. Prostate. 2005;62:374–379. doi: 10.1002/pros.20125. [DOI] [PubMed] [Google Scholar]
- 22.Hoffman RM. Patient-derived orthotopic xenografts: better mimic of metastasis than subcutaneous xenografts. Nat Rev Cancer. 2015;15:451–452. doi: 10.1038/nrc3972. [DOI] [PubMed] [Google Scholar]
- 23.Hoffman RM. Orthotopic metastatic mouse models for anticancer drug discovery and evaluation: a bridge to the clinic. Invest New Drugs. 1999;17:343–359. doi: 10.1023/a:1006326203858. [DOI] [PubMed] [Google Scholar]
- 24.Gavet O, Pines J. Progressive activation of CyclinB1-Cdk1 coordinates entry to mitosis. Dev Cell. 2010;18:533–543. doi: 10.1016/j.devcel.2010.02.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Terskikh A, Fradkov A, Ermakova G, Zaraisky A, Tan P, Kajava AV, Zhao X, Lukyanov S, Matz M, Kim S. "Fluorescent timer": protein that changes color with time. Science. 2000;290:1585–1588. doi: 10.1126/science.290.5496.1585. [DOI] [PubMed] [Google Scholar]
- 26.Chen MR, Yang S, Niu WP, Li ZY, Meng LF, Wu ZX. A novel fluorescent timer based on bicistronic expression strategy in Caenorhabditis elegans. Biochem Biophys Res Commun. 2010;395:82–86. doi: 10.1016/j.bbrc.2010.03.143. [DOI] [PubMed] [Google Scholar]
- 27.Dona E, Barry JD, Valentin G, Quirin C, Khmelinskii A, Kunze A, Durdu S, Newton LR, Fernandez-Minan A, Huber W. Directional tissue migration through a self-generated chemokine gradient. Nature. 2013;503:285–289. doi: 10.1038/nature12635. [DOI] [PubMed] [Google Scholar]
- 28.Khmelinskii A, Keller PJ, Bartosik A, Meurer M, Barry JD, Mardin BR, Kaufmann A, Trautmann S, Wachsmuth M, Pereira G. Tandem fluorescent protein timers for in vivo analysis of protein dynamics. Nat Biotechnol. 2012;30:708–714. doi: 10.1038/nbt.2281. [DOI] [PubMed] [Google Scholar]
- 29.Tryfonidou MA, Lunstrum GP, Hendriks K, Riemers FM, Wubbolts R, Hazewinkel HA, Degnin CR, Horton WA. Novel type II collagen reporter mice: New tool for assessing collagen 2alpha1 expression in vivo and in vitro. Dev Dyn. 2011;240:663–673. doi: 10.1002/dvdy.22569. [DOI] [PubMed] [Google Scholar]
- 30.Li F, Sonveaux P, Rabbani ZN, Liu S, Yan B, Huang Q, Vujaskovic Z, Dewhirst MW, Li CY. Regulation of HIF-1alpha stability through S-nitrosylation. Mol Cell. 2007;26:63–74. doi: 10.1016/j.molcel.2007.02.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Macdonald PJ, Chen Y, Mueller JD. Chromophore maturation and fluorescence fluctuation spectroscopy of fluorescent proteins in a cell-free expression system. Anal Biochem. 2012;421:291–298. doi: 10.1016/j.ab.2011.10.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Shaner NC, Steinbach PA, Tsien RY. A guide to choosing fluorescent proteins. Nat Methods. 2005;2:905–909. doi: 10.1038/nmeth819. [DOI] [PubMed] [Google Scholar]
- 33.Vordermark D, Shibata T, Brown JM. Green fluorescent protein is a suitable reporter of tumor hypoxia despite an oxygen requirement for chromophore formation. Neoplasia. 2001;3:527–534. doi: 10.1038/sj.neo.7900192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Moriyama EH, Niedre MJ, Jarvi MT, Mocanu JD, Moriyama Y, Subarsky P, Li B, Lilge LD, Wilson BC. The influence of hypoxia on bioluminescence in luciferase-transfected gliosarcoma tumor cells in vitro. Photochem Photobiol Sci. 2008;7:675–680. doi: 10.1039/b719231b. [DOI] [PubMed] [Google Scholar]
- 35.Horsman MR, Mortensen LS, Petersen JB, Busk M, Overgaard J. Imaging hypoxia to improve radiotherapy outcome. Nat Rev Clin Oncol. 2012;9:674–687. doi: 10.1038/nrclinonc.2012.171. [DOI] [PubMed] [Google Scholar]
- 36.Rohwer N, Cramer T. Hypoxia-mediated drug resistance: novel insights on the functional interaction of HIFs and cell death pathways. Drug Resist Updat. 2011;14:191–201. doi: 10.1016/j.drup.2011.03.001. [DOI] [PubMed] [Google Scholar]
- 37.Unwith S, Zhao H, Hennah L, Ma D. The potential role of HIF on tumour progression and dissemination. Int J Cancer. 2015;136:2491–2503. doi: 10.1002/ijc.28889. [DOI] [PubMed] [Google Scholar]
- 38.Vaupel P, Kelleher DK, Thews O. Modulation of tumor oxygenation. Int J Radiat Oncol Biol Phys. 1998;42:843–848. doi: 10.1016/s0360-3016(98)00324-1. [DOI] [PubMed] [Google Scholar]
- 39.Danhier F, Danhier P, Magotteaux N, De Preter G, Ucakar B, Karroum O, Jordan B, Gallez B, Preat V. Electron paramagnetic resonance highlights that the oxygen effect contributes to the radiosensitizing effect of paclitaxel. PLoS One. 2012;7:e40772. doi: 10.1371/journal.pone.0040772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lee CT, Boss MK, Dewhirst MW. Imaging tumor hypoxia to advance radiation oncology. Antioxid Redox Signal. 2014;21:313–337. doi: 10.1089/ars.2013.5759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Martinive P, Defresne F, Bouzin C, Saliez J, Lair F, Gregoire V, Michiels C, Dessy C, Feron O. Preconditioning of the tumor vasculature and tumor cells by intermittent hypoxia: implications for anticancer therapies. Cancer Res. 2006;66:11736–11744. doi: 10.1158/0008-5472.CAN-06-2056. [DOI] [PubMed] [Google Scholar]
- 42.Dewhirst MW. Relationships between cycling hypoxia, HIF-1, angiogenesis and oxidative stress. Radiat Res. 2009;172:653–665. doi: 10.1667/RR1926.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Danhier P, De Saedeleer CJ, Karroum O, De Preter G, Porporato PE, Jordan BF, Gallez B, Sonveaux P. Optimization of tumor radiotherapy with modulators of cell metabolism: toward clinical applications. Semin Radiat Oncol. 2013;23:262–272. doi: 10.1016/j.semradonc.2013.05.008. [DOI] [PubMed] [Google Scholar]
- 44.Samanta D, Gilkes DM, Chaturvedi P, Xiang L, Semenza GL. Hypoxia-inducible factors are required for chemotherapy resistance of breast cancer stem cells. Proc Natl Acad Sci U S A. 2014;111:E5429–E5438. doi: 10.1073/pnas.1421438111. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 45.Semenza GL. Targeting HIF-1 for cancer therapy. Nat Rev Cancer. 2003;3:721–732. doi: 10.1038/nrc1187. [DOI] [PubMed] [Google Scholar]
- 46.Hoffman RM, Yang M. Subcellular imaging in the live mouse. Nat Protoc. 2006;1:775–782. doi: 10.1038/nprot.2006.109. [DOI] [PubMed] [Google Scholar]
- 47.Hoffman RM, Yang M. Color-coded fluorescence imaging of tumor-host interactions. Nat Protoc. 2006;1:928–935. doi: 10.1038/nprot.2006.119. [DOI] [PubMed] [Google Scholar]
- 48.Hoffman RM. The multiple uses of fluorescent proteins to visualize cancer in vivo. Nat Rev Cancer. 2005;5:796–806. doi: 10.1038/nrc1717. [DOI] [PubMed] [Google Scholar]