Abstract
AIM: To develop the single chain variable fragment of MG7 murine anti-human gastric cancer monoclonal antibody using the phage display technology for obtaining a tumor-targeting mediator.
METHODS: mRNA was isolated from MG7 producing murine hybridoma cell line and converted into cDNA. The variable fragments of heavy and light chain were amplified separately and assembled into ScFv with a specially constructed DNA linker by PCR. The ScFvs DNA was ligated into the phagmid vector pCANTAB5E and the ligated sample was transformed into competent E. Coli TG1. The transformed cells were infected with M13K07 helper phage to form MG7 recombinant phage antibody library. The volume and recombinant rate of the library were evaluated by means of bacterial colony count and restriction analysis. After two rounds of panning with gastric cancer cell line KATOIII of highly expressing MG7-binding antigen, the phage clones displaying ScFv of the antibody were selected by ELISA from the enriched phage clones. The antigen binding affinity of the positive clone was detected by competition ELISA. HB2151 E.coli was transfected with the positive phage clone demonstrated by competition ELISA for production of a soluble form of the MG7 ScFv. ELISA assay was used to detect the antigen-binding affinity of the soluble MG7 ScFv. Finally, the relative molecular mass of soluble MG7 ScFv was measured by SDS-PAGE.
RESULTS: The V-H, V-L and ScFv DNAs were about 340 bp, 320 bp and 750 bp, respectively. The volume of the library was up to 2 × 106 and 8 of 11 random clones were recombinants. Two phage clones could strongly compete with the original MG7 antibody for binding to the antigen expressed on KATOIII cells. Within 2 strong positive phage clones, the soluble MG7 ScFv from one clone was found to have the binding activity with KATOIII cells. SDS-PAGE showed that the relative molecular weight of soluble MG7 ScFv was 32.
CONCLUSION: The MG7 ScFv was successfully produced by phage antibody technology, which may be useful for broadening the scope of application of the antibody.
Keywords: antibodies, neoplasms/biosynthesis; antibodies, monoclonal; stomach neoplasms/immunology; bacteriophages/genetics
INTRODUCTION
In our previous studies, MG7 hybridoma cell line had been successfully prepared by immunization of mouse with KATOIII gastric cancer cells and hybridization of the B cells from the spleen of the immunized mouse with the murine myeloma cell line SP 2/0. This hybridoma cell line generates a kind of monoclonal antibody against gastric cancer which can specifically recognize an ascertained gastric cancer associated antigen[1]. MG7 antibody was confirmed to be of great value and good potency in the targeting gene therapy of gastric cancer due to the overexpression of its corresponding antigen in a large proportion of patients with gastric cancer. But owing to its murine origin, like many other similar antibodies, MG7 antibody can elicit human anti-mouse immunoreaction and thus its use in clinical practice is restricted[2,3]. One of the efficient solutions to this problem is to remove the constant region of antibody which makes main contribution to the immunogenicity of the murine antibody to human being. It has been proved that antibody devoid of constant region still maintains its capacity of specific antigen-binding affinity[4-10]. Additionally, antibody without constant region, termed ScFv, is a small molecule and comprises 1/6 of its original antibody in molecular mass. Therefore, ScFv can more readily penetrate into the solid tumor in vivo and be easily cleared up from the normal tissue. In the early 90’s, the emergence of recombinant phage library represented a great breakthrough in the antibody technology which provides an economical means to prepare the ScFv/Fab of any desired antibody[11-19]. In the present study, the MG7 recombinant phage antibody derived from MG7 hybridoma was constructed and screened to prepare the MG7 ScFv which might help establish an efficient strategy of targeting gene therapy in gastric cancer.
MATERIALS AND METHODS
Detection of antigen-binding affinity of MG7 antibody
MG7 hybridoma cells and KATOIII cells were cultured with RPMI 1640 (purchased from Gibco) supplemented with heat-inactivated 100 mL·L-1 fetal bovine serum at 37 °C under 50 mL·L-1 CO2. MG7 hybridoma cells were harvested at log phase and stored at -70 °C with aliquot of 106 for RNA isolation. Supernatant was collected for detection of antigen-binding affinity of MG7 antibody by ELISA. KATOIII cells in log phase were transferred into a 96 wellplate and immobilized on the wall by centrifiguation at 1000 × g for 10 min, finally fixed by 0.25 mL·L-1 glutaraldehyde. Supernatant of 0.2 mL was applied to each well and incubated at 4 °C overnight, and 0.1 mL HRP-labeled goat anti-mouse (HRP-GAM) Ig was added into each well. The absorbance value (A) at 492 nm of reactant in each well was measured after incubation for 1 hour at 37 °C and staining with OPD.
Construction of MG7 recombinant phage antibody library
According to the protocol of svtotal RNA isolation system and polyAT tract mRNA isolation system (purchased from Promega), mRNA was isolated from MG7 hybridoma cells and quantified by gel electrophoresis for following reverse transcription reaction. Subsequently, reverse transcription reaction was performed with 0.3 µg mRNA, 2 U reverse transcripase (purchased from Promega) mixed together for incubation of 1 hour under 37 °C. PCR was conducted with a mixture of 10 µg product of reverse transcription, 2 U Taq DNA polymerase and 2 µL V H/VL primers mix (purchased from Promega) in a total volume of 50 µL. The procedure of PCR was arranged in the following order: 95 °C × 5 min; 94 °C × 1 min, 55 °C × 2 min, 72 °C × 2 min and 30 cycles; 72 °C × 10 min. After precise quantification of PCR product by gel electrophoresis, 50 ng of VH and VL product was respectively mixed with 50 ng linker primer and 1 µL Taq DNA polymerase to perform PCR (94 °C× 1 min, 63 °C × 4 min, 7 cycles). Subsequently, 50 ng RS primers (purchased from Promega) underwent another PCR (94 °C × 1 min, 55 °C × 2 min, 72 °C × 2 min and 30 cycles; 72 °C × 10 min). Two µL Sfi I and 0.5 µg ScFv product were added into a sterile 0.5 mL microtube and incubated at 50 °C for 4 hours. After being purified by PCR purification kit, 0.5 µg Sfi I digested ScFv product mixed with 2 µL Not I was incubated at 37 °C for 4 hours and purified again for later use. ScFv (150 ng) and pCANTAB5E (250 ng) mixed with 2 µL T4 DNA ligase was incubated at 16 °C for 16 hours. Ligated product was transformed into TG1 cell. Transformed product with aliquot of 100 µL was placed onto SOBAG plates and incubated overnight at 37 °C to form bacterial clones.
Evaluation of volume and recombinant rate of phage antibody library
Colony count was adopted to exhibit the total number of clones formed on the SOBAG plates. Eleven clones were randomly singled out from the SOBAG plates and passaged into 5 mL 2 × YT-AG medium for an incubation of 12 hours at 37 °C. Plasmid from each clone was respectively isolated and digested by Eco RI and Hin dIII. Gel electrophoresis was conducted with restriction digested product to examine the recombinant phagmid.
Panning and enrichment of MG7 recombinant phage antibody
The initial recombinant phage antibody library was incubated for 1 hour at 37 °C with shaking at 250 r·min-1, and helper phage M13KO7 was added and incubated for another hour at 37 °C with shaking at 250 r·min-1[20]. The culturing product was spinned at 1000 × g for 10 min to precipiate the cells. Then the entire sample was gently resuspended in 10 mL 2 × YT-AK medium. After an overnight incubation at 37 °C with shaking at 250 r·min-1, the culturing product was spinned at 1000 × g for 20 min and the supernatant which contained the recombinant phages was collected. Then, 2 mL PEG/NaCl was added and placed on ice for 45 min for precipitation of recombinant phage clones. It was spinned at 10000 × g for 20 min at 4 °C and the pellet was resuspended in 16mL 2 × YT medium diluted with 14 mL blocking buffer containing 0.1 g·L-1 sodium azide and incubated at room temperature for 15 min. Twenty mL of the diluted recombinant phage was then added to the flask which was coated with KATOIII cells and well blocked. The flask was incubated for 2 hours at 37 °C, washed 10 times with PBS plus another 10 times with PBS containing 1 mL·L-1 Tween20. Ten mL log-phase TG1 cells were added to the flask and incubated with shaking at 37 °C for 1 hour for reinfection. After two rounds of panning, reinfected TG1 cells with bound phages directly in the panning flask were plated for colony isolation.
Screening for MG7 recombinant phages
Recombinant phages were rescued from individual clones and screened for MG7 binding by ELISA. Microtiter wells were coated with KATOIII cells. Bound phages were detected by incubation with a 1:5000 dilution of conjugate (Pharmacia Biotech). And the detection was achieved by addition of TMB substrate. Clones reacted to KATOIII cells were referred to as positives.
Competitive test of positive selected MG7 recombinant phages
Microtiter wells were coated with KATOIII cells as mentioned above . The supernatant of the selected positive MG7 recombinant phages was applied into each well (100 mL·well-1) and then incubated for 1 hour at 37 °C. After disposing of the supernatant, MG7 antibody (100 mg·L-1, 50 µL·well-1) was added and incubated for 1 hour at 37 °C. PBST was used to wash 5 times, and HRP-GAM Ig (1:1000 diluted, 50 µL·well-1) was added and developed by TMB to measure the absorbance value at 450 nm. The inhibiting ratio of selected positive MG7 recombinant phages with MG7 antibody for binding of KATOIII cells was calculated by the following formula: Inhibiting ratio = 1- (value of sample/value of control) × 100%
Detection of antigen-binding affinity of the soluble MG7 ScFv
The positive phages were transfected into E.coli HB2151 cells for the production of a soluble form of the MG7 ScFv. Five mL culturing product of transfected E.coli HB2151 cells with overnight induction of 1 mmol·L-1 isopropyl β-D-thiogalactopyanoside (IPTG) was centrifugated at 1000 × g to collect the sediment and the supernatant (containing extracellular soluble ScFvs). The sediment was given osmotic shock to prepare periplasmic extracts. Microtiter wells were coated with KATOIII cells, and ELISA test was made twice to detect the antigen-binding affinity of soluble MG7 ScFv.
Measurement of the relative molecular weight of soluble MG7 ScFv
Periplasmic extracts from transfected E.coli HB2151 cells induced by IPTG was adopted to measure the relative molecular weight of soluble MG7 ScFv by SDS-PAGE.
RESULTS
Antigen-binding affinity of MG7 antibody
The ELISA showed that the A492 absorbance of reactant with presence of MG7 antibody was up to 0.65 (0.208 in control).
Amplification of VH, VL and ScFv gene
On electrophoresis, VH product formed a band at 350 bp and VL at 320 bp, and ScFv was successfully spliced together to form a fragment of 750 bp, as shown in Figure 1.
Figure 1.
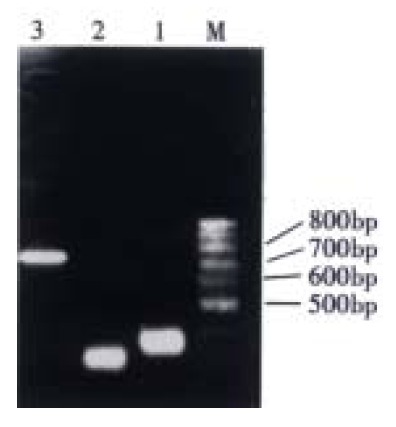
RT-PCR of VH, VL and ScFv fragment of MG7 antibody. 1: VH; 2: VL; 3: ScFv; M: 100 bp ladder
Volume of MG7 phage antibody library
Colony counts showed that MG7 phage antibody library consisted of 2 × 106 clones.
Recombinant rate of MG7 recombinant phage antibody library
Eight of 11 random clones were found to release a 2.1 kb DNA fragment and confirmed to be recombinant phagmid by restriction analysis and gel electophoresis (Figure 2). The recombinant rate was 72.7%.
Figure 2.
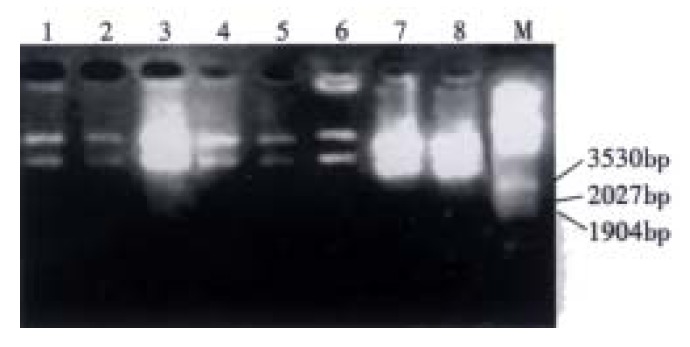
Enzymatic analysis of MG7 recombinant phage antibody library with Eco RI and Hin dIII. 1-8: Recombinant clones from library; M: λ/Eco RI and Hin dIII
Screening of MG7 positive recombinant phages
Using ELISA assay, we yielded six strains of positive clones which had a good reaction with KATOIII cells (Table 1).
Table 1.
ELISA results of screening from enriched phage displayed antibody library
| Screening |
Number of positive clones (A- value) |
Neg. ctrl | |||||
| 1 | 2 | 3 | 4 | 5 | 6 | ||
| First round | 0.495 | 0.508 | 0.488 | 0.805 | 0.845 | 0.580 | 0.157 |
| Second round | 0.543 | 0.606 | 0.560 | 0.840 | 0.796 | 0.758 | 0.185 |
Results of competitive ELISA
Two strong positive clones were found to inhibit the binding of MG7 antibody and KATOIII cells with the inhibiting ratio of 26.1% and 30%, respectively.
Antigen-binding affinity of soluble MG7 ScFv
By means of ELISA assay, one of the strong positive clones exhibited the capacity of binding with KATOIII cells (Table 2).
Table 2.
ELISA results of the soluble MG7 ScFv for binding with KATOIII cells
| ELISA |
Number of strong positive clones (A value) |
Neg. ctrl | |
| 1 | 2 | ||
| First round | 0.776 | 0.287 | 0.201 |
| Second round | 0.802 | 0.346 | 0.223 |
The relative molecular weight of soluble MG7 ScFv
From Figure 3, an extra band on the lane of sample was visualized at Mr 32, as compared with the negative control. The relative molecular weight of soluble MG7 ScFv was 31.
Figure 3.
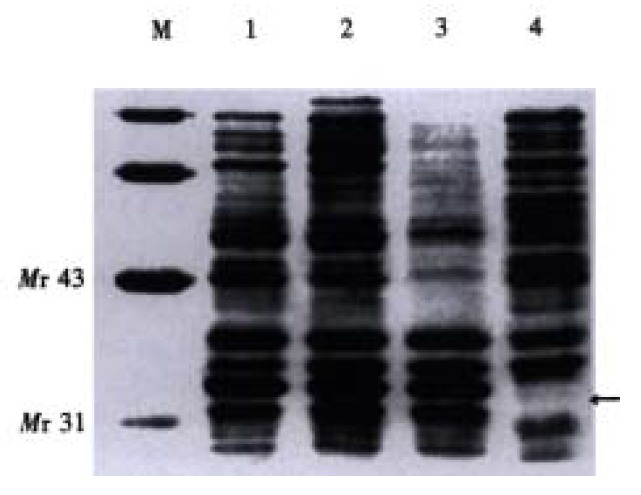
Measurement of the relative molecular weight of soluble MG7 ScFv. 1-3: Periplasmic extracts; 4: Neg. ctrl; M: Low molecular mass protein marker
DISCUSSION
The phage antibody technique is one of the most remarkable achievements in antibody technology. With this technique, the repertoire of VH and VL genes are amplified and joined together by PCR and finally inserted into phagmid[20]. After transformation into E.coli, phage with the fusion of exogenous ScFv and pIII protein exposed on the surface was released from E.coli with the aid of the helper phage M13KO7. This technique uniforms the phenotype of ScFv to its genotype. By immunosorbance of the immobilized antigen, phage with functional ScFv can be bound and enriched. Subsequently, the desired phage harboring functional ScFv gene can be selected from the enriched phage antibody library by ELISA. The resultant ScFv can be solubly expressed in E.coli HB2151. The primary structural information of ScFv of antibody is accessible by DNA sequence of phagmid from the bound phage. Therefore, phage antibody has become an optimal measure to develop the ScFv of desired antibody[21-26].
It is well known that the immune system will be triggered and activated in response to the presence of certain antigens in patients with some kinds of diseases, such as tumor, infective diseases and autoimmune diseases[27-32]. The immune system will produce abundant B lymphocyte clones which can yield and secrete antibody directed against the disease associated antigens in these patients. Therefore, the B lymphocyte population isolated from PBMC of these patients can be used as an ideal material source for construction of the recombinant phage antibody library[29-31]. Additionally, the B lymphocyte isolated from PBMC of immunized animals with given antigen is an alternative material source[20]. Besides the B lymphocyte population from patients or immunized animals, many kinds of antibody-producing hybridomas are also suitable as a kind of material source for construction of the recombinant phage antibody library[6,19]. Owing to the unraveling of biological functions over many antigen recognized by antibody from hybridomas, hybridomas are more favorable as material source for construction of recombinant phage antibody library.
In order to understand the quality of MG7 hybridoma as a material source for construction of MG7 recombinant phage antibody library, we detected the antigen-binding activity of MG7 antibody in present study. ELISA assay showed that the A 492 nm value of reactant with presence of MG7 antibody was 0.65 which was over twice higher than that with absence of MG7 antibody (0.208 only). It demonstrated that MG7 hybridoma could secrete functional antibody and could be used as the source of mRNA to amplify the VH and VL genes of MG7 antibody. Colony count and restriction analysis were conducted for evaluating the volume and quality of MG7 recombinant antibody library. The large volume of MG7 recombinant antibody library (2 × 106) and high recombinant rate (72.7%) confirmed that MG7 phage antibody library comprised sufficient repertoire of recombinant clones for further research. ELISA assay and SDS-PAGE showed that the soluble MG7 ScFv had antigen-binding activity and was Mr 31. Taken together, we have successfully constructed the MG7 recombinant phage antibody library and prepared the phage-displayed/soluble MG7 ScFv.
Gastric cancer is a highly prevalent neoplasm and is the first killer among various malignancies. In advanced cases, many current therapeutic approaches, including surgery combined with chemotherapy, appear to be palliative. These therapeutic approaches can not be targeted to and completely annihilate individual tumor cells, which leads to the failure of preventing metastasis and recurrence of many tumors. Besides, some kinds of therapeutic approaches, such as chemotherapy, can cause damage to both the tumor cells and normal tissue cells. Thus, introduction of a new way of targeting therapy for tumor is desperately needed to overcome these obstacles with the conventional approaches, such as surgery and chemotherapy[33-44]. Targeting therapy for tumors in the last decade has become a highlight in the field of tumor therapy. This therapy mediated by antibody still remains as a promising curative modality among the ways of tumor therapy and attracts worldwide attention[45-50].
Developing ScFv of the MG7 is of great significance in both early diagnosis and treatment of gastric cancer. For instance, MG7 ScFv fused with avidin can be used as a reagent in immuno-PCR for early diagnosis of gastric cancer. Additionally, a new immunotoxin with curative effect on gastric cancer can be developed by fusing the MG7 ScFv and A subunit of ricin. MG7 ScFv can direct the A subunit of ricin to MG7 positive gastric cancer cells. Thus, the construction of MG7 phage antibody library and subsequent preparation of MG7 ScFv may be a step forward in seeking an efficient way for targeting therapy for gastric cancer.
Footnotes
Edited by Ma JY
References
- 1.Fan DM, Zhang XY, Chen XT, Qiao TD, Chen BJ. Preparation and immunohistologic identification of mAbs against a poor differenced gastric cancer line MKN-46.9. Jiefangjun Yixue Zazhi. 1988;13:12–15. [Google Scholar]
- 2.Klimka A, Matthey B, Roovers RC, Barth S, Arends JW, Engert A, Hoogenboom HR. Human anti-CD30 recombinant antibodies by guided phage antibody selection using cell panning. Br J Cancer. 2000;83:252–260. doi: 10.1054/bjoc.2000.1226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Watkins NA, Ouwehand WH. Introduction to antibody engineering and phage display. Vox Sang. 2000;78:72–79. doi: 10.1159/000031154. [DOI] [PubMed] [Google Scholar]
- 4.Zhai W, Davies J, Shang DZ, Chan SW, Allain JP. Human recombinant single-chain antibody fragments, specific for the hypervariable region 1 of hepatitis C virus, from immune phage-display libraries. J Viral Hepat. 1999;6:115–124. doi: 10.1046/j.1365-2893.1999.00146.x. [DOI] [PubMed] [Google Scholar]
- 5.de Greeff A, van Alphen L, Smith HE. Selection of recombinant antibodies specific for pathogenic Streptococcus suis by subtractive phage display. Infect Immun. 2000;68:3949–3955. doi: 10.1128/iai.68.7.3949-3955.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Long MC, Jager S, Mah DC, Jebailey L, Mah MA, Masri SA, Nagata LP. Construction and characterization of a novel recombinant single-chain variable fragment antibody against Western equine encephalitis virus. Hybridoma. 2000;19:1–13. doi: 10.1089/027245700315743. [DOI] [PubMed] [Google Scholar]
- 7.Johns M, George AJ, Ritter MA. In vivo selection of sFv from phage display libraries. J Immunol Methods. 2000;239:137–151. doi: 10.1016/s0022-1759(00)00152-6. [DOI] [PubMed] [Google Scholar]
- 8.Mao S, Gao C, Lo CH, Wirsching P, Wong CH, Janda KD. Ph-age-display library selection of high-affinity human single-chain antibodies to tumor-associated carbohydrate antigens sialyl Lewisx and Lewisx. Proc Natl Acad Sci USA. 1999;96:6953–6958. doi: 10.1073/pnas.96.12.6953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kupsch JM, Tidman NH, Kang NV, Truman H, Hamilton S, Patel N, Newton Bishop JA, Leigh IM, Crowe JS. Isolation of human tumor-specific antibodies by selection of an antibody phage library on melanoma cells. Clin Cancer Res. 1999;5:925–931. [PubMed] [Google Scholar]
- 10.Franconi R, Roggero P, Pirazzi P, Arias FJ, Desiderio A, Bitti O, Pashkoulov D, Mattei B, Bracci L, Masenga V, et al. Functional expression in bacteria and plants of an scFv antibody fragment against tospoviruses. Immunotechnology. 1999;4:189–201. doi: 10.1016/s1380-2933(98)00020-7. [DOI] [PubMed] [Google Scholar]
- 11.Yi K, Chung J, Kim H, Kim I, Jung H, Kim J, Choi I, Suh P, Chung H. Expression and characterization of anti-NCA-95 scFv (CEA 79 scFv) in a prokaryotic expression vector modified to contain a Sfi I and Not I site. Hybridoma. 1999;18:243–249. doi: 10.1089/027245799315899. [DOI] [PubMed] [Google Scholar]
- 12.McCall AM, Adams GP, Amoroso AR, Nielsen UB, Zhang L, Horak E, Simmons H, Schier R, Marks JD, Weiner LM. Isolation and characterization of an anti-CD16 single-chain Fv fragment and construction of an anti-HER2/neu/anti-CD16 bispecific scFv that triggers CD16-dependent tumor cytolysis. Mol Immunol. 1999;36:433–445. doi: 10.1016/s0161-5890(99)00057-7. [DOI] [PubMed] [Google Scholar]
- 13.Winthrop MD, DeNardo SJ, DeNardo GL. Development of a hyperimmune anti-MUC-1 single chain antibody fragments phage display library for targeting breast cancer. Clin Cancer Res. 1999;5:3088s–3094s. [PubMed] [Google Scholar]
- 14.Stadler BM. Antibody production without animals. Dev Biol Stand. 1999;101:45–48. [PubMed] [Google Scholar]
- 15.Topping KP, Hough VC, Monson JR, Greenman J. Isolation of human colorectal tumour reactive antibodies using phage display technology. Int J Oncol. 2000;16:187–195. doi: 10.3892/ijo.16.1.187. [DOI] [PubMed] [Google Scholar]
- 16.Adams GP, Schier R. Generating improved single-chain Fv molecules for tumor targeting. J Immunol Methods. 1999;231:249–260. doi: 10.1016/s0022-1759(99)00161-1. [DOI] [PubMed] [Google Scholar]
- 17.Lekkerkerker A, Logtenberg T. Phage antibodies against human dendritic cell subpopulations obtained by flow cytometry-based selection on freshly isolated cells. J Immunol Methods. 1999;231:53–63. doi: 10.1016/s0022-1759(99)00140-4. [DOI] [PubMed] [Google Scholar]
- 18.van Kuppevelt TH, Dennissen MA, van Venrooij WJ, Hoet RM, Veerkamp JH. Generation and application of type-specific anti-heparan sulfate antibodies using phage display technology. Further evidence for heparan sulfate heterogeneity in the kidney. J Biol Chem. 1998;273:12960–12966. doi: 10.1074/jbc.273.21.12960. [DOI] [PubMed] [Google Scholar]
- 19.Rodenburg CM, Mernaugh R, Bilbao G, Khazaeli MB. Production of a single chain anti-CEA antibody from the hybridoma cell line T84.66 using a modified colony-lift selection procedure to detect antigen-positive ScFv bacterial clones. Hybridoma. 1998;17:1–8. doi: 10.1089/hyb.1998.17.1. [DOI] [PubMed] [Google Scholar]
- 20.Hoogenboom HR, de Bruïne AP, Hufton SE, Hoet RM, Arends JW, Roovers RC. Antibody phage display technology and its applications. Immunotechnology. 1998;4:1–20. doi: 10.1016/s1380-2933(98)00007-4. [DOI] [PubMed] [Google Scholar]
- 21.Songsivilai S, Dharakul T. Genetically engineered single-chain Fvs of human immunoglobulin against hepatitis C virus nucleocapsid protein derived from universal phage display library. Asian Pac J Allergy Immunol. 1998;16:31–41. [PubMed] [Google Scholar]
- 22.Chowdhury PS, Viner JL, Beers R, Pastan I. Isolation of a high-affinity stable single-chain Fv specific for mesothelin from DNA-immunized mice by phage display and construction of a recombinant immunotoxin with anti-tumor activity. Proc Natl Acad Sci USA. 1998;95:669–674. doi: 10.1073/pnas.95.2.669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Desai SA, Wang X, Noronha EJ, Kageshita T, Ferrone S. Characterization of human anti-high molecular weight-melanoma-associated antigen single-chain Fv fragments isolated from a phage display antibody library. Cancer Res. 1998;58:2417–2425. [PubMed] [Google Scholar]
- 24.Teunissen SW, Stassen MH, Pruijn GJ, van Venrooij WJ, Hoet RM. Characterization of an anti-RNA recombinant autoantibody fragment (scFv) isolated from a phage display library and detailed analysis of its binding site on U1 snRNA. RNA. 1998;4:1124–1133. doi: 10.1017/s1355838298980633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Takekoshi M, Maeda F, Tachibana H, Inoko H, Kato S, Takakura I, Kenjyo T, Hiraga S, Ogawa Y, Horiki T, et al. Human monoclonal anti-HCMV neutralizing antibody from phage display libraries. J Virol Methods. 1998;74:89–98. doi: 10.1016/s0166-0934(98)00072-x. [DOI] [PubMed] [Google Scholar]
- 26.Burioni R, Plaisant P, Bugli F, Solforosi L, Delli Carri V, Varaldo PE, Fadda G. A new subtraction technique for molecular cloning of rare antiviral antibody specificities from phage display libraries. Res Virol. 1998;149:327–330. doi: 10.1016/s0923-2516(99)89014-1. [DOI] [PubMed] [Google Scholar]
- 27.Okamoto N, Kennedy SD, Barron-Casella EA, Casella JF, Inoko H, Kickler TS. Identification of a human heavy chain antibody fragment directed against human platelet alloantigen 1a by phage display library. Tissue Antigens. 1998;51:156–163. doi: 10.1111/j.1399-0039.1998.tb02960.x. [DOI] [PubMed] [Google Scholar]
- 28.Jackson H, Bacon L, Pedley RB, Derbyshire E, Field A, Osbourn J, Allen D. Antigen specificity and tumour targeting efficiency of a human carcinoembryonic antigenspecific scFv and affin-ity-matured derivatives. Br J Cancer. 1998;78:181–188. doi: 10.1038/bjc.1998.462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kroll H, Kiefel V, Santoso S. Clinical aspects and typing of platelet alloantigens. Vox Sang. 1998;74 Suppl 2:345–354. doi: 10.1111/j.1423-0410.1998.tb05441.x. [DOI] [PubMed] [Google Scholar]
- 30.Maruyama T, Rodriguez LL, Jahrling PB, Sanchez A, Khan AS, Nichol ST, Peters CJ, Parren PW, Burton DR. Ebola virus can be effectively neutralized by antibody produced in natural human infection. J Virol. 1999;73:6024–6030. doi: 10.1128/jvi.73.7.6024-6030.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Owens GP, Williamson RA, Burgoon MP, Ghausi O, Burton DR, Gilden DH. Cloning the antibody response in humans with chronic inflammatory disease: immunopanning of subacute sclerosing panencephalitis (SSPE) brain sections with antibody phage li-braries prepared from SSPE brain enriches for antibody recog-nizing measles virus antigens in situ. J Virol. 2000;74:1533–1537. doi: 10.1128/jvi.74.3.1533-1537.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Barban V, Fraysse-Corgier S, Paranhos-Baccala G, Petit M, Manin C, Berard Y, Prince AM, Mandrand B, Meulien P. Identification of a human epitope in hepatitis C virus (HCV) core protein using a molecularly cloned antibody repertoire from a non-symptomatic, anti-HCV-positive patient. J Gen Virol. 2000;81:461–469. doi: 10.1099/0022-1317-81-2-461. [DOI] [PubMed] [Google Scholar]
- 33.Wan Z, Wang H, Jiang S. Selection of human anti-HAV McAb from a phage antibody library. Chin J Biotechnol. 1998;14:173–178. [PubMed] [Google Scholar]
- 34.Lu XP, Li BJ, Chen SL, Lu B, Jiang NY. Effect of chemotherapy or targeting chemotherapy on apoptosis of colorectal carcinoma. Shijie Huaren Xiaohua Zazhi. 1999;7:332–334. [Google Scholar]
- 35.Liu HF, Liu WW, Fang DC. Induction of apoptosis in human gastric carcinoma cell line SGC-7901 by anti-Fas monoclonal antibody. Shijie Huaren Xiaohua Zazhi. 1999;7:476–478. [Google Scholar]
- 36.Ning XY, Yang DH. Research and progress is in vivo gene therapy for primary liver cancer. Shijie Huaren Xiaohua Zazhi. 2000;8:89–90. [Google Scholar]
- 37.Chen JP, Lin C, Xu CP, Zhang XY, Fu M, Deng YP, Wei Y, Wu M. Transduction efficiency, biologic effects and mechanism of recombinant RA538, antisense C-myc adenovirus on different cell lines. Shijie Huaren Xiaohua Zazhi. 2000;8:266–270. [Google Scholar]
- 38.Guo SY, Gu QL, Liu BY, Zhu ZG, Yin HR, Lin YZ. Experimental study on the treatment of gastric cancer by TK gene combined with mIL-2 gene. Shijie Huaren Xiaohua Zazhi. 2000;8:974–978. [Google Scholar]
- 39.Pan X, Pan W, Ni CR, Ke CW, Cao GW, Qi ZT. Killing effect of tetracycline-controlled expression of DT/VEGF system on liver cell cancer. Shijie Huaren Xiaohua Zazhi. 2000;8:867–873. [Google Scholar]
- 40.Leng JJ, Chen YQ, Leng XS. Genetic therapy for pancreatic neoplasms. Shijie Huaren Xiaohua Zazhi. 2000;8:916–918. [Google Scholar]
- 41.Pan X, Pan W, Ke CW, Zhang B, Cao GW, Qi ZT. Tetracycline controlled DT/VEGF system gene therapy mediated by aden-ovirus vector. Shijie Huaren Xiaohua Zazhi. 2000;8:1121–1126. [Google Scholar]
- 42.Wang FS, Wu ZZ. Current situation in studies of gene therapy for liver cirrhosis and liver fibrosis. Shijie Huaren Xiaohua Zazhi. 2000;8:371–373. [Google Scholar]
- 43.Pan X, Ke CW, Pan W£¬He X, Cao GW, Qi ZT. Killing effect of DT/VEGF system on gastric carcinoma cell. Shijie Huaren Xiaohua Zazhi. 2000;8:393–396. [Google Scholar]
- 44.Cao GW, Qi ZT, Pan X, Zhang XQ, Miao XH, Feng Y, Lu XH, Kuriyama S, Du P. Gene therapy for human colorectal carcinoma using human CEA promoter contro led bacterial ADP-ribosylating toxin genes human CEA: PEA & amp; DTA gene transfer. World J Gastroenterol. 1998;4:388–391. doi: 10.3748/wjg.v4.i5.388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lu XP, Li BJ, Chen SL, Lu B, Jiang NY. Anti-CEA monoclonal antibody targeting therapy for colorectal carcinoma. Shijie Huaren Xiaohua Zazhi. 1999;7:329–331. [Google Scholar]
- 46.Engelstädter M, Bobkova M, Baier M, Stitz J, Holtkamp N, Chu TH, Kurth R, Dornburg R, Buchholz CJ, Cichutek K. Targeting human T cells by retroviral vectors displaying antibody domains selected from a phage display library. Hum Gene Ther. 2000;11:293–303. doi: 10.1089/10430340050016030. [DOI] [PubMed] [Google Scholar]
- 47.Wu YD, Song XQ, Zhou DN, Hu XH, Gan YQ, Li ZG, Liao P. Expermental and clinical study on targeting treatment of liver cancer using radionuclide anti-AFP antibody-MMC double bomb. Shijie Huaren Xiaohua Zazhi. 1999;7:387–390. [Google Scholar]
- 48.Zhang J, Liu YF, Yang SJ, Sun ZW, Qiao Q, Zhang SZ. Con-struction and expression of mouse/humanized scFv and their fusion to humanized mutant TNFα against hepatocellular carcinoma. Shijie Huaren Xiaohua Zazhi. 2000;8:616–620. [Google Scholar]
- 49.Chen ZN, Bian HJ, Jiang JL. Kecent progress inanti-hepatoma monoclonal antibody and its application. Huaren Xiaohua Zazhi. 1998;6:461–462. [Google Scholar]
- 50.Romanczuk H, Galer CE, Zabner J, Barsomian G, Wadsworth SC, O'Riordan CR. Modification of an adenoviral vector with biologically selected peptides: a novel strategy for gene delivery to cells of choice. Hum Gene Ther. 1999;10:2615–2626. doi: 10.1089/10430349950016654. [DOI] [PubMed] [Google Scholar]


