INTRODUCTION
Hepatocellular carcinoma (HCC) is the most common cause of death from cancer in China. The mechanisms of hepatocarcinogenesis are not yet known clearly. p16INK4a gene, the multiple tumor suppressor gene 1 (MTS1), encodes p16 protein, which acts as an inhibitor by binding directly to CDK4 and CDK6 and preventing its association with a cyclin. It was supposed to exert negative control on cell proliferation. p15INK4b gene, multiple tumor suppressor gene 2 (MTS2), is a homologue of p16INK4a and has a similar role in control of cell proliferation. Both of them were mapped to chromosome 9p21 region[1,2]. Although deletion or mutation of p16INK4a occurred in melanoma, biliary tract cancers, gastric carcinoma, hepatocarcinoma, and alterations of p15INK4b were shown in primary lung cancers, acute leukemia, biliary tract cancers and bladder tumors, there has been no report about whether p15INK4b gene alterated in primary hepatocellular carcinoma[3-9]. In the present study, exon 1, exon 2, exon 3 of p16INK4a and p15INK4b exon 2 in 35 HCC, 35 corresponding adjacent noncancerous liver cirrhosis were analyzed for somatic mutation with PCR-SSCP and one case of aberrant SSCP DNA was cloned and sequenced.
MATERIALS AND METHODS
Specimens and extraction
Tissue specimens used in the study were paraffin embedded and stored in Department of Pathology, the First Affiliated Hospital, West China University of Medical Sciences from 1991-1993. The 35 samples of human primary hepatocarcinoma and 35 corresponding adjacent noncancerous liver cirrhosis were stained with HE and examined under microscope. More than 70% sections used in PCR were hepatocarcinoma sections. And more than 80% cirrhosis sections were the noncancerous liver cirrhosis sections. DNA was extracted from 1-3 sections (10 μm) of paraffin embedded tissue blocks with xylene, ethanol and phenol method, and dissolved in 50 μL of distilled water[10].Ten samples of normal human blood DNA were extracted with standard method. The concentration of DNA was determined with spectrophotometer.
PCR
The exons of p16INK4a gene and exon 2 of p15INK4b gene were amplified using the following primers (Table 1):
Table 1.
Primers for p16INK4a and p15INK4b genes analysis
| Gene | Primers | Fragment length (bp) | Ref |
| p16INK4a exon 1 | 5'GGGAGCAGCATGGAGCCCG3' (sense) | 204 | [6] |
| 5'AGTCGCCCGCCATCCCCT3' (antisense) | |||
| p16INK4a intron 1 and exon 2 | 5'GGAAATTGGAAACTGGAAGC3' (sense) | 168 | [1] |
| 5'GCTGCCCATCATCATGACCT3' (antisense) | |||
| p16INK4a exon 2 and exon 3 | 5'GGCAGGTCATGATGATGGGC3' (sense) | 362 | [1] |
| 5'TCTGAGCTTTGGAAGCTCT3' (antisense) | |||
| p15INK4b exon 2 | 5'GGCCGGCATCTCCCATACCTG3' (sense) | 345 | [9] |
| 5'TGTGGGCGGCTGGGAACCTG3' (antisense) |
The PCR reaction was performed as follows: 200 ng DNA from paraffin embedded tissue or 100 ng DNA from normal human blood cells, 200 μmol/L each dATP, dGTP, dCTP and dTTP, 20 pmol primers, 1.5 u of Taq DNA polymerase (Sino-American Biotechnology Company) with a buffer provided by the manufacturer, in a total reaction volume of 25 μL. The thermal cycle profile was 1 min at 94 °C, 1 min at 66 °C (p16 exon 1, 204 bp) or 57 °C (p16 exon 2, 168 bp) or 55 °C (p16 exon 2, exon 3, 362 bp) or 68 °C (p15 exon 2), 2 min at 72 °C, 40 cycles. The PCR reaction mixture for p16INK4a exon1 (204 bp) exon 2 and exon 3 (362 bp) contained 5% dimethyl sulfoxide.
SSCP
PCR products were directly subjected to silver-staining SSCP analysis according to the method of Peng et al[1]. Ten μL of PCR products were denatured in 30 μL of 98% formamide, 10 mmol/L NaOH, 20 mmol/L EDTA, 0.05% (w/v) bromphenol blue and 0.05% (w/v) xylene cyanol, at 98 °C for 5 min. The samples were immediately loaded on an 8% polyacrylamide gel and run at 1.25 v/cm in 1 × TBE in 4 °C for approximately 14 h. Ten μL of PCR products of p16INK4a exon 2 and exon 3 (362 bp) was digested with Sma I. It generated two fragments (114 bp, 248 bp) which were then subjected to SSCP analysis. After electrophoresis, the gels were fixed, and stained with silver.
PCR cloning and sequencing
The gels were stained with silver. The staining was stopped by immersing the gel in 5% acetic acid, 16% methyl alcohol for 30 min. The gel was rinsed for 1 h in distilled water (changed each 5 min). The abnormal migration single strand DNA band of p15INK4b exon 2 was cut from gel and put into the same volume of distilled water. The gels were incubated in 37 °C water bath for 4 h, centrifuged at 10000 r/min for 5 min, then the gel was discarded. The supernatant was extracted with phenol/chloroform, and chloroform. The DNA from approximately 90 mg gel was placed in the PCR mixture for amplification under the same conditions as the initial PCR.
Following reamplification by PCR, the PCR products were isolated on 1.8% agrosegel. The 345 bp DNA band was cut from the gel and purified with Glassmilk Isolation Kit (made in our Lab). The DNA was blunted and phosphorated with E. coli-Klenow fragment and T4 polynucleotide kinase, ligated to Sma I site of pUC118. The ligation reaction was transformed into E. coli-JM109. The recombinants were screened by minipreparation of plasmid and digesting with restriction endonuclease. The DNA from the clone was screened by PCR-SSCP to identify the clone containing the shift single strand and was sequenced on a ABI 377 DNA sequencer by CyberSyn BJ. Company.
RESULTS
Polymorphism of the p16INK4a gene intron 1 and exon 2
Mutation of p16INK4a gene was analyzed in 31 of the 35 patients with HCC and 8 healthy blood donors. The PCR amplified 168 bp fragment of p16INK4a intron 1 and exon 2 containing 15 nucleotides within exon 2 and 153 nucleotides of its 5' flanking sequence within intron 1. Three patterns (A, B, B') of p16INK4a intron 1 and exon 2 (168 bp) at SSCP analysis were observed in hepatocellular carcinoma and corresponding adjacent noncancerous cirrhosis (Figure 1). The B pattern (48%, 15/31) outnumbered the B'pattern (26%, 8/31). A pattern was the least (13%, 3/31). Two patterns (B, B') at SSCP analysis were observed in healthy human blood cells. The B'pattern (62.5%, 5/8) out numbered the B pattern (37.3%, 3/8). The PCR amplified 362 bp fragment covered exon 2, exon 3 and intron 2 sequences. The PCR products were cleaved into two fragments 114 bp and 248 bp by digesting with Sma I. Neither band shift nor polymorphism at SSCP analysis was observed. The PCR amplified 204 bp fragment containing exon 1 and 19 nucleotides of 5' flanking upstream sequence and 41 nucleotides of 3' flanking downstream sequence. No band shift was detected in all of the samples.
Figure 1.
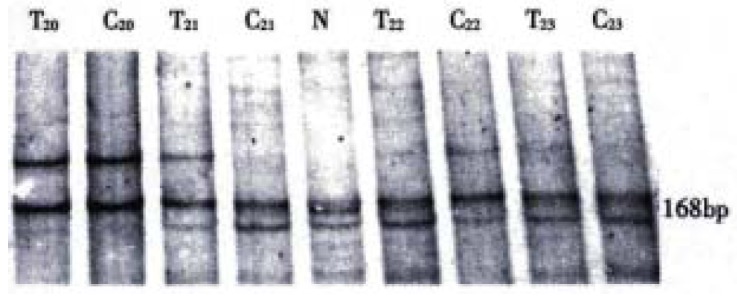
PCR-SSCP analysis of p16 gene intron 1 and exon 2. At the SSCP analysis T20 and C20 showed as A pattern; T23 and C23 showed as B pattern; T21 and C21 showed as B' and B pattern, respectively; T22 and C22 showed as B and B' pattern, respectively. T = Human hepatocarcinoma; C = Adjacent non-cancerous liver cirrhosis; N = Normal human leucocyte
SSCP analysis of p15INK4b exon 2
A 345 bp fragment containing p15INK4b gene exon 2 and 60 nucleotides of its 5' flanking sequence within intron 1 was amplified by PCR from all of the hepatocellular carcinomas, adjacent noncancerous cirrhosis and normal blood cells. No evidence of allele deletion was detected (Figure 2). On repeating SSCP analysis, one case of adjacent noncancerous cirrhosis showed an abnormal migration single strand (Figure 3). But no additional shift band was found in either the corresponding tumor tissue from the same patient or the other hepatocarcinoma and cirrhosis tissues. The identical migration single strand was detected in all of the 10 normal human blood cells. No evidence of polymorphism was found.
Figure 2.
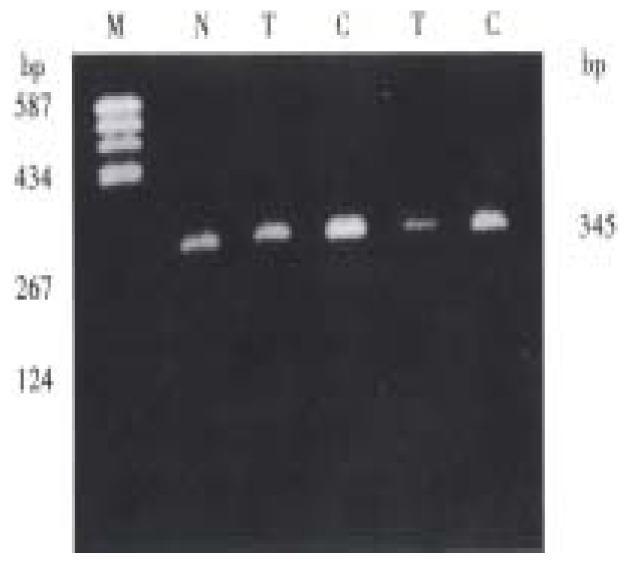
PCR product analysis of p15 gene exon 2 in human hepatocarcinoma on agarose gel.M = pBR322/Hea III; N = Normal human leukocyte; T = Human hepatocarcinoma; C = Adjacent non-cancerous liver cirrhosis
Figure 3.
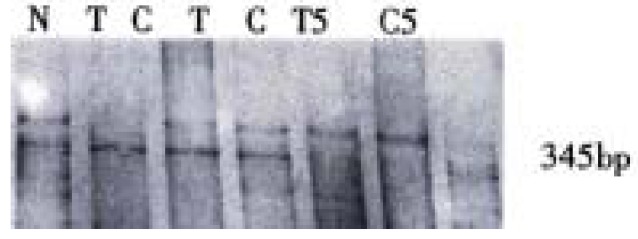
PCR-SSCP analysis of p15 gene exon 2. N = Normal human leukocyte; T = Human hepatocarcinoma; C = Adjacent non-cancerous liver cirrhosis
Cloning and sequencing of the abnormal single strand
The abnormal migration single strand DNA of C5 (adjacent noncancerous cirrhosis) was purified and cloned in Sma I site of pUC118. The recombinant plasmid, pP15E2, containing the p15INK4a exon 2 insert, was selected after minipreparation. The restriction endonuclease map analysis by Hind III, EcoR I, Bgl I revealed that pP15E2 plasmid contained a 345 bp insert (Figure 4) and the insert was placed in antisense orientation. PCR-SSCP analysis of the pP15E2 showed an identical abnormal migration single strand with that of C5 (Data not shown). Sequencing analysis of the insert of pP15E2 indicated that its sequence was identical to that of p15INK4b exon 2 and its upstream 60 nucleotides reported by Kamb (Figure 5).
Figure 4.
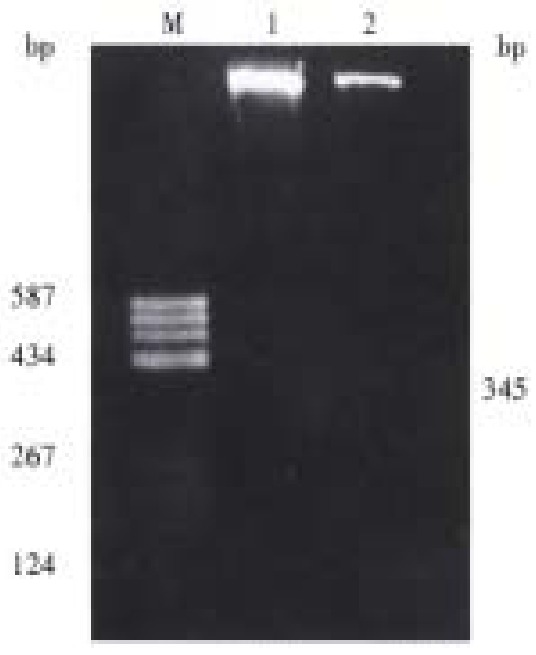
Restriction enzyme analysis of recombinant plasmid which contains aberrant single strand of p15 gene exon 2. M = pBR322/Hea III; 1 = Vector (pUC118) digested with EcoR I and Hind III; 2 = pP15E2 digested with EcoR I and Hind III
Figure 5.

The sequence of p15 gene exon 2 in pP15E2 recombinant plasmid.
DISCUSSION
The p16INK4a gene has been confirmed to be a tumor-suppressor gene by the analysis of p16 gene knock-out mice and its abnormalities have been reported in various kinds of primary cancers and cell lines, such as malignant melanomas, gliomas, glioblastomas, and esophageal squamous-cell carcinomas[1,12-14]. But there have been relatively few studies concerning the alteration of p16INK4a gene in hepatocellullar carcinoma. Kita et al[15] reported only three (5%) intragenic mutations of p16INK4a in primary HCC. Chaubert et al[6] found four patients carried hemizygous germ-line point mutations of the p16INK4a gene, suggesting the existence of familial HCC involving this gene. Hui et al[16] found higher proportion of HCCs may fail to express p16INK4a at the protein level. The general conclusion is that alterations of p16INK4a gene are infrequent in HCC. Our study showedpolymorphism of p16INK4a intron 1 and exon 2 at SSCP pattern. The A pattern was only preasented in HCC patients, not in the healthy blood donors. It is worthy to study whether people with the A pattern has HCC susceptibility. Five patients showed different SSCP patterns in tumor and noncancerous cirrhosis, most of which (4/5) ocurred in large HCC (no statistical significance was revealed). It suggested that intragenic mutation may occur during the progression of HCC, and advanced HCC, but not in the early stages of HCC.
There is argument on whether the inactivation of p15INK4b contributes to the carcinogenesis. Okamato et al[7] observed that non-small-cell lung cancer showed homozygous deletions of p15INK4b (23%), somatic mutation (12%) in exon 2, G→A and C→A polymorphism (8%) within the noncoding sequence of 23 nucleotides and 27 nucleotides of 5' of exon 2, respectively. The latter was named 15Int1-27A gene pattern. But Russin et al[17] detected only the 15Int1-27A polymorphism (13%), no mutation in non-small-cell lung cancers. Sill et al[8] reported 15Int1-27A polymorphism (13%), no somatic mutation in 80 acute leukemia. Orlow et al[1] observed deletions of p15INK4b in primary bladder tumors (8%). Yoshida reported no somatic mutation of p15INK4b gene in biliary tract cancers[4]. The general consensus is that frequency of mutation of p15INK4b in progression of cancer is very uncommon. In the present study no intragenic mutation of p15INK4b exon 2 was detected. Although one case of adjacent non-cancerous liver cirrhosis showed abnormal migration single strand, the cloning and sequencing of the aberrant SSCP DNA showed the sequence is identical to wild type p15INK4b exon 2 and 60 nucleotides upstream of exon 2. It suggested that intragenic mutation of p15INK5b exon 2 was an uncommon event in progression of HCC. This result is in agreement to the infrequent mutation of p16INK4a occcuring in HCC, and also similar to the studies in AML, bladder cancers and biliary tract cancers[4,8,9]. Whether the high frequency of p15INK4b somatic mutation in non-small cell lung cancers is related to the different types of tumor deserves further investigation.
Footnotes
Dr. Yang Qin, graduated from Beijing Medical University as a postgraduate in 1987, associate professor of Institute of Biochemistry and Molecular Biology, West China University of Medical Sciences, as a research fellow supported by Wellcome Trust Fellowship, in MRC Virology Unit, Institute of Virology, Glasgow University in UK during 1993-1994 and as a research fellow in Virology Laboratory, School of Medicine, Catholic University of Louvain in Belgium between 1994 -1995, majoring in molecular biology of cancer and molecular virology, having 21 papers published.
Project supported by the National Natural Science Foundation of China, NO. 39670702
Edited by Zhu LH
proofread by Sun SM
References
- 1.Kamb A, Gruis NA, Weaver-Feldhaus J, Liu Q, Harshman K, Tavtigian SV, Stockert E, Day RS, Johnson BE, Skolnick MH. A cell cycle regulator potentially involved in genesis of many tumor types. Science. 1994;264:436–440. doi: 10.1126/science.8153634. [DOI] [PubMed] [Google Scholar]
- 2.Hannon GJ, Beach D. p15INK4b is a potential effector of TGF-beta-induced cell cycle arrest. Nature. 1994;371:257–261. doi: 10.1038/371257a0. [DOI] [PubMed] [Google Scholar]
- 3.Nobori T, Miura K, Wu DJ, Lois A, Takabayashi K, Carson DA. Deletions of the cyclin-dependent kinase-4 inhibitor gene in multiple human cancers. Nature. 1994;368:753–756. doi: 10.1038/368753a0. [DOI] [PubMed] [Google Scholar]
- 4.Yoshida S, Todoroki T, Ichikawa Y, Hanai S, Suzuki H, Hori M, Fukao K, Miwa M, Uchida K. Mutations of p16Ink4/CDKN2 and p15INK4b/MTS2 genes in biliary tract cancers. Cancer Res. 1995;55:2756–2760. [PubMed] [Google Scholar]
- 5.Wu MS, Shun CT, Sheu JC, Wang HP, Wang JT, Lee WJ, Chen CJ, Wang TH, Lin JT. Overexpression of mutant p53 and c-erbB-2 proteins and mutations of the p15 and p16 genes in human gastric carcinoma: with respect to histological subtypes and stages. J Gastroenterol Hepatol. 1998;13:305–310. doi: 10.1111/j.1440-1746.1998.01560.x. [DOI] [PubMed] [Google Scholar]
- 6.Chaubert P, Gayer R, Zimmermann A, Fontolliet C, Stamm B, Bosman F, Shaw P. Germ-line mutations of the p16INK4(MTS1) gene occur in a subset of patients with hepatocellular carcinoma. Hepatology. 1997;25:1376–1381. doi: 10.1002/hep.510250613. [DOI] [PubMed] [Google Scholar]
- 7.Okamoto A, Hussain SP, Hagiwara K, Spillare EA, Rusin MR, Demetrick DJ, Serrano M, Hannon GJ, Shiseki M, Zariwala M. Mutations in the p16INK4/MTS1/CDKN2, p15INK4b/MTS2, and p18 genes in primary and metastatic lung cancer. Cancer Res. 1995;55:1448–1451. [PubMed] [Google Scholar]
- 8.Sill H, Aguiar RC, Schmidt H, Hochhaus A, Goldman JM, Cross NC. Mutational analysis of the p15 and p16 genes in acute leukaemias. Br J Haematol. 1996;92:681–683. doi: 10.1046/j.1365-2141.1996.340858.x. [DOI] [PubMed] [Google Scholar]
- 9.Orlow I, Lacombe L, Hannon GJ, Serrano M, Pellicer I, Dalbagni G, Reuter VE, Zhang ZF, Beach D, Cordon-Cardo C. Deletion of the p16 and p15 genes in human bladder tumors. J Natl Cancer Inst. 1995;87:1524–1529. doi: 10.1093/jnci/87.20.1524. [DOI] [PubMed] [Google Scholar]
- 10.Wright DK, Manon MM. Sample preparation from paraffin-embedded tissues. In: Michael A Inns Eds. PCR Protocols A Guide to Methods and Applications.San Diego; Academic Press Inc; 1992. pp. 153–158. [Google Scholar]
- 11.Peng H, Du M, Ji J, Isaacson PG, Pan L. High-resolution SSCP analysis using polyacrylamide agarose composite gel and a background-free silver staining method. Biotechniques. 1995;19:410–414. [PubMed] [Google Scholar]
- 12.Serrano M, Lee H, Chin L, Cordon-Cardo C, Beach D, DePinho RA. Role of the INK4a locus in tumor suppression and cell mortality. Cell. 1996;85:27–37. doi: 10.1016/s0092-8674(00)81079-x. [DOI] [PubMed] [Google Scholar]
- 13.Schmidt EE, Ichimura K, Reifenberger G, Collins VP. CDKN2 (p16/MTS1) gene deletion or CDK4 amplification occurs in the majority of glioblastomas. Cancer Res. 1994;54:6321–6324. [PubMed] [Google Scholar]
- 14.Mori T, Miura K, Aoki T, Nishihira T, Mori S, Nakamura Y. Frequent somatic mutation of the MTS1/CDK4I (multiple tumor suppressor/cyclin-dependent kinase 4 inhibitor) gene in esophageal squamous cell carcinoma. Cancer Res. 1994;54:3396–3397. [PubMed] [Google Scholar]
- 15.Kita R, Nishida N, Fukuda Y, Azechi H, Matsuoka Y, Komeda T, Sando T, Nakao K, Ishizaki K. Infrequent alterations of the p16INK4a gene in liver cancer. Int J Cancer. 1996;67:176–180. doi: 10.1002/(SICI)1097-0215(19960717)67:2<176::AID-IJC4>3.0.CO;2-Q. [DOI] [PubMed] [Google Scholar]
- 16.Hui AM, Sakamoto M, Kanai Y, Ino Y, Gotoh M, Yokota J, Hirohashi S. Inactivation of p16INK4 in hepatocellular carcinoma. Hepatology. 1996;24:575–579. doi: 10.1002/hep.510240319. [DOI] [PubMed] [Google Scholar]
- 17.Rusin MR, Okamoto A, Chorazy M, Czyzewski K, Harasim J, Spillare EA, Hagiwara K, Hussain SP, Xiong Y, Demetrick DJ, et al. Intragenic mutations of the p16(INK4), p15(INK4B) and p18 genes in primary non-small-cell lung cancers. Int J Cancer. 1996;65:734–739. doi: 10.1002/(SICI)1097-0215(19960315)65:6<734::AID-IJC4>3.0.CO;2-#. [DOI] [PubMed] [Google Scholar]


