INTRODUCTION
It has been discovered that hepatitis C virus (HCV) presents considerable nucleotide variation and has many genotypes. At least twelve, 5 of which prevalent, i.e.: I/1a, II/1b, III/2a, IV/2b, and V/3a types. The different genotypes of HCV may possess some retationship with regional distribution, clinical manifestation, response to treatment and prognosis of HCV infection, thus to study the genotypes of HCV further in different areas is of practical value. We detected HCV genotypes in 94 patients with hepatitis C and in 6 patients with primary hepatocarcinoma by PCR assay with four kinds of type-specific primers with a view to study the distribution of HCV genotypes in hepatitis C in Shenyang area and their clinical significances.
MATERIALS AND METHODS
Patients
One hundred serum samples with positive anti-HCV and HCV RNA were received from patients who were in clinic or in ward of our department from July 1993-June 1996. Among the 94 patients with hepatitis C, there were 9 acute hepatitis, 73 chronic hepatitis (50 mild degree, 14 moderate and 9 severe ones), 12 active posthepatitic cirrhosis. Mean age 38.8 years ± 15.6 years, male 77 and female 17. Forty-nine of them (52.1%) had history of blood or blood product transfusion. Superinfection or coinfection with B hepatitis was excluded, 2 cirrhosis were confirmed pathologically after operation. Six patients with primary hepatocarcin oma were diagnosed by serum AFP, B -ultrasound, CT and MRI. One had surgical biopsy.
HCV genotyping primers
HCV genotyping primers in core region were synthesized by the Bioengineering Research Center in Shanghai of Chinese Academy of Sciences.The nucleotide sequences of primers were as follows: u-niversal primers: P1 sense 5’CGCGCGACTAGGAAGACTTC3’ (nt 139-158); P2 antisense 5’ATGTACCCCATGAGGTCGCT3’ ( nt 391-410 ), type specific primers: P3 sense 5’AGGAAGACTTCCGAGCGGTC3’ (nt 148-167); P4 anti-sense 5’TGCCTTGGGGATAGGCTGAC3’ (nt 185-204) (HCV-I); P5 antisense 5’GAGCCATCCTGCCCACCCCA3’ (nt 2 72-291) (HCV-II); P6 antisense 5’CCAAGAGGGACGGGAACCTC3’ (nt 302-321) (HCV-III); P7 antisense 5’ ACCCTCGTTTCCGTACAGAG 3’ ( nt 251-270 ) (HCV-IV).
Methods
Serum HCV RNA was extracted using one-step method of guanidinium thiocyanate. For detecting HCV genotypes, Okamoto’s[1] method was used but modified. Major procedures were: (1) reverse transcription and first PCR amplification: in extracted RNA, we added 5 units of avian myeloblastosis virus reverse transcript ase (AMV-RT) (Promega), 20 units of inhibitor of RNAase (RNasin) (Promega), 2 U Taq enzyme (Promega), 2 mM dNTP 3 μL, 10 × AMV- RT buffer 3 μL and universal primers ( P1, P2 ) 0.8 μmol/L. Total volume was 30 μL.After reverse transcription at 42 °C for 20 min, first PCR was performed for 35 cycles. Each cycle included denaturation at 94 °C for 1 min, annealing at 55 °C for 1.5 min and extension at 72 °C for 2 min. (2) The second PCR amplification: in 5 μL products of the first PCR, we added 3 μL 10 × AMV-RT buffer, 2 mM dNTP 3 μL, 2 U Taq enzyme and 0.8 μmol/L of type-specific primers (P3, P4,5,6,7). Total volume was 30 μL. PCR conditions were: denaturation at 94 °C for 1 min, annealing at 60 °C for 1 min, extension at 72 °C 1.5 min, for 30 cycles, then another extension at 72 °C for 5 min. In every amplification, positive control of HCV RNA serum and negative control of normal blood sample were provided. (3) Ten microliters products of the second PCR were subjected to electrophoresis on 6% polyacrylamide gel, stained with ethidium bromide, and observed under ultraviolet ray. Each HCV genotype was characterized by different nucleotide lengths: 57 bp for type I, 144 bp for type II, 174 bp for type III and 1.23 bp for type IV.
RESULTS
One hundred serum samples were genetically classified into 3 types: type II in 58 samples (58%), type III in 27 ( 27% ) and mixed type (II/III) in 14( 14% ), only 1 sample ( 1% ) remained unclassified (Figure 1).
Figure 1.
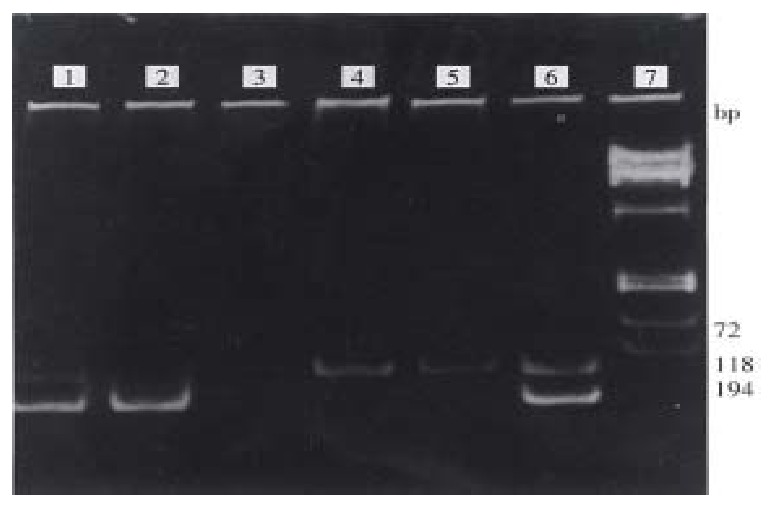
The results in HCV genotyping. 1, 6: II/III mixed type. 2: HCV-II genotype. 3: Negative control. 4,5: HCV-III genotype. 7: Φx 174 DNA / Hae III marker.
In 94 patients with hepatitis C, the distribution of HCV genotypes in patients with hepatitis C was not identical. The data were analyzed with R × C Table test, χ2 = 28.9, P < 0.01. In acute hepatitis, in mild, moderate and severe chronic hepatitis, cirrhosis, the infectious rates of type II were 55.6%, 36.0%, 71.4%, 88.9% and 91.7%, respectively, but those of type III were 22.2%, 46.0%, 7.14%, 0% and 8.3%, respectively. In moderate an d severe chronic hepatitis and cirrhosis, the proportion of type II was significantly elevated in comparison with that in mild chronic hepatitis (P < 0.05 or P < 0.01), but the proportion of type III was significantly lower than that in mild ones (P < 0.05). Six patients with hepatocarcinoma were all infected by genotype II HCV (Table 1). The genotype distribution of HCV in post-transfusional and sporadic hepatitis C was identical. The results were analyzed with R × C Table χ2 test, χ2 = 3.74, P > 0.05 (Table 2).
Table 1.
HCV genotypes of hepatitis C and hepatocarcinoma
| Clinical status | Cases |
HCV II |
HCV III |
Mixed (II/III) |
Unknown |
||||
| Cases | (%) | Cases | (%) | Cases | (%) | Cases | (%) | ||
| Acute hepatitis | 9 | 5 | (55.6) | 2 | (22.2) | 2 | (22.2) | ||
| Chronic hepatitis | |||||||||
| Mild degree | 50 | 18 | (36.0) | 23 | (46.0) | 9 | (18.0) | ||
| Moderate degree | 14 | 10 | (71.4)a | 1 | (7.14)a | 2 | (14.3) | 1 | (7.14) |
| Severe degree | 9 | 8 | (88.9)b | 1 | (11.1) | ||||
| Cirrhosis of liver | 12 | 11 | (91.7)b | 1 | (8.3) | ||||
| Hepatocarcinoma | 6 | 6 | (100.0)a | ||||||
| Total | 100 | 58 | (58) | 27 | (27) | 14 | (14) | 1 | (1) |
P < 0.05,
P < 0 .01, compared with mild chronic hepatitis, by χ2 test.
Table 2.
HCV genotypes of post-transfusional and sporadic hepatitis C
| Groups | Cases |
HCV II |
HCV III |
Mixed (II/III) |
Unknown |
||||
| Cases | (%) | Cases | (%) | Cases | (%) | Cases | (%) | ||
| Post-transfusional hepatitis | 49 | 26 | (53.1) | 12 | (24.5) | 10 | (20.4) | 1 | (2.0) |
| Sporadic hepatitis | 45 | 26 | (57.8) | 15 | (33.3) | 4 | (8.9) | ||
DISCUSSION
There are some differences in distribution of HCV genotypes in different regions. In America, infection of HCV-I genotype is predominated, but in China and Japan, HCV-II genotype is dominant over HCV-III genotype. Du et al[2] reported that infection by type II was predominant in Beijing, Wuhan, Guangzhou, Xi’an, Chongqing, Guangxi areas. Among these areas the highest infection rate of type III was in Beijing (19%), the infection rates of mixed type II/III were the highest in Guangzhou (9%). Our study showed in Shenyang that the infection rate of type II ( 58% ) > type III ( 27% ) and mixed type II/III (14%), the latter two were higher in Shenyang than those in other parts of China. Therefore, future prevention and treatment strategy should be directed towards type II HCV mainly, but not neglecting type III and type II/III. In our study, no HCV-I and HCV-IV were found. One serum sample was positive for HCV RNA by repeated PCR assay, but it could not be classified after the two tests, indicating there might be other HCV genotype in our area.
Clinical manifestations of hepatitis C infected by HCV type II are usually more severe, those of HCV type III are milder. The HCV genotypes seem to be closely related to the clinical condition and degree of severity of hepatitis C. All our 6 patients with hepatoca rcinoma were found to carry HCV type II which indicated that infection of type II may be related to occurrence of hepatocarcinoma[3].
No significant difference was found in the distribution of HCV genotypes in post-transfusional and sporadic hepatitis C, showing HCV genotypes may be not related to routes of infection. The infection rate ( 20.9% ) of mixed type ( II/III ) in post-transfusional hepatitis was higher than that in sporadic hepatitis ( 9.8% ), indicating repeated blood or blood product transfusion may be contributory.
Footnotes
Edited by Xie-Ning Wu
Proofread by Qi-Hong Miao
References
- 1.Okamoto H, Sugiyama Y, Okada S, Kurai K, Akahane Y, Sugai Y, Tanaka T, Sato K, Tsuda F, Miyakawa Y. Typing hepatitis C virus by polymerase chain reaction with type-specific primers: application to clinical surveys and tracing infectious sources. J Gen Virol. 1992;73(Pt 3):673–679. doi: 10.1099/0022-1317-73-3-673. [DOI] [PubMed] [Google Scholar]
- 2.Du SC, Tao QM, Zhu L, Liu JX, Wang H, Sun Y. Genotyping study of hepatitis C virus by restriction fragment length polymorphsim in its 5'NC region. Zhonghua Yixue Zazhi. 1993;1:7–9. [Google Scholar]
- 3.Yang JM, Liu WW, Luo YH. Genotypic investigation of hepatitis C virus in patient with primary hepatic carcinoma, liver cirrhosis and hepatitis. Zhonghua Chuanranbing Zazhi. 1995;1:1–3. [Google Scholar]


