Abstract
Fragile X syndrome (FXS), the most-frequently inherited form of intellectual disability and the most-prevalent single-gene cause of autism, results from a lack of fragile X mental retardation protein (FMRP), an RNA-binding protein that acts, in most cases, to repress translation. Multiple pharmacological and genetic manipulations that target receptors, scaffolding proteins, kinases and translational control proteins can rescue neuronal morphology, synaptic function and behavioural phenotypes in FXS model mice, presumably by reducing excessive neuronal translation to normal levels. Such rescue strategies might also be explored in the future to identify the mRNAs that are critical for FXS pathophysiology.
Since its initial description as an X-linked heritable form of mental insufficiency1 and the subsequent demonstration that patients exhibit a constriction at the tip of the X chromosome (indicating a region of chromosomal fragility)2, fragile X syndrome (FXS) has become recognized as the most-prevalent form of inherited cognitive impairment. Moreover, as the diagnosis of autism spectrum disorder (ASD) has become more sophisticated, it has become evident that individuals with ASD and FXS have several characteristics in common, such as avoidance of eye contact, repetitive behaviours and reduced social interactions3. Indeed, FXS is now considered to lie within the autism spectrum and to be the most-common single-gene cause of ASD.
The cause of both FXS and the X-chromosome restriction noted above is an expansion of 200 or more CGG repeats in the fragile X mental retardation 1 (FMR1) gene, which causes its methylation and inactivation4. The loss of FMR protein (FMRP), the product of FMR1, alters synapse function and morphology, with profound effects on higher brain function. FMRP is thought to be ubiquitous in cells; in neurons, it is found in dendrites, at the base of synapses, in axons and in the soma, and it shuttles in and out of the nucleus5. In the absence of FMRP, global protein synthesis in the hippocampus is elevated by ~15–20%6–8. It is almost certain that this excessive protein synthesis is a major contributor to the pathophysiology in FXS. In addition, the elevation of protein synthesis in the absence of FMRP means that synaptic activation is unable to further increase protein production, and it is likely that this absence of stimulus-induced protein synthesis plays an important part in disease pathophysiology9–11.
It seems axiomatic that the key to understanding the molecular basis of FXS will be to identify the mRNAs under the control of FMRP and to define the mechanism (or mechanisms) by which it represses their translation. These tasks have been made daunting by the fact that the brain contains a highly complex population of mRNAs, which in many cases are the products of alternative splicing and/or RNA editing12,13. Moreover, various mRNAs are over-represented in certain cell types (for example, in neurons versus glia) and even in certain locations within cells (such as in the cell soma versus the dendrites). This compartmentalization of mRNAs adds to the difficulty in determining which mRNA targets of FMRP are important for any given biological phenomenon. Parsing these and other biological variables is certainly a formidable challenge, but their importance for understanding FXS is reflected in the continuous appearance of essential new information in the literature, reflecting the intense research interest in this area.
Although both pharmacological and genetic approaches have been successful in mitigating several pathophysiological features of FXS in animal models of the disorder, emergent genetic rescue experiments (those in which FXS is corrected when both Fmr1 and a second gene are lacking) in mice have been particularly important for dissecting key facets of the disease. Ten studies have reported such genetic rescues of Fmr1-knockout (KO) mice6,8,14–21 (TABLE 1), eight of which investigated protein synthesis and found that it was restored to wild-type levels6,8,14,15,17–19. If resetting translational homeostasis is central to these and perhaps all rescue mechanisms, then it follows that there must be specific mRNAs whose translation is both elevated in FXS and restored to wild-type levels in the genetic rescue experiments. Identifying these mRNAs and understanding their functional roles is likely to have profound implications for the understanding and treatment of FXS.
Table 1.
Manipulations that reset translational homeostasis and rescue phenotypes in FXS mice
| Target | Rescue strategy | Normalization of excessive protein synthesis? | Normalization of mGluR-induced LTD? | Behavioural phenotypes normalized | Refs |
|---|---|---|---|---|---|
| mGluR5 | Genetic reduction and mGluR5 antagonist | Yes | Yes | Extinction memory, startle response, locomotor activity and audiogenic seizures | 6,76 |
| GABAB receptor | receptor GABAB antagonist | Yes | ND | Audiogenic seizures, repetitive behaviour, locomotor activity and motor learning | 80 |
| PIKE | Genetic reduction | Yes | Yes | Repetitive behaviours and nest building | 18 |
| p110β subunit of PI3K | Genetic reduction | Yes | ND | Goal-directed decision making, behavioural flexibility, nest building and repetitive behaviour | 19 |
| HOMER1A | Genetic reduction | Yes | No | Audiogenic seizures and anxiety behaviour | 17 |
| TSC2 | Genetic reduction | Yes | Yes | Context discrimination | 14 |
| S6K1 | Genetic reduction | Yes | Yes | Recognition memory, behavioural flexibility, social novelty and motor learning | 15 |
| CPEB | Genetic reduction | Yes | Yes | Audiogenic seizures, anxiety behaviour, nest building and spontaneous alteration | 8 |
| RAS | Farnesyltransferase inhibitor | Yes | Yes | Audiogenic seizures | 88 |
| MEK1 | MEK1 inhibitor | Yes | ND | Audiogenic seizures | 77 |
| MNK1, MNK2 and p-eIF4E | Genetic reduction of MNK1 and/or MNK2, MNK inhibitor, and genetic reduction of p-eIF4E | Yes | Yes | Social novelty, social interaction, anxiety behaviour and audiogenic seizures | 16 |
| MMP9 | Genetic reduction and MMP9 inhibitor | ND | Yes | Anxiety behaviour, locomotor activity and social novelty | 20 |
| CB1R | Genetic reduction and CB1R antagonist | ND | ND | Recognition memory, anxiety behaviour, nociception and audiogenic seizures | 21 |
CB1R, cannabinoid 1 receptor; CPEB, cytoplasmic polyadenylation element binding protein; FXS, fragile X syndrome; LTD, long term depression; MEK1, MAPK/ERK kinase 1; mGluR, metabotropic glutamate receptor; MMP9, matrix metalloproteinase 9; MNK, MAP kinase-interacting serine/threonine-protein kinase; ND, not determined; PI3K, phosphoinositide 3-kinase; p-eIF4E, phosphorylated eukaryotic initiation factor 4E; PIKE, PI3K enhancer; S6K1, p70 S6 kinase 1; TSC2, tuberous sclerosis complex 2.
In this Review, we discuss recent advances in FXS research, with a particular focus on the underlying mechanisms by which FMRP controls mRNA translation. In support of the idea that FXS is a disease of translation gone awry, we describe evidence that alteration of the translational landscape can reverse pathophysiologies associated with the syndrome. Finally, we provide a roadmap for future investigations into how restoration of translational homeostasis in the FXS brain can be leveraged into new therapies for treating the disease.
Translational control by FMRP
Nearly two decades ago, data suggested that FMRP was most likely to be a repressor of translation22, a conclusion that was supported by the findings of many subsequent studies in humans7, mice11,23,24 and Drosophila melanogaster25 (although it is important to note that some studies suggest that FMRP can also activate translation; discussed below). Translation involves three broad steps: initiation, elongation and termination26. Initiation of translation begins with the association of eukaryotic initiation factor 4F (eIF4F) — a complex of eIF4E, eIF4G and eIF4A — with the 7-methyl-guanosine residue that ‘caps’ the 5′ ends of mRNAs. The eIF4F complex helps to position the 40S ribosomal subunit on the 5′ end of the mRNA. This promotes 40S subunit ‘scanning’ of the 5′ untranslated region (UTR) until the initiation codon (which usually is the first AUG) is recognized, after which the 40S subunit is joined by the 60S ribosomal subunit to form an 80S monosome that is capable of elongating the nascent polypeptide chain. Finally, termination of translation occurs when the 80S monosome dissociates from the mRNA at the termination codon, thereby releasing the completed polypeptide.
Initiation is by far the most complex step of translation and is generally considered to be the rate-liming step; hence, it is subject to many forms of regulation27. However, it is worth noting that microRNAs (miRNAs), which have an important role in regulating translation and act primarily at initiation, also regulate elongation28. Similarly, emerging evidence indicates that FMRP acts at both initiation and elongation, and also works in conjunction with miRNAs.
Regulation of translation initiation
Studies have suggested that FMRP may regulate translation initiation through its interactions with the cap-binding translation factor eIF4E and cytoplasmic FMRP-interacting protein 1 (CYFIP1). Repression of the cap-dependent translation process described above takes place when eIF4E-binding proteins (4E-BPs) bind to eIF4E. Derepression (and initiation of translation) occurs when the 4E-BPs are phosphorylated by mammalian target of rapamycin complex 1 (mTORC1; see FIG. 1), which permits eIF4E to associate with eIF4G29. eIF4G, in turn, indirectly recruits the 40S ribosomal subunit to the 5′ end of the mRNA. When bound to eIF4G, eIF4E can be phosphorylated by MAP kinase-interacting serine/threonine-protein kinase 1 (MNK1; also known as MKNK1) and/ or MNK2 (also known as MKNK2), which can modulate translation30 (FIG. 1). FMRP binds to CYFIP1 (REFS 31,32), which is both a component of the WAVE (Wiskott–Aldrich syndrome protein family verprolin homologue) regulatory complex, which promotes actin remodelling, and a non-canonical 4E-BP33. Thus, the activity of the FMRP–CYFIP1 complex, through an as-yet-unknown mechanism, can switch between translational control and actin remodelling34. It has been shown that activation of either brain-derived neurotrophic factor (BDNF) or group 1 metabotropic glutamate receptor (mGluR) signalling can release CYFIP1 from eIF4E (but not CYFIP1 from FMRP) to promote translation33,35. This activation is consistent with the observation that the involvement of the FMRP–CYFIP1 complex in long-lasting synaptic plasticity is controlled by MNK1 and/or MNK2 (REF. 36). In Fmr1-KO mice, interactions between eIF4E and eIF4G are increased17,37, as is eIF4E phosphorylation16. Finally, FMRP can also regulate initiation indirectly by suppressing the translation of components of the mTORC1 signalling pathway (see below). Thus, FMRP can directly and indirectly regulate translation initiation.
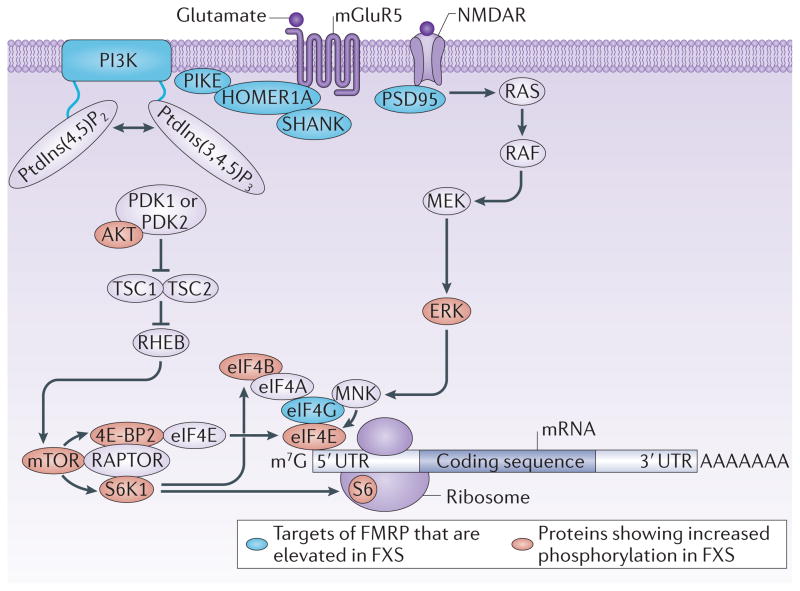
Figure 1. Translational control pathways that are dysregulated in FXS.
Normally, stimulation of cell surface receptors, including NMDA receptors (NMDARs) and group 1 metabotropic glutamate receptors (mGluRs; such as mGluR5), results in the activation of the phosphoinositide 3-kinase (PI3K)23,94, mammalian target of rapamycin complex 1 (mTORC1; which comprises mTOR bound to regulatory-associated protein of mTOR (RAPTOR))37 and extracellular signal-regulated kinase (ERK)77 signalling pathways in neurons. mTORC1 phosphorylates eukaryotic initiation factor 4E (eIF4E) binding proteins (4E-BPs), including 4E-BP2, the predominant 4E-BP isoform in the mouse brain, which derepresses eIF4E to promote ‘cap’-dependent translation. mTORC1 also phosphorylates and activates p70 S6 kinase 1 (S6K1), which phosphorylates ribosomal protein S6 and eIF4B. Phosphorylation of eIF4B by S6K1 stimulates the helicase activity of eIF4A to promote cap-dependent translation. mTORC1-dependent translation is triggered upstream by a signalling pathway involving PI3K, 3-phosphoinositide-dependent protein kinase 1 (PDK1) and/or PDK2, AKT, tuberous sclerosis 1 (TSC1) and/ or TSC2 and RAS homologue enhanced in brain (RHEB). ERK phosphorylates and activates MAP kinase-interacting serine/threonine-protein kinases (MNKs), which phosphorylate eIF4E to promote translation. This ERK-dependent translation is triggered upstream by a pathway involving RAS, RAF and MAPK/ERK kinase 1 (MEK1). mGluR5 signals to the PI3K–AKT–mTORC1 pathway via HOMER1A, SH3 and multiple ankyrin repeat domains (SHANK) proteins and PI3K enhancer (PIKE), and NMDARs signal to ERK via postsynaptic density protein 95 (PSD95). The protein levels of several targets of the RNA-binding protein fragile X mental retardation protein (FMRP)67,68 are increased in fragile X syndrome (FXS). The increased expression of these proteins results in basally elevated PI3K, mTORC1 and ERK signalling and thus increased translation. m7G, 7-methyl-guanosine; PtdIns(4,5)P2, phosphatidylinositol-4,5-bisphosphate; PtdIns(3,4,5)P3, phosphatidylinositol-3,4,5-trisphosphate; UTR, untranslated region. Adapted from REF. 5, Nature Publishing Group.
Cooperation with miRNAs and other factors
FMRP has also been suggested to regulate translation through its interaction with miRNAs. Several studies have revealed both biochemical and functional interactions among FMRP, miRNAs, the Argonaute (also known as eIF2C) protein of the RNA-induced silencing complex (RISC)38–43, Dicer and miRNA precursors44. In D. melanogaster, Fmr1 is physically associated with miRNAs, and loss-of-function mutations suggest that Fmr1 modulates miRNA expression to control neuronal development40,42. For example, steady-state levels of miR-124a were reduced in Fmr1-mutant flies, and the effects of miR-124a on dendritic arborization were shown to be dependent on Fmr1 (REF. 42). In mice, FMRP is associated with the RISC and/or miRNAs — such as miR-125a, miR-125b and miR-132 — that cooperate to regulate the protein synthesis that is important for determining dendritic spine morphology38,41. It is possible that FMRP may co-opt the RISC and/or miRNAs to repress synthesis of GluN2A (an NMDA receptor subunit), as interactions between the GluN2A mRNA 3′ UTR and miR-125b have been reported38. However, the extent to which the binding site of FMRP and those for miRNAs in this region overlap is not known.
Surprisingly, recent work has shown that FMRP is not only a negative regulator of translation but can also enhance translation, depending on the proximity of the FMRP-binding sites within the mRNA to the RNA helicase Moloney leukaemia virus 10 (MOV10) and the presence or absence of GC-rich secondary structures in the mRNA45. FMRP binds directly to G-quartet structures46–49, which provide a motif that drives mRNA localization to dendrites50, although the role of these interactions in translation is not known. G-rich sequences in the 3′ UTR of the mRNA of the important synapse component postsynaptic density protein 95 (PSD95; also known as DLG4)51 occur within regions that are binding sites for miR-125a and FMRP41,52. Thus, it is possible that FMRP and associated factors may cooperate to regulate the accessibility of miRNA target sequences that are embedded within the secondary structure of the mRNA53. The presence of such interactions between FMRP and miRNAs would predict dysregulation of miRNAs in FXS: indeed, this has been recently reported in Fmr1-KO mice54 and human FXS induced pluripotent stem cell (iPSC)-derived neurons55.
FMRP phosphorylation also plays an important part in the bidirectional regulation of mRNA translation in neurons41,56–59. Phosphorylated FMRP can repress translation via recruitment of the RISC, whereas mGluR-induced dephosphorylation of FMRP by protein phosphatase 2A (PP2A) stimulates translation and concomitant release of the RISC41. A second study has also reported that phosphorylated FMRP is necessary for miRNA-mediated repression of translation60. Translational repression can also be removed by mGluR-induced ubiquitylation and degradation of FMRP locally within dendrites and at synapses9,57, and the dephosphorylation of FMRP can enhance its ubiquitylation57.
Stalling polyribosomes to regulate elongation
Experiments from several laboratories have demonstrated that most FMRP co-sediments with polyribosomes when analysed by sucrose gradient ultracentrifugation61–63. Such co-sedimentation suggests, but does not prove, that FMRP is directly associated with the translational apparatus (although it is important to note that some laboratories found that FMRP sediments to the translating ribonucleoprotein (RNP) part of a sucrose gradient, which suggests an altogether different mode of FMRP activity (see below)61,62). This polysome co-sedimentation suggests that FMRP might inhibit translation at the level of polypeptide elongation (also called ribosome transit)63. A subsequent in vivo study gave strong credence to this hypothesis. Using crosslinking and immunoprecipitation (CLIP) — a technique in which ultraviolet (UV) light is used to induce covalent crosslinking between proteins and the mRNAs to which they are bound followed by RNP immunoprecipitation and high-throughput sequencing64,65 — it was shown that FMRP binds most frequently to the coding regions of mRNAs, with fewer binding sites within the 5′ and 3′ UTRs (the sites most-often bound by other RNA-binding proteins). Thus, in contrast to the findings of in vitro RNA–protein binding studies46,66, the in vivo CLIP study of FMRP67 suggests that FMRP binds to specific mRNAs in a cis-element-independent manner (although another study did detect some cis-element-specific binding sites for FMRP68). These data suggest that, by binding to the coding regions of target mRNAs, FMRP could act as a roadblock to impede ribosome transit and thus slow polypeptide elongation (FIG. 2). . Further corroborating evidence from structural analysis of D. melanogaster Fmrp showed that it interacts with the ribosome via Ribosomal protein L5 (REF. 69), which again implies that it could alter ribosome function to limit its ability to elongate polypeptides.
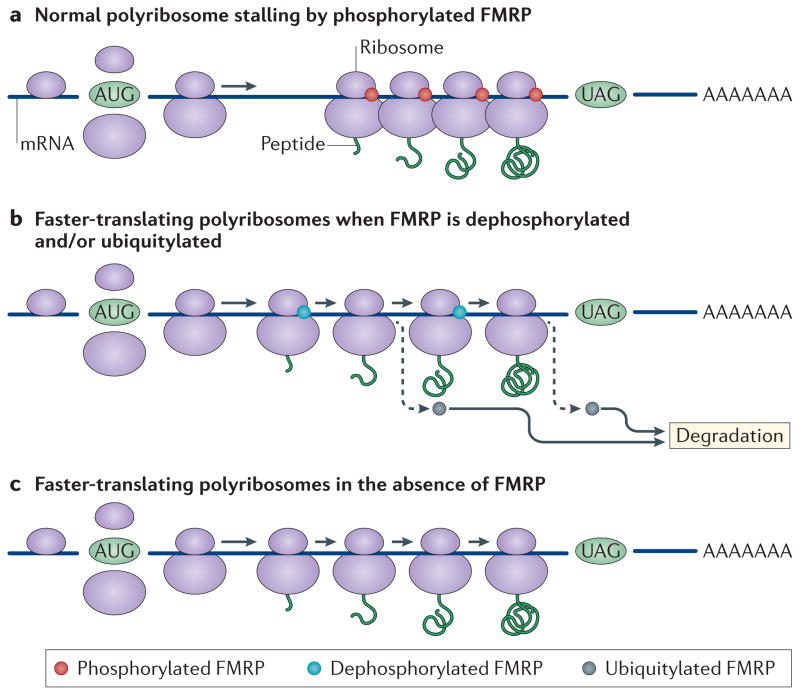
Figure 2. FMRP may stall polyribosomes to reduce the rate of translation elongation.
a | It is hypothesized that phosphorylated fragile X mental retardation protein (FMRP) associates with actively translating ribosomes and causes them to stall and accumulate on the mRNA molecule, slowing elongation (AUG and UAG are the initiation and termination codons, respectively). b | FMRP-regulated translation increases when FMRP is dephosphorylated56,57, ubiquitylated9,57 and eventually destroyed via proteasomal degradation. c | In fragile X syndrome, the absence of FMRP results in faster translation of FMRP target mRNAs8. Adapted from REF. 5, Nature Publishing Group.
A recent study directly measured ribosome transit in the presence or absence of FMRP8. The investigators prepared forebrain lysates from wild-type and Fmr1-KO mice. These were supplemented with hippuristanol, a drug that inhibits the initiation-factor helicase eIF4A70 and blocks new rounds of translation initiation, allowing ribosomes that are already associated with mRNAs to continue translation. When the incorporation of radioactive amino acids into elongating polypeptides was determined over time, Fmr1-KO brain lysates were found to elongate polypeptides approximately 40–50% faster than wild-type lysates, demonstrating that FMRP does indeed slow or stall ribosomes.
Rescue of phenotypes in FXS model mice
The observations described above not only demonstrate the complexity of FMRP function but also suggest that there are multiple ways by which FMRP activity might be either modulated or mimicked, thereby providing a potentially rich source of therapies to treat FXS. Animal models of FXS, particularly those that use genetically altered mice, have been especially important for assessing such possible interventions.
Targeting mGluR5
FMRP acts downstream of mGluRs, including mGluR5 (also known as GRM5), a G protein-coupled receptor that populates the postsynaptic membrane71. Activation of mGluR5 triggers kinase signalling cascades that lead to long-term depression (LTD), a long-lasting form of synaptic plasticity. mGluR-induced LTD requires both protein synthesis and FMRP expression; in the absence of FMRP, LTD is exaggerated, resulting in weaker synaptic connections between pre- and postsynaptic regions72. These weak synaptic connections probably contribute to the reduced cognitive function observed in individuals with FXS71.
The observations of exaggerated mGluR-induced LTD in hippocampal slices of Fmr1-KO mice led to the mGluR theory of FXS, which posits that many syndrome-associated phenotypes could be attributable to increased signalling through these receptors72. Thus, downregulation of mGluR signalling would be predicted to mitigate many of the brain abnormalities that characterize this disease. Initial studies to test this hypothesis relied on drugs such as MPEP (2-methyl-6-(phenylethynyl)-pyridine), which inhibit mGluR5 activation and do indeed suppress some FXS-like seizure phenotypes in mice73,74. However, the most compelling evidence for the importance of mGluR5 in FXS comes from a mouse model in which one allele of the gene encoding mGluR5 is disrupted. When mated with Fmr1-KO mice, the resulting offspring, which exhibit a 50% reduction in levels of mGluR5 expression, display a remarkably robust rescue of several FXS-associated phenotypes6. Subsequent studies have confirmed various aspects of these initial observations75–77.
Although the work described above implies that an excess of mGluR5-mediated LTD at excitatory synapses may contribute to FXS, other studies have shown that stimulating inhibitory GABA-regulated synapses with GABA receptor agonists, such as benzodiazepine or arbaclofen, also ameliorates several FXS-associated phenotypes in mice78–81 and D. melanogaster75,82. Arbaclofen has been reported to have some success in treating individuals with FXS83. It is important to emphasize that many human trials of FXS suffer from numerous confounding factors, including difficulties in selecting appropriate outcome measures, strong placebo effects and drug tolerance84,85. These studies do not invalidate the mGluR theory, but they do illustrate the difficulty in translating results from animal models into the clinic. To confirm the mGluR hypothesis, it will be essential to identify other molecules in the mGluR signalling pathway that, when either depleted or overexpressed, reduce pathophysiologies associated with FXS.
In Fmr1-KO mice, mGluR-induced LTD is independent of protein synthesis, presumably because the levels of proteins required for LTD are already elevated. Thus, a resetting of translation in the brain — that is, restoration of translational homeostasis — might correct the abnormal mGluR5-mediated synaptic plasticity (and related aberrant behavioural phenotypes) in FXS model mice. Indeed, although there is likely to be more to FXS pathophysiology than altered protein synthesis (BOX 1), studies based on restoring balanced protein synthesis have been remarkably successful and may portend new classes of drugs to treat individuals with FXS (BOX 1; see also BOX 2 for unexpected results on the involvement of tuberous sclerosis complex (TSC) in FXS).
Targeting translational control molecules
p70 S6 kinase 1 (S6K1; also known as RPS6KB1) is a substrate for mTORC1 that phosphorylates a diverse set of substrates, including the initiation factor eIF4B (FIG. 1), which stimulates the helicase activity of eIF4A86. In addition, S6K1 phosphorylates and inactivates eukaryotic elongation factor 2 (eEF2) kinase, which normally phosphorylates and inactivates eEF2 to inhibit translation elongation. Thus, activation of S6K1 can promote translation through its effects on both initiation and elongation. Fmr1-KO mice exhibit increased S6K1 signalling, as evidenced by elevated phosphorylation of the mTORC1 site on S6K1 (REF. 37), increased phosphorylation of ribosome protein S6 and eIF4B, and increased levels of eEF2 (REF. 15). Genetic reduction of S6K1 in Fmr1-KO mice prevents the increased phosphorylation of S6 and eIF4B and the increased levels of eEF2, and this correlates with the resetting of protein synthesis to levels comparable to those in wild-type mice15. In addition, reducing S6K1 levels in Fmr1-KO mice prevents the immature dendritic spine morphology, enhanced mGluR-induced LTD and multiple behavioural phenotypes seen in these mice15. These results support the idea that dysregulated protein synthesis is the key causal factor in FXS and suggest that S6K1 can be targeted to reset translational homeostasis to stabilize neurological function in the disease.
As described above, eIF4E is a substrate for MNK1 and/or MNK2 (FIG. 1) and, when phosphorylated, can increase translation30. Recently, it was shown that haploinsufficiency for either MNK1 or MNK2 prevents increased eIF4E phosphorylation, excessive protein synthesis, immature dendritic spine morphology, enhanced mGluR-induced LTD and several aberrant behaviours in Fmr1-KO mice16. These findings were mimicked when the Fmr1-KO mice were crossed with mice in which Ser209 of eIF4E (the residue that is phosphorylated by MNK1 and MNK2) is mutated to alanine, rendering it incapable of being phosphorylated by these kinases16. These findings also support the idea that targeting molecules that regulate translation can prevent multiple phenotypes in FXS model mice. Notably, MNK1 and/or MNK2 activation and eIF4E phosphorylation are regulated by extracellular signal-regulated kinase 1 (ERK1) and/or ERK2 (REF. 87), which are key components of the signalling pathway downstream of mGluRs and other receptors that mediate protein synthesis-dependent synaptic plasticity. Inhibitors of ERK1 and/or ERK2 signalling corrected the excess basal protein synthesis and reduced the susceptibility of Fmr1-KO mice to audiogenic seizures88, as did lovastatin, a cholesterol-lowering drug that decreases RAS–ERK signalling88. Interestingly, lovasta-tin has also been used to correct impairments in plasticity and behaviour in a mouse model of neurofibromatosis type 1 (NF1), a developmental disorder that is associated with intellectual disability89.
Cytoplasmic polyadenylation element-binding protein (CPEB) associates with the CPE in the 3′ UTR of its target mRNAs and stimulates translation by promoting poly(A)-tail lengthening90. CPEB controls translation in many biological systems, including neurons and the brain91,92. Although global translation is not altered in the brains of mice that lack CPEB, excessive translation as well as enhanced mGluR-induced LTD, dendrite spine morphology and numerous behavioural phenotypes are rescued in Fmr1-KO mice with a genetic reduction of CPEB (FIG. 3). CPEB often promotes cytoplasmic polyadenylation, and it is possible that this mRNA-processing event might be involved in controlling the elevated protein synthesis in the brains of FMRP-deficient mice8. Once again, these results indicate that rebalancing protein synthesis can rescue FXS-associated pathophysiologies.
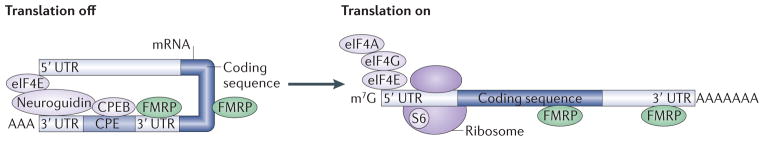
Figure 3. Involvement of CPEB in mediating FMRP activity.
Cytoplasmic polyadenylation element-binding protein (CPEB) associates with CPEs in the 3′ untranslated region (UTR) of target mRNAs and stimulates translation by promoting lengthening of the poly(A) tail. Neuroguidin is a eukaryotic initiation factor 4E (eIF4E)-binding protein (4E-BP) that binds to CPEB to prevent this CPE-dependent translation. It is known that excessive translation in fragile X syndrome (FXS) model mice is normalized by genetic reduction of CPEB, although the mechanisms through which the two proteins interact are currently unclear. In the left panel, CPEB is shown associated with fragile X mental retardation protein (FMRP) and neuroguidin, which in turn interacts with the ‘cap’-binding factor eIF4E. This configuration of factors would be hypothesized to silence mRNAs that are bound by both FMRP and CPEB. As shown in the right panel, group 1 metabotropic glutamate receptor (mGluR; not shown) activation could lead to poly(A)-tail elongation and dissociation of neuroguidin from eIF4E, thereby allowing for the assembly of the eIF4F complex (which consists of eIF4E, eIF4G and eIF4A) on the cap and the initiation of translation. In this scenario, FMRP remains bound to the mRNA and S6K1 phosphorylates the initiation factors and ribosomal protein S6 on the 40S subunit to stimulate translation8. m7G, 7-methyl-guanosine.
Targeting molecules upstream and downstream of translation
FMRP controls the synaptic synthesis of two key components of the phosphoinositide 3-kinase (PI3K) signalling complex: the catalytic subunit, p110β (also known as PIK3CB), and PI3K enhancer (PIKE)23. PIKE67 and p110β68,93 mRNAs are targets of FMRP, and the translation and expression of both are elevated in the brains of Fmr1-KO mice23,37. Enhanced basal PI3K activity and the absence of mGluR-dependent activation of p110β-associated PI3K have been observed in both Fmr1-KO mice and human FXS cells23,94. Treatment of cultured neurons and synaptic fractions from Fmr1-KO mice23, as well as cultured cells from patients with FXS94,95, with either class 1a PI3K inhibitors or p110β subunit-selective inhibitors ameliorates excess protein synthesis and other FXS-associated cellular phenotypes. More-recent work has shown that diverse FXS-associated phenotypes, including impaired signalling and protein synthesis, abnormal spine morphology, impaired synaptic plasticity and behavioural abnormalities, can be rescued by genetic knockdown of either p110β or PIKE in the mouse model18,19. Genetic reduction of the D. melanogaster orthologue of PIKE, Centaurin 1A (CenG1A), abolished the excessive PI3K signalling and impairments in neuronal development and short-term memory that are seen in the fly model of FXS18. Importantly, acute silencing of p110β-associated PI3K activity in adult FXS model mice rescued FXS-associated phenotypes, including higher-order cognitive impairments19. These studies suggest that targeting the PI3K signalling complex associated with mGluRs may provide an alternative strategy to dampen excess signalling and restore translational homeostasis. Because FMRP directly represses the synthesis of the PI3K signalling complex, which is overactive in FXS, targeting PI3K signalling would be predicted to correct the excess protein synthesis that is downstream of the mGluR–PI3K signalling complex.
Metabotropic receptors are linked to PI3K signalling by a long isoform of the postsynaptic scaffold protein Homer. In Fmr1-KO mice, co-immunoprecipitation between long Homer and mGluR5 is reduced96, suggesting that there may be an altered balance between the levels of the long isoform of Homer, which promotes PI3K signalling, and of the short isoform of Homer, which disrupts PI3K signalling. Interestingly, in mice in which Fmr1 is knocked out and there is also a genetic reduction of short Homer, mGluR–Homer scaffolds are restored and the impaired mGluR signalling and excessive protein synthesis seen in Fmr1-KO mice are rescued (FIG. 1). Moreover, rescue of altered network hyper-excitability, sensitivity to acoustic stimulation and behavioural anomalies was also observed in mice in which both Fmr1 and Homer1a were knocked out17 (TABLE 1). However, the exaggerated mGluR-induced LTD phenotype present in Fmr1-KO mice was not rescued, nor was the translational dysregulation of FMRP target mRNAs. This study suggests that restoration of Homer scaffolds can rescue many but not all FXS-associated phenotypes. More work is needed to understand the mechanism by which Homer scaffolds are disrupted in FXS and the possible inter-relationship with the exaggerated PI3K signalling. It is possible that elevated PI3K signalling leads to disruption of Homer scaffolds and the uncoupling of mGluR5 to PI3K.
Elevated expression and activity of the extracellular protein matrix metalloproteinase 9 (MMP9) contributes to impairments in dendritic spine morphology and altered neuronal signalling in FXS. FXS-associated phenotypes can be rescued in Fmr1-KO mice either through the genetic reduction of MMP9 (REF. 20) or by treatment with minocycline, which lowers MMP9 activity97. Minocycline has also been reported to have success in ameliorating symptoms in individuals with FXS98. MMP9 mRNA is a target of FMRP, which regulates its translation at synapses99, and increased translation of MMP9 in Fmr1-KO mice is prevented by inhibiting MNK1 and/or MNK2 and phosphorylation of eIF4E16. In the absence of FMRP, excessive MMP9 activity may contribute to exaggerated PI3K–mTORC1 signalling: indeed, in Fmr1-KO mice, several defects, including dendritic spine defects, exaggerated mGluR-induced LTD, behavioural impairments, macroorchidism and increased basal phosphorylation of PI3K–mTORC1 pathway components, were abolished by genetic deletion of MMP9 (REF. 20). That elevated MMP9 activity exacerbates PI3K signalling suggests that loss of FMRP exerts indirect effects on PI3K signalling beyond the translational dysregulation of FMRP target mRNAs that directly regulate PI3K, such as PIKE and p110β. Altered function of the endocannabinoid system may also play a part in FXS. For example, cannabinoid 1 receptor (CB1R)-dependent regulation of synaptic strength due to mGluR5 activation is altered in Fmr1-KO mice100. Moreover, it was shown that both genetic reduction of CB1R and treatment with CB1R antagonists normalize behavioural impairments, susceptibility to audiogenic seizures and upregulation of the mTORC1 signalling pathway in Fmr1-KO mice21. Thus, MMP9 and CB1R are linked to altered translational control and may be viable therapeutic targets for resetting translational homeostasis in FXS.
Other therapeutic targets have been examined, and interventions affecting several of these have rescued FXS-associated phenotypes in mouse models, although it remains unclear whether excessive protein synthesis due to loss of FMRP is rescued in these animals. These targets include p21-activated kinase (PAK)101, striatum-enriched protein-tyrosine phosphatase (STEP; also known as PTPN5)102, glycogen synthase kinase 3 (REFS 103–105) and amyloid precursor protein (APP)106. Each of these approaches is thought to restore function by targeting distal mechanisms that may not directly result from loss of FMRP-mediated regulation of protein synthesis.
Key FMRP target mRNAs
Several different approaches, including both in vitro and in vivo analyses, have been used to identify FMRP target mRNAs. The three canonical RNA-binding motifs present in FMRP, an RGG (Arg-Gly-Gly) box and two KH (heterogeneous nuclear RNP K homology) domains, have been the focus of these studies. One study46 used in vitro RNA selection (also known as SELEX (selected evolution of ligands by exponential enrichment)) to demonstrate that the RGG box of FMRP binds to a G-quartet structure. Several RNAs harbour such a G-quartet, including those encoding the structural protein microtubule-associated pro-tein 1B (MAP1B), the axon-guidance molecule semaphorin 3F and the voltage-gated potassium channel Kv3.1 (also known as KCNC1). To get a better sense of the range of RNAs bound by FMRP (irrespective of whether they contain a G-quartet), another study107 performed RIP–ChIP (co-immunoprecipitation of RNA followed by microarray analysis) with an FMRP antibody using mouse brain lysates. This revealed that 432 mRNAs were co-precipitated with FMRP and that more than half of them were aberrantly translated in Fmr1-KO mice. Interestingly, approximately 70% of these RNAs contained G-quartets, which would seem to strongly validate the findings of the in vitro selection experiments. However, RNA immunoprecipitation can result in re-association of RNA–protein interactions in lysed cells108, suggesting that methods to directly analyse RNA–protein interactions in vivo would provide more-accurate information.
To circumvent this potential problem, a more-recent study67 used UV–CLIP and RNA sequencing, which is a rigorous method that avoids nonspecific interactions because the RNAs and proteins are covalently linked before cell lysis and immunoprecipitation64. In this experiment, the FMRP-associated RNAs showed little overlap with those of the previous study107. Moreover, FMRP crosslinked to coding regions of RNA in a cis-element-independent manner and thus did not bind to G-quartets even in RNAs that contained them. Of course, it is possible that G-quartets are not efficiently UV-crosslinked to proteins, and thus may indeed be bona fide sites of FMRP interaction. In this study, FMRP crosslinked to 842 distinct brain mRNAs67, several of which encode cell-scaffolding proteins (including MAP1B, tau and myosin)26. FMRP was also shown to bind to mRNAs encoding proteins that mediate synaptic function and cell signalling, such as calcium/ calmodulin-dependent protein kinase II (CaMKII), GluRs and many RHO GTPases, guanine nucleotide exchange factors (GEFs) and other GTPases. Other signalling factor mRNAs, such as tyrosine kinases and protein tyrosine phosphatases, were also found to be bound by FMRP. In addition, FMRP was shown to associate with mRNAs encoding translation initiation factors (including Argonaute and eIF4G), translation elongation factors (such as eEF2), several potassium-gated channels and even some presynaptic components, such as Piccolo and Bassoon. Interestingly, 117 of the mRNAs that FMRP was shown to associate with have been linked to autism, including phosphatase and tensin homologue (PTEN), TSC2 and SH3 and multiple ankyrin repeat domains 3 (SHANK3)67.
These studies have therefore revealed that there are many potential targets of FMRP that might be affected by loss of the protein in FXS. Comparisons of tissue or cultured neurons from wild-type and Fmr1-KO mouse brains have revealed many mRNAs that seem to be regulated at the translational level, and these have been investigated extensively by western blotting8,23,37,109, polysome sucrose gradient ultracentrifugation41,67 and mass spectrometry110. However, although significant changes in the levels of several of these mRNAs are associated with various diseases, such as cancer, autism and schizophrenia111, the increase in their levels in either cells or tissues that lack FMRP is relatively small (overall, the levels of most proteins increase by approximately 15–20%). Thus, it is reasonable to ask how such a small increase can cause such a dramatic disease. Could it be that the amalgamation of the effects of all of the dysregulated mRNAs causes FXS, or are there a few key mRNAs whose translation changes in a more-dramatic way, thus making them more-directly responsible for the disease state5? Investigation of this issue is paramount because it has important implications for understanding FXS aetiology and may also provide a focus for the development of new treatments for the disease.
Perspective
One of the most vexing questions in FXS research is why normal brain function is so dependent on FMRP-regulated translation. Similarly, it is important to understand how resetting translational homeostasis in Fmr1-KO mice by genetic deletion or pharmacological inhibition of receptors, scaffolding proteins, protein kinases and translational control proteins reverses such a range of morphological, synaptic and behavioural phenotypes (TABLE 1). To address these issues, we must not only determine the identities of the mRNAs whose translation is altered in FXS model mice but also identify the mRNAs whose translation is rescued when genetic and pharmacological approaches are used to restore translational homeostasis (FIG. 4). It will then be important to determine the importance of these mRNAs and to decipher the molecular mechanism (or mechanisms) by which their translation is rescued. Upregulation of PI3K18,19,23,94 and mTORC1–S6K1 (REFS 95,112) signalling, as well as excessive protein synthesis, has been reported in non-neuronal cells, human blood cells and post-mortem brain tissue from individuals with FXS94,95. To determine their relevance to human disease, these findings (and those made in the Fmr1-KO mice) must be confirmed by analysing neurons derived from patients with FXS. iPSC-derived neurons from individuals with FXS have been generated113,114 and used to screen for compounds that increase FMRP expression115. Similar approaches should be used to screen for compounds that reset translational homeostasis in iPSC-derived neurons from individuals with FXS.
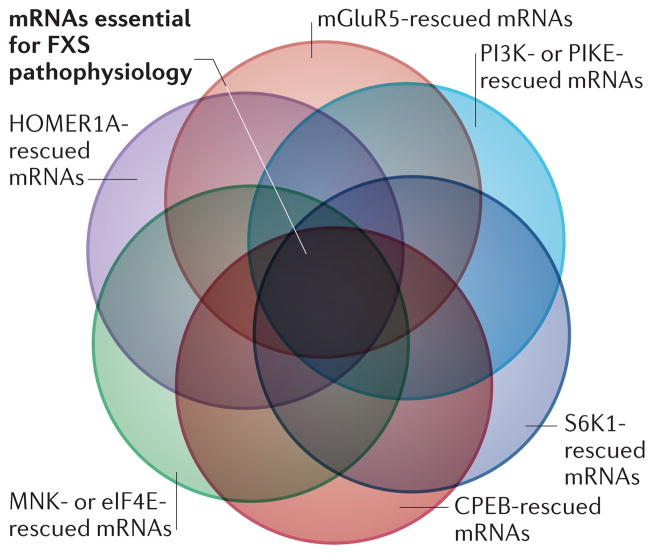
Figure 4. Identification of dysregulated mRNAs that are rescued by pharmacological or genetic manipulations in FXS model mice.
Venn diagram in which the circles represent populations of mRNAs (currently unknown) whose aberrant translation in fragile X mental retardation 1 (Fmr1)-knockout mice is rescued when the following are either inhibited with pharmacological agents or genetically ablated: HOMER1A; metabotropic glutamate receptor 5 (mGluR5); phosphoinositide 3-kinase (PI3K) and/ or PI3K enhancer (PIKE); p70 S6 kinase 1 (S6K1); cytoplasmic polyadenylation element-binding protein (CPEB); and MAP kinase-interacting serine/threonine-protein kinase (MNK) and/or eukaryotic initiation factor 4E (eIF4E). It is proposed that there is a subset of mRNAs that are rescued under all conditions. These mRNAs are likely to be essential for normal neuronal function and have the potential to be novel therapeutic targets. FXS, fragile X syndrome.
As discussed above, multiple genetic manipulations and pharmacological treatments targeting molecules such as mGluR5, HOMER1A, PI3K, S6K1, RAS, MAPK/ERK kinase 1 (MEK1; also known as MAP2K1), ERK, MNK1 and/or MNK2, and CPEB can rescue FXS model mice. Because all of these molecules converge on the translational apparatus (FIGS 1,3), it seems likely that the translation of a common set of mRNAs is rescued in these various paradigms and that these mRNAs almost certainly have a profound impact on FXS pathophysiology (FIG. 4). It is likely that the identification of these mRNAs will build on results showing that FMRP represses translation at the level of ribosome transit (polypeptide elongation)8,67. For example, determination of ribosome transit times for all mRNAs in an unbiased, whole-genome manner in FXS model mice and in genetic rescue conditions will reveal transcripts that are centrally important for the disease. This approach should not only permit the identification of the mRNA targets of FMRP whose translation is essential for normal brain function but also suggest new therapies to treat the syndrome based on alteration of the translational landscape.
Box 1. Non-canonical actions of FMRP: regulation of ion channels.
Fragile X mental retardation protein (FMRP) has been shown to regulate the translation, expression and activity of the voltage-gated potassium channels Kv4.2 (also known as KCND2)116–118 and Kv3.1b119, as well as potassium/sodium hyper-polarization-activated cyclic nucleotide-gated ion channel 1 (HCN1)120. Interestingly, FMRP also interacts directly with and regulates several ion channels in a manner that is independent of its canonical function in translation. For example, FMRP binds to the sodium-activated potassium channel Slack (also known as KCNT1), and this interaction causes opening of the channel121. FMRP also interacts with the β4 subunit of BK (big potassium) channels, which results in excessive action potential broadening during repetitive neuronal activity, enhanced presynaptic calcium influx and increased release of neurotransmitters122. Recently, a missense mutation in the amino terminus of FMRP in a human patient with intellectual disability and seizures was shown to impair the interaction between FMRP and BK channels as well as other aspects of presynaptic function123. Finally, FMRP was reported to directly interact with the calcium channel Cav2.2 (also known as CACNA1B) carboxy-terminal domain, the region that not only targets the channel for proteasome-mediated destruction but also controls synaptic exocytosis124. Thus, it is possible that the lack of FMRP in fragile X syndrome could result in abnormalities in neuronal excitability, firing and homeostatic plasticity that are independent of its role in regulating translation.
Box 2. TSC and FXS: two wrongs make a right.
Tuberous sclerosis complex (TSC) is a neurodevelopmental disorder caused by mutations in TSC1 (encodes hamartin) and TSC2 (REF. 125). Inactivation of TSC1 and/ or TSC2 is known to activate mammalian target of rapamycin complex 1 (mTORC1) signalling via the small G protein RAS homologue enhanced in brain (RHEB)126; thus, one would predict that TSC model mice, like fragile X mental retardation 1 (Fmr1)-knockout (KO) mice, would exhibit excessive translation. However, Tsc2 heterozygous KO mice were shown to exhibit depressed translation in the hippocampus. Astonishingly, when TSC model mice were crossed with Fmr1-KO mice, several of the phenotypes that were present in both Fmr1-KO mice and Tsc2 heterozygous KO mice, including altered levels of protein synthesis, were returned to normal14. Although the precise molecular mechanisms responsible for the rescue of excessive translation and fragile X syndrome (FXS)-associated phenotypes by genetically reducing levels of TSC2 are unknown, it is tempting to speculate that the levels of TSC2, the mRNA of which is a target of FMR protein (FMRP)67, are elevated in the neurons of Fmr1-KO mice. Regardless of the mechanism (or mechanisms), the rescue of phenotypes displayed by Fmr1-KO mice by the genetic reduction of TSC2 is consistent with the idea that resetting translational homeostasis is critical for amelioration of FXS pathophysiologies.
Acknowledgments
This work was supported by Fragile X Syndrome Research Center grant HD082013 from the US National Institutes of Health.
Glossary
- Alternative splicing
A form of nuclear pre-mRNA splicing in which different exons can be included into the mature mRNA
- RNA editing
A post-transcriptional event in which ribonucleotides are modified
- for example
from adenosine to inosine or from cytosine to uridine
- RNA-induced silencing complex
(RISC) A complex of a microRNA and Argonaute (plus associated factors, such as GW182) that silences mRNA expression by inhibiting translation and/or causing mRNA instability
- G-quartet
A guanosine-containing quadruplex structure in RNA that is thought to be a binding site for fragile X mental retardation protein
- Induced pluripotent stem cell
(iPSC) A type of stem cell that is produced by transducing a differentiated cell with certain transcription factors. iPSCs have the potential to be converted into any terminally differentiated cell type
- Polyribosomes
Functional units of protein synthesis made of several ribosomes attached along the length of an mRNA molecule
- Cis
A chemical term denoting a structure in which, for example, two atoms reside on the same side of a particular structure (in contrast to trans, which refers to atoms on different sides of a structure). In molecular biology, cis refers to a sequence of DNA or RNA that often serves as a binding site for a trans-acting factor, such as a protein
- Long-term depression
(LTD) A form of synaptic plasticity that results in a long-lasting decrease in the strength of synaptic transmission. LTD is exaggerated in fragile X mental retardation 1-knockout mice
Footnotes
Competing interests statement
The authors declare no competing interests.
References
- 1.Martin JP, Bell JA. Pedigree of mental defect showing sex-linkage. J Neurol Psychiatry. 1943;6:154–157. doi: 10.1136/jnnp.6.3-4.154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lubs HA. A marker X chromosome. Am J Hum Genet. 1969;21:231–244. [PMC free article] [PubMed] [Google Scholar]
- 3.Santoro MR, Bray SM, Warren ST. Molecular mechanisms of fragile X syndrome: a twenty-year perspective. Annu Rev Pathol. 2012;7:219–245. doi: 10.1146/annurev-pathol-011811-132457. [DOI] [PubMed] [Google Scholar]
- 4.Nelson DL, Orr HT, Warren ST. The unstable repeats—three evolving faces of neurological disease. Neuron. 2013;77:825–843. doi: 10.1016/j.neuron.2013.02.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Darnell JC, Klann E. The translation of translational control by FMRP: therapeutic targets for FXS. Nat Neurosci. 2013;16:1530–1536. doi: 10.1038/nn.3379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dolen G, et al. Correction of fragile X syndrome in mice. Neuron. 2007;56:955–962. doi: 10.1016/j.neuron.2007.12.001. Describes the first genetic rescue of multiple FXS pathophysiologies accomplished with reduction of mGluR5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Qin M, et al. Altered cerebral protein synthesis in fragile X syndrome: studies in human subjects and knockout mice. J Cereb Blood Flow Metab. 2013;33:499–507. doi: 10.1038/jcbfm.2012.205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Udagawa T, et al. Genetic and acute CPEB1 depletion ameliorate fragile X pathophysiology. Nat Med. 2013;19:1473–1477. doi: 10.1038/nm.3353. Demonstrates that genetic reduction of CPEB1 prevents multiple FXS-associated phenotypes. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hou L, et al. Dynamic translational and proteasomal regulation of fragile X mental retardation protein controls mGluR-dependent long-term depression. Neuron. 2006;51:441–454. doi: 10.1016/j.neuron.2006.07.005. [DOI] [PubMed] [Google Scholar]
- 10.Muddashetty RS, Kelic S, Gross C, Xu M, Bassell GJ. Dysregulated metabotropic glutamate receptor-dependent translation of AMPA receptor and postsynaptic density-95 mRNAs at synapses in a mouse model of fragile X syndrome. J Neurosci. 2007;27:5338–5348. doi: 10.1523/JNEUROSCI.0937-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Weiler IJ, et al. Fragile X mental retardation protein is necessary for neurotransmitter-activated protein translation at synapses. Proc Natl Acad Sci USA. 2004;101:17504–17509. doi: 10.1073/pnas.0407533101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fernandez-Moya SM, Bauer KE, Kiebler MA. Meet the players: local translation at the synapse. Front Mol Neurosci. 2014;7:84. doi: 10.3389/fnmol.2014.00084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hutten S, Sharangdhar T, Kiebler M. Unmasking the messenger. RNA Biol. 2014;11:992–997. doi: 10.4161/rna.32091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Auerbach BD, Osterweil EK, Bear MF. Mutations causing syndromic autism define an axis of synaptic pathophysiology. Nature. 2011;480:63–68. doi: 10.1038/nature10658. Shows that genetic reduction of TSC2 ameliorates FXS pathophysiologies. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bhattacharya A, et al. Genetic removal of p70 S6 kinase 1 corrects molecular, synaptic, and behavioral phenotypes in fragile X syndrome mice. Neuron. 2012;76:325–337. doi: 10.1016/j.neuron.2012.07.022. Reveals that genetic removal of S6K1 prevents multiple FXS-associated phenotypes. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gkogkas CG, et al. Pharmacogenetic inhibition of eIF4E-dependent Mmp9 mRNA translation reverses fragile X syndrome-like phenotypes. Cell Rep. 2014;9:1742–1755. doi: 10.1016/j.celrep.2014.10.064. Shows that genetic reduction and pharmacological inhibition of MNK1 and MNK2, and genetic reduction of phosphorylated eIF4E, can correct several FXS pathophysiologies. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ronesi JA, et al. Disrupted Homer scaffolds mediate abnormal mGluR5 function in a mouse model of fragile X syndrome. Nat Neurosci. 2012;15:431–440. doi: 10.1038/nn.3033. Demonstrates that disrupting HOMER1A prevents multiple FXS pathophysiologies. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gross C, et al. Increased expression of the PI3K enhancer PIKE mediates deficits in synaptic plasticity and behavior in fragile X syndrome. Cell Rep. 2015;11:727–736. doi: 10.1016/j.celrep.2015.03.060. Describes how reducing elevated PIKE levels in models of FXS rescues associated phenotypes. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gross C, et al. Selective role of the catalytic PI3K subunit p110b in impaired higher-order cognition in fragile x syndrome. Cell Rep. 2015;11:681–688. doi: 10.1016/j.celrep.2015.03.065. Shows that genetic reduction of the PI3K subunit p110β ameliorates FXS-associated phenotypes. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sidhu H, Dansie LE, Hickmott PW, Ethell DW, Ethell IM. Genetic removal of matrix metalloproteinase 9 rescues the symptoms of fragile X syndrome in a mouse model. J Neurosci. 2014;34:9867–9879. doi: 10.1523/JNEUROSCI.1162-14.2014. Demonstrates that genetically reducing MMP9 prevents FXS pathophysiologies. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Busquets-Garcia A, et al. Targeting the endocannabinoid system in the treatment of fragile X syndrome. Nat Med. 2013;19:603–607. doi: 10.1038/nm.3127. Shows that genetic reduction, as well as pharmacological inhibition, of CB1R prevents and rescues FXS-associated pathophysiologies. [DOI] [PubMed] [Google Scholar]
- 22.Feng Y, et al. Translational suppression by trinucleotide repeat expansion at FMR1. Science. 1995;268:731–734. doi: 10.1126/science.7732383. [DOI] [PubMed] [Google Scholar]
- 23.Gross C, et al. Excess phosphoinositide 3-kinase subunit synthesis and activity as a novel therapeutic target in fragile X syndrome. J Neurosci. 2010;30:10624–10638. doi: 10.1523/JNEUROSCI.0402-10.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Grosset C, et al. A mechanism for translationally coupled mRNA turnover: interaction between the poly(A) tail and a c-fos RNA coding determinant via a protein complex. Cell. 2000;103:29–40. doi: 10.1016/s0092-8674(00)00102-1. [DOI] [PubMed] [Google Scholar]
- 25.Zhang YQ, et al. Protein expression profiling of the Drosophila fragile X mutant brain reveals up-regulation of monoamine synthesis. Mol Cell Proteom. 2005;4:278–290. doi: 10.1074/mcp.M400174-MCP200. [DOI] [PubMed] [Google Scholar]
- 26.Groppo R, Richter JD. Translational control from head to tail. Curr Opin Cell Biol. 2009;21:444–451. doi: 10.1016/j.ceb.2009.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hinnebusch AG. The scanning mechanism of eukaryotic translation initiation. Annu Rev Biochem. 2014;83:779–812. doi: 10.1146/annurev-biochem-060713-035802. [DOI] [PubMed] [Google Scholar]
- 28.Fabian MR, Sonenberg N, Filipowicz W. Regulation of mRNA translation and stability by microRNAs. Annu Rev Biochem. 2010;79:351–379. doi: 10.1146/annurev-biochem-060308-103103. [DOI] [PubMed] [Google Scholar]
- 29.Gingras AC, et al. Hierarchical phosphorylation of the translation inhibitor 4E-BP1. Genes Dev. 2001;15:2852–2864. doi: 10.1101/gad.912401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Waskiewicz AJ, et al. Phosphorylation of the cap-binding protein eukaryotic translation initiation factor 4E by protein kinase Mnk1 in vivo. Mol Cell Biol. 1999;19:1871–1880. doi: 10.1128/mcb.19.3.1871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Schenck A, et al. CYFIP/Sra-1 controls neuronal connectivity in Drosophila and links the Rac1 GTPase pathway to the fragile X protein. Neuron. 2003;38:887–898. doi: 10.1016/s0896-6273(03)00354-4. [DOI] [PubMed] [Google Scholar]
- 32.Schenck A, Bardoni B, Moro A, Bagni C, Mandel JL. A highly conserved protein family interacting with the fragile X mental retardation protein (FMRP) and displaying selective interactions with FMRP-related proteins FXR1P and FXR2P. Proc Natl Acad Sci USA. 2001;98:8844–8849. doi: 10.1073/pnas.151231598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Napoli I, et al. The fragile X syndrome protein represses activity-dependent translation through CYFIP1, a new 4E-BP. Cell. 2008;134:1042–1054. doi: 10.1016/j.cell.2008.07.031. [DOI] [PubMed] [Google Scholar]
- 34.De Rubeis S, et al. CYFIP1 coordinates mRNA translation and cytoskeleton remodeling to ensure proper dendritic spine formation. Neuron. 2013;79:1169–1182. doi: 10.1016/j.neuron.2013.06.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Genheden M, et al. BDNF stimulation of protein synthesis in cortical neurons requires the MAP kinase-interacting kinase MNK1. J Neurosci. 2015;35:972–984. doi: 10.1523/JNEUROSCI.2641-14.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Panja D, et al. Two-stage translational control of dentate gyrus LTP consolidation is mediated by sustained BDNF–TrkB signaling to MNK. Cell Rep. 2014;9:1430–1445. doi: 10.1016/j.celrep.2014.10.016. [DOI] [PubMed] [Google Scholar]
- 37.Sharma A, et al. Dysregulation of mTOR signaling in fragile X syndrome. J Neurosci. 2010;30:694–702. doi: 10.1523/JNEUROSCI.3696-09.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Edbauer D, et al. Regulation of synaptic structure and function by FMRP-associated microRNAs miR-125b and miR-132. Neuron. 2010;65:373–384. doi: 10.1016/j.neuron.2010.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Jin P, Alisch RS, Warren ST. RNA and microRNAs in fragile X mental retardation. Nat Cell Biol. 2004;6:1048–1053. doi: 10.1038/ncb1104-1048. [DOI] [PubMed] [Google Scholar]
- 40.Jin P, et al. Biochemical and genetic interaction between the fragile X mental retardation protein and the microRNA pathway. Nat Neurosci. 2004;7:113–117. doi: 10.1038/nn1174. [DOI] [PubMed] [Google Scholar]
- 41.Muddashetty RS, et al. Reversible inhibition of PSD-95 mRNA translation by miR-125a, FMRP phosphorylation and mGluR signaling. Mol Cell. 2011;42:673–688. doi: 10.1016/j.molcel.2011.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Xu XL, Li Y, Wang F, Gao FB. The steady-state level of the nervous-system-specific microRNA-124a is regulated by dFMR1 in Drosophila. J Neurosci. 2008;28:11883–11889. doi: 10.1523/JNEUROSCI.4114-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Yang Y, et al. The bantam microRNA is associated with Drosophila fragile X mental retardation protein and regulates the fate of germline stem cells. PLoS Genet. 2009;5:e1000444. doi: 10.1371/journal.pgen.1000444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Cheever A, Ceman S. Translation regulation of mRNAs by the fragile X family of proteins through the microRNA pathway. RNA Biol. 2009;6:175–178. doi: 10.4161/rna.6.2.8196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kenny PJ, et al. MOV10 and FMRP regulate AGO2 association with microRNA recognition elements. Cell Rep. 2014;9:1729–1741. doi: 10.1016/j.celrep.2014.10.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Darnell JC, et al. Fragile X mental retardation protein targets G quartet mRNAs important for neuronal function. Cell. 2001;107:489–499. doi: 10.1016/s0092-8674(01)00566-9. [DOI] [PubMed] [Google Scholar]
- 47.Menon L, Mader SA, Mihailescu MR. Fragile X mental retardation protein interactions with the microtubule associated protein 1B RNA. RNA. 2008;14:1644–1655. doi: 10.1261/rna.1100708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Menon L, Mihailescu MR. Interactions of the G quartet forming semaphorin 3F RNA with the RGG box domain of the fragile X protein family. Nucleic Acids Res. 2007;35:5379–5392. doi: 10.1093/nar/gkm581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Schaeffer C, et al. The fragile X mental retardation protein binds specifically to its mRNA via a purine quartet motif. EMBO J. 2001;20:4803–4813. doi: 10.1093/emboj/20.17.4803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Subramanian M, et al. G-quadruplex RNA structure as a signal for neurite mRNA targeting. EMBO Rep. 2011;12:697–704. doi: 10.1038/embor.2011.76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Zalfa F, et al. A new function for the fragile X mental retardation protein in regulation of PSD-95 mRNA stability. Nat Neurosci. 2007;10:578–587. doi: 10.1038/nn1893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.John B, et al. Human microRNA targets. PLoS Biol. 2004;2:e363. doi: 10.1371/journal.pbio.0020363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Stefanovic S, Bassell GJ, Mihailescu MR. G quadruplex RNA structures in PSD-95 mRNA: potential regulators of miR-125a seed binding accessibility. RNA. 2015;21:48–60. doi: 10.1261/rna.046722.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Liu T, et al. A microRNA profile in Fmr1 knockout mice reveals microRNA expression alterations with possible roles in fragile X syndrome. Mol Neurobiol. 2014;51:1053–1063. doi: 10.1007/s12035-014-8770-1. [DOI] [PubMed] [Google Scholar]
- 55.Halevy T, Czech C, Benvenisty N. Molecular mechanisms regulating the defects in fragile X syndrome neurons derived from human pluripotent stem cells. Stem Cell Rep. 2015;4:37–46. doi: 10.1016/j.stemcr.2014.10.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Ceman S, et al. Phosphorylation influences the translation state of FMRP-associated polyribosomes. Hum Mol Genet. 2003;12:3295–3305. doi: 10.1093/hmg/ddg350. [DOI] [PubMed] [Google Scholar]
- 57.Nalavadi VC, Muddashetty RS, Gross C, Bassell GJ. Dephosphorylation-induced ubiquitination and degradation of FMRP in dendrites: a role in immediate early mGluR-stimulated translation. J Neurosci. 2012;32:2582–2587. doi: 10.1523/JNEUROSCI.5057-11.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Narayanan U, et al. FMRP phosphorylation reveals an immediate-early signaling pathway triggered by group I mGluR and mediated by PP2A. J Neurosci. 2007;27:14349–14357. doi: 10.1523/JNEUROSCI.2969-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Niere F, Wilkerson JR, Huber KM. Evidence for a fragile X mental retardation protein-mediated translational switch in metabotropic glutamate receptor-triggered Arc translation and long-term depression. J Neurosci. 2012;32:5924–5936. doi: 10.1523/JNEUROSCI.4650-11.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Li Y, Tang W, Zhang LR, Zhang CY. FMRP regulates miR196a-mediated repression of HOXB8 via interaction with the AGO2 MID domain. Mol Biosyst. 2014;10:1757–1764. doi: 10.1039/c4mb00066h. [DOI] [PubMed] [Google Scholar]
- 61.Corbin F, et al. The fragile X mental retardation protein is associated with poly(A)+ mRNA in actively translating polyribosomes. Hum Mol Genet. 1997;6:1465–1472. doi: 10.1093/hmg/6.9.1465. [DOI] [PubMed] [Google Scholar]
- 62.Khandjian EW, et al. Biochemical evidence for the association of fragile X mental retardation protein with brain polyribosomal ribonucleoparticles. Proc Natl Acad Sci USA. 2004;101:13357–13362. doi: 10.1073/pnas.0405398101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Stefani G, Fraser CE, Darnell JC, Darnell RB. Fragile X mental retardation protein is associated with translating polyribosomes in neuronal cells. J Neurosci. 2004;24:9272–9276. doi: 10.1523/JNEUROSCI.2306-04.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Licatalosi DD, et al. HITS-CLIP yields genome-wide insights into brain alternative RNA processing. Nature. 2008;456:464–469. doi: 10.1038/nature07488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Ule J, et al. CLIP identifies Nova-regulated RNA networks in the brain. Science. 2003;302:1212–1215. doi: 10.1126/science.1090095. [DOI] [PubMed] [Google Scholar]
- 66.Darnell JC, et al. Kissing complex RNAs mediate6interaction between the fragile-X mental retardation protein KH2 domain and brain polyribosomes. Genes Dev. 2005;19:903–918. doi: 10.1101/gad.1276805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Darnell JC, et al. FMRP stalls ribosomal translocation on mRNAs linked to synaptic function and autism. Cell. 2011;146:247–261. doi: 10.1016/j.cell.2011.06.013. Identifies over 800 FMRP target mRNAs and demonstrates that FMRP stalls polysomes on several of these target mRNAs. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Ascano M, Jr, et al. FMRP targets distinct mRNA sequence elements to regulate protein expression. Nature. 2012;492:382–386. doi: 10.1038/nature11737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Chen E, Sharma MR, Shi X, Agrawal RK, Joseph S. Fragile X mental retardation protein regulates translation by binding directly to the ribosome. Mol Cell. 2014;54:407–417. doi: 10.1016/j.molcel.2014.03.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Bordeleau ME, et al. Functional characterization of IRESes by an inhibitor of the RNA helicase eIF4A. Nat Chem Biol. 2006;2:213–220. doi: 10.1038/nchembio776. [DOI] [PubMed] [Google Scholar]
- 71.Dolen G, Bear MF. Role for metabotropic glutamate receptor 5 (mGluR5) in the pathogenesis of fragile X syndrome. J Physiol. 2008;586:1503–1508. doi: 10.1113/jphysiol.2008.150722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Bear MF, Huber KM, Warren ST. The mGluR theory of fragile X mental retardation. Trends Neurosci. 2004;27:370–377. doi: 10.1016/j.tins.2004.04.009. [DOI] [PubMed] [Google Scholar]
- 73.Chuang SC, et al. Prolonged epileptiform discharges induced by altered group I metabotropic glutamate receptor-mediated synaptic responses in hippocampal slices of a fragile X mouse model. J Neurosci. 2005;25:8048–8055. doi: 10.1523/JNEUROSCI.1777-05.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Yan QJ, Rammal M, Tranfaglia M, Bauchwitz RP. Suppression of two major fragile X syndrome mouse model phenotypes by the mGluR5 antagonist MPEP. Neuropharmacology. 2005;49:1053–1066. doi: 10.1016/j.neuropharm.2005.06.004. [DOI] [PubMed] [Google Scholar]
- 75.Chang S, et al. Identification of small molecules rescuing fragile X syndrome phenotypes in Drosophila. Nat Chem Biol. 2008;4:256–263. doi: 10.1038/nchembio.78. [DOI] [PubMed] [Google Scholar]
- 76.Michalon A, et al. Chronic pharmacological mGlu5 inhibition corrects fragile X in adult mice. Neuron. 2012;74:49–56. doi: 10.1016/j.neuron.2012.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Osterweil EK, Krueger DD, Reinhold K, Bear MF. Hypersensitivity to mGluR5 and ERK1/2 leads to excessive protein synthesis in the hippocampus of a mouse model of fragile X syndrome. J Neurosci. 2010;30:15616–15627. doi: 10.1523/JNEUROSCI.3888-10.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Bhakar AL, Dolen G, Bear MF. The pathophysiology of fragile X (and what it teaches us about synapses) Annu Rev Neurosci. 2012;35:417–443. doi: 10.1146/annurev-neuro-060909-153138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Braat S, Kooy RF. Insights into GABAergic system deficits in fragile X syndrome lead to clinical trials. Neuropharmacology. 2014;88:48–54. doi: 10.1016/j.neuropharm.2014.06.028. [DOI] [PubMed] [Google Scholar]
- 80.Henderson C, et al. Reversal of disease-related pathologies in the fragile X mouse model by selective activation of GABAB receptors with arbaclofen. Sci Transl Med. 2012;4:152ra128. doi: 10.1126/scitranslmed.3004218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Heulens I, D’Hulst C, Van Dam D, De Deyn PP, Kooy RF. Pharmacological treatment of fragile X syndrome with GABAergic drugs in a knockout mouse model. Behav Brain Res. 2012;229:244–249. doi: 10.1016/j.bbr.2012.01.031. [DOI] [PubMed] [Google Scholar]
- 82.Gatto CL, Pereira D, Broadie K. GABAergic circuit dysfunction in the Drosophila fragile X syndrome model. Neurobiol Dis. 2014;65:142–159. doi: 10.1016/j.nbd.2014.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Berry-Kravis EM, et al. Effects of STX209 (arbaclofen) on neurobehavioral function in children and adults with fragile X syndrome: a randomized, controlled, phase 2 trial. Sci Transl Med. 2012;4:152ra127. doi: 10.1126/scitranslmed.3004214. [DOI] [PubMed] [Google Scholar]
- 84.Gomez-Mancilla B, et al. Development of mavoglurant and its potential for the treatment of fragile X syndrome. Expert Opin Investig Drugs. 2014;23:125–134. doi: 10.1517/13543784.2014.857400. [DOI] [PubMed] [Google Scholar]
- 85.Jacquemont S, et al. The challenges of clinical trials in fragile X syndrome. Psychopharmacology (Berl ) 2014;231:1237–1250. doi: 10.1007/s00213-013-3289-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Raught B, et al. Phosphorylation of eucaryotic translation initiation factor 4B Ser422 is modulated by S6 kinases. EMBO J. 2004;23:1761–1769. doi: 10.1038/sj.emboj.7600193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Waskiewicz AJ, Flynn A, Proud CG, Cooper JA. Mitogen-activated protein kinases activate the serine/threonine kinases Mnk1 and Mnk2. EMBO J. 1997;16:1909–1920. doi: 10.1093/emboj/16.8.1909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Osterweil EK, et al. Lovastatin corrects excess protein synthesis and prevents epileptogenesis in a mouse model of fragile X syndrome. Neuron. 2013;77:243–250. doi: 10.1016/j.neuron.2012.01.034. Demonstrates that a drug widely prescribed for the treatment of high cholesterol can reduce excessive protein synthesis and prevent audiogenic seizures in a mouse model of FXS. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Li W, et al. The HMG-CoA reductase inhibitor lovastatin reverses the learning and attention deficits in a mouse model of neurofibromatosis type 1. Curr Biol. 2005;15:1961–1967. doi: 10.1016/j.cub.2005.09.043. [DOI] [PubMed] [Google Scholar]
- 90.Ivshina M, Lasko P, Richter JD. Cytoplasmic polyadenylation element binding proteins in development, health, and disease. Annu Rev Cell Dev Biol. 2014;30:393–415. doi: 10.1146/annurev-cellbio-101011-155831. [DOI] [PubMed] [Google Scholar]
- 91.Wu L, et al. CPEB-mediated cytoplasmic polyadenylation and the regulation of experience-dependent translation of α-CaMKII mRNA at synapses. Neuron. 1998;21:1129–1139. doi: 10.1016/s0896-6273(00)80630-3. [DOI] [PubMed] [Google Scholar]
- 92.Zearfoss NR, Alarcon JM, Trifilieff P, Kandel E, Richter JD. A molecular circuit composed of CPEB-1 and c-Jun controls growth hormone-mediated synaptic plasticity in the mouse hippocampus. J Neurosci. 2008;28:8502–8509. doi: 10.1523/JNEUROSCI.1756-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Miyashiro KY, et al. RNA cargoes associating with FMRP reveal deficits in cellular functioning in Fmr1 null mice. Neuron. 2003;37:417–431. doi: 10.1016/s0896-6273(03)00034-5. [DOI] [PubMed] [Google Scholar]
- 94.Gross C, Bassell GJ. Excess protein synthesis in FXS patient lymphoblastoid cells can be rescued with a p110β-selective inhibitor. Mol Med. 2012;18:336–345. doi: 10.2119/molmed.2011.00363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Kumari D, et al. Identification of fragile X syndrome-specific molecular markers in human fibroblasts: a useful model to test the efficacy of therapeutic drugs. Hum Mutat. 2014;35:1485–1494. doi: 10.1002/humu.22699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Giuffrida R, et al. A reduced number of metabotropic glutamate subtype 5 receptors are associated with constitutive homer proteins in a mouse model of fragile X syndrome. J Neurosci. 2005;25:8908–8916. doi: 10.1523/JNEUROSCI.0932-05.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Bilousova TV, et al. Minocycline promotes dendritic spine maturation and improves behavioural performance in the fragile X mouse model. J Med Genet. 2009;46:94–102. doi: 10.1136/jmg.2008.061796. [DOI] [PubMed] [Google Scholar]
- 98.Leigh MJ, et al. A randomized double-blind, placebo-controlled trial of minocycline in children and adolescents with fragile X syndrome. J Dev Behav Pediatr. 2013;34:147–155. doi: 10.1097/DBP.0b013e318287cd17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Janusz A, et al. The fragile X mental retardation protein regulates matrix metalloproteinase 9 mRNA at synapses. J Neurosci. 2013;33:18234–18241. doi: 10.1523/JNEUROSCI.2207-13.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Tang AH, Alger BE. Homer protein-metabotropic glutamate receptor binding regulates endocannabinoid signaling and affects hyperexcitability in a mouse model of fragile X syndrome. J Neurosci. 2015;35:3938–3945. doi: 10.1523/JNEUROSCI.4499-14.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Dolan BM, et al. Rescue of fragile X syndrome phenotypes in Fmr1 KO mice by the small-molecule PAK inhibitor FRAX486. Proc Natl Acad Sci USA. 2013;110:5671–5676. doi: 10.1073/pnas.1219383110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Goebel-Goody SM, et al. Genetic manipulation of STEP reverses behavioral abnormalities in a fragile X syndrome mouse model. Genes Brain Behav. 2012;11:586–600. doi: 10.1111/j.1601-183X.2012.00781.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Mines MA, Jope RS. Glycogen synthase kinase-3: a promising therapeutic target for fragile X syndrome. Front Mol Neurosci. 2011;4:35. doi: 10.3389/fnmol.2011.00035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Franklin AV, et al. Glycogen synthase kinase-3 inhibitors reverse deficits in long-term potentiation and cognition in fragile X mice. Biol Psychiatry. 2014;75:198–206. doi: 10.1016/j.biopsych.2013.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Guo W, et al. Inhibition of GSK3β improves hippocampus-dependent learning and rescues neurogenesis in a mouse model of fragile X syndrome. Hum Mol Genet. 2012;21:681–691. doi: 10.1093/hmg/ddr501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Westmark CJ, et al. Reversal of fragile X phenotypes by manipulation of AβPP/Aβ levels in Fmr1KO mice. PLoS ONE. 2011;6:e26549. doi: 10.1371/journal.pone.0026549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Brown V, et al. Microarray identification of FMRP-associated brain mRNAs and altered mRNA translational profiles in fragile X syndrome. Cell. 2001;107:477–487. doi: 10.1016/s0092-8674(01)00568-2. [DOI] [PubMed] [Google Scholar]
- 108.Mili S, Steitz JA. Evidence for reassociation of RNA-binding proteins after cell lysis: implications for the interpretation of immunoprecipitation analyses. RNA. 2004;10:1692–1694. doi: 10.1261/rna.7151404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Zalfa F, et al. The fragile X syndrome protein FMRP associates with BC1 RNA and regulates the translation of specific mRNAs at synapses. Cell. 2003;112:317–327. doi: 10.1016/s0092-8674(03)00079-5. [DOI] [PubMed] [Google Scholar]
- 110.Liao L, Park SK, Xu T, Vanderklish P, Yates JR., 3rd Quantitative proteomic analysis of primary neurons reveals diverse changes in synaptic protein content in fmr1 knockout mice. Proc Natl Acad Sci USA. 2008;105:15281–15286. doi: 10.1073/pnas.0804678105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Pasciuto E, Bagni C. SnapShot: FMRP mRNA targets and diseases. Cell. 2014;158:1446–1446.e1. doi: 10.1016/j.cell.2014.08.035. [DOI] [PubMed] [Google Scholar]
- 112.Hoeffer CA, et al. Altered mTOR signaling and enhanced CYFIP2 expression levels in subjects with fragile X syndrome. Genes Brain Behav. 2012;11:332–341. doi: 10.1111/j.1601-183X.2012.00768.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Doers ME, et al. iPSC-derived forebrain neurons from FXS individuals show defects in initial neurite outgrowth. Stem Cells Dev. 2014;23:1777–1787. doi: 10.1089/scd.2014.0030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Sheridan SD, et al. Epigenetic characterization of the FMR1 gene and aberrant neurodevelopment in human induced pluripotent stem cell models of fragile X syndrome. PLoS ONE. 2011;6:e26203. doi: 10.1371/journal.pone.0026203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Kumari D, et al. High-throughput screening to identify compounds that increase fragile X mental retardation protein expression in neural stem cells differentiated from fragile X syndrome patient-derived induced pluripotent stem cells. Stem Cells Transl Med. 2015;4:800–808. doi: 10.5966/sctm.2014-0278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Gross C, Yao X, Pong DL, Jeromin A, Bassell GJ. Fragile X mental retardation protein regulates protein expression and mRNA translation of the potassium channel Kv4.2. J Neurosci. 2011;31:5693–5698. doi: 10.1523/JNEUROSCI.6661-10.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Lee HY, et al. Bidirectional regulation of dendritic voltage-gated potassium channels by the fragile X mental retardation protein. Neuron. 2011;72:630–642. doi: 10.1016/j.neuron.2011.09.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Routh BN, Johnston D, Brager DH. Loss of functional A-type potassium channels in the dendrites of CA1 pyramidal neurons from a mouse model of fragile X syndrome. J Neurosci. 2013;33:19442–19450. doi: 10.1523/JNEUROSCI.3256-13.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Strumbos JG, Brown MR, Kronengold J, Polley DB, Kaczmarek LK. Fragile X mental retardation protein is required for rapid experience-dependent regulation of the potassium channel Kv3.1b. J Neurosci. 2010;30:10263–10271. doi: 10.1523/JNEUROSCI.1125-10.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Brager DH, Akhavan AR, Johnston D. Impaired dendritic expression and plasticity of h-channels in the fmr1–/y mouse model of fragile X syndrome. Cell Rep. 2012;1:225–233. doi: 10.1016/j.celrep.2012.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Brown MR, et al. Fragile X mental retardation protein controls gating of the sodium-activated potassium channel Slack. Nat Neurosci. 2010;13:819–821. doi: 10.1038/nn.2563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Deng PY, et al. FMRP regulates neurotransmitter release and synaptic information transmission by modulating action potential duration via BK channels. Neuron. 2013;77:696–711. doi: 10.1016/j.neuron.2012.12.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Myrick LK, et al. Independent role for presynaptic FMRP revealed by an FMR1 missense mutation associated with intellectual disability and seizures. Proc Natl Acad Sci USA. 2015;112:949–956. doi: 10.1073/pnas.1423094112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Ferron L, Nieto-Rostro M, Cassidy JS, Dolphin AC. Fragile X mental retardation protein controls synaptic vesicle exocytosis by modulating N-type calcium channel density. Nat Commun. 2014;5:3628. doi: 10.1038/ncomms4628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Tsai P, Sahin M. Mechanisms of neurocognitive dysfunction and therapeutic considerations in tuberous sclerosis complex. Curr Opin Neurol. 2011;24:106–113. doi: 10.1097/WCO.0b013e32834451c4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Hoeffer CA, Klann E. mTOR signaling: at the crossroads of plasticity, memory and disease. Trends Neurosci. 2010;33:67–75. doi: 10.1016/j.tins.2009.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]