Abstract
Hereditary Spastic Paraplegia (HSP) is a syndrome characterised by lower limb spasticity, occurring alone or in association with other neurological manifestations, such as cognitive impairment, seizures, ataxia or neuropathy. HSP occurs worldwide, with different populations having different frequencies of causative genes. The Greek population has not yet been characterised. The purpose of this study was to describe the clinical presentation and molecular epidemiology of the largest cohort of HSP in Greece, comprising 54 patients from 40 families. We used a targeted next-generation sequencing (NGS) approach to genetically assess a proband from each family. We made a genetic diagnosis in >50% of cases and identified 11 novel variants. Variants in SPAST and KIF5A were the most common causes of autosomal dominant HSP, whereas SPG11 and CYP7B1 were the most common cause of autosomal recessive HSP. We identified a novel variant in SPG11, which led to disease with later onset and may be unique to the Greek population and report the first nonsense mutation in KIF5A. Interestingly, the frequency of HSP mutations in the Greek population, which is relatively isolated, was very similar to other European populations. We confirm that NGS approaches are an efficient diagnostic tool and should be employed early in the assessment of HSP patients.
Introduction
Hereditary spastic Paraplegia (HSP) refers to a heterogeneous group of disorders characterised by slowly progressive limb spasticity and weakness.1 Spasticity can occur in isolation (pure HSP), or can be complicated by additional features including learning disability, seizures, neuropathy or ataxia (complicated HSP). Approximately two-thirds of patients have a family history of the disorder, with the remainder occurring sporadically.2
All modes of inheritance are known to cause HSP and at least 45 genetic types are known, although a smaller number of genes account for the majority of cases.3 Despite the abundance of genes involved in HSP, many of the genes impact on the same molecular pathways. These include axonal transport (KIF5A, KIF1A), endoplasmic reticulum formation (ATL1, SPAST, REEP1), endosomal and lysosomal trafficking (SPG11, AP4B1), and mitochondrial function (SPG7, mATP6, HSPD1).2
The clinical heterogeneity of HSP and the diversity of genetic causes can make it difficult to achieve a genetic diagnosis in many patients. In most clinical settings, a small number of genes can be sequenced by conventional Sanger methods owing to the cost and low frequency of mutations in many genes. Overall, >50% of autosomal dominant (AD) HSP cases and 70% of autosomal recessive (AR) HSP cases never receive a genetic diagnosis.4 HSP occurs in all populations, and there are regional variations in the frequency of causative genes. However, no study has evaluated the genetic spectrum of HSP in Greece. In this study, we present the largest Greek cohort of HSP patients, comprising 54 patients from 40 families.
The development of massively parallel sequencing has revolutionized genetics, making it possible to simultaneously sequence thousands of genes, faster and at lower cost than traditional Sanger sequencing.5 There are a number of different NGS technologies, including whole-genome, exome, targeted exome and panel-based sequencing. Although sequencing a whole-genome provides unparalleled genetic information, the interpretation of these variants is difficult and time consuming. In addition, when only single probands are available for sequencing, genome or exome sequencing may reveal thousands of potentially pathogenic variants. Targeted exome sequencing and panel-based sequencing offer some advantages owing to cost savings and the ease and speed of data interpretation.
In this study, we used a combination of targeted exome sequencing and panel sequencing to characterise the mutational spectrum of HSP in probands from 40 Greek HSP families, providing insights into the genetics of HSP in this population for the first time.
Materials and Methods
Patient recruitment
This study included consecutive patients referred with suspected HSP to the Neurogenetics Unit, 1st Department of Neurology, University of Athens Medical School, Eginition Hospital, over an 18-year period (1995–2013). The Athens Neurogenetics Unit is the only unit of its kind in Greece and has uniquely offered molecular diagnostic testing for adult patients with neurogenetic disorders within the framework of the Greek public health service. It receives referrals for clinical assessment and/or molecular genetic testing in patients with suspected neurogenetic disorders from all regions of the country. In the case of HSP, the Unit has not offered diagnostic molecular testing, but has assessed patients clinically and stored DNA for future analysis. Patients included in the present study originate from various regions of Greece, were in most cases referred by other neurologists in the Department and gave informed consent for molecular genetic testing.
Targeted exome sequencing
Nine patients were sequenced on an Illumina Trusight Exome panel, which targets 2731 genes known to cause human disease. These include 34 genes associated with HSP (see Supplementary Table). The complete list of targeted genes is available online from http://support.illumina.com/downloads/trusight_exome_product_files.html.
DNA samples were diluted to 5 ng/μl and were then tagged and fragmented according to the Trusight Enrichment DNA Sample Preparation Guide. Libraries were sequenced with an Illumina HiSeq 2500. Sequences were aligned using BWA and variants called with GATK. Mean target coverage was 42, and 87% of bases were read 10 times. There was an average of 7065 variants called per sample.
Truseq custom amplicon panel sequencing
Thirty-one patients were sequenced using a custom HSP amplicon panel, which was designed to target the coding regions of the following genes implicated in HSP: ATL1, SPAST, BSCL2, HSPD1, GJC, KIAA0196, KIF5A, NIPA1, PLP, PNPLA6, REEP1, RTN2, SPG11, SPG7, CYP7B1, FA2H. DNA was diluted to 250 ng and procedures followed the Truseq Custom Amplicon Library Preparation Guide. Libraries were sequenced on an Illumina Miseq and variants were called on the Illumina BaseSpace platform. Mean coverage of the Custom Panel was 592 reads per targeted base. A graphical representation of coverage per patient and coverage per gene may be found in Supplementary Figure 1.
Sanger sequencing
All potentially pathogenic variants identified by next-generation sequencing (NGS) were confirmed by conventional Sanger sequencing. The exon carrying the variant was PCR amplified using flanking intronic primers (primer sequences available on request). The PCR product was purified and then sequenced in both directions using Big Dye Terminator version 3.1 Cycle Sequencing Kit (Applied Biosystems, Foster City, CA, USA). Sequencing products were purified and read on an ABI 3730 DNA Analyzer (Applied Biosystems). Sequences were analysed using Seqscape 3 software (Applied Biosystems).
Variants are described with reference to the following transcripts: SPAST: NM_014946; NG_008730.1, KIF5A: NM_004984; NG_008155.1, REEP1: NM_001164732.1; NG_013037.1, ATL1: NM_015915; NG_009028.1, CYP7B1: NM_004820.3; NG_008338.1, SPG11: NM_025137; NG_008885.1, SPG7: NM_003119; NG_008082.1, GJC2: NM_020435; NG_011838.1, ZFYVE26: NM_015346; NG_011836.1. All variants detected have been deposited in the Leiden Open Variation Database found at http://databases.lovd.nl/shared/genes (Patient IDs 39349–39359 and 46990–47000).
Results
Clinical characterisation
We identified 54 patients from 40 families affected by HSP. The clinical descriptions of the patients and families can be found in Table 1. Thirty-six patients (67%) presented with pure HSP and 18 (33%) with complicated HSP.
Table 1. Clinical description of families and patients.
| Family | Inheritance | Individual | Sex | Phenotype | Age at onset | Urinary symptoms | LL sensory disturbance | Neuropathy | Cognitive impairment | TCC | WML | Cerebellar signs | Additional features |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | X-linked | HSP1 | M | Pure | 27 | + | − | − | − | − | − | − | |
| 1 | X-linked | HSP3 | M | Pure | 28 | + | + | − | − | − | − | − | |
| 1 | X-linked | HSP4 | M | Pure | 20 | − | + | − | − | − | − | − | |
| 2 | AD | HSP8 | F | Pure | NA | − | − | − | − | − | − | − | |
| 3 | AD | HSP9 | M | Pure | 40 | + | − | − | − | − | − | − | |
| 4 | Sporadic | HSP11 | M | Pure | 60 | + | + | − | − | − | − | − | |
| 5 | AD | HSP12 | M | Pure | 35 | − | − | − | − | − | − | − | |
| 6 | AR | HSP13 | M | Complicated | 60 | − | + | + | − | − | − | + | |
| 7 | AD | HSP14 | M | Pure | 14 | − | − | − | − | − | − | − | |
| 8 | Unknown | HSP16 | M | Complicated | 40 | + | + | − | + | − | − | − | |
| 9 | AD | HSP17 | M | Complicated | 40 | − | − | − | + | − | − | − | |
| 10 | Sporadic | HSP19 | M | Pure | 9 m | − | − | − | − | − | − | − | |
| 11 | Unknown | HSP20 | F | Pure | 63 | − | − | − | − | − | − | − | |
| 12 | Sporadic | HSP21 | M | Complicated | 10 | − | − | + | − | − | − | − | Sensorimotor axonal neuropathy |
| 13 | Sporadic | HSP22 | M | Pure | 24 | − | − | − | − | − | − | − | |
| 14 | AD | HSP27 | M | Pure | 44 | + | − | − | − | − | − | − | |
| 15 | Sporadic | HSP28 | F | Pure | 37 | + | − | − | − | − | − | − | |
| 16 | Unknown | HSP29 | M | Complicated | 39 | + | + | + | + | − | + | − | |
| 17 | Sporadic | HSP30 | M | Pure | 24 | + | + | − | − | − | − | − | |
| 18 | Sporadic | HSP31 | M | Pure | 34 | − | − | − | − | − | − | − | |
| 19 | AD | HSP32 | F | Pure | 52 | − | − | − | − | − | − | − | |
| 20 | AR | HSP33 | M | Complicated | 8 | − | + | − | − | − | − | − | Sensorineural hearing loss |
| 20 | AR | HSP34 | M | Complicated | 10 | − | + | − | − | − | − | + | Sensorineural hearing loss |
| 20 | AR | HSP35 | F | Complicated | 13 | − | + | − | − | − | + | + | Sensorineural hearing loss |
| 21 | AD | HSP37 | F | Pure | NA | − | − | − | − | − | − | − | |
| 21 | AD | HSP40 | M | Complicated | 6 | − | + | + | − | − | − | − | Sensory axonal polyneuropathy |
| 22 | Sporadic | HSP41 | F | Pure | 27 | − | + | − | − | − | − | − | Parents 3rd cousins |
| 23 | AR | HSP45 | M | Pure | 19 | − | − | − | − | − | − | − | |
| 24 | AD | HSP46 | M | Pure | 45 | − | − | − | − | − | + | − | |
| 25 | AR | HSP47 | F | Pure | 41 | − | − | − | − | − | + | − | |
| 25 | AR | HSP59 | M | Pure | NA | − | − | − | − | − | − | − | |
| 26 | AR | HSP48 | F | Complicated | 15 | − | − | − | + | + | + | + | Dysarthria |
| 27 | AD | HSP49 | M | Pure | 46 | − | − | − | + | − | − | + | |
| 27 | AD | HSP83 | F | Pure | NA | − | − | − | − | − | − | − | |
| 27 | AD | HSP86 | F | Pure | 40 | − | + | − | − | − | − | − | |
| 27 | AD | HSP87 | M | Complicated | 73 | − | + | − | − | − | − | − | Chorea |
| 27 | AD | HSP88 | M | Complicated | 59 | − | − | − | − | − | − | − | Chorea |
| 27 | AD | HSP89 | M | Complicated | 4 | − | − | − | − | − | − | − | Epilepsy |
| 27 | AD | HSP90 | M | Pure | 16 | − | − | − | − | − | − | − | |
| 27 | AD | HSP91 | M | Pure | 18 | − | − | − | − | − | − | − | |
| 28 | AD | HSP50 | M | Pure | 22 | − | − | − | − | − | − | − | |
| 29 | AD | HSP54 | F | Pure | 45 | − | − | − | − | − | − | − | |
| 30 | AD | HSP55 | M | Pure | NA | − | − | − | − | − | − | − | |
| 31 | AD | HSP62 | M | Pure | 8 | − | + | − | − | − | − | − | |
| 32 | Sporadic | HSP64 | M | Complicated | 12 | + | − | − | + | + | + | + | |
| 33 | AD | HSP65 | M | Pure | 10 | − | + | − | − | − | − | − | |
| 34 | AD | HSP66 | F | Complicated | 18 m | − | + | − | + | − | − | − | |
| 34 | AD | HSP70 | M | Complicated | 18 m | − | + | − | + | − | − | − | |
| 35 | Sporadic | HSP67 | M | Complicated | 5 | − | + | + | − | − | − | − | Sensorimotor axonal neuropathy |
| 36 | AD | HSP68 | F | Pure | 25 | − | + | − | − | − | − | − | |
| 37 | Sporadic | HSP69 | M | Pure | 49 | − | + | − | − | − | + | − | |
| 38 | AD | HSP71 | F | Pure | 50 | − | − | − | − | − | − | − | |
| 39 | Sporadic | HSP73 | F | Complicated | 15 | − | − | − | + | + | + | − | Ichthyosis |
| 40 | Sporadic | HSP60 | M | Pure | NA | − | − | − | − | − | − | − |
Abbreviations: AD, autosomal dominant; AR, autosomal recessive; Complicated, HSP with additional neurological features such as ataxia, neuropathy, cognitive impairment; M, male; F, female; NA, information not available; Pure, lower/upper limb slowly progressive spasticity ± back pain; +, present; −, absent; TCC, thinning of the corpus callosum; WML, white matter lesions.
Pure forms: the majority of pure HSP occurred in families with an AD mode of inheritance (55%), followed by sporadic inheritance (31%) and AR inheritance (7%). X-linked dominant inheritance occurred in one family (3%). Just over two-thirds of the patients with pure HSP were men. The mean age of onset was 32 years. The most frequent symptoms described in addition to spastic paraplegia were lower limb sensory disturbance (28%) and urinary urgency/frequency (19%). Cerebellar signs and cognitive impairment occurred rarely. White matter lesions on MRI were described in three patients.
Complicated forms: the most frequent mode of inheritance in families with a complicated HSP phenotype was AD (38%), sporadic (30%) and AR (23%). The mode of inheritance was unknown in 15%. There was a significant excess of men with a complicated phenotype (77%) compared with women (23%). The mean age at onset was 23 years. The most frequently reported additional symptoms were lower limb sensory disturbance (61%), cognitive impairment (44%), cerebellar signs (28%) and neuropathy (28%). Urinary symptoms occurred in 17%. Three patients had thinning of the corpus callosum on MRI and four patients had white matter lesions.
Molecular characterisation
We performed genetic analysis on one proband from each HSP family (40 total), using the Truseq custom HSP panel (targeting 16 of the most frequently mutated HSP genes) or Trusight Exome. MLPA analysis was not performed. This may underestimate the frequency of some genes owing to genomic deletions.
A genetic diagnosis was made in 6/9 patients using the Trusight Exome and 15/31 patients using the Truseq custom HSP panel.
A genetic diagnosis was achieved in 13 of the 30 patients with pure HSP (43%) and in 8 of the 10 patients with complicated HSP (80%). Where a clear AR pattern of inheritance was given, a genetic diagnosis could be made in all cases. Single heterozygous variants found in known AR HSP genes are given in Supplementary Table 1.
Novel variants
Of the 21 potentially pathogenic variants we identified, 11 were novel. These are listed in Table 2. These include two nonsense mutations in SPAST, c.430C>T and c.575T>A, and one in KIF5A c.2590C>T. We also identified a novel frameshift mutation in SPAST, c.1591dupC. Of the remaining seven novel missense variants, all but one are absent in available databases, including the ExAC browser,6 the Exome Variant Server and an internal database of over 600 exomes. All novel variants are predicted to be pathogenic by at least two in silico tools and all occur at conserved residues.
Table 2. Novel mutations detected with frequency data and in silico prediction score.
| Gene | Nucleotide change | Predicted AA change | Mutation type | Freq. (ExAC) | Freq. (EVS) | Freq. (internal db) | SIFT | Polyphen | Mutation taster | GERP++ | Family segregation |
|---|---|---|---|---|---|---|---|---|---|---|---|
| SPAST | c.430C>T | p.(Q144*) | Nonsense | 0 | 0 | 0 | 1 | NA | 1 | 4.6 | Yes |
| SPAST | c.575T>A | p.(L192*) | Nonsense | 0 | 0 | 0 | 1 | NA | 1 | 5.77 | NA |
| SPAST | c.1508G>C | p.(R503P) | Missense | 0 | 0 | 0 | 1 | 1 | 1 | 5.13 | NA |
| SPAST | c.1591dupC | p.(Q531Pfs*9) | Frameshift | 0 | 0 | 0 | SIFTindel: Damaging | 5.74 | n/a | ||
| KIF5A | c.745C>G | p.(L249V) | Missense | 0 | 0 | 0 | 1 | 0.631 | 0.999 | 1.15 | NA |
| KIF5A | c.2590C>T | p.(R864*) | Nonsense | 0 | 0 | 0 | 1 | NA | 1 | 4.9 | NA |
| REEP1 | c.166G>A | p.(D56N) | Missense | 0 | 0 | 0 | 1 | 0.98 | 0.63 | 5.07 | Yes |
| SPG11 | c.2278T>C | p.(C760R) | Missense | 0 | 0 | 0 | 0.99 | 0.998 | 1 | 3.82 | NA |
| SPG11 | c.5381T>C | p.(L1794P) | Missense | 0.00004 | 0.00015 | 0 | 0.99 | 1 | 1 | 5.8 | NA |
| CYP7B1 | c.1304T>C | p.(L435P) | Missense | 0 | 0 | 0 | 1 | 1 | 1 | 5.55 | NA |
| CYP7B1 | c.1322C>T | p.(P441L) | Missense | 0 | 0 | 0 | 1 | 0.99 | 0.48 | 5.09 | NA |
Only the SPG11 c.5381T>C variant has a recorded frequency on control databases, with a reported frequency of five in 120 880 alleles (0.00004136) on the ExAC browser.6 No homozygotes have been reported. This variant has maximally pathogenic scores on all three in silico tools and occurs at a highly conserved residue. It occurs in combination with another deleterious SPG11 variant in three probands. In all cases the variant occurs with a frameshift or nonsense mutation and in one family, the same compound heterozygote mutations, c.[5470C>T] [5381T>C] occur in two affected brothers. An unaffected brother carried only the c. 5381T>C variant. For these reasons, we believe that the c. 5381T>C variant in SPG11 may be pathogenic when it occurs in trans with another pathogenic SPG11 variant or perhaps in the homozygous state, and it is possible that this is a unique finding in the Greek population.
Autosomal dominant mutations
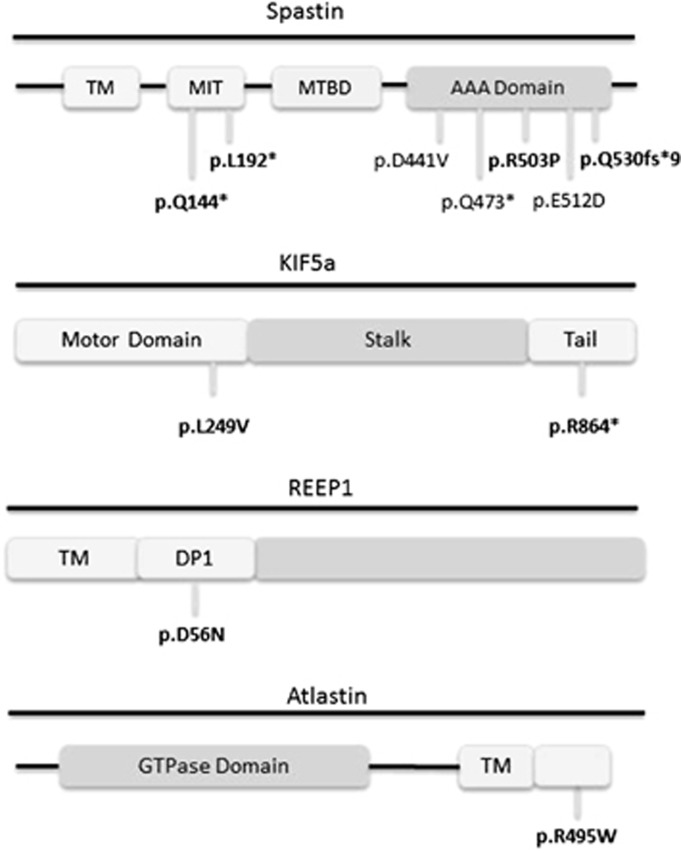
SPAST/SPG4
Variants in SPAST were responsible for the majority of AD HSP in our cohort (see Table 3, Figure 1). We identified seven patients with potentially pathogenic variants in SPAST, of which four were novel and three previously described as pathogenic. The age of onset of HSP in patients with SPAST variants was highly variable ranging from early childhood to 50 years. We observed a trend for an earlier age of onset in truncating mutations at the 5′ end of the gene (HSP62 and HSP66). All but one SPAST patient had a pure HSP phenotype, although sensory disturbance was common. One patient with early childhood onset had mild cognitive impairment in addition to spastic paraplegia (HSP66–c.430C>T variant) and another was found to have white matter lesions on MRI (HSP46–c.1536G>C variant).
Table 3. Autosomal dominant HSP genes.
| Individual | Inheritance | Gene | Nucleotide change | Predicted AA change | Phenotype | AAO | Additional features |
|---|---|---|---|---|---|---|---|
| HSP27 | AD | SPAST | c.1591dupC | p.(Q531Pfs*9) | Pure HSP | 44 | Urinary urgency |
| HSP31 | Sporadic | SPAST | c.1508G>C | p.(R503P) | Pure HSP | 34 | |
| HSP46 | AD | SPAST | c.1536G>C | p.(E512D) | Pure HSP | 45 | Periventricular and subcortical white matter lesions |
| HSP62 | AD | SPAST | c.575T>A | p.(L192*) | Pure HSP | 8 | Sensory disturbance (UL/LL) |
| HSP65 | AD | SPAST | c.1322A>T | p.(D441V) | Pure HSP | 10 | Sensory disturbance (LL) |
| HSP66 | AD | SPAST | c.430C>T | p.(Q144*) | Complicated HSP | 18 m | MCI, sensory disturbance, pes cavus |
| HSP71 | AD | SPAST | c.1417C>T | p.(Q473*) | Pure HSP | 50 | |
| HSP21 | Sporadic | KIF5A | c.745C>G | p.(L249V) | Complicated HSP | 10 | Sensorimotor axonal neuropathy |
| HSP67 | Sporadic | KIF5A | c.2590C>T | p.(R864*) | Complicated HSP | 5 | Sensorimotor axonal neuropathy, LL hemiatrophy, pes cavus |
| HSP50 | AD | REEP1 | c.166G>A | p.(D56N) | Pure HSP | 22 | Pes cavus |
| HSP55 | AD | ATL1 | c.1483C>T | p.(R495W) | Pure HSP | N/A |
Abbreviations: AAO, age at onset; MCI, mild cognitive impairment; LL, lower limb; UL, upper limb.
Figure 1.
Schematic representations of the spastin, KIF5A, REEP1 and atlastin proteins. Novel mutations are shown in bold. TM: transmembrane, MIT: microtubule interacting domain, MTBD: microtubule-binding domain, AAA: ATPase-associated with diverse cellular activity.
KIF5A
KIF5A was the second most common AD HSP gene in our cohort. We identified two patients with KIF5A variants, both of which were novel. The c.2590C>T mutation is the first nonsense mutation identified in KIF5A to date. Both patients presented in early childhood with HSP complicated by a sensorimotor axonal neuropathy, the typical phenotype of SPG10.
A single patient with a novel REEP1 variant and a single patient with a previously described pathogenic ATL1 variant were also identified.
Autosomal recessive mutations
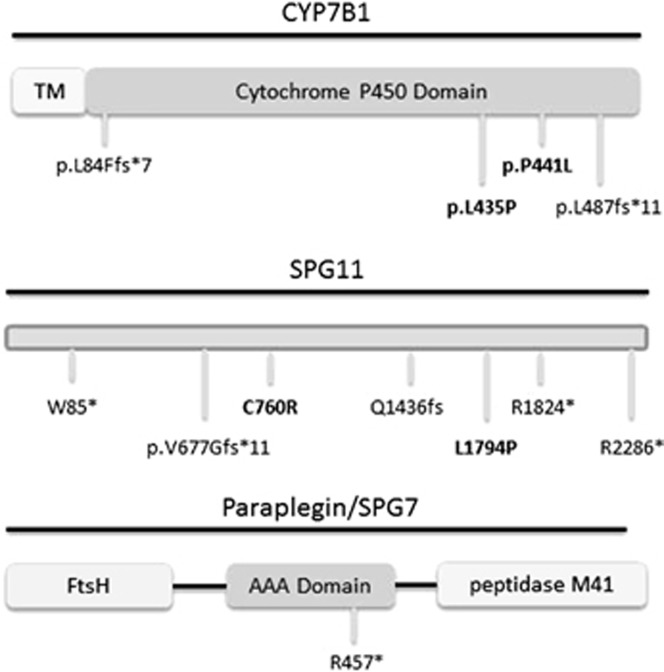
SPG11
We identified five probands with variants in SPG11. Two patients (HSP73 and HSP48) had a typical presentation of SPG11 associated HSP with a complicated phenotype, thinning of the corpus callosum and onset below age 20 (see Table 4,Figure 2). These patients had a novel homozygous variant, c.2278T>C, and two known pathogenic variants c.[6856C>T (;) 2030_2034del], respectively.
Table 4. Autosomal recessive HSP genes.
| Individual | Inheritance | Gene | Nucleotide change | Predicted AA change | Phenotype | AAO | Additional features | Family segregation |
|---|---|---|---|---|---|---|---|---|
| HSP33 | AR | CYP7B1 | c.250delC | p.(L84Ffs*7) | Complicated HSP | 8 | Hearing loss, sensory disturbance (LL), symmetric frontoparietal WML | Same mutation found in two affected siblings |
| HSP45 | AR | CYP7B1 | c.1322C>T | p.(P441L) | Pure HSP | 19 | − | NA |
| HSP41 | Sporadic | CYP7B1 | c.[1304T>C (;) 1460dupT] | p.(L435P) (;) p.(L487Ffs*11) | Pure HSP | 27 | Sensory disturbance (LL) | |
| HSP73 | Sporadic | SPG11 | c.2278T>C | p.(C760R) | Complicated HSP | 15 | MCI, ichthyosis, TCC, WML | NA |
| HSP29 | Unclear | SPG11 | c.[255G>A (;) 5381T>C] | p.(W85*) (;) p.(L1794P) | Complicated HSP | 39 | Cognitive impairment, hearing loss, mild sensory axonal neuropathy, ventricular/cortical sulci enlargement, WML | N/A |
| HSP13 | AR | SPG11 | c.[4307_4308del (;) 5381T>C] | p.(Q1436Rfs*7) (;) p.(L1794P) | Complicated HSP | 60 | UL hypereflexia, LL hyporeflexia, dysdiadochokinesis | N/A |
| HSP47 | AR | SPG11 | c.[5470C>T] [5381T>C] | p.(R1824*); p.(L1794P) | Pure HSP | 41 | Periventricular, centrum semiovale WML | Same mutations found in affected brother, unaffected brother has only L1974P variant |
| HSP48 | AR | SPG11 | c.[6856C>T (;) 2030_2034del] | p.(R2286*) (;) p.(V677Gfs*11) | Complicated HSP | 15 | Cognitive impairment, hearing loss, dysarthria, TCC, periventricular WML | NA |
| HSP16 | Unclear | SPG7 | c.1369C>T | p.(R457*) | Complicated HSP | 40 | Urinary urgency, distal UL weakness, rest tremor, MCI, sensory disturbance (LL), UL hypereflexia | NA |
Abbreviations: AAO, age at onset; MCI, mild cognitive impairment; LL, lower limb; TCC, thinning of the corpus callosum; UL, upper limb; WML, white matter lesions.
Figure 2.
Schematic representations of the SPG11, CYP7B1 and paraplegin proteins. Novel mutations are in shown in bold. TM: transmembrane domain, FtsH: filamentous temperature sensitive H.
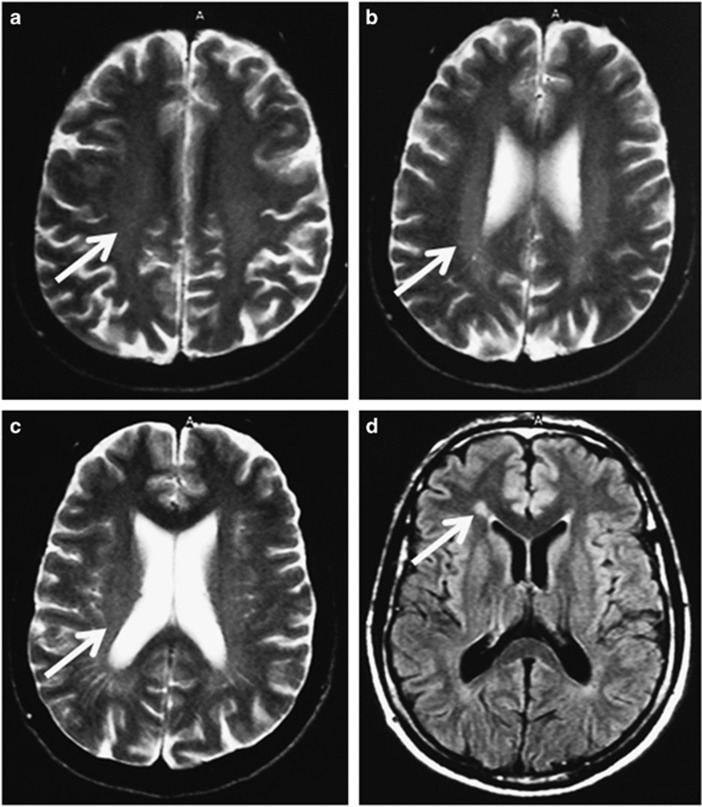
Interestingly, the three patients who had the c. 5381T>C variant in SPG11 in addition to another deleterious variant had a significantly older age at onset. HSP29 developed symptoms at age 39. He had a complicated phenotype comprising spastic paraplegia, bilateral sensorineural deafness, diminished vibration sensation in the upper and lower limbs and mild sensory axonal neuropathy on Nerve Conduction Studies. MRI revealed mild ventricular and cortical sulci enlargement with diffuse periventricular T2 hyperintensity and a typical ‘ears of the lynx' appearance at the frontal horn of the right lateral ventricle (see Figure 3). We identified a novel nonsense mutation c.255G>A in exon 1 of SPG11 in addition to the c. 5381T>C variant, which we believe may be pathogenic. DNA was not available from other family members to assess if these variants occurred in cis or in trans.
Figure 3.
(a–c) axial T2 images showing diffuse periventricular hyperintensity (arrows), along with ventricular and cortical sulci enlargement; (d) axial FLAIR image showing early ‘flame-' or ‘ears-of-the-lynx-' formation (arrow).
HSP13 developed symptoms at age 60. He had two affected siblings. His parents were unaffected to our knowledge, implying an AR mode of inheritance. He presented with progressive spastic paraplegia, hypereflexia in the upper limbs with hyporeflexia in the lower limbs. There was dysdiadochokinesis. We identified two variants in SPG11, a frameshift mutation in exon 25 (c.4307_4308del) and the c. 5381T>C variant in exon 30.
HSP47 had a pure HSP phenotype, and developed symptoms at age 41. MRI revealed periventricular and centrum semiovale white matter lesions. The same c.[5470C>T] [5381T>C] variants were found in the proband and her affected brother. An unaffected brother carried only the c. 5381T>C variant, proving that the mutations occur in trans in these patients.
CYP7B1
We identified three probands with variants in CYP7B1/SPG5a. One patient with a homozygous frameshift variant (c.250delC), had onset of spastic paraplegia at age 8 with hearing loss and sensory disturbance in the lower limbs. Two siblings, who were similarly affected, were found to carry the same homozygous variant. An affected sister, with onset of symptoms age 13 years, was found to have symmetric frontoparietal white matter lesions and gaze-evoked nystagmus. All three siblings had prominent impairment of vibration sensation and an affected brother also had ataxia. This finding confirms previous reports7 that mutations in CYP7B1 may be associated with sensorineural deafness.
Patient HSP45 was found to have a homozygous novel mutation in CYP7B1 (c.1322C>T). This patient with pure HSP had onset of symptoms at age 19. Two siblings had later age of onset in the 30 and 40 s, although DNA was not available for testing.
Patient HSP41 was found to have the c.[1304T>C (;) 1460dupT] variants in CYP7B1. Although the c.1304T>C variant is novel, the c.1460dupT variant is known to cause HSP.8 This patient, with a pure HSP phenotype, had onset of symptoms at age 27. Vibration and proprioception in the lower limbs was impaired, whereas imaging of the neuroaxis was normal.
SPG7
One patient (HSP16) was found to have a homozygous c.1369C>T variant in SPG7. This variant has previously been reported to cause HSP in compound heterozygous state.9 This patient, with onset of symptoms at age 40, was found to have mild distal upper limb weakness, rest tremor, lower limb sensory symptoms and mild cognitive impairment. Although hand tremor has been reported in up to 10% of patients with SPG4, this is the first report of rest tremor occurring in a case of SPG7.10
X-linked mutations
One patient was found to carry a small deletion in ABCD1 (c.1174_1178del: p.(Leu392SfsTer7), HSP4). This patient had been reported to have a pure HSP phenotype with possibly dominant inheritance. When the family history was reviewed, it was clear that males were affected severely, but females only mildly, and there was no male-to-male transmission of the disease, suggesting X-linked inheritance. The proband had onset of symptoms at age 20 with progressive spastic paraplegia, urinary symptoms and sensory abnormalities in the lower limbs. Brain MRI was normal and thoracic spine MRI at age 55 revealed thinning of the thoracic spinal. Two cousins were similarly affected, and were also found to carry the same mutation. After the mutation was identified, very long chain fatty acids were tested in the proband and found to be within the range for adrenoleukodystrophy.
Discussion
In this paper we describe, for the first time, the clinical and genetic spectrum of HSP in Greece. We provide a clinical description of 54 cases from 40 families. Each of these families was genetically screened using a NGS approach. We made a genetic diagnosis in 21 families (52.5%). Our cohort consisted of a heterogeneous population with pure and complicated, sporadic and familial HSP. When variants in SPAST are excluded, our approach rendered a genetic diagnosis in 13 of 33 patients (39%), which is superior to the 25% diagnostic rate found by Kumar et al.,8 who used a similar NGS panel to screen SPAST-negative patients in Australia.
Our study describes for the first time the mutational spectrum of HSP in Greece. The frequency of gene mutations in HSP varies considerably across populations, therefore a comprehensive assessment of mutation frequency is essential to characterise the population and guide accurate genetic testing. We show that point mutations in SPAST are by far the most common cause of AD HSP in the Greek population, which is consistent with other studies. KIF5A variants were the second most common cause of AD HSP, and were the most frequent cause of complicated, early childhood onset HSP, consistent with previous reports estimating the frequency of KIF5A mutations in AD HSP at 10%.11 In addition, we report the first nonsense mutation in KIF5A (c.2590C>T) in a patient with a typical SPG10 phenotype consisting of early-onset spastic paraplegia with sensorimotor axonal neuropathy. To date, most pathogenic KIF5A variants have been detected in the motor domain and are expected to impair protein function through a dominant-negative effect.12 However, two KIF5A variants outside the motor domain have been found,13, 14 and the mechanism with which these cause disease has not been identified.15 We therefore suggest that further functional work is required before KIF5A variants outside the motor domain can be excluded, particularly in patients with typical SPG10 or axonal CMT2 phenotypes.
ATL1 and REEP1 were relatively rare causes of AD HSP in our population, which is surprising in comparison with other Western European populations. No mutations in NIPA1 were identified.
SPG11 was the commonest cause of AR HSP, again in accordance with previous findings. Interestingly, we identified a novel c.5381T>C variant, which, when in combination with another deleterious SPG11 variant, leads to a syndrome of complicated HSP with relatively late onset. This variant appears to be unique to the Greek cohort under study. The majority of SPG11 variants identified to date lead to premature protein termination or splicing defects. However, at least two missense mutations have already been identified in families with a typical SPG11 phenotype.16, 17 Clearly, further genetic and functional work will be required to determine whether and how missense mutations in SPG11 lead to disease.
CYP7B1 variants were the second most common cause of AR HSP, and we confirm that sensorineural deafness can occur as part of the spectrum of CYP7B1-associated HSP.
Surprisingly, we identified a family with a pathogenic deletion in ABCD1 mimicking HSP and presenting with what was initially thought to be dominant inheritance. It has previously been shown that adrenoleukodystrophy can mimic HSP and even present with an apparently AR mode of inheritance.18 It is essential to identify patients with adrenoleukodystrophy as they require careful monitoring of adrenal function and are potentially treatable with stem cell transplant should cerebral adrenoleukodystrophy develop.19
We did not detect potentially pathogenic variants in 19 probands. Sixteen of these were sequenced using the Truseq custom HSP panel and three were sequenced using the Trusight Exome. Of these 19, 9 had an AD family history of HSP, 8 had sporadic inheritance and the family history was unknown in 2. Seventeen had a pure HSP phenotype, with only two patients having a complicated phenotype. This raises interesting questions as to whether there are many more genes causing HSP that have not yet been identified. It will be necessary to perform whole-exome or whole-genome analysis on a large cohort of phenotypically similar pure HSP cases to address this.
This study shows that a NGS approach can be an efficient way to make a genetic diagnosis in a heterogeneous population naive to genetic testing. With one test, more than half of patients can be given a genetic diagnosis, allowing for resources to be concentrated on investigating gene-negative patients and families. NGS approaches may be particularly relevant in resource poor settings, where biochemical and radiological testing may not be readily available or may be prohibitively expensive. This study confirms that NGS approaches are efficient and cost-effective and should be employed in the first line of investigation in HSP patients.
Acknowledgments
The authors would like to thank all the patients in this study for their essential help with this work. This study was supported by the Medical Research Council, The Leonard Wolfson Experimental Neurology Centre, The Wellcome Trust and The Brain Research Trust. This study was also supported by the National Institute for Health Research University College London Hospitals Biomedical Research Centre.
The authors declare no conflict of interest.
Footnotes
Supplementary Information accompanies this paper on European Journal of Human Genetics website (http://www.nature.com/ejhg)
Supplementary Material
References
- Fink JK: Hereditary spastic paraplegia: clinico-pathologic features and emerging molecular mechanisms. Acta Neuropathol 2013; 126: 307–328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lo Giudice T, Lombardi F, Santorelli FM, Kawarai T, Orlacchio A: Hereditary spastic paraplegia: Clinical-genetic characteristics and evolving molecular mechanisms. Exp Neurol 2014; 261: 518–539. [DOI] [PubMed] [Google Scholar]
- Noreau A, Dion PA, Rouleau GA: Molecular aspects of hereditary spastic paraplegia. Exp Cell Res 2014; 325: 18–26. [DOI] [PubMed] [Google Scholar]
- Ruano L, Melo C, Silva MC, Coutinho P: The global epidemiology of hereditary ataxia and spastic paraplegia: a systematic review of prevalence studies. Neuroepidemiology 2014; 42: 174–183. [DOI] [PubMed] [Google Scholar]
- Buermans HPJ, den Dunnen JT: Next generation sequencing technology: advances and applications. Biochim Biophys Acta 2014; 1842: 1932–1941. [DOI] [PubMed] [Google Scholar]
- ExAC. Exome Aggregation Consortium. Available at http://exac.broadinstitute.org (last accessed 23 April 2015).
- Arnoldi A, Crimella C, Tenderini E et al: Clinical phenotype variability in patients with hereditary spastic paraplegia type 5 associated with CYP7B1 mutations. Clin Genet 2012; 81: 150–157. [DOI] [PubMed] [Google Scholar]
- Kumar KR, Blair NF, Vandebona H et al: Targeted next generation sequencing in SPAST-negative hereditary spastic paraplegia. J Neurol 2013; 260: 2516–2522. [DOI] [PubMed] [Google Scholar]
- Klebe S, Depienne C, Gerber S et al: Spastic paraplegia gene 7 in patients with spasticity and/or optic neuropathy. Brain 2012; 135: 2980–2993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Bot ST, van den Elzen RTM, Mensenkamp AR et al: Hereditary spastic paraplegia due to SPAST mutations in 151 Dutch patients: new clinical aspects and 27 novel mutations. J Neurol Neurosurg Psychiatry 2010; 81: 1073–1078. [DOI] [PubMed] [Google Scholar]
- Goizet C, Boukhris A, Mundwiller E et al: Complicated forms of autosomal dominant hereditary spastic paraplegia are frequent in SPG10. Hum Mutat 2009; 30: E376–E385. [DOI] [PubMed] [Google Scholar]
- Liu Y-T, Laurá M, Hersheson J et al: Extended phenotypic spectrum of KIF5A mutations: from spastic paraplegia to axonal neuropathy. Neurology 2014; 83: 612–619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crimella C, Baschirotto C, Arnoldi A et al: Mutations in the motor and stalk domains of KIF5A in spastic paraplegia type 10 and in axonal Charcot-Marie-Tooth type 2. Clin Genet 2012; 82: 157–164. [DOI] [PubMed] [Google Scholar]
- Lo Giudice M, Neri M, Falco M et al: A missense mutation in the coiled-coil domain of the KIF5A gene and late-onset hereditary spastic paraplegia. Arch Neurol 2006; 63: 284–287. [DOI] [PubMed] [Google Scholar]
- Ebbing B, Mann K, Starosta A et al: Effect of spastic paraplegia mutations in KIF5A kinesin on transport activity. Hum Mol Genet 2008; 17: 1245–1252. [DOI] [PubMed] [Google Scholar]
- Denora PS, Schlesinger D, Casali C et al: Screening of ARHSP-TCC patients expands the spectrum of SPG11 mutations and includes a large scale gene deletion. Hum Mutat 2009; 30: E500–E519. [DOI] [PubMed] [Google Scholar]
- Crimella C, Arnoldi A, Crippa F et al: Point mutations and a large intragenic deletion in SPG11 in complicated spastic paraplegia without thin corpus callosum. J Med Genet 2009; 46: 345–351. [DOI] [PubMed] [Google Scholar]
- Zhan Z-X, Liao X-X, Du J et al: Exome sequencing released a case of X-linked adrenoleukodystrophy mimicking recessive hereditary spastic paraplegia. Eur J Med Genet 2013; 56: 375–378. [DOI] [PubMed] [Google Scholar]
- Mitchell R, Nivison-Smith I, Anazodo A et al: Outcomes of haematopoietic stem cell transplantation for inherited metabolic disorders: a report from the Australian and New Zealand Children's Haematology Oncology Group and the Australasian Bone Marrow Transplant Recipient Registry. Pediatr Transplant 2013; 17: 582–588. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.