Figure 2. Mice treated with DMH1 reduce primary mammary tumor growth.
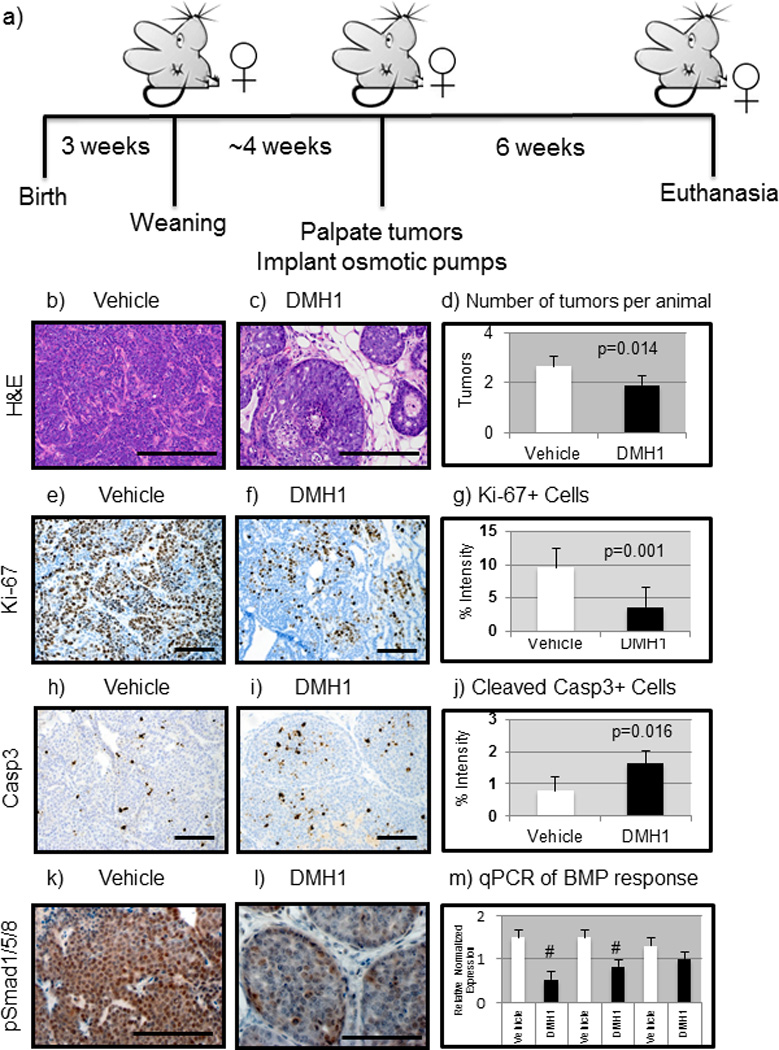
a) FVBn mice expressing the MMTV.PyVmT constitutively expressed oncogene were implanted with slow release (6 weeks) osmotic pumps after first palpation of tumors (~7weeks). b) Vehicle (DMSO only, n=12) treated tumors were stained with H&E which indicated advanced adenocarcinoma morphology. c) DMH1 treated animals (n=12) revealed a much less advanced adenocarcinoma appearance. d) The number of tumors per animal greater than .5cm in diameter were counted at sacrifice and revealed a statistically significant (P=0.014) reduction with DMH1 treatment (1.92) compared to vehicle treated controls (2.66). e) IHC for Ki-67 to indicate actively proliferating cells revealed strongly proliferative cells in vehicle treated animals. f) DMH1 treated tumors revealed less actively stained Ki-67+ cells. g) IHC for the proliferation marker Ki-67 showed significant (P=0.001) reduction in proliferating tumor cells (vehicle=9.52, DMH1=3.61). h) IHC for activation of apoptosis via cleavage of caspase 3 revealed little apoptosis in vehicle treated primary tumors. i) DMH1 treated tumors displayed significantly higher levels of activated caspase 3 by IHC. j) Quantitation of IHC for apoptosis maker cleaved-caspase3 showed significant (P=0.016) increase in apoptotic tumor cells (vehicle=.79, DMH1=1.62). k) IHC for active BMP signaling with pSmad1/5/8 demonstrated that primary tumors are strongly positive for BMP signaling. l) DMH1 treated tumors had reduced staining for pSmad1/5/8 indicating reduced yet not absent BMP signaling. m) EpCAM+ tumor cells isolated by FACS had RNA isolated and SYBR qPCR was performed for BMP transcriptional targets, which demonstrated a significant reduction in Id1 and Smad6. Normalized expression for qPCR was performed in comparison to GAPDH expression. Quantification of IHC was performed using NIH ImageJ software to detect pixels positive for stain in a 20X field of view. A minimum of four tumors with at least five representative images were used in analysis. # Indicate statistical significance (P=<0.05) by performing a student’s T-test. Error bars indicate SEM.
