Abstract
CRISPR-Cas9 nucleases are primarily guided by RNA-DNA interactions but also require Cas9-mediated recognition of a protospacer adjacent motif (PAM). While potentially advantageous for specificity, extended PAM sequences limit the targeting range of Cas9 orthologues for genome editing. One possible strategy to relieve this restriction is to relax specificities for certain positions within the PAM. Here we used molecular evolution to modify the NNGRRT PAM specificity of Staphylococcus aureus Cas9 (SaCas9). One variant we identified, referred to as KKH SaCas9, shows robust genome editing activities at endogenous human target sites with NNNRRT PAMs. Importantly, using GUIDE-seq, we show that both wild-type and KKH SaCas9 induce comparable numbers of off-target effects in human cells. KKH SaCas9 increased the targeting range of SaCas9 by nearly two- to four-fold. Our molecular evolution strategy does not require structural information and therefore should be applicable to a wide range of Cas9 orthologues.
Originally discovered as an essential component of the bacterial clustered regularly interspaced short palindromic repeat (CRISPR) immune system, the CRISPR-associated protein 9 (Cas9) has become a widely used customizable nuclease for genome editing1–3. Cas9 cleavage specificity is directed by two short RNAs known as the crRNA and tracrRNA4, 5, which can be fused into a single guide RNA (sgRNA)4, 6–8. The 5′ end of the sgRNA (derived from the crRNA) can base pair with the target DNA site, thereby permitting straightforward re-programming of site-specific cleavage by the Cas9/sgRNA complex4. However, Cas9 must also recognize a specific protospacer adjacent motif (PAM) that lies proximal to the DNA that base pairs with the sgRNA4, 9–12, a requirement that is needed to initiate sequence-specific recognition13 but that can also constrain the targeting range of these nucleases for genome editing. The broadly used Streptococcus pyogenes Cas9 (SpCas9) recognizes a short NGG PAM4, 14, which occurs once in every 8 bps of random DNA sequence. By contrast, other Cas9 orthologues characterized to date can require longer PAMs12, 15–18. For example, Staphylococcus aureus Cas9 (SaCas9), one of several smaller Cas9 orthologues that are better suited for viral delivery12, 17, 18, recognizes a longer NNGRRT PAM that is expected to occur once in every 32 bps of random DNA. Broadening the targeting range of Cas9 orthologues is important for various applications, including the modification of small genetic elements (e.g., transcription factor binding sites19, 20) or performing allele- specific alterations by positioning sequence differences within the PAM21.
A potential strategy for improving the targeting range of orthogonal Cas9s that recognize extended PAMs is to alter their PAM recognition specificities. In previous work22, we demonstrated the feasibility of changing the PAM specificity of SpCas9 using a combination of structure-guided design and directed evolution performed with a bacterial cell-based selection system. A limitation of this approach is the need to evolve a separate variant for each potential PAM sequence, a challenge that becomes even greater for Cas9 orthologues that specify longer PAMs. An alternative strategy for such orthologues might be to evolve variants that have relaxed or partially relaxed specificities for certain positions within the PAM. The capability to engineer such variants would expand the utility of Cas9 orthologues that specify longer PAM sequences.
We devised an unbiased genetic approach for engineering Cas9 variants with relaxed PAM recognition specificities that does not require structural information. We tested this strategy using SaCas9, for which no structural data was available at the time we initiated these studies. In an initial step, we sought to conservatively estimate the PAM-interacting domain for SaCas9 by sequence comparisons with the structurally well-characterized SpCas923–25. Although SpCas9 and SaCas9 differ substantially at the primary sequence level (Fig. 1a, Supplementary Fig. 1), alignment of both with 10 additional orthologues enabled us to conservatively define a predicted PAM-interacting domain for SaCas9 (Online Methods; Supplementary Figs. 1 and 2).
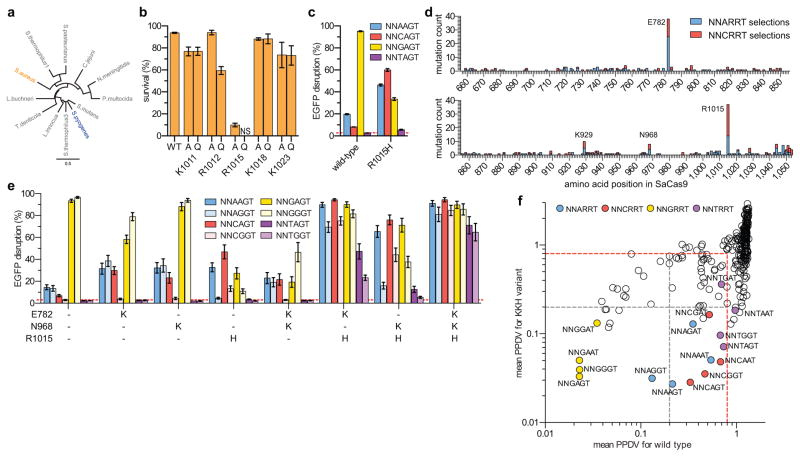
Figure 1.
Selection and assembly of SaCas9 variants with altered PAM specificities (a) Phylogenetic tree of Cas9 orthologues with SpCas9 and SaCas9 highlighted. (b) Activity of SaCas9 variants with single amino acid substitutions assessed in the bacterial positive selection assay (see also Supplementary Fig. 3b). Error bars represent s.e.m., n = 3; NS = no survival. (c) Human cell activity of wild-type and R1015H SaCas9. EGFP disruption activity quantified by flow cytometry; error bars represent s.e.m, n = 3, mean level of background EGFP loss represented by dashed red line (for this and panel e). (d) Total number of substitutions observed at each amino acid position when selecting for SaCas9 variants with altered PAM specificities (see also Supplementary Figs. 4 and 5). Starter mutations at R1015 are not counted. (e) Human cell EGFP disruption activity of variants containing mutations observed when selecting for altered PAM specificities. (f) Mean post-selection PAM depletion value (PPDV) scatterplot of wild-type SaCas9 versus the KKH variant (n = 2, see also Supplementary Fig. 8c). Two libraries with different protospacers and 8 randomized basepairs in place of the PAM were used to determine which PAMs are targetable by each Cas9. Statistically significant depletion indicated by the red dashed line (relative to a dCas9 control, see Supplementary Figs. 8a and 8b), and 5-fold depletion by the grey dashed line.
Because the guanine at the third position in the SaCas9 PAM is the most strictly specified base17, we randomly mutagenized the predicted PI domain and used our previously described bacterial cell- based method22 to attempt to select for mutants capable of cleaving sites with each of the three other possible nucleotides at the 3rd PAM position (i.e., NN[A/C/T]RRT PAMs (NNHRRT); Supplementary Fig. 3a). All but one of the surviving variants from the selections against sites containing NNARRT and NNCRRT PAMs harbored an R1015H mutation (Supplementary Fig. 4), whereas we did not obtain any variants from the selections with NNTRRT PAMs. These results strongly suggested that R1015 might participate in recognition of the guanine at the third position of the SpCas9 PAM. Indeed, in our alignments we found that R1015 of SaCas9 is in the vicinity of SpCas9 R1335 (Supplementary Fig. 2), a residue previously implicated in recognition of the third base position of the PAM22, 24. Consistent with this, we found that mutation of R1015 to an alanine or glutamine substantially decreased SaCas9 activity on a target site containing an NNGRRT PAM (Fig. 1b) when tested in our bacterial selection system (Supplementary Fig. 3b). Alanine or glutamine substitutions of other positively charged residues in the vicinity of R1015 did not have as strong of an effect on SaCas9 activity (Fig. 1b, Supplementary Fig. 2).
Our bacterial-based selection results also suggested that the R1015H mutation might at least partially relax the specificity of SaCas9 at the third position of the PAM. However, we found that the R1015H single mutant had suboptimal activity in our previously described human cell-based EGFP disruption assay26 when tested against sites with any nucleotide at the 3rd position of NNNRRT PAMs (Fig. 1c). Because this suggested that additional mutations might be required to increase or optimize the activity of the R1015H mutant in human cells, we randomly mutagenized a region encompassing the predicted PI domain of an SaCas9 variant that also harbored a R1015Q mutation. We then selected for variants from this library that could cleave target sites with each of the three different NNHRRT PAMs using our bacterial selection system. We used R1015Q because, unlike R1015H, this mutant did not show activity in bacteria (Fig. 1b, and data not shown, respectively). Although no surviving clones were again observed when selecting against NNTRRT PAMs, selections with the R1015Q variant against NNARRT or NNCRRT yielded mutations at E782, K929, N968, and a mutation of the Q at 1015 to H (Supplementary Fig. 5).
Combined with the selection results from wild-type SaCas9, the most frequent missense mutations identified across all selections were E782K, K929R, N968K and R1015H (Fig. 1d, Supplementary Figs. 4 and 5), suggesting that a combination of these mutations might permit efficient cleavage of sites that contain an A or C at the third position of the SaCas9 PAM. We therefore tested SaCas9 variants containing different combinations of these mutations using the human cell-based EGFP disruption assay with sgRNAs targeted to sites harboring each of the 4 bases at the third position of the PAM (i.e. on NNNRRT PAMs) (Fig. 1e, Supplementary Fig. 6). We found that the variants with the triple mutant combinations E782K/N968K/R1015H and E782K/K929R/R1015H were highly active at sites with NNNRRT PAMs (Fig. 1e, Supplementary Fig. 6), whereas the quadruple mutant variant containing all four mutations (E782K/K929R/N968K/R1015H) had generally lower activities on these sites (Supplementary Fig. 6). We chose the E782K/N968K/R1015H (hereafter referred to as the KKH variant) for further characterization, and verified using our human cell-based EGFP disruption assay that all three substitutions comprising the KKH variant were required for activity (Fig. 1e).
To more comprehensively define the PAM specificities of KKH and wild-type SaCas9, we used our previously described bacterial cell-based site-depletion assay22 (Supplementary Fig. 7). This method yields Cas9 PAM specificity profiles by identifying the relative cleavage (and therefore depletion in bacterial cells) of DNA plasmids bearing randomized PAM sequences, quantified as a post-selection PAM depletion value (PPDV). We performed site-depletion experiments with both wild-type and KKH SaCas9 using libraries with two different spacer sequences each with 8 randomized bases in place of the PAM (Supplementary Fig. 7). Control experiments using catalytically inactive SaCas9 showed little depletion of any PAM sequence (Supplementary Fig. 8a), enabling us to establish a threshold for statistically significant depletion as a PPDV of 0.794 (Supplementary Fig. 8b). Previous experiments have shown that PAMs with PPDVs of <0.2 in our bacterial site-depletion assay can be efficiently cleaved in our human cell-based EGFP disruption assay22. With wild-type SaCas9, the most depleted PAMs (based on mean PPDVs obtained from the two libraries) were, as expected, the four NNGRRT PAMs (Fig. 1f and Supplementary Fig. 8c). Notably, other PAMs with mean PPDVs <0.1 included those of the form NNGRRN (Supplementary Fig. 8d), suggesting that for some spacer sequences the last position of the PAM may not be fully specified as a T in our bacterial-based assay (although a previous report demonstrated by an in vitro PAM depletion assay, ChIP-seq and targeting of endogenous human sites that a thymine at the sixth position of the PAM was highly preferred17). By contrast, with the KKH variant, PAMs with mean PPDVs of <0.2 included not only the NNGRRT PAMs but also all four NNARRT, all four NNCRRT, and three of the four NNTRRT PAMs (Fig. 1f, Supplementary Figs. 8c and 8e). These results suggested that KKH SaCas9 appears to have a broadened PAM targeting range relative to its wild-type counterpart.
To assess the robustness of the KKH SaCas9 variant in human cells, we tested its activity on 55 different endogenous gene target sites containing a variety of NNNRRT PAMs (Fig. 2a). The KKH variant showed efficient activity with a mean mutagenesis frequency of 24.7% across all sites, with 80% of sites (44 of 55 sites) showing greater than 5% disruption. Analysis of KKH SaCas9 activity across all 55 sites revealed ordered preferences for the 3rd position of the PAM (NN[G>A=C>T]RRT; Fig. 2b) as well as the 4th/5th positions of the PAM (NNN[AG>GG>GA>AA]T; Fig. 2c). Consistent with this, we observed differences among the 16 possible combinations of the 3rd/4th/5th positions of an NNNRRT PAM (Supplementary Fig. 9a). KKH SaCas9 functioned efficiently on spacer lengths ranging from 21–23 nucleotides (Fig. 2d), spacer sequences with variable GC content (Supplementary Fig. 9b), and PAMs with variable GC content (Supplementary Fig. 9c). Sequence logos derived from sites cleaved with low, medium, and high efficiencies (0–10%, 10–30%, and >30% mean mutagenesis frequencies, respectively) revealed little sequence preference across the entire target site other than at the 4th and 5th positions of the NNNRRT PAM, and perhaps a slight preference for guanine at the 2nd PAM position on sites cleaved with high efficiencies (Supplementary Fig. 9d).
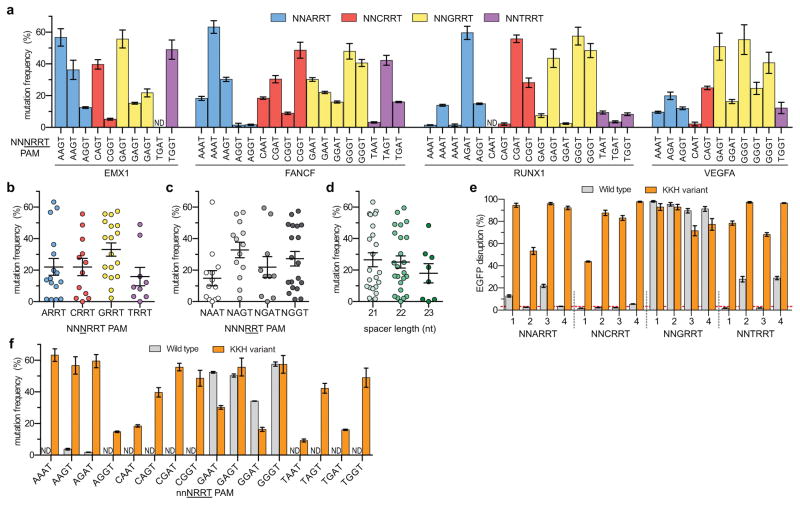
Figure 2.
Activity of the SaCas9 KKH variant targeted to endogenous sites in human cells (a) Mutagenesis frequencies across 55 different sites bearing NNNRRT PAMs induced by KKH SaCas9, determined by T7E1 assay. Error bars represent s.e.m., n = 3, ND, not detectable by T7E1 assay. (b) KKH variant preference for the third position of the PAM. Mean activities from data in panel a are shown for this and panels b and c. (c) KKH variant preference for the fourth and fifth positions of the PAM. (d) Spacer length preference of the KKH SaCas9 variant. (e) Comparison of the human cell EGFP disruption activity of wild-type and KKH SaCas9 targeted to various sites containing NNNRRT PAMs. EGFP disruption quantified by flow cytometry; error bars represent s.e.m, n = 3, mean level of background EGFP loss represented by dashed red line. (f) Mutagenesis frequencies of wild-type SaCas9 against one site for each of the 16 possible NNNRRT sites from panel a (sites with the highest KKH activity were selected). Error bars represent s.e.m., n = 3, ND, not detectable by T7E1 assay.
To demonstrate that the KKH variant enables modification of PAMs that cannot be targeted by wild-type SaCas9, we performed direct comparisons of these nucleases in human cells on sites bearing various NNNRRT PAMs. Assessment of 16 sites using our EGFP disruption assay and 16 endogenous human gene targets (Figs. 2e and 2f, respectively) showed that KKH SaCas9 robustly modified target sites bearing NNNRRT PAMs whereas wild-type SaCas9 efficiently targeted only sites with NNGRRT PAMs. For all 24 sites with NNHRRT PAMs, the KKH variant induced substantially higher rates of mutagenesis than wild-type SaCas9. On the eight sites with NNGRRT PAMs, KKH SaCas9 induced comparable or slightly lower levels of mutagenesis compared with wild-type (Figs. 2e and 2f). These results collectively demonstrate that the KKH variant can cleave sites with NNNRRT PAMs, thereby enabling targeting of sites with NNHRRT PAMs that currently cannot be efficiently altered by wild-type SaCas9 in human cells.
To assess the impact of the KKH mutations on the genome-wide specificity of SaCas9, we used the GUIDE-seq (Genome-wide Unbiased Identification of DSBs Enabled by sequencing) method to directly compare the off-target profiles of wild-type and KKH SaCas9 with the same sgRNAs27. When tested with sgRNAs targeted to six endogenous human gene sites containing NNGRRT PAMs, we observed that wild-type and KKH SaCas9 induced nearly identical GUIDE-seq tag integration rates and on-target cleavage frequencies for all six sites (Supplementary Figs. 10a and 10b, respectively). Furthermore, wild-type and KKH SaCas9 induce mutations at similar numbers of off-target sites with each of the six sgRNAs (Figs. 3a and 3b). Off-target sites for the KKH variant generally adhered to the NNNRRT PAM motif, and off-target sites for wild-type SaCas9 adhered to an NNGRR[T>G] motif (Fig. 3b). With one of the sgRNAs, which induced the highest number of off-target sites among the six sgRNAs tested, we observed a similar number of off-target sites with wild-type and KKH SaCas9. However, the off-target sites were only partially overlapping between wild-type and KKH SaCas9, as might be expected given their different PAM specificities (Figs. 3b and 3c). Although we would not advocate the use of the KKH variant for targeting sites with NNGRRT PAMs (because wild-type SaCas9 can show higher on-target activities than KKH for these sites), these results suggest that KKH SaCas9 mostly cleaves off-target sites with the expected PAMs and generally induces numbers of off-target sites comparable to those observed with wild-type SaCas9.
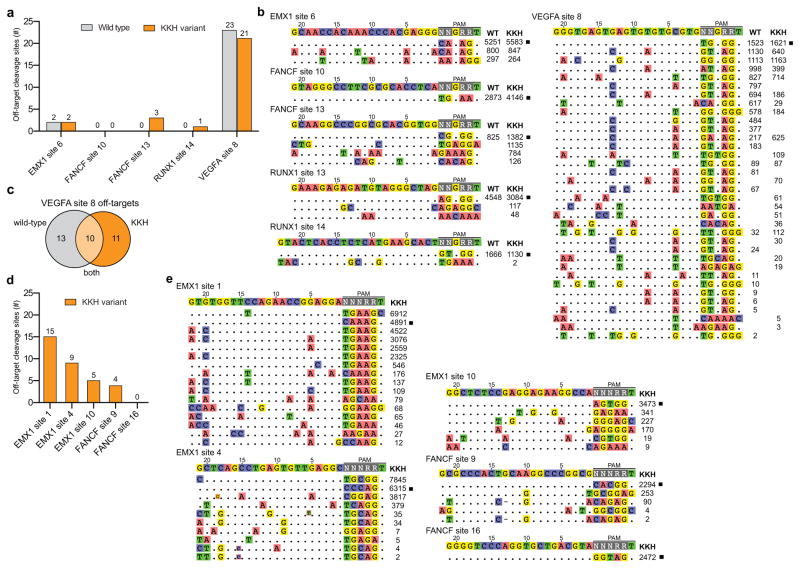
Figure 3.
Genome-wide specificity profiles of wild-type and KKH SaCas9 (a) and (b) Direct comparison of wild-type and KKH SaCas9 targeted to sites containing NNGRRT PAMs, represented by total number of off-targets (panel a) and mismatches observed at each off-target site (panel b). For panels b and e, GUIDE-seq read counts at each site are indicated; on-target sequences are marked with a black box; mismatched positions within off-target sites are highlighted; sequences have been corrected for cell-type specific SNPs; sites with potential sgRNA or DNA bulge nucleotides are indicated by a small red-bordered base or a dash, respectively. (c) Venn diagram highlighting the differences in off-target site cleavage by wild-type and KKH SaCas9 at VEGFA site 8. (d) and (e) Specificity profile of the KKH variant targeted to sites containing NNHRRT PAMs, represented by total number of off-targets (panel d) and mismatches observed at each off-target site (panel e).
To further examine the genome-wide specificity of KKH SaCas9, we tested five additional sgRNAs targeted to sites containing NNHRRT PAMs (Figs. 3d and 3e). Off-target sites detected by GUIDE-seq were generally low in number (comparable to the numbers observed with wild-type SpCas9 and SpCas9 variants in previously published experiments22, 27), displayed potential DNA- and RNA-bulged off-targets28 and contained expected PAM sequences. Taken together, our experiments demonstrate that the genome-wide specificities of wild-type and KKH SaCas9 are similar and generally show low numbers of off-target mutations in human cells as judged by GUIDE-seq.
Although wild-type SaCas9 remains the most optimal choice for targeting NNGRRT PAMs, the KKH SaCas9 variant we describe here can robustly target sites with NNARRT and NNCRRT PAMs and has a reasonable success rate for sites with NNTRRT PAMs. Thus, we conservatively estimate that the KKH variant increases the targeting range of SaCas9 by nearly two- to four-fold in random DNA sequence (see Supplementary Note 1), thereby improving the prospects for more broadly utilizing SaCas9 in a variety of different applications that require highly precise targeting. Using GUIDE-seq, we demonstrated that KKH SaCas9 induces similar numbers of off-target mutations as wild-type SaCas9 when targeted to the same sites that contain NNGRRT PAMs. Also, KKH SaCas9 induces only a small number of off-target mutations when targeted to sites bearing NNHRRT PAMs. Although KKH SaCas9 recognizes a modified PAM sequence relative to wild-type SaCas9, our findings are not entirely surprising given that the total combined length of the protospacer and PAM is still long enough with the KKH variant (24 to 26 bps) to be reasonably orthogonal to the human genome. Furthermore, it is possible that modifying PAM recognition can improve specificity by altering the energetics of Cas9/sgRNA interaction with its target site (similar to the previously proposed mechanisms for improved specificities of truncated sgRNAs29 or the D1135E SpCas9 mutant22).
Our results suggested that R1015 in wild-type SaCas9 contacts the G in the third PAM position, a finding confirmed by structures of SaCas9 bound to target DNA sites that were published as we were finalizing this work for submission30. We speculate that the R1015H substitution removes this contact and relaxes specificity at the third position; however, loss of the R1015 to G contact could also conceivably reduce the energy associated with target site binding, which may explain why the R1015H mutation alone is not sufficient for robust activity at NNNRRT sites in human cells. Because the E782K and N968K substitutions both add positive charge, it is possible that they may make non-specific interactions with the DNA phosphate backbone to compensate energetically for the loss of the R1015 to guanine contact. The recently published SaCas9 structure30 supports this hypothesis: E782 is in the vicinity of the target strand DNA-backbone (near the spacer/PAM junction) and N968 is near the non-target strand backbone within the PAM.
The genetic approach described here does not require structural information and therefore should be applicable to many other Cas9 orthologues. The only requirement to evolve Cas9 nucleases with broadened PAM specificities is that they function in our bacterial-based selection. Although previous studies demonstrated that PAM recognition can be altered by swapping the PAM-interacting domains of highly related Cas9 orthologues25, it remains to be determined whether this strategy is generalizable or effective when using more divergent orthologues. By contrast, the evolution strategies we have described here and in a previous study22 can, in principle, be used to engineer PAM recognition specificities beyond those encoded within naturally occurring Cas9 orthologues. We envision that our overall strategy can be employed to expand the targeting range and extend the utility of the numerous Cas9 orthologues that exist in nature.
METHODS
Plasmids and oligonucleotides
A list and sequences of plasmids used in this study can be found in Supplementary Note 2. Oligonucleotides are listed in Supplementary Table 1 and sgRNA target sites are listed in Supplementary Table 2. A selection of the new plasmids in this study will be deposited with the nonprofit plasmid repository Addgene: http://www.addgene.org/crispr-cas (see Supplementary Information).
Bacterial Cas9/sgRNA expression plasmids were used to express both a human codon optimized version of SaCas9 and the sgRNA, each expressed under a separate T7 promoter. Bacterial expression plasmids used in the selections were derived from BPK210122 whereas those used in the site-depletion assay were modified to express a sgRNA with a shortened repeat:anti-repeat sequence (see below). All sgRNAs in these bacterial expression plasmids included two guanines at the 5′ end of the spacer sequence for proper expression from the T7 promoter.
To generate libraries of SaCas9 variants, amino acids M657-G1053 of SaCas9 were randomly mutagenized using Mutazyme II (Agilent Technologies) at a frequency of ~5.5 mutations/kilobase. Both wild-type and R1015Q SaCas9 were used as starting template for mutagenesis, resulting in two libraries with estimated complexities of greater than 6×106 clones.
Positive selection plasmids were assembled by ligating oligonucleotide duplexes encoding target sites into XbaI/SphI-digested p11-lacY-wtx131. For the site-depletion experiments, two separate libraries containing different spacer sequences were generated. For each library, an oligonucleotide containing 8 randomized nucleotides adjacent to the spacer sequence (in place of the PAM) was complexed with a bottom strand primer and filled in using Klenow(-exo) (refer to Supplementary Table 1). The resulting product was digested with EcoRI and ligated into EcoRI/SphI-digested p11-lacY-wtx1. Estimated complexities of the two site-depletion libraries were greater than 4×106 clones.
For human cell experiments, human codon-optimized wild-type and variant SaCas9s were expressed from a plasmid containing a CAG promoter. sgRNA expression plasmids (containing a U6 promoter) were generated by ligating oligonucleotide duplexes encoding the spacer sequence into BsmBI digested VVT122 or BPK2660 (containing the full length 120 nt crRNA:tracrRNA sgRNA or a 84 nt shortened repeat:anti-repeat version, respectively). All sgRNAs used in this study for human expression included one guanine at the 5′ end of the spacer to ensure proper expression from the U6 promoter, and also used a shortened sgRNA (Supplementary Fig. 11) similar to that previously described17.
Bioinformatic analysis of Cas9 orthologue sequences
Similar to alignments performed in previous studies15, 17, 24, Cas9 orthologues similar to both SpCas9 and SaCas9 were aligned using ClustalW2 (http://www.ebi.ac.uk/Tools/msa/clustalw2/). The resulting phylogenetic tree and protein alignment were visualized using Geneious version 8.1.6 and ESPript (http://espript.ibcp.fr/ESPript/ESPript/).
Bacterial-based positive selection assay
The bacterial positive selection assays were performed as previously described22. Briefly, Cas9/sgRNA plasmids were transformed into E. coli BW25141(λDE3)32 containing a positive selection plasmid. Transformations were plated on both non-selective (chloramphenicol) and selective (chloramphenicol + 10 mM arabinose) conditions. Cas9 cleavage of the selection plasmid was estimated by calculating the percent survival: (# of colonies on selective plates/# of colonies on non-selective plates)×100. To select for SaCas9 variants capable of recognizing alternative PAMs, the wild-type and R1015Q libraries with mutagenized PI domains were transformed into competent E. coli BW25141(λDE3) containing positive selection plasmids with NNAAGT, NNAGGT, NNCAGT, NNCGGT, NNTAGT, or NNTGGT PAMs. Approximately 1×105 clones were screened by plating on selective conditions, and surviving colonies containing SaCas9 variants presumed to cleave the selection plasmid were mini-prepped (MGH DNA Core). All variants were re-screened individually in the positive selection assay, and those with greater than ~20% survival were sequenced to determine the mutations required for recognition of the alternate PAM.
Bacterial-based site-depletion assay
The site-depletion experiments were performed as previously described22. Briefly, the randomized PAM libraries were electroporated into competent E. coli BW25141(λDE3) containing either wild-type, catalytically inactive (D10A/H557A), or KKH variant SaCas9/sgRNA plasmids. Greater than 1×105 colonies were plated on chloramphenicol/carbenicillin plates, and selection plasmids with PAMs resistant to Cas9 targeting contained within the surviving colonies were isolated by maxiprep (Qiagen). The region of the plasmid containing the spacer sequence and PAM was PCR-amplified using the primers listed in Supplementary Table 1. The KAPA HTP library preparation kit (KAPA BioSystems) was used to generate a dual-indexed Tru-seq Illumina sequencing library using ~500 ng purified PCR product from each site-depletion condition prior to an Illumina MiSeq high-throughput sequencing run at the Dana- Farber Cancer Institute Molecular Biology Core. The data from the site-depletion experiments was analyzed as previously described22, with the exception that the script was modified to analyze 8 randomized nucleotides. The counts of all possible 8 nt strings for each site-depletion treatment can be found in Supplementary Table 3. Cas9 ability to recognize PAMs was determined by calculating the post-selection PAM depletion value (PPDV) of any given PAM: the ratio of the post-selection frequency of that PAM to the pre-selection library frequency. A control experiment using catalytically inactive SaCas9 was used to establish that a PPDV of 0.794 represents statistically significant depletion relative to the input library.
Human cell culture and transfection
U2OS cells obtained from our collaborator T. Cathomen (Freiburg) and U2OS.EGFP cells harboring a single integrated copy of an EGFP-PEST reporter gene33 were cultured in Advanced DMEM medium (Life Technologies) with 10% FBS, penicillin/streptomycin, and 2 mM GlutaMAX (Life Technologies) at 37°C with 5% CO2. Cell line identities were validated by STR profiling (ATCC) and deep sequencing, and cells were tested bi-weekly for mycoplasma contamination. U2OS.EGFP culture medium was additionally supplemented with 400 μg/mL G418. Cells were co-transfected with 750 ng Cas9 plasmid and 250 ng sgRNA plasmid using the DN-100 program of a Lonza 4D-nucleofector following the manufacturer’s instructions.
Human cell EGFP disruption assay
EGFP disruption experiments were performed as previously described26, 33. Approximately 52 hours post- transfection, a Fortessa flow cytometer (BD Biosciences) was used to measure EGFP fluorescence in transfected U2OS.EGFP cells. Negative control transfections of Cas9 and empty U6 promoter plasmids were used to establish background EGFP loss at ~2.5% for all experiments (represented as a red dashed line in figures).
T7E1 assay
T7E1 assays were performed as previously described33 to quantify Cas9-induced mutagenesis at endogenous loci in human cells. Approximately 72 hours post-transfection, genomic DNA was isolated using the Agencourt DNAdvance Genomic DNA Isolation Kit (Beckman Coulter Genomics). Target loci were PCR-amplified from ~100 ng of genomic DNA using the primers listed in Supplementary Table 1. Following an Agencourt Ampure XP clean-up step (Beckman Coulter Genomics), ~200 ng purified PCR product was denatured and hybridized prior to digestion with T7E1 (New England Biolabs). Following a second clean-up step, mutagenesis frequencies were quantified using a Qiaxcel capillary electrophoresis instrument (Qiagen).
GUIDE-seq experiments
GUIDE-seq experiments were performed and analyzed as previously described27. Briefly, U2OS cells were transfected as described above with Cas9 and sgRNA plasmids, as well as 100 pmol of a phosphorylated, phosphorothioate-modified double-stranded oligodeoxynucleotide (dsODN) with an embedded NdeI site. Restriction fragment length polymorphism (RFLP) analyses were performed to determine frequency of dsODN-tag integration frequencies22, 27, and T7E1 assays were performed to quantify on-target Cas9 mutagenesis frequencies. dsODN tag-specific amplification and library preparation27 was performed prior to high-throughput sequencing using an Illumina MiSeq Sequencer. When mapping potential off-target sites, the cut-off for alignment to the on-target spacer sequence was set at 8 mismatches for 21 nucleotide spacers, 9 mismatches for 22 nucleotide spacers, and 10 mismatches for 23 nucleotide spacers. Off-target sites with potential DNA- or RNA-bulges28 were identified by manual alignment.
Supplementary Material
Acknowledgments
We thank D. Edgell for providing the bacterial strain and precursor plasmids related to the bacterial selections, and J. Angstman for comments on the manuscript. This work was supported by a National Institutes of Health (NIH) Director’s Pioneer Award (DP1 GM105378) and NIH R01 GM107427 to J.K.J., the Jim and Ann Orr Research Scholar Award (to J.K.J.), and a Natural Sciences and Engineering Research Council of Canada Postdoctoral Fellowship (to B.P.K.). New reagents described in this work will be deposited with the non-profit plasmid distribution service Addgene (http://www.addgene.org/crispr-cas).
Footnotes
Accession codes: GUIDE-seq data has been deposited to the NCBI sequence read archive (SRA) under study accession SRP064571
Author Contributions
B.P.K. and M.S.P. performed all bacterial and human cell experiments, and N.T.N. assisted with GUIDE- seq experiments. S.Q.T., V.V.T., and Z.Z. analyzed the site-depletion and GUIDE-seq data. B.P.K. and J.K.J directed the research, interpreted experiments, and wrote the manuscript with input from all authors.
Competing Financial Interests
J.K.J. is a consultant for Horizon Discovery. J.K.J. has financial interests in Editas Medicine, Hera Testing Laboratories, Poseida Therapeutics, and Transposagen Biopharmaceuticals. J.K.J.’s interests were reviewed and are managed by Massachusetts General Hospital and Partners HealthCare in accordance with their conflict of interest policies. A patent application has been filed describing the KKH SaCas9 variant.
Ed sum
Molecular evolution broadens the targeting range of the Staphylococcus aureus Cas9 for genome editing in human cells.
References
- 1.Hsu PD, Lander ES, Zhang F. Development and applications of CRISPR-Cas9 for genome engineering. Cell. 2014;157:1262–1278. doi: 10.1016/j.cell.2014.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Sander JD, Joung JK. CRISPR-Cas systems for editing, regulating and targeting genomes. Nat Biotechnol. 2014;32:347–355. doi: 10.1038/nbt.2842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Doudna JA, Charpentier E. Genome editing. The new frontier of genome engineering with CRISPR-Cas9. Science. 2014;346:1258096. doi: 10.1126/science.1258096. [DOI] [PubMed] [Google Scholar]
- 4.Jinek M, et al. A programmable dual-RNA-guided DNA endonuclease in adaptive bacterial immunity. Science. 2012;337:816–821. doi: 10.1126/science.1225829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Deltcheva E, et al. CRISPR RNA maturation by trans-encoded small RNA and host factor RNase III. Nature. 2011;471:602–607. doi: 10.1038/nature09886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Jinek M, et al. RNA-programmed genome editing in human cells. Elife. 2013;2:e00471. doi: 10.7554/eLife.00471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mali P, et al. RNA-guided human genome engineering via Cas9. Science. 2013;339:823–826. doi: 10.1126/science.1232033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cong L, et al. Multiplex genome engineering using CRISPR/Cas systems. Science. 2013;339:819–823. doi: 10.1126/science.1231143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mojica FJ, Diez-Villasenor C, Garcia-Martinez J, Almendros C. Short motif sequences determine the targets of the prokaryotic CRISPR defence system. Microbiology. 2009;155:733–740. doi: 10.1099/mic.0.023960-0. [DOI] [PubMed] [Google Scholar]
- 10.Shah SA, Erdmann S, Mojica FJ, Garrett RA. Protospacer recognition motifs: mixed identities and functional diversity. RNA Biol. 2013;10:891–899. doi: 10.4161/rna.23764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sapranauskas R, et al. The Streptococcus thermophilus CRISPR/Cas system provides immunity in Escherichia coli. Nucleic Acids Res. 2011;39:9275–9282. doi: 10.1093/nar/gkr606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Horvath P, et al. Diversity, activity, and evolution of CRISPR loci in Streptococcus thermophilus. J Bacteriol. 2008;190:1401–1412. doi: 10.1128/JB.01415-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sternberg SH, Redding S, Jinek M, Greene EC, Doudna JA. DNA interrogation by the CRISPR RNA-guided endonuclease Cas9. Nature. 2014;507:62–67. doi: 10.1038/nature13011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jiang W, Bikard D, Cox D, Zhang F, Marraffini LA. RNA-guided editing of bacterial genomes using CRISPR-Cas systems. Nat Biotechnol. 2013;31:233–239. doi: 10.1038/nbt.2508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fonfara I, et al. Phylogeny of Cas9 determines functional exchangeability of dual-RNA and Cas9 among orthologous type II CRISPR-Cas systems. Nucleic Acids Res. 2014;42:2577–2590. doi: 10.1093/nar/gkt1074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Esvelt KM, et al. Orthogonal Cas9 proteins for RNA-guided gene regulation and editing. Nat Methods. 2013;10:1116–1121. doi: 10.1038/nmeth.2681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ran FA, et al. In vivo genome editing using Staphylococcus aureus Cas9. Nature. 2015;520:186–191. doi: 10.1038/nature14299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhang Y, et al. Processing-independent CRISPR RNAs limit natural transformation in Neisseria meningitidis. Mol Cell. 2013;50:488–503. doi: 10.1016/j.molcel.2013.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Vierstra J, et al. Functional footprinting of regulatory DNA. Nat Methods. 2015 doi: 10.1038/nmeth.3554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Canver MC, et al. BCL11A enhancer dissection by Cas9-mediated in situ saturating mutagenesis. Nature. 2015 doi: 10.1038/nature15521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Courtney DG, et al. CRISPR/Cas9 DNA cleavage at SNP-derived PAM enables both in vitro and in vivo KRT12 mutation-specific targeting. Gene Ther. 2015 doi: 10.1038/gt.2015.82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kleinstiver BP, et al. Engineered CRISPR-Cas9 nucleases with altered specificities. Nature. 2015;523:481–485. doi: 10.1038/nature14592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jinek M, et al. Structures of Cas9 Endonucleases Reveal RNA-Mediated Conformational Activation. Science. 2014 doi: 10.1126/science.1247997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Anders C, Niewoehner O, Duerst A, Jinek M. Structural basis of PAM-dependent target DNA recognition by the Cas9 endonuclease. Nature. 2014;513:569–573. doi: 10.1038/nature13579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Nishimasu H, et al. Crystal Structure of Cas9 in Complex with Guide RNA and Target DNA. Cell. 2014 doi: 10.1016/j.cell.2014.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Fu Y, et al. High-frequency off-target mutagenesis induced by CRISPR-Cas nucleases in human cells. Nat Biotechnol. 2013;31:822–826. doi: 10.1038/nbt.2623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tsai SQ, et al. GUIDE-seq enables genome-wide profiling of off-target cleavage by CRISPR-Cas nucleases. Nat Biotechnol. 2015;33:187–197. doi: 10.1038/nbt.3117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lin Y, et al. CRISPR/Cas9 systems have off-target activity with insertions or deletions between target DNA and guide RNA sequences. Nucleic Acids Res. 2014;42:7473–7485. doi: 10.1093/nar/gku402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Fu Y, Sander JD, Reyon D, Cascio VM, Joung JK. Improving CRISPR-Cas nuclease specificity using truncated guide RNAs. Nat Biotechnol. 2014;32:279–284. doi: 10.1038/nbt.2808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Nishimasu H, et al. Crystal Structure of Staphylococcus aureus Cas9. Cell. 2015;162:1113–1126. doi: 10.1016/j.cell.2015.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chen Z, Zhao H. A highly sensitive selection method for directed evolution of homing endonucleases. Nucleic Acids Res. 2005;33:e154. doi: 10.1093/nar/gni148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kleinstiver BP, Fernandes AD, Gloor GB, Edgell DR. A unified genetic, computational and experimental framework identifies functionally relevant residues of the homing endonuclease I-BmoI. Nucleic Acids Res. 2010;38:2411–2427. doi: 10.1093/nar/gkp1223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Reyon D, et al. FLASH assembly of TALENs for high-throughput genome editing. Nat Biotechnol. 2012;30:460–465. doi: 10.1038/nbt.2170. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.