Abstract
Activating mutations of oncogenic RAS genes are frequently detected in human cancers. The studies in genetically engineered mouse models (GEMMs) reveal that Kras-activating mutations predispose mice to early onset tumors in the lung, pancreas, and gastrointestinal tract. Nevertheless, most of these tumors do not have metastatic phenotypes. Metastasis occurs when tumors acquire additional genetic changes in other cancer driver genes. Studies on clinical specimens also demonstrated that KRAS mutations are present in premalignant tissues and that most of KRAS mutant human cancers have co-mutations in other cancer driver genes, including TP53, STK11, CDKN2A, and KMT2C in lung cancer; APC, TP53, and PIK3CA in colon cancer; and TP53, CDKN2A, SMAD4, and MED12 in pancreatic cancer. Extensive efforts have been devoted to develop therapeutic agents that target enzymes involved in RAS posttranslational modifications, that inhibit downstream effectors of RAS signaling pathways, and that kill RAS mutant cancer cells through synthetic lethality. Recent clinical studies have revealed that sorafenib, a pan-RAF and VEGFR inhibitor, has impressive benefits for KRAS mutant lung cancer patients. Combination therapy of MEK inhibitors with either docetaxel, AKT inhibitors, or PI3K inhibitors also led to improved clinical responses in some KRAS mutant cancer patients. This review discusses knowledge gained from GEMMs, human cancer cells, and patient-related studies on RAS-mediated tumorigenesis and anti-RAS therapy. Emerging evidence demonstrates that RAS mutant cancers are heterogeneous because of the presence of different mutant alleles and/or co-mutations in other cancer driver genes. Effective subclassifications of RAS mutant cancers may be necessary to improve patients' outcomes through personalized precision medicine.
Keywords: RAS genes, neoplasms, adenocarcinoma, animal models, clinical trial, antineoplastic agents
Introduction
RAS proteins are small G proteins that cycle between active GTP-bound and inactive GDP-bound forms and function as molecular switches for signal transductions initiated in the cell membrane [1,2]. Synthesized in cytosol, RAS proteins are transferred to the inner leaflet of the plasma membrane, where they interact with diverse membrane receptors and execute signal transduction in a variety of signaling pathways that govern cell growth, proliferation, differentiation, and death. Activation of upstream growth factor receptors, such as epidermal growth factor receptor (EGFR), insulin-like growth factor 1 receptor, and platelet-derived growth factor receptor (PDGFR), results in the assembly of adaptor proteins Grb2 and the Son of Sevenless (SOS) complex. SOS is one of the guanine nucleotide exchange factors (GEFs) that activate RAS by promoting binding of RAS with GTP via catalysis of the release of GDP from RAS [3,4]. Intrinsic GTPase activity enhanced by GTPase-activating proteins (GAPs) [5] converts GTP to GDP, leading to inactive GDP-bound RAS (Fig. 1). RAS mutations that diminish GTPase activity or decrease GDP-binding capacity render RAS in constitutively active GTP-bound status. In the absence of a RAS mutation, increased RAS activity in human cancer cells frequently results from RAS gene amplifications [6,7] and overexpression [8], an increase in activity of upstream signals from tyrosine kinase growth factor receptors such as HER2 and EGFR [4,9], or/and altered expression of microRNAs such as let-7 [10,11].
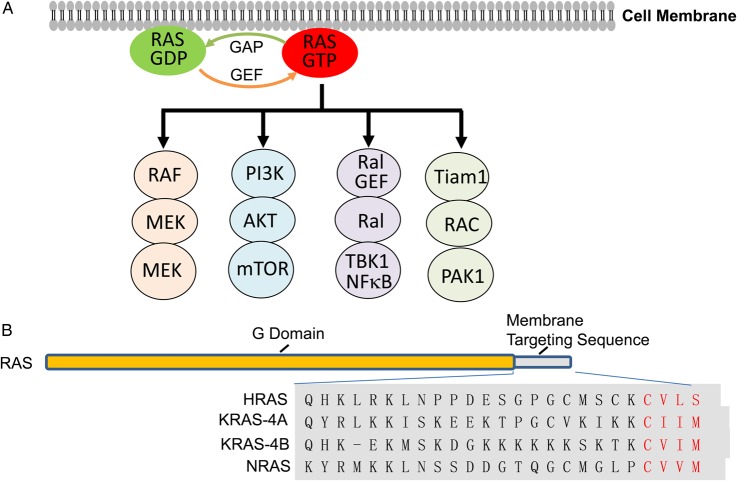
Figure 1.
Diagrams of RAS proteins and RAS signaling pathways (A) Major RAS signaling pathways. RAS GEF activated by upstream growth factor receptors promotes binding of RAS with GTP via catalysis of the release of GDP from RAS, leading to the activation of downstream pathways (see details in other review articles [18,19]). Intrinsic GTPase activity enhanced by GAPs converts GTP to GDP, leading to inactive GDP-bound RAS. RAS mutations that cause the loss of GTPase activity render RAS in a persistent GTP-bound status. (B) Structures of RAS proteins. RAS proteins consist of G domain (amino acids 1–164) that has 93%–99% conserved sequences among RAS proteins and functions as GTPase, and membrane targeting sequences (amino acids 165–188/189) that is highly variable. The C-terminal CAAX motif required for farnesylation is marked red.
RAS activation leads to stimulation of a wide range of downstream signaling pathways, most notably the RAF/mitogen-activated protein kinase (MAPK) kinase (MEK)/ERK [12,13], phosphoinositide 3-kinase (PI3K)/AKT/mammalian target of rapamycin (mTOR), RalGEF/RAL [14,15], and Tiam1/RAC [16,17] (Fig. 1) (see details in other review articles [18,19]). GTP-RAS binds directly to and activates RAF [12,13,20], the catalytic subunit of PI3K p110 [21,22], Ral guanine nucleotide exchange factors (RalGEF) [23,24], and RAC GEFs such as Tiam1 and Vav [16,25]. The signaling cascades initiated by these RAS-interacting proteins form networks through crosstalk and feedback interactions, which have been shown to play critical roles in the initiation and progression of malignancies [14,26–28]. Because activating mutations in RAS genes are among the most frequently observed oncogenic mutations in human cancers, RAS signaling and anti-RAS therapeutic agents have been intensively investigated. However, RAS proteins are regarded as non-druggable with small molecule inhibitors because of their high affinity for GTP and their simple protein structures. Thus, extensive efforts have been made to develop therapeutic agents that modulate posttranscriptional modification and/or plasma membrane localization of RAS proteins [29,30], that intervene in downstream signal transductions, and that induce synthetic lethality in RAS mutant cancer cells [31]. Recently, small molecules have been reported to bind irreversibly to the mutant KRAS (G12C) protein [32], or to interfere with RAS/SOS [33,34] or RAS-effector protein interactions [35]. Nevertheless, effective anti-RAS treatment is not yet available clinically. This review discusses knowledge gained from genetically engineered mouse models (GEMMs), human cancer cell lines, clinical studies about RAS-mediated signaling in tumorigenesis, and the development of anti-RAS therapy. It is likely that RAS mutant cancers are heterogeneous and different therapeutic strategies may be required for different subclasses of RAS mutant cancers.
GEMMs with RAS Mutations
Mammalian cells have three RAS genes (HRAS, KRAS, and NRAS) that encode four highly homologous RAS proteins, because the KRAS gene encodes two splicing isoforms: a major KRAS-4B and a minor KRAS-4A. These proteins have highly identical sequences in the first 164 amino residues containing the G domain for GTP binding and hydrolysis. The remaining 24/25 C-terminal residues are highly variable among isoforms and critical for membrane localization (Fig. 1). KRAS-4B, HRAS, and NRAS are ubiquitously expressed, whereas KRAS-4A is expressed mainly in renal, hepatic, and gastrointestinal tissues [36,37]. Evidence has shown that these highly conserved RAS isoforms carry out similar but indispensable functions that govern cell growth, differentiation, proliferation, apoptosis, tumorigenesis, and tumor progression [38]. Gene knockout studies reveal that Hras or Nras knockout mice, and even the Hras and Nras double knockout mice, are viable and show no obvious abnormalities [39,40]. In contrast, knockout of the Kras gene is embryonically lethal [41]. Although some of the differences may be derived from differences in their expression patterns [42,43], it has been reported that different RAS isoforms have different biological functions and different effects on tumor progression [42–46].
Expression of mutant Kras12D alone in murine embryonic fibroblasts (MEFs) induced enhanced proliferation and partial transformation accompanied with an elevation of CDK2 and CDK4. However, elevated ERK and AKT phosphorylation is not observed in MEFs expressing endogenous Kras12D [47]. Moreover, expression of endogenous oncogenic Kras12D during murine embryos results in widespread morphological aberrations and early embryonic lethality [47]. Expression of endogenous Kras12V also frequently results in embryonic lethality, although some mice may reach adulthood and develop lung adenomas and adenocarcinomas [48]. Conditional expression of these mutant Kras alleles in the lung, pancreas, and gastrointestinal tract induces preneoplastic epithelial hyperplasias, adenomas, pancreatic intraepithelial neoplasia (PanIN), and adenocarcinomas [47–51]. The majority of these Kras-driven tumors do not have an invasive or metastatic phenotype, although progression to invasive and metastatic cancers is detected at low frequencies [51,52]. However, the progression to highly invasive and metastatic cancers is dramatically enhanced by the presence of mutations in other cancer driver genes, including defects in Tp53 [53–55], Stk11 [56–58], Ink4a/Arf [53,59], Smad4 [60], Pten [61], Tgfbr2 [62], and Runx3 [63]; and activation of Wnt/beta-catenin signaling [64]. For example, Kras-activating mutations in lung epithelial cells predispose mice to early onset of lung adenocarcinoma [49,50,65]. Nevertheless, such tumors do not have an invasive or metastatic phenotype. Metastasis occurs when additional genetic changes, such as Tp53 mutations or Stk11 deletion, are introduced [54,57].
Efforts were made to delineate the downstream effectors and pathways that are required for RAS-mediated tumorigenesis in GEMMs. Several genes or pathways have been identified as participants in Kras-driven lung and pancreatic cancers, because their ablation or deletion prevents or reduces mutant Kras-induced tumors. Among them are PI3K/PDK1 [26,66], PI3K/RAC1 [67–70], RAF/MEK/ERK [71,72], TBK1/IKK/NFκB [73–75], IL-6/STAT3 [76,77], YAP [78], Foxm1/NFκB [79], anabolic glucose metabolism [80], GM-CSF-mediated recruitment of myeloid cells [81], ERK/RHOA/focal adhesion kinase (FAK) network [82], fibroblast activating protein (FAP) [83], and Myc [84].
For example, PI3K signaling was found to be necessary in Kras-induced malignant transformations in pancreatic cancer [85] and lung cancer [66,86]. Pancreas-specific inactivation of Pik3ca, but not Pik3cb, prevented the occurrence of all types of malignant lesions induced by expression of mutant Kras, including PanIN and acinar-to-ductal metaplasia [69,85]. Pik3ca ablation and chronic inhibition led to up-regulation of AKT signaling, possibly resulting from compensatory activity from other PI3K isoforms. In contrast, Pik3ca ablation significantly diminished both Rac1 activity and expression in Kras mutant pancreatic cells, accompanied by significant inhibition of Kras-activated Rac1 guanine exchange factors Tiam1 and Vav1 [69]. Pancreas-specific ablation of Rac1 has the same phenotype as Pik3ca ablation in Kras mutant mice [68,69], indicating the PI3K/RAC axis plays an important role in Kras-driven pancreatic tumor development. Similarly, Stat3 phosphorylation is found to occur at multiple stages of Kras12D-driven pancreatic tumorigenesis but not in normal pancreatic tissue [76,77]. Disruption of the IL-6 gene and conditional inactivation of Stat3 in the pancreas reduced PanIN and pancreatic ductal adenocarcinoma (PDAC) formation in mutant Kras12D mice [76,77], indicating that Stat3 activity is required for the development of the early premalignant pancreatic lesions, acinar-to-ductal metaplasia, and PanIN, and for the progression from PanIN to invasive PDAC [77]. A recent study showed that TBK1/IKK regulates autocrine cytokines CCL5 and IL-6, which contributes to Kras-driven tumorigenesis [74].
Treatment Response in GEMMs with RAS-driven Tumors
In a doxycycline-inducible Hras12V-driven mouse melanoma model with null Ink4a, withdrawal of doxycycline resulted in Hras12V down-regulation, marked apoptosis in the tumor cells and host-derived endothelial cells, and histological regression of primary and explanted tumors [87], demonstrating that the mutant Hras12V is required in both the initiation and maintenance of solid tumors. The same results were observed in lung tumors [88] and pancreatic tumors [89,90] induced by doxycycline-regulated Kras12D. Withdrawal of doxycycline in transgenic mice expressing Kras4b12D in type II pneumocytes, with or without deficiencies in either the Tp53 gene or the Ink4a/Arf locus, led to rapid regression of Kras4b12D-mediated tumors [88]. Although the p53 gene or Ink4a/Arf deficiencies dramatically accelerate tumor initiation and progression, removal of doxycycline caused a rapid regression of tumor burdens, implying that continued production of mutant Kras is necessary to maintain the viability of tumor cells, regardless of the presence of other cancer drivers.
In Kras-driven mouse pancreatic cancer models, treatment with MEK inhibitors (AZD6244, GDC-0973) or PI3K inhibitors (BEZ235, GDC-0941, BKM120) (Fig. 2) alone induced only partial tumor growth suppression, which did not significantly prolong overall survival (OS). Treatment with a MEK1 inhibitor resulted in cytostatic effects accompanied by sustained activation of the PI3K/AKT/mTOR pathway and receptor tyrosine kinases EGFR, HER2, PDGFR, and AXL [90]. Similar results were observed in Kras12D-driven lung cancer in mice [91]. Treatment with a dual pan-PI3K and mTOR inhibitor BEZ235 is able to substantially suppress the growth of Pik3ca1047R-induced lung tumors but not Kras12D-driven lung tumors [91], suggesting that PI3K may be required for Kras-induced tumorigenesis but less crucial for tumor maintenance. Nevertheless, simultaneously targeting both MEK and PI3K pathways led to marked synergy in shrinking these Kras mutant cancers, resulting in a significant survival advantage when compared with controls [90,92,93]. Partial inhibition of Kras12D-driven lung tumorigenesis was also observed in treatment with the MEK inhibitor selumetinib (AZD6244) and the TBK1/IKK/JAK inhibitor CYT387 [74]. The synergistic combination's effects on Kras12D-driven lung tumors were also reported for combination therapy of farnesyl and geranylgeranyl diphosphate synthases inhibitor lipophilic bisphosphonates plus mTOR inhibitor rapamycin [94]; or combination of MEK inhibitor selumetinib plus the BCL2/BCL-XL inhibitor ABT-263 (navitoclax) [95].
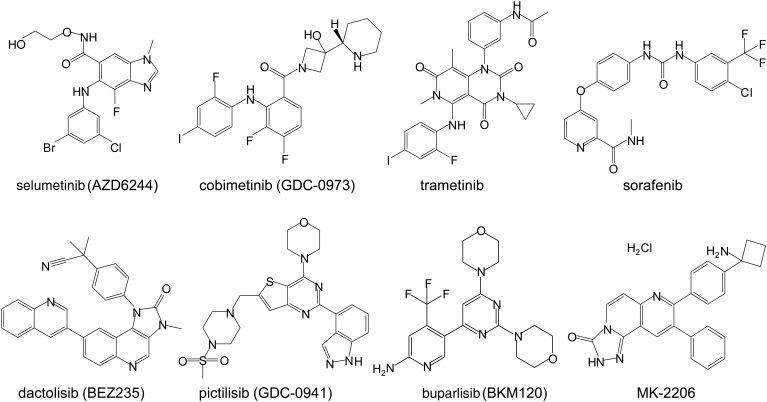
Figure 2.
Chemical structures of small molecule inhibitors targeting RAS downstream effectors MEK inhibitors: selumetinib, cobimetinib, and trametinib; RAF inhibitor: sorafenib; PI3K inhibitors: dactolisib, pictilisib, and buparlisib; and AKT inhibitor: MK-2206.
A study comparing treatment responses of Kras12D-driven lung cancer with concomitant loss of either Tp53 or Stk11 revealed that loss of either gene markedly impaired the response of Kras mutant cancers to docetaxel monotherapy [96]. Nevertheless, Kras/Tp53 mutant tumors, which had increased MEK/ERK signaling, were sensitive to the combination therapy of docetaxel plus selumetinib. In contrast, Kras/Stk11 mutant tumors, which had activation of AKT and SRC, were resistant to this combination therapy [96]. Similarly, Kras and Kras/Tp53 mutant lung tumors were found to be susceptible to Myc inhibition with dominant-negative Myc [84] or with the bromodomain and extra-terminal bromodomain inhibitor JQ1 [97], whereas Kras/Stk11 mutant mouse cancer cells and human lung cancer cells were resistant to the JQ1 treatment [97]. Knockdown of STK11 in human KRAS mutant lung cancer cells sensitive to JQ1 caused resistance to this inhibitor, indicating the causal relationship between STK11 deficiency and JQ1 resistance. In contrast, Kras/Stk11 mutant mouse tumors responded to the treatment with the drug phenformin which affects metabolisms [98], and were highly sensitive to the inhibition of deoxythymidylate kinase [99], suggesting that the presence of co-mutations of another cancer driver gene may have a dramatic impact on responses to anticancer drugs.
Although both the ablation of oncogenic Kras expression and the combination of MEK and PI3K/mTOR inhibitors can induce complete regression of Kras-driven tumors, resistance to Kras ablation or to the combination therapy has been observed in relapsed tumors [100,101]. Some of these relapsed tumors had increased activity in mitochondria, autophagy, and lysosomes functions, had reduced glycolytic activity, and were sensitive to oxidative phosphorylation inhibitors such as oligomycin [100]. Yap1 amplification is another mechanism of resistance identified in the relapsed tumors after Kras12D ablation, indicating that Yap1 activation may lead to a bypass of Kras dependency [101]. In a systemic screening with 15,294 open reading frames, YAP1's overexpression was identified in surviving cells after inducible shRNA-mediated KRAS knockdown in the human KRAS-dependent colon cancer cell line HCT116, and in acquired resistance to Kras suppression in a Kras-driven murine lung cancer model [102]. In addition, constitutive AKT activation also caused resistance to Kras ablation [90], indicating multiple mechanisms underlying resistance to anti-RAS therapy.
Treatment Response in RAS Mutant Human Cancer Cells
Studies in human cancer cell lines demonstrated that KRAS gene expression is required for survival of some KRAS mutant cancer cells. Disrupting KRAS by homologous recombination in KRAS mutant colon cancer cell lines DLD-1 and HCT116 resulted in inhibition of MYC expression and suppression of tumor cell growth both in vitro and in vivo in nude mice [103], implicating the expression of mutant KRAS is crucial for growth of these tumor cells. Knockdown of KRAS with shRNAs in a panel of KRAS mutant lung and pancreatic cancer cell lines has revealed that KRAS mutant cancer cell lines can be classified into KRAS-dependent and -independent groups, based on the requirement of KRAS expression for cell survivals [104]. Many of KRAS-dependent cells exhibit a classic epithelial morphology and gene expression signature, expressing prominent E-cadherin, whereas KRAS-independent cell lines have epithelial mesenchymal transition phenotype, expressing little or no E-cadherin, but expressing the mesenchymal marker vimentin. SYK, RON kinases, and integrin b6 were high in KRAS-dependent cell lines and were required for epithelial differentiation and cell survival in KRAS-dependent cells [104].
Synthetic lethality screening with siRNA libraries in cell lines with or without mutant RAS genes has revealed a number of genes whose knockdown may selectively induce lethality in cells with mutant RAS genes. Reported synthetic lethal partners for oncogenic RAS include PLK1, a serine/threonine protein kinase that regulates cell mitosis [105]; the transcription factor WT1 [106]; TBK1, an IκB kinase that regulates the stability of IκB [107]; SYK [104]; and CDK4 [108]. The synthetic lethality interactions of these targets with oncogenic RAS suggest that the inhibitors targeting to these molecules might selectively kill RAS mutant cancer cells. Indeed, small molecule inhibitors that disrupt mitosis, including paclitaxel and the PLK1 inhibitor BI-2536 [109], were found to be synthetic lethal in RAS mutant cells [105]. Synthetic lethality screening is also employed to identify combination therapy for investigational drugs targeting RAS downstream pathways. Using a pooled 5000 shRNA library targeting 1200 ‘druggable’ genes, Bcl-XL was identified as a synthetic lethal partner for selumetinib in KRAS mutant colon cancer cell lines HCT116 and SW620 [95]. A combination of selumetinib and ABT-263 induced substantial apoptosis in ∼50% of KRAS mutant cancer cell lines derived from colon, lung, and pancreatic cancers, particularly in those with epithelial signature [95]. The synergistic effects of the combination are also demonstrated in vivo in xenograft tumors and in Kras12D-driven mice lung tumors [95]. Of note, a study using shRNA library screening for genes that may have synthetic lethal interactions with the oncogenic KRAS gene in the colon cancer cell line DLD-1 has led to the identification of ∼370–1600 mutant KRAS synthetic lethal genes, depending on the stringency of statistical analyses [105], demonstrating the diversity of biological processes or pathways regulated by KRAS and the possible challenges that may be encountered by employing the synthetic lethality approach to identify therapeutic targets.
The synthetic lethality approach has also been investigated in the identification of anticancer agents for RAS mutant cancer cells by screening chemical libraries [110–112]. Several of such anticancer agents have been reported, including triphenyl tetrazolium and a sulfinyl cytidine derivative that showed ∼6-fold selectivity for cell lines containing mutant KRAS [111]; erastin which exhibited lethal selectivity in human tumor cells harboring mutations in the HRAS, KRAS, or BRAF oncogene by acting on mitochondrial voltage-dependent anion channels and inducing oxidative cell death [112,113]; lanperisone [114]; and oncrasin-1 [115]. Erastin [113], lanperisone [114], and oncrasin compounds [116,117] all induced cell killing effects in RAS mutant tumor cells by inducing oxidative stress, although through different underlying mechanisms.
Through chemical library screening on cells with or without a mutant KRAS gene [115] and lead compound optimization [118–120], we recently developed a novel compound designated oncrasin-72 (NSC743380) that is highly active [median growth inhibitory concentration (IC50) between 10 nM and 1 μM] in vitro in 30 of 102 cancer cell lines tested [118,121], including many KRAS mutant cancer cell lines [115,119,121]. Mechanistic characterization revealed that NSC743380 and its analogs induced apoptosis in sensitive cancer cells [115,118,119], inhibited phosphorylation of the C-terminal domain of RNA Pol II [120,122], induced sustained JNK activation by inhibiting its dephosphorylation [119], induced reactive oxygen species (ROS) accumulation [117], inhibited STAT3 phosphorylation, and suppressed cyclin D1 expression [118], suggesting that NSC743380 modulates multiple cancer-related targets. Blocking NSC743380-induced ROS generation with antioxidants dramatically abolished its apoptosis-inducing ability but had minimal effect on its inhibition of STAT3, suggesting that STAT3 inhibition is not caused by ROS production. In contrast, knockdown of STAT3 by siRNA induced ROS generation and suppressed tumor cell growth [121], suggesting that STAT3 inhibition might be upstream of ROS induction. Interestingly, the activation of RAS signaling pathways has been reported to up-regulate the overall cellular antioxidant capacity [123]. The interactions among RAS, STAT3, and redox pathways have been discussed in another review [124]. In vivo studies have shown that the intravenous or intraperitoneal administration of NSC743380 caused complete tumor regression or significant growth suppression in several xenograft tumor models [118,121], indicating that NSC743380 has promising in vivo activity. More recently, we improved NSC743380's stability and safety through the synthesis and evaluation of its prodrugs. Oncrasin-266 spontaneously releases NSC743380 in physiological solutions in vitro and in vivo, has improved stability and pharmacokinetics, and is better tolerated in mice at a higher dose level (150–300 mg/kg, i.p.) than NSC743380 [125], suggesting that the prodrug is a favorable candidate for further development.
Nevertheless, the correlations between NSC743380's anticancer activity and KRAS mutations in the NCI-60 cell lines and in the 50 lung cancer cell lines tested were not significant [118,121], because some KRAS mutant cancer cell lines were resistant while some KRAS wild-type cancer cell lines were sensitive to NSC743380. Our recent study revealed that the expression of a sulfotransferase (SULT), SULT1A1, in cancer cells is required for NSC743380's anticancer activity and that the expression of SULT1A1 is capable of predicting the responses to NSC743380 [126]. SULT1A1 is a biotransformation enzyme that bioactivates several pro-carcinogens [127–133] and some anticancer drugs, such as tamoxifen [134]. Identification of SULT1A1 as a predictive biomarker for NSC743380 sheds light on mechanisms of selectivity and the possible toxicity of this compound [135]. The process of identifying this predictive biomarker underscores the importance of activity characterization in a large set of molecularly annotated cancer cell lines and rigorous validation of causal relationships between the sensitivity and the biomarker.
Studies on Clinical Specimens and Clinical Trials
KRAS mutations are frequently found in human adenocarcinomas of the pancreas (70%–90%) [136–138], colon (50%) [139,140], and lung (35%) [141–143]. Based on human cancer gene mutation datasets retrieved from www.cBioPortal.org, KRAS mutations are also frequently detected in multiple myeloma (22%) and cancers of the ovary (15%), uterine (18%), and stomach (16%). NRAS mutations are frequently detected in melanoma (30%), multiple myeloma (18%), colorectal cancer (10%), and thyroid cancer (8%). In contrast, HRAS mutations occur in low frequencies (5% or less) in cancers of the bladder, head/neck, and uterine. The majority of genetic alterations in these oncogenic RAS genes are missense mutations in codons 12, 13, and 61. The most common mutant alleles for KRAS are 12D, 12C, 12V, 12R, and 13D [144,145]. Evidence has shown that different KRAS mutant alleles may have different clinical impacts on the prognosis of lung [146,147], colon [148–150], and pancreatic [151] cancers, although the alleles associated with poor clinical outcomes are not consistent in these studies. Molecular characterization of clinical specimens from patients who participated in prospective phase II biomarker-integrated approaches of targeted therapy for lung cancer elimination revealed that the expressions of cell cycle regulators PLK1, CCNB1, and CCNE1 were lower in KRAS12C and KRAS12V mutant tumors, but were higher in the remaining KRAS mutant tumors, when compared with KRAS wild-type cancer [146]. Analysis of NSCLC cell lines revealed that cancer cells with mutant KRAS12D had activated PI3K and MEK signaling, whereas those with mutant KRAS12C or KRAS12V had activated Ral signaling and decreased growth factor-dependent AKT activation. Moreover, ectopic expression of KRAS12D or KRAS12C in TP53 knockdown human bronchial epithelial cells (HBECsiP53) had different effects on AKT activation and RalA/B expression [146], suggesting that different mutant alleles may have different preferences in activating downstream pathways.
Consistent with the roles of Kras mutations in tumor initiation in mouse models, analysis of clinical specimens revealed that KRAS mutations are frequently detected in human hyperplastic/metaplastic pancreatic acinar-ductal cells [152–154] and colorectal adenomas [155–157]. KRAS alterations may represent an early event in pancreatic ductal tumorigenesis, whereas TP53 gene changes may represent an event required for the malignancy progression of ductal tumors from lower to higher grades [158]. Whole-genome analysis on pancreatic cancer revealed that, although KRAS is mutated in ∼95% of PDACs, PDACs can be classified into four classes based on patterns of chromosomal rearrangements: stable, locally rearranged, scattered, and unstable [138]. These results demonstrated that mutations in the cooperative pathways or cancer drivers may further differentiate KRAS mutant cancers into subgroups.
The data retrieved from whole-genome sequencing analyses for lung, pancreatic, and colorectal cancers at The Cancer Genome Atlas (TCGA) databases (http://www.cbioportal.org) revealed that the majority of KRAS mutant cancers have co-mutations in other cancer driver genes (Fig. 3). The common co-mutations detected in lung cancer are TP53, SKT11, CDKN2A, and KMT2C; in colon cancer, APC, TP53, and PIK3CA; and in pancreatic cancer, TP53, CDKN2A, SMAD4, and MED12. A recent study using immunohistochemical analysis showed that LKB1 (encoded by STK11) is lost in 30% of KRAS mutant lung adenocarcinoma. KRAS mutant NSCLC patients with concurrent LKB1 loss had a high number of metastatic sites at the time of diagnosis, and had a high incidence of extra-thoracic metastases [159]. Interestingly, concurrent mutations in KRAS and STK11 in human cancer cells resulted in susceptibility to the depletion of the coatomer complex I subunits [160], which are required for lysosomal maturation and CDC42-mediated transformation. More recently, an integrative analysis of genomic, transcriptomic, and proteomic data from early stage and chemo-refractory lung adenocarcinoma demonstrated that KRAS mutant lung cancer can be classified into three subgroups by co-occurring genetic alterations in STK11, TP53, and CDKN2A/B [161]. These three groups have distinct clinical outcomes and treatment responses. KRAS/STK11 cells showed increased vulnerability to HSP90 inhibitors. These results strongly indicate that concurrent mutations in KRAS and STK11 genes may represent a subgroup of KRAS mutant tumors that differ from other KRAS mutant cancers in treatment responses.
Figure 3.
Molecular heterogeneity in KRAS mutant adenocarcinomas Status of KRAS mutations and co-mutations in other cancer driver genes in 230 lung adenocarcinomas (A); 90 pancreatic adenocarcinoma (B); and 220 colorectal adenocarcinoma (C) retrieved from The Cancer Genome Atlas (TCGA) databases at the website http://www.cbioportal.org. Each vertical line represents a tumor. The graph shows mutations in the top seven cancer driver genes in lung, pancreatic, and colorectal adenocarcinomas. Red, amplification; blue, homozygous deletion; green, missense mutation; black, truncating mutation; blown, inframe mutation. Note most of KRAS mutant cancers have co-mutations in other cancer driver genes.
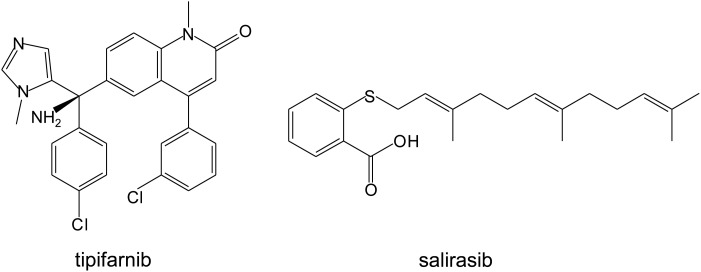
Because posttranslational modifications of RAS proteins are required for them to be translocated to plasma membrane to execute their biologic functions, efforts have been made to target enzymes involved in these posttranslational modifications, including farnesylation at the cysteine residue of the C-terminal CAAX motif, removal of the AAX peptide, and methylation of farnesyl-cysteine at the C-terminal [162]. For HRAS and NRAS, palmitoylation on cysteine residues near the C-terminal is also required for RAS re-localization to the membrane. For KRAS-4B, a polybasic domain located at the C-terminal serves as the second signal for membrane localization [163,164] (Fig. 1). Because farnesylation of RAS is critical to its biologic function, farnesyltransferase inhibitors (FTIs) have been intensively investigated in clinical studies. Several phase II and phase III clinical trials showed that the FTIs tipifarnib and salirasib (Fig. 4), either alone or in combination therapy, did not have significant activity in lung, pancreatic, and colorectal cancers [165–168].
Figure 4.
Chemical structures of therapeutic agents that modulate RAS subcellular localizations
Targeting the RAS downstream pathways, particularly the RAF/MEK/ERK and PI3K/AKT/mTOR pathways [18,169], has also been investigated for treatment of RAS mutant cancers in clinics. A clinical trial with biomarker-integrated targeted therapy for lung cancer has revealed that sorafenib, a pan-RAF and VEGFR inhibitor [170], has impressive benefits for KRAS mutant patients [171]. However, selective inhibition of BRAF with a dominant-negative construct [172] in mice or with BRAF-selective inhibitors such as vemurafenib in patients [173,174] promoted the development and/or progression of RAS mutant cancers, possibly because of the activation of other RAF isoforms, such as RAF1. MEK inhibitors selumetinib and trametinib have been investigated for treatment of KRAS mutant tumors in a few clinical trials. A biomarker-derived multi-arm phase II trial revealed that selumetinib monotherapy failed to achieve its primary end point, with a response rate of 11% [175]. Nevertheless, combination therapy of MEK inhibitors with either docetaxel [176], AKT inhibitors [177], or PI3K inhibitors [178] led to improved clinical responses. In a randomized multicenter phase II study, docetaxel plus selumetinib treatment for patients with advanced KRAS mutant NSCLC resulted in an objective response rate of 37%, with a median OS of 94 months and median progression-free survival (PFS) of 53 months, whereas the patients who received docetaxel alone had an OS and PFS of 52 and 21 months, respectively [176]. Combination of selumetinib with AKT inhibitor MK-2206 resulted in an objective response of 3/13 (23%) of KRAS mutant NSCLC patients [177], while combination of trametinib with PI3K inhibitor buparlisib led to objective responses in KRAS mutant ovarian cancer and NSCLC [178]. However, a combination of trametinib with gemcitabine for treatment of metastatic pancreatic cancer [179], or selumetinib with irinotecan for treatment of KRAS-mutated colorectal cancer [180], did not improve PFS or overall response rate when compared with gemcitabine or irinotecan plus placebo. Together, these studies demonstrate that targeting multiple RAS signaling pathways may provide benefits to a subgroup of RAS mutant cancer patients, and that effective strategies to stratify patients for precision therapy will be required to improve efficacies.
Conclusions and Perspectives
Studies in GEMMs have demonstrated that Ras gene mutations are sufficient to initiate tumorigenesis, although the presence of additional genetic alterations in other cancer driver genes is often required for progression to invasive and metastatic cancers [51,52]. These studies have led to the identification of several genes whose abnormalities cooperatively promote the initiation and progression of Ras mutant tumors. Similarly, several genes whose ablations or inhibition prevent and/or reduce Ras-mediated tumorigenesis were reported, suggesting that they may serve as potential therapeutic targets for treating RAS mutant cancers. Studies in human cancers demonstrated that RAS gene mutations are frequently detected in premalignant tissue specimens and that most KRAS mutant cancers have co-mutations of other cancer driver genes. Moreover, emerging evidence has demonstrated that KRAS mutant cancers are heterogeneous in terms of signaling/metabolic aberrations and responses to treatments. The variations in co-mutations, mutant alleles of RAS genes, and origins of tumor cells can all cause such heterogeneity. Thus, effective classification of RAS mutant cancers will be required to improve anti-RAS therapy through personalized precision medicine. Because crosstalk and feedback activations are commonly observed in RAS-mediated signaling pathways, and because most solid tumors carry multiple concomitantly activated oncogenes or inactivated tumor suppressor genes [181], simultaneously targeting multiple cancer-associated pathways is likely required for effective anti-RAS therapy. Our experience with selective cell killing of NSC743380 in SULT1A1+ cells [126] indicates that even though some passenger mutations in tumor cells may not contribute to tumorigenesis, they may have an impact on treatment response because of altered drug metabolism. Therefore, they may be used as biomarkers to identify responders.
The initiation and progression of KRAS mutant tumors are also drastically affected by tumor microenvironments. Several tumor microenvironment factors have been identified as crucial in Kras-mediated tumor initiation and progressions in GEMMs. Among them are GM-CSF-mediated recruitment of myeloid cells [81], the IL-6/STAT3 signaling pathway [76,77], the ERK/RHOA/FAK network [82], and FAP [83]—a member of the serine protease family selectively expressed in stromal fibroblasts of epithelial cancers [182]. Genetic deletion and pharmacologic inhibition of FAP resulted in inhibition of Kras12D-driven lung tumors in mice, possibly through indirect inhibition of tumor cell growth by modulating extracellular matrix/integrin-mediated singling [83]. These results demonstrate the feasibility of modulating the tumor microenvironment in the treatment of RAS mutant cancer, and highlight the necessity of incorporating factors of tumor microenvironment into the design of future anti-RAS therapies.
Funding
This work was supported in part by the National Institutes of Health grant R01CA190628; Specialized Program of Research Excellence (SPORE) grant CA070907 developmental award; The University of Texas MD Anderson Cancer Center support grant CA-016672; and by endowed funds to The University of Texas MD Anderson Cancer Center, including the Lung Cancer Moon Shots Program, Stading Lung Cancer Research Fund, the Homer Flower Research Fund, and the Goldman Sachs Philanthropy Fund.
References
- 1.Bar-Sagi D, Hall A. Ras and Rho GTPases: a family reunion. Cell 2000, 103: 227–238. [DOI] [PubMed] [Google Scholar]
- 2.Colicelli J. Human RAS superfamily proteins and related GTPases. Sci STKE 2004, 2004: RE13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Egan SE, Giddings BW, Brooks MW, Buday L, Sizeland AM, Weinberg RA. Association of Sos Ras exchange protein with Grb2 is implicated in tyrosine kinase signal transduction and transformation. Nature 1993, 363: 45–51. [DOI] [PubMed] [Google Scholar]
- 4.Buday L, Downward J. Epidermal growth factor regulates p21ras through the formation of a complex of receptor, Grb2 adapter protein, and Sos nucleotide exchange factor. Cell 1993, 73: 611–620. [DOI] [PubMed] [Google Scholar]
- 5.Bernards A, Settleman J. GAP control: regulating the regulators of small GTPases. Trends Cell Biol 2004, 14: 377–385. [DOI] [PubMed] [Google Scholar]
- 6.Hoa M, Davis SL, Ames SJ, Spanjaard RA. Amplification of wild-type K-ras promotes growth of head and neck squamous cell carcinoma. Cancer Res 2002, 62: 7154–7156. [PubMed] [Google Scholar]
- 7.Filmus JE, Buick RN. Stability of c-K-ras amplification during progression in a patient with adenocarcinoma of the ovary. Cancer Res 1985, 45: 4468–4472. [PubMed] [Google Scholar]
- 8.Coleman WB, Throneburg DB, Grisham JW, Smith GJ. Overexpression of c-K-ras, c-N-ras and transforming growth factor beta co-segregate with tumorigenicity in morphologically transformed C3H 10T1/2 cell lines. Carcinogenesis 1994, 15: 1005–1012. [DOI] [PubMed] [Google Scholar]
- 9.Ehrhardt A, David MD, Ehrhardt GR, Schrader JW. Distinct mechanisms determine the patterns of differential activation of H-Ras, N-Ras, K-Ras 4B, and M-Ras by receptors for growth factors or antigen. Mol Cell Biol 2004, 24: 6311–6323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Johnson SM, Grosshans H, Shingara J, Byrom M, Jarvis R, Cheng A, Labourier E et al. RAS is regulated by the let-7 microRNA family. Cell 2005, 120: 635–647. [DOI] [PubMed] [Google Scholar]
- 11.Kumar MS, Erkeland SJ, Pester RE, Chen CY, Ebert MS, Sharp PA, Jacks T. Suppression of non-small cell lung tumor development by the let-7 microRNA family. Proc Natl Acad Sci USA 2008, 105: 3903–3908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Moodie SA, Willumsen BM, Weber MJ, Wolfman A. Complexes of Ras.GTP with Raf-1 and mitogen-activated protein kinase kinase. Science 1993, 260: 1658–1661. [DOI] [PubMed] [Google Scholar]
- 13.Vojtek AB, Hollenberg SM, Cooper JA. Mammalian Ras interacts directly with the serine/threonine kinase Raf. Cell 1993, 74: 205–214. [DOI] [PubMed] [Google Scholar]
- 14.Lim KH. Divergent roles for RalA and RalB in malignant growth of human pancreatic carcinoma cells. Curr Biol 2006, 16: 2385–2394. [DOI] [PubMed] [Google Scholar]
- 15.Martin TD, Chen XW, Kaplan RE, Saltiel AR, Walker CL, Reiner DJ, Der CJ. Ral and Rheb GTPase activating proteins integrate mTOR and GTPase signaling in aging, autophagy, and tumor cell invasion. Mol Cell 2014, 53: 209–220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lambert JM, Lambert QT, Reuther GW, Malliri A, Siderovski DP, Sondek J, Collard JG et al. Tiam1 mediates Ras activation of Rac by a PI(3)K-independent mechanism. Nat Cell Biol 2002, 4: 621–625. [DOI] [PubMed] [Google Scholar]
- 17.Malliri A, van der Kammen RA, Clark K, van der Valk M, Michiels F, Collard JG. Mice deficient in the Rac activator Tiam1 are resistant to Ras-induced skin tumours. Nature 2002, 417: 867–871. [DOI] [PubMed] [Google Scholar]
- 18.Takashima A, Faller DV. Targeting the RAS oncogene. Expert Opin Ther Targets 2013, 17: 507–531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Pylayeva-Gupta Y, Grabocka E, Bar-Sagi D. RAS oncogenes: weaving a tumorigenic web. Nat Rev Cancer 2011, 11: 761–774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhang XF, Settleman J, Kyriakis JM, Takeuchi-Suzuki E, Elledge SJ, Marshall MS, Bruder JT et al. Normal and oncogenic p21ras proteins bind to the amino-terminal regulatory domain of c-Raf-1. Nature 1993, 364: 308–313. [DOI] [PubMed] [Google Scholar]
- 21.Rodriguez-Viciana P, Warne PH, Dhand R, Vanhaesebroeck B, Gout I, Fry MJ, Waterfield MD et al. Phosphatidylinositol-3-OH kinase as a direct target of Ras. Nature 1994, 370: 527–532. [DOI] [PubMed] [Google Scholar]
- 22.Pacold ME, Suire S, Perisic O, Lara-Gonzalez S, Davis CT, Walker EH, Hawkins PT et al. Crystal structure and functional analysis of Ras binding to its effector phosphoinositide 3-kinase gamma. Cell 2000, 103: 931–943. [DOI] [PubMed] [Google Scholar]
- 23.Spaargaren M, Bischoff JR. Identification of the guanine nucleotide dissociation stimulator for Ral as a putative effector molecule of R-ras, H-ras, K-ras, and Rap. Proc Natl Acad Sci USA 1994, 91: 12609–12613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hofer F, Fields S, Schneider C, Martin GS. Activated Ras interacts with the Ral guanine nucleotide dissociation stimulator. Proc Natl Acad Sci USA 1994, 91: 11089–11093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Caloca MJ, Zugaza JL, Matallanas D, Crespo P, Bustelo XR. Vav mediates Ras stimulation by direct activation of the GDP/GTP exchange factor Ras GRP1. EMBO J 2003, 22: 3326–3336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Eser S, Reiff N, Messer M, Seidler B, Gottschalk K, Dobler M, Hieber M et al. Selective requirement of PI3K/PDK1 signaling for Kras oncogene-driven pancreatic cell plasticity and cancer. Cancer Cell 2013, 23: 406–420. [DOI] [PubMed] [Google Scholar]
- 27.Wellbrock C, Karasarides M, Marais R. The RAF proteins take centre stage. Nat Rev Mol Cell Biol 2004, 5: 875–885. [DOI] [PubMed] [Google Scholar]
- 28.Martin TD, Samuel JC, Routh ED, Der CJ, Yeh JJ. Activation and involvement of Ral GTPases in colorectal cancer. Cancer Res 2011, 71: 206–215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.van der Hoeven D, Cho KJ, Ma X, Chigurupati S, Parton RG, Hancock JF. Fendiline inhibits K-Ras plasma membrane localization and blocks K-Ras signal transmission. Mol Cell Biol 2013, 33: 237–251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Cho KJ, van der Hoeven D, Hancock JF. Inhibitors of K-Ras plasma membrane localization. Enzymes 2013, 33 Pt A: 249–265. [DOI] [PubMed] [Google Scholar]
- 31.Fang B. Development of synthetic lethality anticancer therapeutics. J Med Chem 2014, 57: 7859–7873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ostrem JM, Peters U, Sos ML, Wells JA, Shokat KM. K-Ras(G12C) inhibitors allosterically control GTP affinity and effector interactions. Nature 2013, 503: 548–551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Maurer T, Garrenton LS, Oh A, Pitts K, Anderson DJ, Skelton NJ, Fauber BP et al. Small-molecule ligands bind to a distinct pocket in Ras and inhibit SOS-mediated nucleotide exchange activity. Proc Natl Acad Sci USA 2012, 109: 5299–5304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Winter JJ, Anderson M, Blades K, Brassington C, Breeze AL, Chresta C, Embrey K et al. Small molecule binding sites on the Ras: SOS complex can be exploited for inhibition of Ras activation. J Med Chem 2015, 58: 2265–2274. [DOI] [PubMed] [Google Scholar]
- 35.Shima F, Yoshikawa Y, Ye M, Araki M, Matsumoto S, Liao J, Hu L et al. In silico discovery of small-molecule Ras inhibitors that display antitumor activity by blocking the Ras-effector interaction. Proc Natl Acad Sci USA 2013, 110: 8182–8187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Pells S, Divjak M, Romanowski P, Impey H, Hawkins NJ, Clarke AR, Hooper ML et al. Developmentally regulated expression of murine K-ras isoforms. Oncogene 1997, 15: 1781–1786. [DOI] [PubMed] [Google Scholar]
- 37.Fiorucci G, Hall A. All three human ras genes are expressed in a wide range of tissues. Biochim Biophys Acta 1988, 950: 81–83. [DOI] [PubMed] [Google Scholar]
- 38.Downward J. Targeting RAS signalling pathways in cancer therapy. Nat Rev Cancer 2003, 3: 11–22. [DOI] [PubMed] [Google Scholar]
- 39.Umanoff H, Edelmann W, Pellicer A, Kucherlapati R. The murine N-ras gene is not essential for growth and development. Proc Natl Acad Sci USA 1995, 92: 1709–1713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Esteban LM, Vicario-Abejon C, Fernandez-Salguero P, Fernandez-Medarde A, Swaminathan N, Yienger K, Lopez E et al. Targeted genomic disruption of H-ras and N-ras, individually or in combination, reveals the dispensability of both loci for mouse growth and development. Mol Cell Biol 2001, 21: 1444–1452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Johnson L, Greenbaum D, Cichowski K, Mercer K, Murphy E, Schmitt E, Bronson RT et al. K-ras is an essential gene in the mouse with partial functional overlap with N-ras. Genes Dev 1997, 11: 2468–2481. [Erratum appears in Genes Dev 1997, 11: 3277]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Nakamura K, Ichise H, Nakao K, Hatta T, Otani H, Sakagami H, Kondo H et al. Partial functional overlap of the three Ras genes in mouse embryonic development. Oncogene 2008, 27: 2961–2968. [DOI] [PubMed] [Google Scholar]
- 43.Potenza N, Vecchione C, Notte A, De Rienzo A, Rosica A, Bauer L, Affuso A et al. Replacement of K-Ras with H-Ras supports normal embryonic development despite inducing cardiovascular pathology in adult mice. EMBO Rep 2005, 6: 432–437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Koera K, Nakamura K, Nakao K, Miyoshi J, Toyoshima K, Hatta T, Otani H et al. K-ras is essential for the development of the mouse embryo. Oncogene 1997, 15: 1151–1159. [DOI] [PubMed] [Google Scholar]
- 45.Walsh AB, Bar-Sagi D. Differential activation of the Rac pathway by Ha-Ras and K-Ras. J Biol Chem 2001, 276: 15609–15615. [DOI] [PubMed] [Google Scholar]
- 46.Billadeau D, Liu P, Jelinek D, Shah N, LeBien TW, Van Ness B. Activating mutations in the N- and K-ras oncogenes differentially affect the growth properties of the IL-6-dependent myeloma cell line ANBL6. Cancer Res 1997, 57: 2268–2275. [PubMed] [Google Scholar]
- 47.Tuveson DA, Shaw AT, Willis NA, Silver DP, Jackson EL, Chang S, Mercer KL et al. Endogenous oncogenic K-ras(G12D) stimulates proliferation and widespread neoplastic and developmental defects. Cancer Cell 2004, 5: 375–387. [DOI] [PubMed] [Google Scholar]
- 48.Guerra C, Mijimolle N, Dhawahir A, Dubus P, Barradas M, Serrano M, Campuzano V et al. Tumor induction by an endogenous K-ras oncogene is highly dependent on cellular context. Cancer Cell 2003, 4: 111–120. [DOI] [PubMed] [Google Scholar]
- 49.Jackson EL, Willis N, Mercer K, Bronson RT, Crowley D, Montoya R, Jacks T et al. Analysis of lung tumor initiation and progression using conditional expression of oncogenic K-ras. Genes Dev 2001, 15: 3243–3248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Johnson L, Mercer K, Greenbaum D, Bronson RT, Crowley D, Tuveson DA, Jacks T. Somatic activation of the K-ras oncogene causes early onset lung cancer in mice. Nature 2001, 410: 1111–1116. [DOI] [PubMed] [Google Scholar]
- 51.Hingorani SR, Petricoin EF, Maitra A, Rajapakse V, King C, Jacobetz MA, Ross S et al. Preinvasive and invasive ductal pancreatic cancer and its early detection in the mouse. Cancer Cell 2003, 4: 437–450. [DOI] [PubMed] [Google Scholar]
- 52.Kim CF, Jackson EL, Kirsch DG, Grimm J, Shaw AT, Lane K, Kissil J et al. Mouse models of human non-small-cell lung cancer: raising the bar. Cold Spring Harb Symp Quant Biol 2005, 70: 241–250. [DOI] [PubMed] [Google Scholar]
- 53.Bardeesy N, Aguirre AJ, Chu GC, Cheng KH, Lopez LV, Hezel AF, Feng B et al. Both p16(Ink4a) and the p19(Arf)-p53 pathway constrain progression of pancreatic adenocarcinoma in the mouse. Proc Natl Acad Sci USA 2006, 103: 5947–5952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Jackson EL, Olive KP, Tuveson DA, Bronson R, Crowley D, Brown M, Jacks T. The differential effects of mutant p53 alleles on advanced murine lung cancer. Cancer Res 2005, 65: 10280–10288. [DOI] [PubMed] [Google Scholar]
- 55.O'Dell MR, Huang JL, Whitney-Miller CL, Deshpande V, Rothberg P, Grose V, Rossi RM et al. Kras(G12D) and p53 mutation cause primary intrahepatic cholangiocarcinoma. Cancer Res 2012, 72: 1557–1567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Ji H, Ramsey MR, Hayes DN, Fan C, McNamara K, Kozlowski P, Torrice C et al. LKB1 modulates lung cancer differentiation and metastasis. Nature 2007, 448: 807–810. [DOI] [PubMed] [Google Scholar]
- 57.Carretero J, Shimamura T, Rikova K, Jackson AL, Wilkerson MD, Borgman CL, Buttarazzi MS et al. Integrative genomic and proteomic analyses identify targets for Lkb1-deficient metastatic lung tumors. Cancer Cell 2010, 17: 547–559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Morton JP, Jamieson NB, Karim SA, Athineos D, Ridgway RA, Nixon C, McKay CJ et al. LKB1 haploinsufficiency cooperates with Kras to promote pancreatic cancer through suppression of p21-dependent growth arrest. Gastroenterology 2010, 139: 586–597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Aguirre AJ, Bardeesy N, Sinha M, Lopez L, Tuveson DA, Horner J, Redston MS et al. Activated Kras and Ink4a/Arf deficiency cooperate to produce metastatic pancreatic ductal adenocarcinoma. Genes Dev 2003, 17: 3112–3126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Bardeesy N, Cheng KH, Berger JH, Chu GC, Pahler J, Olson P, Hezel AF et al. Smad4 is dispensable for normal pancreas development yet critical in progression and tumor biology of pancreas cancer. Genes Dev 2006, 20: 3130–3146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Hill R, Calvopina JH, Kim C, Wang Y, Dawson DW, Donahue TR, Dry S et al. PTEN loss accelerates KrasG12D-induced pancreatic cancer development. Cancer Res 2010, 70: 7114–7124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Ijichi H, Chytil A, Gorska AE, Aakre ME, Fujitani Y, Fujitani S, Wright CV et al. Aggressive pancreatic ductal adenocarcinoma in mice caused by pancreas-specific blockade of transforming growth factor-beta signaling in cooperation with active Kras expression. Genes Dev 2006, 20: 3147–3160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Lee YS, Lee JW, Jang JW, Chi XZ, Kim JH, Li YH, Kim MK et al. Runx3 inactivation is a crucial early event in the development of lung adenocarcinoma. Cancer Cell 2013, 24: 603–616. [DOI] [PubMed] [Google Scholar]
- 64.Pacheco-Pinedo EC, Durham AC, Stewart KM, Goss AM, Lu MM, DeMayo FJ, Morrisey EE. Wnt/beta-catenin signaling accelerates mouse lung tumorigenesis by imposing an embryonic distal progenitor phenotype on lung epithelium. J Clin Invest 2011, 121: 1935–1945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Meuwissen R, Linn SC, van der Valk M, Mooi WJ, Berns A. Mouse model for lung tumorigenesis through Cre/lox controlled sporadic activation of the K-Ras oncogene. Oncogene 2001, 20: 6551–6558. [DOI] [PubMed] [Google Scholar]
- 66.Castellano E, Sheridan C, Thin MZ, Nye E, Spencer-Dene B, Diefenbacher ME, Moore C et al. Requirement for interaction of PI3-kinase p110alpha with RAS in lung tumor maintenance. Cancer Cell 2013, 24: 617–630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Kissil JL, Walmsley MJ, Hanlon L, Haigis KM, der Kim CF, Sweet-Cordero A, Eckman MS et al. Requirement for Rac1 in a K-ras induced lung cancer in the mouse. Cancer Res 2007, 67: 8089–8094. [DOI] [PubMed] [Google Scholar]
- 68.Heid I, Lubeseder-Martellato C, Sipos B, Mazur PK, Lesina M, Schmid RM, Siveke JT. Early requirement of Rac1 in a mouse model of pancreatic cancer. Gastroenterology 2011, 141: 719–730. [DOI] [PubMed] [Google Scholar]
- 69.Wu CY, Carpenter ES, Takeuchi KK, Halbrook CJ, Peverley LV, Bien H, Hall JC et al. PI3K regulation of RAC1 is required for KRAS-induced pancreatic tumorigenesis in mice. Gastroenterology 2014, 147: 1405–1416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Zhou C, Licciulli S, Avila JL, Cho M, Troutman S, Jiang P, Kossenkov AV et al. The Rac1 splice form Rac1b promotes K-ras-induced lung tumorigenesis. Oncogene 2013, 32: 903–909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Trejo CL, Juan J, Vicent S, Sweet-Cordero A, McMahon M. MEK1/2 inhibition elicits regression of autochthonous lung tumors induced by KRASG12D or BRAFV600E. Cancer Res 2012, 72: 3048–3059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Collisson EA, Trejo CL, Silva JM, Gu S, Korkola JE, Heiser LM, Charles RP et al. A central role for RAF->MEK->ERK signaling in the genesis of pancreatic ductal adenocarcinoma. Cancer Discov 2012, 2: 685–693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Ling J, Kang Y, Zhao R, Xia Q, Lee DF, Chang Z, Li J et al. KrasG12D-induced IKK2/β/NF-κB activation by IL-1α and p62 feedforward loops is required for development of pancreatic ductal adenocarcinoma. Cancer Cell 2012, 21: 105–120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Zhu Z, Aref AR, Cohoon TJ, Barbie TU, Imamura Y, Yang S, Moody SE et al. Inhibition of KRAS-driven tumorigenicity by interruption of an autocrine cytokine circuit. Cancer Discov 2014, 4: 452–465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Chien Y, Kim S, Bumeister R, Loo YM, Kwon SW, Johnson CL, Balakireva MG et al. RalB GTPase-mediated activation of the IkappaB family kinase TBK1 couples innate immune signaling to tumor cell survival. Cell 2006, 127: 157–170. [DOI] [PubMed] [Google Scholar]
- 76.Lesina M, Kurkowski MU, Ludes K, Rose-John S, Treiber M, Kloppel G, Yoshimura A et al. Stat3/Socs3 activation by IL-6 transsignaling promotes progression of pancreatic intraepithelial neoplasia and development of pancreatic cancer. Cancer Cell 2011, 19: 456–469. [DOI] [PubMed] [Google Scholar]
- 77.Corcoran RB, Contino G, Deshpande V, Tzatsos A, Conrad C, Benes CH, Levy DE et al. STAT3 plays a critical role in KRAS-induced pancreatic tumorigenesis. Cancer Res 2011, 71: 5020–5029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Zhang W, Nandakumar N, Shi Y, Manzano M, Smith A, Graham G, Gupta S et al. Downstream of mutant KRAS, the transcription regulator YAP is essential for neoplastic progression to pancreatic ductal adenocarcinoma. Sci Signal 2014, 7: ra42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Wang IC, Ustiyan V, Zhang Y, Cai Y, Kalin TV, Kalinichenko VV. Foxm1 transcription factor is required for the initiation of lung tumorigenesis by oncogenic Kras(G12D.). Oncogene 2014, 33: 5391–5396. [DOI] [PubMed] [Google Scholar]
- 80.Ying H, Kimmelman AC, Lyssiotis CA, Hua S, Chu GC, Fletcher-Sananikone E, Locasale JW et al. Oncogenic Kras maintains pancreatic tumors through regulation of anabolic glucose metabolism. Cell 2012, 149: 656–670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Pylayeva-Gupta Y, Lee KE, Hajdu CH, Miller G, Bar-Sagi D. Oncogenic Kras-induced GM-CSF production promotes the development of pancreatic neoplasia. Cancer Cell 2012, 21: 836–847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Konstantinidou G, Ramadori G, Torti F, Kangasniemi K, Ramirez RE, Cai Y, Behrens C et al. RHOA-FAK is a required signaling axis for the maintenance of KRAS-driven lung adenocarcinomas. Cancer Discov 2013, 3: 444–457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Santos AM, Jung J, Aziz N, Kissil JL, Pure E. Targeting fibroblast activation protein inhibits tumor stromagenesis and growth in mice. J Clin Invest 2009, 119: 3613–3625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Soucek L, Whitfield JR, Sodir NM, Masso-Valles D, Serrano E, Karnezis AN, Swigart LB et al. Inhibition of Myc family proteins eradicates KRas-driven lung cancer in mice. Genes Dev 2013, 27: 504–513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Baer R, Cintas C, Dufresne M, Cassant-Sourdy S, Schonhuber N, Planque L, Lulka H et al. Pancreatic cell plasticity and cancer initiation induced by oncogenic Kras is completely dependent on wild-type PI 3-kinase p110alpha. Genes Dev 2014, 28: 2621–2635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Gupta S, Ramjaun AR, Haiko P, Wang Y, Warne PH, Nicke B, Nye E et al. Binding of ras to phosphoinositide 3-kinase p110alpha is required for ras-driven tumorigenesis in mice. Cell 2007, 129: 957–968. [DOI] [PubMed] [Google Scholar]
- 87.Chin L, Tam A, Pomerantz J, Wong M, Holash J, Bardeesy N, Shen Q et al. Essential role for oncogenic Ras in tumour maintenance. Nature 1999, 400: 468–472. [DOI] [PubMed] [Google Scholar]
- 88.Fisher GH, Wellen SL, Klimstra D, Lenczowski JM, Tichelaar JW, Lizak MJ, Whitsett JA et al. Induction and apoptotic regression of lung adenocarcinomas by regulation of a K-Ras transgene in the presence and absence of tumor suppressor genes. Genes Dev 2001, 15: 3249–3262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Collins MA, Bednar F, Zhang Y, Brisset JC, Galban S, Galban CJ, Rakshit S et al. Oncogenic Kras is required for both the initiation and maintenance of pancreatic cancer in mice. J Clin Invest 2012, 122: 639–653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Pettazzoni P, Viale A, Shah P, Carugo A, Ying H, Wang H, Genovese G et al. Genetic events that limit the efficacy of MEK and RTK inhibitor therapies in a mouse model of KRAS-driven pancreatic cancer. Cancer Res 2015, 75: 1091–1101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Engelman JA, Chen L, Tan X, Crosby K, Guimaraes AR, Upadhyay R, Maira M et al. Effective use of PI3K and MEK inhibitors to treat mutant Kras G12D and PIK3CA H1047R murine lung cancers. Nat Med 2008, 14: 1351–1356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Junttila MR, Devasthali V, Cheng JH, Castillo J, Metcalfe C, Clermont AC, Otter DD et al. Modeling targeted inhibition of MEK and PI3 kinase in human pancreatic cancer. Mol Cancer Ther 2015, 14: 40–47. [DOI] [PubMed] [Google Scholar]
- 93.Alagesan B, Contino G, Guimaraes AR, Corcoran RB, Deshpande V, Wojtkiewicz GR, Hezel AF et al. Combined MEK and PI3K inhibition in a mouse model of pancreatic cancer. Clin Cancer Res 2015, 21: 396–404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Xia Y, Liu YL, Xie Y, Zhu W, Guerra F, Shen S, Yeddula N et al. A combination therapy for KRAS-driven lung adenocarcinomas using lipophilic bisphosphonates and rapamycin. Sci Transl Med 2014, 6: 263ra161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Corcoran RB, Cheng KA, Hata AN, Faber AC, Ebi H, Coffee EM, Greninger P et al. Synthetic lethal interaction of combined BCL-XL and MEK inhibition promotes tumor regressions in KRAS mutant cancer models. Cancer Cell 2013, 23: 121–128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Chen Z, Cheng K, Walton Z, Wang Y, Ebi H, Shimamura T, Liu Y et al. A murine lung cancer co-clinical trial identifies genetic modifiers of therapeutic response. Nature 2012, 483: 613–617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Shimamura T, Chen Z, Soucheray M, Carretero J, Kikuchi E, Tchaicha JH, Gao Y et al. Efficacy of BET bromodomain inhibition in Kras-mutant non-small cell lung cancer. Clin Cancer Res 2013, 19: 6183–6192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Shackelford DB, Abt E, Gerken L, Vasquez DS, Seki A, Leblanc M, Wei L et al. LKB1 inactivation dictates therapeutic response of non-small cell lung cancer to the metabolism drug phenformin. Cancer Cell 2013, 23: 143–158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Liu Y, Marks K, Cowley GS, Carretero J, Liu Q, Nieland TJ, Xu C et al. Metabolic and functional genomic studies identify deoxythymidylate kinase as a target in LKB1-mutant lung cancer. Cancer Discov 2013, 3: 870–879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Viale A, Pettazzoni P, Lyssiotis CA, Ying H, Sanchez N, Marchesini M, Carugo A et al. Oncogene ablation-resistant pancreatic cancer cells depend on mitochondrial function. Nature 2014, 514: 628–632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Kapoor A, Yao W, Ying H, Hua S, Liewen A, Wang Q, Zhong Y et al. Yap1 activation enables bypass of oncogenic Kras addiction in pancreatic cancer. Cell 2014, 158: 185–197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Shao DD, Xue W, Krall EB, Bhutkar A, Piccioni F, Wang X, Schinzel AC et al. KRAS and YAP1 converge to regulate EMT and tumor survival. Cell 2014, 158: 171–184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Shirasawa S, Furuse M, Yokoyama N, Sasazuki T. Altered growth of human colon cancer cell lines disrupted at activated Ki-ras. Science 1993, 260: 85–88. [DOI] [PubMed] [Google Scholar]
- 104.Singh A, Greninger P, Rhodes D, Koopman L, Violette S, Bardeesy N, Settleman J. A gene expression signature associated with ‘K-Ras addiction’ reveals regulators of EMT and tumor cell survival. Cancer Cell 2009, 15: 489–500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Luo J, Emanuele MJ, Li D, Creighton CJ, Schlabach MR, Westbrook TF, Wong KK et al. A genome-wide RNAi screen identifies multiple synthetic lethal interactions with the Ras oncogene. Cell 2009, 137: 835–848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Vicent S, Chen R, Sayles LC, Lin C, Walker RG, Gillespie AK, Subramanian A et al. Wilms tumor 1 (WT1) regulates KRAS-driven oncogenesis and senescence in mouse and human models. J Clin Invest 2010, 120: 3940–3952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Barbie DA, Tamayo P, Boehm JS, Kim SY, Moody SE, Dunn IF, Schinzel AC et al. Systematic RNA interference reveals that oncogenic KRAS-driven cancers require TBK1. Nature 2009, 462: 108–112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Puyol M, Martin A, Dubus P, Mulero F, Pizcueta P, Khan G, Guerra C et al. A synthetic lethal interaction between K-Ras oncogenes and Cdk4 unveils a therapeutic strategy for non-small cell lung carcinoma. Cancer Cell 2010, 18: 63–73. [DOI] [PubMed] [Google Scholar]
- 109.Steegmaier M, Hoffmann M, Baum A, Lenart P, Petronczki M, Krssak M, Gurtler U et al. BI 2536, a potent and selective inhibitor of polo-like kinase 1, inhibits tumor growth in vivo. Curr Biol 2007, 17: 316–322. [DOI] [PubMed] [Google Scholar]
- 110.Kaelin WG., Jr The concept of synthetic lethality in the context of anticancer therapy. Nat Rev Cancer 2005, 5: 689–698. [DOI] [PubMed] [Google Scholar]
- 111.Torrance CJ, Agrawal V, Vogelstein B, Kinzler KW. Use of isogenic human cancer cells for high-throughput screening and drug discovery. Nat Biotech 2001, 19: 940–945. [DOI] [PubMed] [Google Scholar]
- 112.Dolma S, Lessnick SL, Hahn WC, Stockwell BR. Identification of genotype-selective antitumor agents using synthetic lethal chemical screening in engineered human tumor cells. Cancer Cell 2003, 3: 285–296. [DOI] [PubMed] [Google Scholar]
- 113.Yagoda N, von Rechenberg M, Zaganjor E, Bauer AJ, Yang WS, Fridman DJ, Wolpaw AJ et al. RAS-RAF-MEK-dependent oxidative cell death involving voltage-dependent anion channels. Nature 2007, 447: 864–868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Shaw AT, Winslow MM, Magendantz M, Ouyang C, Dowdle J, Subramanian A, Lewis TA et al. Selective killing of K-ras mutant cancer cells by small molecule inducers of oxidative stress. Proc Natl Acad Sci USA 2011, 108: 8773–8778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Guo W, Wu S, Liu J, Fang B. Identification of a small molecule with synthetic lethality for K-ras and protein kinase C iota. Cancer Res 2008, 68: 7403–7408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Guo W, Wei X, Wu S, Wang L, Peng H, Wang J, Fang B. Antagonistic effect of flavonoids on NSC-741909-mediated antitumor activity via scavenging of reactive oxygen species. Eur J Pharmacol 2010, 649: 51–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Wei X, Guo W, Wu S, Wang L, Huang P, Liu J, Fang B. Oxidative stress in NSC-741909-induced apoptosis of cancer cells. J Transl Med 2010, 8: 37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Guo W, Wu S, Wang L, Wei X, Liu X, Wang J, Lu Z et al. Antitumor activity of a novel oncrasin analogue is mediated by JNK activation and STAT3 inhibition. PLoS One 2011, 6: e28487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Wei X, Guo W, Wu S, Wang L, Lu Y, Xu B, Liu J et al. Inhibiting JNK dephosphorylation and induction of apoptosis by novel anticancer agent NSC-741909 in cancer cells. J Biol Chem 2009, 284: 16948–16955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Wu S, Wang L, Guo W, Liu X, Liu J, Wei X, Fang B. Analogues and derivatives of oncrasin-1, a novel inhibitor of the C-terminal domain of RNA polymerase II and their antitumor activities. J Med Chem 2011, 54: 2668–2679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Liu X, Guo W, Wu S, Wang L, Wang J, Dai B, Kim ES et al. Antitumor activity of a novel STAT3 inhibitor and redox modulator in non-small cell lung cancer cells. Biochem Pharmacol 2012, 83: 1456–1464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Guo W, Wu S, Wang L, Wang R, Wei L, Liu J, Fang B. Interruption of RNA processing machinery by a small compound 1-[(4-chlorophenyl) methyl]-1H-indole-3-carboxaldehyde (oncrasin-1). Mol Cancer Ther 2009, 8: 441–448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Young TW, Mei FC, Yang G, Thompson-Lanza JA, Liu J, Cheng X. Activation of antioxidant pathways in ras-mediated oncogenic transformation of human surface ovarian epithelial cells revealed by functional proteomics and mass spectrometry. Cancer Res 2004, 64: 4577–4584. [DOI] [PubMed] [Google Scholar]
- 124.Fang B. Genetic interactions of STAT3 and anticancer drug development. Cancers 2014, 6: 494–525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Wu S, Wang L, Huang X, Cao M, Hu J, Li H, Zhang H et al. Prodrug oncrasin-266 improves the stability, pharmacokinetics, and safety of NSC-743380. Bioorg Med Chem 2014, 22: 5234–5240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Huang X, Cao M, Wang L, Wu S, Liu X, Li H, Zhang H et al. Expression of sulfotransferase SULT1A1 in cancer cells predicts susceptibility to the novel anticancer agent NSC-743380. Oncotarget 2015, 6: 345–354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Svendsen C, Meinl W, Glatt H, Alexander J, Knutsen HK, Hjertholm H, Rasmussen T et al. Intestinal carcinogenesis of two food processing contaminants, 2-amino-1-methyl-6-phenylimidazo[4,5-b]pyridine and 5-hydroxymethylfurfural, in transgenic FVB min mice expressing human sulfotransferases. Mol Carcinog 2012, 51: 984–992. [DOI] [PubMed] [Google Scholar]
- 128.Suzuki Y, Umemura T, Ishii Y, Hibi D, Inoue T, Jin M, Sakai H et al. Possible involvement of sulfotransferase 1A1 in estragole-induced DNA modification and carcinogenesis in the livers of female mice. Mutation Res 2012, 749: 23–28. [DOI] [PubMed] [Google Scholar]
- 129.Gamage NU, Duggleby RG, Barnett AC, Tresillian M, Latham CF, Liyou NE, McManus ME et al. Structure of a human carcinogen-converting enzyme, SULT1A1. Structural and kinetic implications of substrate inhibition. J Biol Chem 2003, 278: 7655–7662. [DOI] [PubMed] [Google Scholar]
- 130.Arlt VM, Glatt H, Muckel E, Pabel U, Sorg BL, Schmeiser HH, Phillips DH. Metabolic activation of the environmental contaminant 3-nitrobenzanthrone by human acetyltransferases and sulfotransferase. Carcinogenesis 2002, 23: 1937–1945. [DOI] [PubMed] [Google Scholar]
- 131.Banoglu E. Current status of the cytosolic sulfotransferases in the metabolic activation of promutagens and procarcinogens. Curr Drug Metab 2000, 1: 1–30. [DOI] [PubMed] [Google Scholar]
- 132.Michejda CJ, Kroeger Koepke MB. Carcinogen activation by sulfate conjugate formation. Adv Pharmacol 1994, 27: 331–363. [DOI] [PubMed] [Google Scholar]
- 133.Kroeger-Koepke MB, Koepke SR, Hernandez L, Michejda CJ. Activation of a beta-hydroxyalkylnitrosamine to alkylating agents: evidence for the involvement of a sulfotransferase. Cancer Res 1992, 52: 3300–3305. [PubMed] [Google Scholar]
- 134.Mercer KE, Apostolov EO, da Costa GG, Yu X, Lang P, Roberts DW, Davis W et al. Expression of sulfotransferase isoform 1A1 (SULT1A1) in breast cancer cells significantly increases 4-hydroxytamoxifen-induced apoptosis. Int J Mol Epidemiol Genet 2010, 1: 92–103. [PMC free article] [PubMed] [Google Scholar]
- 135.Eldridge SR, Cover J, Morris J, Fang B, Horn TL, Elsass KE, Hamre JR et al. Characterization of acute biliary hyperplasia in Fisher 344 rats administered the indole-3-carbinol analog, NSC-743380. Toxicol Appl Pharmacol 2014, 281: 303–309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Almoguera C, Shibata D, Forrester K, Martin J, Arnheim N, Perucho M. Most human carcinomas of the exocrine pancreas contain mutant c-K-ras genes. Cell 1988, 53: 549–554. [DOI] [PubMed] [Google Scholar]
- 137.Smit VT, Boot AJ, Smits AM, Fleuren GJ, Cornelisse CJ, Bos JL. KRAS codon 12 mutations occur very frequently in pancreatic adenocarcinomas. Nucleic Acids Res 1988, 16: 7773–7782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Waddell N, Pajic M, Patch AM, Chang DK, Kassahn KS, Bailey P, Johns AL et al. Whole genomes redefine the mutational landscape of pancreatic cancer. Nature 2015, 518: 495–501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Bos JL, Fearon ER, Hamilton SR, Verlaan-de Vries M, van Boom JH, van der Eb AJ, Vogelstein B. Prevalence of ras gene mutations in human colorectal cancers. Nature 1987, 327: 293–297. [DOI] [PubMed] [Google Scholar]
- 140.Vogelstein B, Fearon ER, Hamilton SR, Kern SE, Preisinger AC, Leppert M, Nakamura Y et al. Genetic alterations during colorectal-tumor development. N Engl J Med 1988, 319: 525–532. [DOI] [PubMed] [Google Scholar]
- 141.Cancer Genome Atlas Research Network. Comprehensive molecular profiling of lung adenocarcinoma. Nature 2014, 511: 543–550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Mills NE, Fishman CL, Rom WN, Dubin N, Jacobson DR. Increased prevalence of K-ras oncogene mutations in lung adenocarcinoma. Cancer Res 1995, 55: 1444–1447. [PubMed] [Google Scholar]
- 143.Aviel-Ronen S, Blackhall FH, Shepherd FA, Tsao MS. K-ras mutations in non-small-cell lung carcinoma: a review. Clin Lung Cancer 2006, 8: 30–38. [DOI] [PubMed] [Google Scholar]
- 144.Bryant KL, Mancias JD, Kimmelman AC, Der CJ. KRAS: feeding pancreatic cancer proliferation. Trends Biochem Sci 2014, 39: 91–100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Vasan N, Boyer JL, Herbst RS. A RAS renaissance: emerging targeted therapies for KRAS-mutated non-small cell lung cancer. Clin Cancer Res 2014, 20: 3921–3930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Ihle NT, Byers LA, Kim ES, Saintigny P, Lee JJ, Blumenschein GR, Tsao A et al. Effect of KRAS oncogene substitutions on protein behavior: implications for signaling and clinical outcome. J Natl Cancer Inst 2012, 104: 228–239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Siegfried JM, Gillespie AT, Mera R, Casey TJ, Keohavong P, Testa JR, Hunt JD. Prognostic value of specific KRAS mutations in lung adenocarcinomas. Cancer Epidemiol Biomarkers Prev 1997, 6: 841–847. [PubMed] [Google Scholar]
- 148.Blons H, Emile JF, Le MK, Julie C, Zaanan A, Tabernero J, Mini E et al. Prognostic value of KRAS mutations in stage III colon cancer: post hoc analysis of the PETACC8 phase III trial dataset. Ann Oncol 2014, 25: 2378–2385. [DOI] [PubMed] [Google Scholar]
- 149.Lee DW, Kim KJ, Han SW, Lee HJ, Rhee YY, Bae JM, Cho NY et al. KRAS mutation is associated with worse prognosis in stage III or high-risk stage II colon cancer patients treated with adjuvant FOLFOX. Ann Surg Oncol 2015, 22: 187–194. [DOI] [PubMed] [Google Scholar]
- 150.Yoon HH, Tougeron D, Shi Q, Alberts SR, Mahoney MR, Nelson GD, Nair SG et al. KRAS codon 12 and 13 mutations in relation to disease-free survival in BRAF-wild-type stage III colon cancers from an adjuvant chemotherapy trial (N0147 alliance). Clin Cancer Res 2014, 20: 3033–3043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Ogura T, Yamao K, Hara K, Mizuno N, Hijioka S, Imaoka H, Sawaki A et al. Prognostic value of K-ras mutation status and subtypes in endoscopic ultrasound-guided fine-needle aspiration specimens from patients with unresectable pancreatic cancer. J Gastroenterol 2013, 48: 640–646. [DOI] [PubMed] [Google Scholar]
- 152.Tada M, Ohashi M, Shiratori Y, Okudaira T, Komatsu Y, Kawabe T, Yoshida H et al. Analysis of K-ras gene mutation in hyperplastic duct cells of the pancreas without pancreatic disease. Gastroenterology 1996, 110: 227–231. [DOI] [PubMed] [Google Scholar]
- 153.Shi C, Hong SM, Lim P, Kamiyama H, Khan M, Anders RA, Goggins M et al. KRAS2 mutations in human pancreatic acinar-ductal metaplastic lesions are limited to those with PanIN: implications for the human pancreatic cancer cell of origin. Mol Cancer Res 2009, 7: 230–236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Yanagisawa A, Ohtake K, Ohashi K, Hori M, Kitagawa T, Sugano H, Kato Y. Frequent c-Ki-ras oncogene activation in mucous cell hyperplasias of pancreas suffering from chronic inflammation. Cancer Res 1993, 53: 953–956. [PubMed] [Google Scholar]
- 155.Hiraoka S, Kato J, Tatsukawa M, Harada K, Fujita H, Morikawa T, Shiraha H et al. Laterally spreading type of colorectal adenoma exhibits a unique methylation phenotype and K-ras mutations. Gastroenterology 2006, 131: 379–389. [DOI] [PubMed] [Google Scholar]
- 156.Barry EL, Baron JA, Grau MV, Wallace K, Haile RW. K-ras mutations in incident sporadic colorectal adenomas. Cancer 2006, 106: 1036–1040. [DOI] [PubMed] [Google Scholar]
- 157.Takayama T, Ohi M, Hayashi T, Miyanishi K, Nobuoka A, Nakajima T, Satoh T et al. Analysis of K-ras, APC, and beta-catenin in aberrant crypt foci in sporadic adenoma, cancer, and familial adenomatous polyposis. Gastroenterology 2001, 121: 599–611. [DOI] [PubMed] [Google Scholar]
- 158.Pellegata NS, Sessa F, Renault B, Bonato M, Leone BE, Solcia E, Ranzani GN. K-ras and p53 gene mutations in pancreatic cancer: ductal and nonductal tumors progress through different genetic lesions. Cancer Res 1994, 54: 1556–1560. [PubMed] [Google Scholar]
- 159.Calles A, Sholl LM, Rodig SJ, Pelton AK, Hornick JL, Butaney M, Lydon C et al. Immunohistochemical loss of LKB1 is a biomarker for more aggressive biology in KRAS-mutant lung adenocarcinoma. Clin Cancer Res 2015, 21: 2851–2860. [DOI] [PubMed] [Google Scholar]
- 160.Kim HS, Mendiratta S, Kim J, Pecot CV, Larsen JE, Zubovych I, Seo BY et al. Systematic identification of molecular subtype-selective vulnerabilities in non-small-cell lung cancer. Cell 2013, 155: 552–566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Skoulidis F, Byers LA, Diao L, Papadimitrakopoulou VA, Tong P, Izzo J, Behrens C et al. Co-occurring genomic alterations define major subsets of KRAS-mutant lung adenocarcinoma with distinct biology, immune profiles, and therapeutic vulnerabilities. Cancer Discov 2015, 5: 860–877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Hancock JF. Ras proteins: different signals from different locations. Nat Rev Mol Cell Biol 2003, 4: 373–384. [DOI] [PubMed] [Google Scholar]
- 163.Cadwallader KA, Paterson H, Macdonald SG, Hancock JF. N-terminally myristoylated Ras proteins require palmitoylation or a polybasic domain for plasma membrane localization. Mol Cell Biol 1994, 14: 4722–4730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Hancock JF, Paterson H, Marshall CJ. A polybasic domain or palmitoylation is required in addition to the CAAX motif to localize p21ras to the plasma membrane. Cell 1990, 63: 133–139. [DOI] [PubMed] [Google Scholar]
- 165.Riely GJ, Johnson ML, Medina C, Rizvi NA, Miller VA, Kris MG, Pietanza MC et al. A phase II trial of Salirasib in patients with lung adenocarcinomas with KRAS mutations. J Thoracic Oncol 2011, 6: 1435–1437. [DOI] [PubMed] [Google Scholar]
- 166.Karp JE, Vener TI, Raponi M, Ritchie EK, Smith BD, Gore SD, Morris LE et al. Multi-institutional phase 2 clinical and pharmacogenomic trial of tipifarnib plus etoposide for elderly adults with newly diagnosed acute myelogenous leukemia. Blood 2012, 119: 55–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Rao S, Cunningham D, de Gramont A, Scheithauer W, Smakal M, Humblet Y, Kourteva G et al. Phase III double-blind placebo-controlled study of farnesyl transferase inhibitor R115777 in patients with refractory advanced colorectal cancer. J Clin Oncol 2004, 22: 3950–3957. [DOI] [PubMed] [Google Scholar]
- 168.Van CE, van de Velde H, Karasek P, Oettle H, Vervenne WL, Szawlowski A, Schoffski P et al. Phase III trial of gemcitabine plus tipifarnib compared with gemcitabine plus placebo in advanced pancreatic cancer. J Clin Oncol 2004, 22: 1430–1438. [DOI] [PubMed] [Google Scholar]
- 169.Rusconi P, Caiola E, Broggini M. RAS/RAF/MEK inhibitors in oncology. Curr Med Chem 2012, 19: 1164–1176. [DOI] [PubMed] [Google Scholar]
- 170.Wilhelm SM, Carter C, Tang L, Wilkie D, McNabola A, Rong H, Chen C et al. BAY 43-9006 exhibits broad spectrum oral antitumor activity and targets the RAF/MEK/ERK pathway and receptor tyrosine kinases involved in tumor progression and angiogenesis. Cancer Res 2004, 64: 7099–7109. [DOI] [PubMed] [Google Scholar]
- 171.Kim ES, Herbst RS, Wistuba II, Lee JJ, Blumenschein GR Jr, Tsao A, Stewart DJ et al. The BATTLE trial: personalizing therapy for lung cancer. Cancer Discov 2011, 1: 44–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Heidorn SJ, Milagre C, Whittaker S, Nourry A, Niculescu-Duvas I, Dhomen N, Hussain J et al. Kinase-dead BRAF and oncogenic RAS cooperate to drive tumor progression through CRAF. Cell 2010, 140: 209–221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Callahan MK, Rampal R, Harding JJ, Klimek VM, Chung YR, Merghoub T, Wolchok JD et al. Progression of RAS-mutant leukemia during RAF inhibitor treatment. N Engl J Med 2012, 367: 2316–2321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Robert C, Arnault JP, Mateus C. RAF inhibition and induction of cutaneous squamous cell carcinoma. Curr Opin Oncol 2011, 23: 177–182. [DOI] [PubMed] [Google Scholar]
- 175.Lopez-Chavez A, Thomas A, Rajan A, Raffeld M, Morrow B, Kelly R, Carter CA et al. Molecular profiling and targeted therapy for advanced thoracic malignancies: a biomarker-derived, multiarm, multihistology phase II basket trial. J Clin Oncol 2015, 33: 1000–1007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Janne PA, Shaw AT, Pereira JR, Jeannin G, Vansteenkiste J, Barrios C, Franke FA et al. Selumetinib plus docetaxel for KRAS-mutant advanced non-small-cell lung cancer: a randomised, multicentre, placebo-controlled, phase 2 study. Lancet Oncol 2013, 14: 38–47. [DOI] [PubMed] [Google Scholar]
- 177.Tolcher AW, Khan K, Ong M, Banerji U, Papadimitrakopoulou V, Gandara DR, Patnaik A et al. Antitumor activity in RAS-driven tumors by blocking AKT and MEK. Clin Cancer Res 2015, 21: 739–748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Bedard PL, Tabernero J, Janku F, Wainberg ZA, Paz-Ares L, Vansteenkiste J, Van CE et al. A phase Ib dose-escalation study of the oral pan-PI3K inhibitor buparlisib (BKM120) in combination with the oral MEK1/2 inhibitor trametinib (GSK1120212) in patients with selected advanced solid tumors. Clin Cancer Res 2015, 21: 730–738. [DOI] [PubMed] [Google Scholar]
- 179.Infante JR, Somer BG, Park JO, Li CP, Scheulen ME, Kasubhai SM, Oh DY et al. A randomised, double-blind, placebo-controlled trial of trametinib, an oral MEK inhibitor, in combination with gemcitabine for patients with untreated metastatic adenocarcinoma of the pancreas. Eur J Cancer 2014, 50: 2072–2081. [DOI] [PubMed] [Google Scholar]
- 180.Hochster HS, Uboha N, Messersmith W, Gold PJ, ONeil BH, Cohen D, Denlinger C et al. Phase II study of selumetinib (AZD6244, ARRY-142886) plus irinotecan as second-line therapy in patients with K-RAS mutated colorectal cancer . Cancer Chemother Pharmacol 2015, 75: 17–23. [Erratum appears in Cancer Chemother Pharmacol 2015, 75: 25. Note: Lenz, H-J [added]]. [DOI] [PubMed] [Google Scholar]
- 181.Vogelstein B, Papadopoulos N, Velculescu VE, Zhou S, Diaz LA Jr, Kinzler KW. Cancer genome landscapes. Science 2013, 339: 1546–1558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Scanlan MJ, Raj BK, Calvo B, Garin-Chesa P, Sanz-Moncasi MP, Healey JH, Old LJ et al. Molecular cloning of fibroblast activation protein alpha, a member of the serine protease family selectively expressed in stromal fibroblasts of epithelial cancers. Proc Natl Acad Sci USA 1994, 91: 5657–5661. [DOI] [PMC free article] [PubMed] [Google Scholar]