Abstract
Oncogenic mutations in Kirsten rat sarcoma viral oncogene homolog (KRAS) occur in 15%–30% of non-small cell lung cancer (NSCLC). However, despite decades of intensive research, there is still no direct KRAS inhibitor with clinically proven efficacy. Considering its association with poor treatment response and prognosis of lung cancer, developing an effective inhibitory approach is urgently needed. Here, we review different strategies currently being explored to target KRAS-mutant NSCLC, discuss opportunities and challenges, and also propose some novel methods and concepts with the promise of clinical application.
Keywords: KRAS, mutations, NSCLC, therapy
Introduction
Non-small cell lung cancer (NSCLC) accounts for >85% of human lung cancer, which is a leading cause of cancer death in both men and women worldwide [1]. The overall survival of patients with advanced or metastatic NSCLC is still dismal, with a median of ∼8–10 months [2]. The identification of targetable genetic alterations such as epidermal growth factor receptor (EGFR) mutations and echinoderm microtubule-associated protein-like 4 (EML4)-anaplastic lymphoma kinase (ALK) translocations has provided unique advantages in treating lung cancer patients [3,4]. Although oncogenic Kirsten rat sarcoma viral oncogene homolog (KRAS) mutation was identified in NSCLC in the 1980s [5], there is still no direct inhibitor in the clinical setting. More importantly, it was found that KRAS mutation impairs response to certain cytotoxic agents as well as targeted therapies such as erlotinib [6,7], whereas targeting KRAS may help sensitize the response to certain chemotherapeutic agents such as docetaxel [8]. Considering KRAS mutation occurs in 15%–30% of NSCLC [9,10] and also correlates with poor clinical prognosis [11,12], finding effective approaches to target KRAS mutation is urgently needed.
Here, we review historical efforts to target mutant KRAS, with specific focus on approaches with promising preclinical/clinical data. We also propose some novel and provocative ideas, as well as an integrated approach for successful targeting.
Approaches of Targeting KRAS-mutant NSCLC
KRAS: its signaling and mutation in NSCLC
KRAS (Kirsten RAS) belongs to the RAS oncogene family, which also includes HRAS and NRAS [9]. Although all RAS activating mutations promote oncogenesis, we now know that mutated isoforms vary among different tissues. In addition, in different cancer types, each isoform can have distinctive codon mutations and amino acid substitutions [13]. In NSCLC, KRAS mutation is the most dominant among all RAS mutations and occurs in 15%–30% of NSCLC cases [9,10], the majority in adenocarcinoma of the lung [1]. Among all KRAS mutations, G12 codon has the highest frequency of mutation in NSCLC (∼92%), followed by Q61 (∼8%) [14]. Among all G12 mutations, the most predominant change is G12C (∼59%), followed by G12D (∼29%) and G12V (∼6%) [14]. Since RAS is a small GTPase, its activity normally cycles between a GTP-bound active state and a GDP-bound inactive state, a process that is facilitated partly by the stimulation of GTP hydrolysis through GAP (GTPase activating protein). However, when RAS protein is mutationally activated, GAP stimulation is impaired due to the formation of persistently GTP-bound RAS [15]. Therefore, although KRAS, like other RAS members, is essential for normal development [16], its activating mutation can be oncogenic due to the consistent activation of its downstream signaling pathways that are involved in deregulated cell proliferation, growth, invasion and metabolism, etc. [9,10].
We now know that RAS protein activates almost a dozen of downstream effectors, resulting in various biological aspects that are crucial for cancer initiation, maintenance, and progression [9,17]. The best validated effectors of RAS-driven oncogenesis include rapidly accelerated fibrosarcoma (RAF)/mitogen/extracellular signal-related kinase (MEK)/extracellular signal-regulated kinase (ERK), phosphatidylinositol-4,5-bisphosphate 3-kinase (PI3K)/AKT/mammalian target of rapamycin (mTOR) and Ral guanine nucleotide dissociation stimulator (RALGDS)/Ras-related protein (RAL) signaling pathways as well as Ras-related C3 botulinum toxin substrate 1 (Rac1) small GTPase, which are now considered as therapeutic targets with various developed inhibitors [14] (Table 1). With the difficulty of developing a direct RAS inhibitor, targeting those downstream effectors has proved to be a rational and reliable approach [8,14,17,29,30]. On the contrary, targeting RAS upstream activator alone such as the receptor tyrosine kinase EGFR has not been proved to be an efficient approach, mainly because the activating mutation of RAS has already resulted in constitutive activation of its downstream signaling pathways. In fact, KRAS mutation predicts decreased response to certain targeted therapy against its upstream activator (e.g. EGFR tyrosine kinase inhibitor) [6,7]. However, the value of combined therapy against both RAS upstream activator and downstream effector should not be neglected. For example, human epidermal growth factor receptor 3 (HER3), also known as Erb-B2 receptor tyrosine kinase 3 (ERBB3), was found transcriptionally induced by MEK inhibitors in KRAS-mutant NSCLC through MYC degradation; and dual inhibition of HER3 (the upstream activator) and MEK (the downstream effector) was consequently found synergistic [31]. Interestingly, a Phase I/IB trial of MEK162 (a MEK inhibitor) in combination with erlotinib in NSCLC harboring KRAS mutation is now ongoing (NCT01859026). In addition, since the proper maturation, prenylation, transportation, and membrane localization are all crucially important for the RAS protein to exert its function as a master driver of multiple downstream signaling pathways [32], approaches to target each of these steps have been investigated, with some more promising than others [32] (Fig. 1).
Table 1.
List of inhibitors targeting mutant RAS or RALs
| Inhibitor | Designated target | Stage of development | Efficacy in patients | Reference |
|---|---|---|---|---|
| SCH-54292 | RAS | Preclinical | NA | [18] |
| TLN-4601 | RAS | Phase II | Lack of efficacy; glioblastoma | [19] |
| Salirasib | RAS | Phase II | PFS: 4.7 months; pancreatic cancer | [20] |
| Deltarasin | RAS | Preclinical | NA | [21] |
| Lonafarnib | Farnesyl transferase | Phase II | OS: 19 months; PFS: 10 months; ovarian cancer | [22] |
| Tipifarnib | Farnesyl transferase | Phase II | OS: 8.3 months; PFS: 6.8 months; gliomas | [23] |
| BKM120 | PI3K | Phase II | OS: 11 months; PFS: 1.9 months; NSCLC | [24] |
| Afuresertib | AKT | Phase I | Lack of efficacy; multiple myeloma | [25] |
| Everolimus | mTOR | FDA approval | OS: recruiting; PFS: 7.8 months; breast cancer | [26] |
| Vemurafenib | RAF | FDA approval | OS: 6.2 months; PFS: 5.3 months; melanoma | [27] |
| Selumetinib | MEK | Phase II | OS: 9.4 months; PFS: 5.3 months; NSCLC | [8] |
| BVD-523 | ERK | Phase II | Recruiting; AML or MDS | [28] |
OS, overall survival; PFS, progression-free survival; AML, acute myelogenous leukemia; MDS, myelodysplastic syndrome.
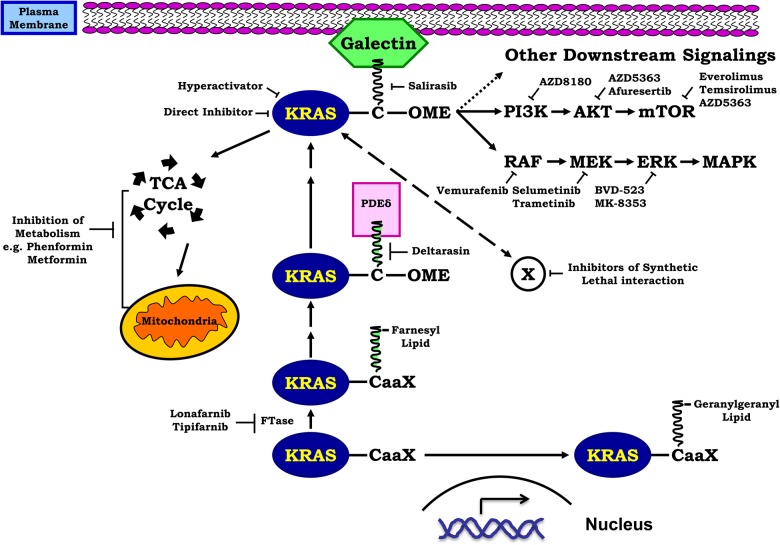
Figure 1.
Approaches of targeting KRAS-mutant NSCLC right after synthesis, inactive KRAS proteins undergo a series of post-translational modifications including prenylation, such as adding a lipid tail by farnesylation at a CaaX tetrapeptide motif on the C-terminus with the help of FTase C: cysteine; aa, two aliphatic residues; and X, a variable residue. Lonafarnib and tipifarnib are FTase inhibitors. However, the blockage of this step can be bypassed through another lipidation process geranylgeranylation via GGTase-I. The transportation of KRAS to the membrane relies on the interaction of its lipid tail with PDEδ, which can be blocked by deltarasin. Salirasib is a farnesyl mimic that competes with KRAS for its docking protein galectin on the plasma membrane. After proper localization, KRAS activates multiple downstream signaling pathways, which can be targeted individually or in combination. Also shown are the direct inhibitor and hyper-activator of KRAS, as well as inhibitors targeting cancer metabolism. Please refer to the text for detail description.
Past endeavors to target KRAS prenylation and membrane localization
Shortly after its synthesis in the cytoplasm, inactive RAS protein undergoes a series of post-translational modifications including prenylation, which, for example, adds a 15-carbon farnesyl lipid tail to its C-terminus with the help of farnesyltransferase (FTase) [32,33]. This process was shown to be important for proper trafficking of RAS protein to the plasma membrane [32] and for mediating its transforming activity [34]. Targeting FTase is therefore an appealing concept. However, specific FTase inhibitors such as lonafarnib and tipifarnib were largely ineffective in clinical trials, likely because RAS protein was found to be able to bypass this inhibition by geranylgeranylation, another process of prenylation [33]. A combination of FTase and geranylgeranyltransferase (GGTase) inhibitors was therefore proposed [33].
Another approach was tested by targeting the RAS docking site galectin using a farnesyl mimic such as salirasib, to compete with active RAS and lead to its degradation [32]. Unfortunately, despite promising preclinical data [35,36], early-phase clinical trials of salirasib monotherapy, especially in KRAS-mutant lung adenocarcinoma, were not successful [37]. Although there is lack of explanation in the literature, this observation could suggest that using farnesyl mimics alone is probably not sufficient to significantly degrade active KRAS via targeting its localization, and/or targeting KRAS membrane localization alone may not be sufficient to result in clinical significance. In fact, several studies have proposed combining salirasib with other strategies for KRAS-mutant cancers [38–40]. Nevertheless, the above-mentioned unsuccessful clinical trials, together with the tremendous challenge of designing a direct RAS inhibitor, make targeting its downstream signaling pathway the most clinically applicable approach. However, new discoveries are emerging.
A novel approach to target KRAS localization
Since the transportation of RAS protein requires PDEδ, Zimmermann et al. [21] discovered through stringent screening a small molecule inhibitor deltarasin that can specifically interrupt the interaction between the lipid tail of KRAS and the central pocket of PDEδ, therefore disrupting the proper trafficking and membrane localization of KRAS. Using pancreatic adenocarcinoma cell lines, they found that deltarasin redistributed KRAS into the cytoplasm instead of aggregating on the cell membrane, therefore attenuating its signaling [21]. Interestingly, deltarasin was found to be more potent in pancreatic cancer cells harboring KRAS mutation and dependent on KRAS signaling, although only a very limited number of cell lines were tested [21].
When we tested deltarasin in NSCLC cell lines, we did not find a statistical difference in IC50 values between KRAS-mutant (five cell lines) vs. wild-type (three cell lines) NSCLC cells [41]. Although limited samples could be the reason for these differences, the requirement for a deltarasin concentration in the range of 3–5 μM by virtually all cell lines suggests that it is necessary for a certain proportion of PDEδ to be occupied by deltarasin before a significant inhibitory effect can be achieved, e.g. 50% as measured by IC50. However, under certain deltarasin concentrations (e.g. 3 μM), more cell lines with mutant KRAS achieved a defined threshold of cell killing (e.g. 20%), suggesting that there could be potential mutant selectivity, although this effect is small [41]. Interestingly, compared with KRAS wild-type cell lines, the KRAS-mutant NSCLC cells in general exhibited much more pronounced apoptosis after being treated with deltarasin, with significant inhibition of both phospho-ERK and phospho-AKT levels [41]. Consistent with the findings of Zimmerman et al. [21], we did not observe any effects of deltarasin on the overall production of KRAS or total RAS [41]. Further studies are needed to explore the potential of deltarasin, for example, using sensitive in vivo models of lung cancer such as KRAS mutation-driven genetically engineered mouse (GEM) models, and a patient-derived xenograft (PDX) model implanted with human NSCLC tissue harboring different KRAS mutations. In addition, the experiment from salirasib suggested that it is probably not surprising to see limited effect of deltarasin alone in vivo and a combination approach is likely necessary to achieve significant clinical effect.
New insights in developing a direct KRAS inhibitor
The greatest hurdles to the successful development of a direct KRAS inhibitor are the very strong affinity between KRAS and GDP/GTP, which is measured in the picomolar range [9,42], and the abundance of both GDP and GTP in the cells [9]. Although there is no direct RAS inhibitor fits all types of KRAS mutation even in the laboratory setting, it seems that we may take advantage of each unique mutation for targeting. This approach has been proved promising in Kevin Shokat's laboratory [43]. These investigators took advantage of the cysteine residue of the KRAS G12C mutation and developed small molecule inhibitors that irreversibly bind to the mutant cysteine, which then subvert the preference of KRAS to favor GDP over GTP by conformational change and selectively inhibit the oncogenic signaling of KRAS G12C [43]. Their work clearly demonstrated that (1) a direct RAS inhibitor can be possibly designed, and (2) the idea of ‘one inhibitor fits all mutations’ may not be appropriate for KRAS mutations. Instead, mutation-specific inhibitors need to be pursued. Since KRAS G12C is one of the most common KRAS mutations in NSCLC, it will be interesting to see how effective these compounds will be after chemical optimization in future assessments in vivo.
MEK inhibition-based combination therapy
Targeting the downstream signaling pathways of oncogenic KRAS is probably the most widely explored approach. Studies have shown that the inhibition of RAF/MEK/ERK signaling seems to be the most important [28], and based on this, multiple RAF, MEK, and ERK inhibitors have been or are currently being tested in clinical trials (Table 1) [29]. Specifically in lung cancer, Janne et al. [8] conducted a Phase II clinical trial that demonstrated a MEK inhibitor selumetinib is effective in recurrent/metastatic NSCLC patients when used in combination with docetaxel. Based on this promising effect, selumetinib is now being evaluated in a Phase III clinical trial in NSCLC (SELECT-1 trial, NCT01933932).
Since KRAS activates dozens of downstream signaling [9], combination therapy is likely the key to its successful inhibition. In a recent large-scale screening study [44], among all inhibitors of downstream effectors, MEK inhibitor conferred the strongest cell killing in KRAS-mutant cancer cell lines [44]. Therefore, a MEK inhibition-based combination therapy is considered the most rational approach. However, since concomitant mutations such as those in liver kinase B1 (LKB1), phosphate and tensin homolog (PTEN), and PI3K catalytic subunit alpha (PIK3CA) may change the response to MEK inhibition [45–47], finding the best combination in certain genetic contexts is important.
Many MEK inhibition-based combination therapies have been tested. The most common have included combination with a PI3K, AKT, or mTOR inhibitor [48–51], largely because RAS activates PI3K, which in turn activates AKT and mTOR signaling [9,28,52]. In addition, up-regulation of PI3K/AKT/mTOR signaling through mutations such as activating mutations in PIK3CA or loss of function in PTEN, as well as up-regulation of HER3 and AKT may confer resistance to MEK inhibitors [31,45–47]. Despite modest enhancements in treatment effect, many of the combination therapies tested such as with PI3K were found to be toxic [53,54]. Discovering novel MEK inhibition-based combinations with minimal toxicity is therefore urgently needed.
Toward this goal, Corcoran et al. [55] performed a synthetic lethality-based screening and found that the BCL-XL inhibitor ABT-263 is synergistic with the MEK inhibitor selumetinib in KRAS-mutant NSCLC and colon cancers. Although selumetinib alone induced Bim, a pro-apoptotic protein, it was bound and inhibited by the anti-apoptotic protein BCL-XL [55]. By also giving the BCL-XL inhibitor ABT-263, the inhibitory effect on Bim was abrogated and robust apoptosis was observed [55]. Since ABT-263 was found to cause Grade III and IV thrombocytopenia in 41% of patients in a recent Phase II study of small cell lung cancer [56], we developed a novel BCL-XL inhibitor, which is more potent but induces much less toxicity [57] and is currently in testing. Similarly, using a kinome-centered synthetic lethality screen, Sun et al. [31] found that targeting HER3 may act synergistically with MEK inhibition in KRAS-mutant cancers including NSCLC.
During cancer progression, there is continuous rewiring of metabolism in the cancer cells to meet the requirement of both energy and metabolic intermediates. Thus, targeting cancer metabolism [58–60], especially in combination with MEK inhibition, could be an appealing strategy [46]. Using KRAS-mutant NSCLC cell lines both in vitro and in vivo, we have demonstrated for the first time that combining selumetinib and phenformin, a drug targeting mitochondrial complex 1, is potentially synergistic [46,61]. This is reminiscent of a similar study using BRAF inhibitor and phenformin in melanoma [62]. These observations support the concept that combination strategies acting on cancer metabolism with a targeted therapy, such as MEK inhibition, is a promising novel approach.
Taking advantage of synthetic lethality
Because of the phenotypic changes collectively known as ‘hallmarks of cancer’ [59,63] that are induced by oncogenic KRAS during cancer initiation, maintenance, and progression, targeting mechanisms underlying these changes (aka ‘non-oncogene addiction’) may provide synthetic killing [64]. The concept behind ‘synthetic lethality’ is to identify potential targets by targeting cells that will only be significantly damaged if they have KRAS mutation [65]. Therefore, this approach not only provides a strategy to target the traditionally considered ‘undruggable’ KRAS, but it also offers mutant selectivity to spare normal cells [65].
Using RNAi library screening, researchers have so far identified a wide range of genes involved in various cellular process that have synthetic lethal interactions with mutant KRAS. Cox et al. [14] provide a nice summary of KRAS synthetic lethal genes. However, two major challenges must be tackled before this method can be widely and effectively exploited: (1) synthetic interactions appear to be context dependent and tissue specific [66,67], therefore, an improved screening method is needed [14]; and (2) some identified targets such as the transcription factor GATA2 are themselves undruggable, therefore, it can be challenging in some situations to find appropriate targeting strategies [68].
Hyperactivating KRAS: a novel approach with potential
It was observed some time ago that the expression and activity levels of an oncogene are important for its function [69–71]. While oncogenic KRAS induces cell proliferation at an endogenous physiological level to promote tumor initiation and growth [70,71], overexpression or hyperactivation could paradoxically inhibit cell growth by inducing senescence [59,70], apoptotic-dependent, or apoptotic-independent cell death [72]. Therefore, the level of oncogenic KRAS is delicately controlled, which is not surprisingly given its crucial importance in regulating cancer cell metabolism, the number of multiple downstream signaling pathways that cross-talk with each other, and other hallmarks of cancer [59,63]. Since directly abrogating oncogenic KRAS activity is challenging, we adopted an alternative approach by hyperactivating KRAS. Using carefully designed KRAS activators, we observed in our pilot studies that these compounds have KRAS-mutant selectivity in NSCLC cell lines. Furthermore, in both xenograft and GEM models, we observed their inhibitory effect on tumor growth (Xu et al. unpublished data). Although potential oncogenic effect from systemic administration of KRAS activator is a reasonable concern for this approach, we did not observe this effect from either our in vitro or in vivo studies. Whether this is due to the duration of KRAS activation and/or the activated level achieved by KRAS activator certainly needs to be further explored. In addition, since concomitant genetic alterations can potentially modify the response of KRAS-mutant NSCLC to various treatments, we are currently investigating whether this will also occur with our KRAS activators.
Summary and Perspective
In this article, we have reviewed different approaches for targeting KRAS mutation in NSCLC as shown in Fig. 1, although many of these are still in the preclinical stage. Targeting mutant KRAS has been proved to be one of the most challenging tasks in cancer research; therefore, while exploring other novel approaches, the combination of different strategies appears most promising. For example, the combination of dual-targeting KRAS downstream signaling (e.g. MEK inhibition) and KRAS localization (e.g. deltarasin) may have potential to achieve better efficacy. While Ostrem et al. [43] demonstrated the possibility of designing a direct inhibitor for each individual KRAS mutation, our observation that phenformin enhances selumetinib sensitivity in KRAS-mutant NSCLC [46,61] also suggests the value of further exploration in the field of targeting cancer metabolism. Finally, the development of better screening approaches, the delivery of genes with synthetic–lethal interactions with KRAS, and fine-tuning oncogenic KRAS activity are all important steps to achieve our final goal of conquering KRAS-mutant NSCLC.
Funding
This work was supported by the grant from National Cancer Institute, National Institutes of Health (NIH; No. 1R01CA193828-01) and the NIH T32 training grant (No. 1T32CA160040-01A1, PI: DMS). J.Z. is an awardee of the T32 training grant.
Acknowledgements
We would like to thank Anthea Hammond for editing the manuscript.
References
- 1.Ettinger DS, Wood DE, Akerley W, Bazhenova LA, Borghaei H, Camidge DR, Cheney RT et al. Non-small cell lung cancer, version 1.2015. J Natl Compr Canc Netw 2014, 12: 1738–1761. [DOI] [PubMed] [Google Scholar]
- 2.Schiller JH, Harrington D, Belani CP, Langer C, Sandler A, Krook J, Zhu J et al. Comparison of four chemotherapy regimens for advanced non-small-cell lung cancer. N Engl J Med 2002, 346: 92–98. [DOI] [PubMed] [Google Scholar]
- 3.Lynch TJ, Bell DW, Sordella R, Gurubhagavatula S, Okimoto RA, Brannigan BW, Harris PL et al. Activating mutations in the epidermal growth factor receptor underlying responsiveness of non-small-cell lung cancer to gefitinib. N Engl J Med 2004, 350: 2129–2139. [DOI] [PubMed] [Google Scholar]
- 4.Soda M, Choi YL, Enomoto M, Takada S, Yamashita Y, Ishikawa S, Fujiwara S et al. Identification of the transforming EML4-ALK fusion gene in non-small-cell lung cancer. Nature 2007, 448: 561–566. [DOI] [PubMed] [Google Scholar]
- 5.Pao W, Girard N. New driver mutations in non-small-cell lung cancer. Lancet Oncol 2011, 12: 175–180. [DOI] [PubMed] [Google Scholar]
- 6.Eberhard DA, Johnson BE, Amler LC, Goddard AD, Heldens SL, Herbst RS, Ince WL et al. Mutations in the epidermal growth factor receptor and in KRAS are predictive and prognostic indicators in patients with non-small-cell lung cancer treated with chemotherapy alone and in combination with erlotinib. J Clin Oncol 2005, 23: 5900–5909. [DOI] [PubMed] [Google Scholar]
- 7.Roberts PJ, Stinchcombe TE. KRAS mutation: should we test for it, and does it matter? J Clin Oncol 2013, 31: 1112–1121. [DOI] [PubMed] [Google Scholar]
- 8.Jänne PA, Shaw AT, Pereira JR, Jeannin G, Vansteenkiste J, Barrios C, Franke FA et al. Selumetinib plus docetaxel for KRAS-mutant advanced non-small-cell lung cancer: a randomised, multicentre, placebo-controlled, phase 2 study. Lancet Oncol 2013, 14: 38–47. [DOI] [PubMed] [Google Scholar]
- 9.Malumbres M, Barbacid M. RAS oncogenes: the first 30 years. Nat Rev Cancer 2003, 3: 459–465. [DOI] [PubMed] [Google Scholar]
- 10.Rodenhuis S, van de Wetering ML, Mooi WJ, Evers SG, van Zandwijk N, Bos JL. Mutational activation of the K-ras oncogene. A possible pathogenetic factor in adenocarcinoma of the lung. N Engl J Med 1987, 317: 929–935. [DOI] [PubMed] [Google Scholar]
- 11.Slebos RJ, Kibbelaar RE, Dalesio O, Kooistra A, Stam J, Meijer CJ, Wagenaar SS et al. K-Ras oncogene activation as a prognostic marker in adenocarcinoma of the lung. N Engl J Med 1990, 323: 561–565. [DOI] [PubMed] [Google Scholar]
- 12.Mitsudomi T, Steinberg SM, Oie HK, Mulshine JL, Phelps R, Viallet J, Pass H et al. Ras gene mutations in non-small cell lung cancers are associated with shortened survival irrespective of treatment intent. Cancer Res 1991, 51: 4999–5002. [PubMed] [Google Scholar]
- 13.Prior IA, Lewis PD, Mattos C. A comprehensive survey of Ras mutations in cancer. Cancer Res 2012, 72: 2457–2467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cox AD, Fesik SW, Kimmelman AC, Luo J, Der CJ. Drugging the undruggable RAS: mission possible? Nat Rev Drug Discov 2014, 13: 828–851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bos JL, Rehmann H, Wittinghofer A. GEFs and GAPs: critical elements in the control of small G proteins. Cell 2007, 129: 865–877. [DOI] [PubMed] [Google Scholar]
- 16.Johnson L, Greenbaum D, Cichowski K, Mercer K, Murphy E, Schmitt E, Bronson RT et al. K-Ras is an essential gene in the mouse with partial functional overlap with N-ras. Genes Dev 1997, 11: 2468–2481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Rajalingam K, Schreck R, Rapp UR, Albert S. Ras oncogenes and their downstream targets. Biochim Biophys Acta 2007, 1773: 1177–1195. [DOI] [PubMed] [Google Scholar]
- 18.Ganguly AK, Pramanik BN, Huang EC, Liberles S, Heimark L, Liu YH, Tsarbopoulos A et al. Detection and structural characterization of Ras oncoprotein-inhibitors complexes by electrospray mass spectrometry. Bioorg Med Chem 1997, 5: 817–820. [DOI] [PubMed] [Google Scholar]
- 19.Mason WP, Belanger K, Nicholas G, Vallieres I, Mathieu D, Kavan P, Desjardins A et al. A phase II study of the Ras-MAPK signaling pathway inhibitor TLN-4601 in patients with glioblastoma at first progression. J Neurooncol 2012, 107: 343–349. [DOI] [PubMed] [Google Scholar]
- 20.Bustinza-Linares E, Kurzrock R, Tsimberidou AM. Salirasib in the treatment of pancreatic cancer. Future Oncol 2010, 6: 885–891. [DOI] [PubMed] [Google Scholar]
- 21.Zimmermann G, Papke B, Ismail S, Vartak N, Chandra A, Hoffmann M, Hahn SA et al. Small molecule inhibition of the KRAS-PDEδ interaction impairs oncogenic KRAS signalling. Nature 2013, 497: 638–642. [DOI] [PubMed] [Google Scholar]
- 22.Bachmann HS, Meier W, du Bois A, Kimmig R, Kuhlmann JD, Siffert W, Sehouli J et al. The FNTB promoter polymorphism rs11623866 as a potential predictive biomarker for lonafarnib treatment of ovarian cancer patients. Br J Clin Pharmacol 2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Haas-Kogan DA, Banerjee A, Poussaint TY, Kocak M, Prados MD, Geyer JR, Fouladi M et al. Phase II trial of tipifarnib and radiation in children with newly diagnosed diffuse intrinsic pontine gliomas. Neuro Oncol 2011, 13: 298–306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Vansteenkiste JF, Canon JL, Braud FD, Grossi F, De Pas T, Gray JE, Su WC et al. Safety and efficacy of buparlisib (BKM120) in patients with PI3K pathway-activated non-small cell lung cancer: results from the phase II BASALT-1 study. J Thorac Oncol 2015, 10: 1319–1327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Spencer A, Yoon SS, Harrison SJ, Morris SR, Smith DA, Brigandi RA, Gauvin J et al. The novel AKT inhibitor afuresertib shows favorable safety, pharmacokinetics, and clinical activity in multiple myeloma. Blood 2014, 124: 2190–2195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yardley DA, Noguchi S, Pritchard KI, Burris HA, Baselga J, Gnant M, Hortobagyi GN et al. Everolimus plus exemestane in postmenopausal patients with HR+ breast cancer: BOLERO-2 final progression-free survival analysis. Adv Ther 2013, 30: 870–884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Larkin J, Ascierto PA, Dreno B, Atkinson V, Liszkay G, Maio M, Mandala M et al. Combined vemurafenib and cobimetinib in BRAF-mutated melanoma. New Engl J Med 2014, 371: 1867–1876. [DOI] [PubMed] [Google Scholar]
- 28.Infante JR, Janku F, Tolcher AW, Patel MR, Sullivan RJ, Flaherty K, Carvajal RD et al. Dose escalation stage of a first-in-class phase I study of the novel oral ERK 1/2 kinase inhibitor BVD-523 (ulixertinib) in patients with advanced solid tumors. J Clin Oncol 2015, 33: 2506. [Google Scholar]
- 29.Samatar AA, Poulikakos PI. Targeting RAS-ERK signalling in cancer: promises and challenges. Nat Rev Drug Discov 2014, 13: 928–942. [DOI] [PubMed] [Google Scholar]
- 30.Montagut C, Settleman J. Targeting the RAF-MEK-ERK pathway in cancer therapy. Cancer Lett 2009, 283: 125–134. [DOI] [PubMed] [Google Scholar]
- 31.Sun C, Hobor S, Bertotti A, Zecchin D, Huang S, Galimi F, Cottino F et al. Intrinsic resistance to MEK inhibition in KRAS mutant lung and colon cancer through transcriptional induction of ERBB3. Cell Rep 2014, 7: 86–93. [DOI] [PubMed] [Google Scholar]
- 32.Baker NM, Der CJ. Cancer: drug for an ‘undruggable’ protein. Nature 2013, 497: 577–578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Berndt N, Hamilton AD, Sebti SM. Targeting protein prenylation for cancer therapy. Nat Rev Cancer 2011, 11: 775–791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Jackson JH, Cochrane CG, Bourne JR, Solski PA, Buss JE, Der CJ. Farnesol modification of Kirsten-Ras exon 4B protein is essential for transformation. Proc Natl Acad Sci USA 1990, 87: 3042–3046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Marom M, Haklai R, Ben-Baruch G, Marciano D, Egozi Y, Kloog Y. Selective inhibition of Ras-dependent cell growth by farnesylthiosalisylic acid. J Biol Chem 1995, 270: 22263–22270. [DOI] [PubMed] [Google Scholar]
- 36.Rotblat B, Ehrlich M, Haklai R, Kloog Y. The Ras inhibitor farnesylthiosalicylic acid (salirasib) disrupts the spatiotemporal localization of active Ras: a potential treatment for cancer. Methods Enzymol 2008, 439: 467–489. [DOI] [PubMed] [Google Scholar]
- 37.Riely GJ, Johnson ML, Medina C, Rizvi NA, Miller VA, Kris MG, Pietanza MC et al. A phase II trial of salirasib in patients with lung adenocarcinomas with KRAS mutations. J Thorac Oncol 2011, 6: 1435–1437. [DOI] [PubMed] [Google Scholar]
- 38.Wolfson E, Schmukler E, Schokoroy ST, Kloog Y, Pinkas-Kramarski R. Enhancing FTS (Salirasib) efficiency via combinatorial treatment. Biol Cell 2015, 107: 130–143. [DOI] [PubMed] [Google Scholar]
- 39.Schmukler E, Wolfson E, Haklai R, Elad-Sfadia G, Kloog Y, Pinkas-Kramarski R. Chloroquine synergizes with FTS to enhance cell growth inhibition and cell death. Oncotarget 2014, 5: 173–184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Charette N, De Saeger C, Horsmans Y, Leclercq I, Stärkel P. Salirasib sensitizes hepatocarcinoma cells to TRAIL-induced apoptosis through DR5 and survivin-dependent mechanisms. Cell Death Dis 2013, 4: e471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zhang J, Xu K, Shin D, Deng X. Targeting KRAS mutant non-small cell lung cancer with deltarasin—a small molecule inhibitor of KRAS-PDEδ interaction ed.: ASCO 2015: Chicago, IL, USA. [Google Scholar]
- 42.John J, Sohmen R, Feuerstein J, Linke R, Wittinghofer A, Goody RS. Kinetics of interaction of nucleotides with nucleotide-free H-ras p21. Biochemistry 1990, 29: 6058–6065. [DOI] [PubMed] [Google Scholar]
- 43.Ostrem JM, Peters U, Sos ML, Wells JA, Shokat KM. K-Ras(G12C) inhibitors allosterically control GTP affinity and effector interactions. Nature 2013, 503: 548–551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Garnett MJ, Edelman EJ, Heidorn SJ, Greenman CD, Dastur A, Lau KW, Greninger P et al. Systematic identification of genomic markers of drug sensitivity in cancer cells. Nature 2012, 483: 570–575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Chen Z, Cheng K, Walton Z, Wang Y, Ebi H, Shimamura T, Liu Y et al. A murine lung cancer co-clinical trial identifies genetic modifiers of therapeutic response. Nature 2012, 483: 613–617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zhang J, Wang D, Qian G, Liu F, Rahman M, Nannapaneni S, Wang X et al. Phenformin combines with selumetinib in targeting KRAS mutant non-small cell lung cancer cells with alternative LKB1 status. J Clin Oncol 2014, 32: Suppl. 2589. [Google Scholar]
- 47.Wee S, Jagani Z, Xiang KX, Loo A, Dorsch M, Yao YM, Sellers WR et al. PI3K pathway activation mediates resistance to MEK inhibitors in KRAS mutant cancers. Cancer Res 2009, 69: 4286–4293. [DOI] [PubMed] [Google Scholar]
- 48.Engelman JA, Chen L, Tan X, Crosby K, Guimaraes AR, Upadhyay R, Maira M et al. Effective use of PI3K and MEK inhibitors to treat mutant Kras G12D and PIK3CA H1047R murine lung cancers. Nat Med 2008, 14: 1351–1356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ku BM, Jho EH, Bae YH, Sun JM, Ahn JS, Park K, Ahn MJ. BYL719, a selective inhibitor of phosphoinositide 3-kinase α, enhances the effect of selumetinib (AZD6244, ARRY-142886) in KRAS-mutant non-small cell lung cancer. Invest New Drugs 2015, 33: 12–21. [DOI] [PubMed] [Google Scholar]
- 50.Qu Y, Wu X, Yin Y, Yang Y, Ma D, Li H. Antitumor activity of selective MEK1/2 inhibitor AZD6244 in combination with PI3K/mTOR inhibitor BEZ235 in gefitinib-resistant NSCLC xenograft models. J Exp Clin Cancer Res 2014, 33: 52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Holt SV, Logie A, Davies BR, Alferez D, Runswick S, Fenton S, Chresta CM et al. Enhanced apoptosis and tumor growth suppression elicited by combination of MEK (selumetinib) and mTOR kinase inhibitors (AZD8055). Cancer Res 2012, 72: 1804–1813. [DOI] [PubMed] [Google Scholar]
- 52.Young A, Lyons J, Miller AL, Phan VT, Alarcón IR, McCormick F. Ras signaling and therapies. Adv Cancer Res 2009, 102: 1–17. [DOI] [PubMed] [Google Scholar]
- 53.LoRusso P, Shapiro G, Pandya SS, Kwak EL, Jones C, Belvin M, Musib LC et al. A first-in-human phase IB study to evaluate the MEK inhibitor GDC-0973, combined with the pan-PI3K inhibitor GDC-0941, in patients with advanced solid tumors. J Clin Oncol 2012, 30: Suppl. 2566. [Google Scholar]
- 54.Speranza G, Kinders RJ, Khin S, Weil MK, Do KT, Horneffer Y, Juwara L et al. Pharmacodynamic biomarker-driven trial of MK-2206, and AKT inhibitor, with AZD6244 (selumeti-nib), a MEK inhibitor, in patients with advanced colorectal carcinoma. J Clin Oncol 2012, 30: Suppl. 3529. [Google Scholar]
- 55.Corcoran RB, Cheng KA, Hata AN, Faber AC, Ebi H, Coffee EM, Greninger P et al. Synthetic lethal interaction of combined BCL-XL and MEK inhibition promotes tumor regressions in KRAS mutant cancer models. Cancer Cell 2013, 23: 121–128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Rudin CM, Hann CL, Garon EB, de Oliveira MR, Bonomi PD, Camidge DR, Chu Q et al. Phase II study of single-agent navitoclax (ABT-263) and biomarker correlates in patients with relapsed small cell lung cancer. Clin Cancer Res 2012, 18: 3163–3169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Park D, Magis AT, Li R, Owonikoko TK, Sica GL, Sun SY, Ramalingam SS et al. Novel small-molecule inhibitors of Bcl-XL to treat lung cancer. Cancer Res 2013, 73: 5485–5496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Galluzzi L, Kepp O, Vander Heiden MG, Kroemer G. Metabolic targets for cancer therapy. Nat Rev Drug Discov 2013, 12: 829–846. [DOI] [PubMed] [Google Scholar]
- 59.Hanahan D, Weinberg RA. Hallmarks of cancer: the next generation. Cell 2011, 144: 646–674. [DOI] [PubMed] [Google Scholar]
- 60.Zhao Y, Butler EB, Tan M. Targeting cellular metabolism to improve cancer therapeutics. Cell Death Dis 2013, 4: e532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Zhang J, Wang D, Nannapaneni S, Liu F, Qian G, Rahman A, Wang X et al. Dual targeting MEK and cancer metabolism to tackle KRAS mutant non-small cell lung cancer with alternative LKB1 status. ed.: Annual GASCO meeting: Atlanta, GA: 2014. [Google Scholar]
- 62.Yuan P, Ito K, Perez-Lorenzo R, Del Guzzo C, Lee JH, Shen CH, Bosenberg MW et al. Phenformin enhances the therapeutic benefit of BRAF(V600E) inhibition in melanoma. Proc Natl Acad Sci USA 2013, 110: 18226–18231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Hanahan D, Weinberg RA. The hallmarks of cancer. Cell 2000, 100: 57–70. [DOI] [PubMed] [Google Scholar]
- 64.Luo J, Solimini NL, Elledge SJ. Principles of cancer therapy: oncogene and non-oncogene addiction. Cell 2009, 136: 823–837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Chan DA, Giaccia AJ. Harnessing synthetic lethal interactions in anticancer drug discovery. Nat Rev Drug Discov 2011, 10: 351–364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Kim HS, Mendiratta S, Kim J, Pecot CV, Larsen JE, Zubovych I, Seo BY et al. Systematic identification of molecular subtype-selective vulnerabilities in non-small-cell lung cancer. Cell 2013, 155: 552–566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Garraway LA, Sellers WR. Lineage dependency and lineage-survival oncogenes in human cancer. Nat Rev Cancer 2006, 6: 593–602. [DOI] [PubMed] [Google Scholar]
- 68.Kumar MS, Hancock DC, Molina-Arcas M, Steckel M, East P, Diefenbacher M, Armenteros-Monterroso E et al. The GATA2 transcriptional network is requisite for RAS oncogene-driven non-small cell lung cancer. Cell 2012, 149: 642–655. [DOI] [PubMed] [Google Scholar]
- 69.Serrano M, Lin AW, McCurrach ME, Beach D, Lowe SW. Oncogenic Ras provokes premature cell senescence associated with accumulation of p53 and p16INK4a. Cell 1997, 88: 593–602. [DOI] [PubMed] [Google Scholar]
- 70.Sarkisian CJ, Keister BA, Stairs DB, Boxer RB, Moody SE, Chodosh LA. Dose-dependent oncogene-induced senescence in vivo and its evasion during mammary tumorigenesis. Nat Cell Biol 2007, 9: 493–505. [DOI] [PubMed] [Google Scholar]
- 71.Tuveson DA, Shaw AT, Willis NA, Silver DP, Jackson EL, Chang S, Mercer KL et al. Endogenous oncogenic K-Ras(G12D) stimulates proliferation and widespread neoplastic and developmental defects. Cancer Cell 2004, 5: 375–387. [DOI] [PubMed] [Google Scholar]
- 72.Overmeyer JH, Maltese WA. Death pathways triggered by activated Ras in cancer cells. Front Biosci -Landmark 2011, 16: 1693–1713. [DOI] [PMC free article] [PubMed] [Google Scholar]