Vukovic et al. report that Hif-1α and Hif-2α are not required for leukemia stem cell maintenance and AML propagation, but they act synergistically to suppress leukemia development in mice. Furthermore, knockout of HIF-2α or pharmacological inhibition of the HIF pathway in human AML cells has no impact on their survival and proliferation under hypoxic conditions.
Abstract
Leukemogenesis occurs under hypoxic conditions within the bone marrow (BM). Knockdown of key mediators of cellular responses to hypoxia with shRNA, namely hypoxia-inducible factor-1α (HIF-1α) or HIF-2α, in human acute myeloid leukemia (AML) samples results in their apoptosis and inability to engraft, implicating HIF-1α or HIF-2α as therapeutic targets. However, genetic deletion of Hif-1α has no effect on mouse AML maintenance and may accelerate disease development. Here, we report the impact of conditional genetic deletion of Hif-2α or both Hif-1α and Hif-2α at different stages of leukemogenesis in mice. Deletion of Hif-2α accelerates development of leukemic stem cells (LSCs) and shortens AML latency initiated by Mll-AF9 and its downstream effectors Meis1 and Hoxa9. Notably, the accelerated initiation of AML caused by Hif-2α deletion is further potentiated by Hif-1α codeletion. However, established LSCs lacking Hif-2α or both Hif-1α and Hif-2α propagate AML with the same latency as wild-type LSCs. Furthermore, pharmacological inhibition of the HIF pathway or HIF-2α knockout using the lentiviral CRISPR-Cas9 system in human established leukemic cells with MLL-AF9 translocation have no impact on their functions. We therefore conclude that although Hif-1α and Hif-2α synergize to suppress the development of AML, they are not required for LSC maintenance.
Oxygen measurement in the BM indicated that normal and malignant hematopoiesis occur under hypoxic conditions (Spencer et al., 2014). Hif-1 and Hif-2 are key mediators of cellular responses to hypoxia and regulate gene expression to facilitate adaptation to low oxygen tension (Semenza, 2014). Oxygen-regulated α-subunits of Hif-1 and Hif-2, namely Hif-1α and Hif-2α, are paralogs that have common and also distinct functions during responses to hypoxia. Several studies investigated the role of Hif-1α (Wang et al., 2011; Velasco-Hernandez et al., 2014) and Hif-2α (Rouault-Pierre et al., 2013) in acute myeloid leukemia (AML; Gezer et al., 2014; Vyas, 2014). HIF-1α or HIF-2α knockdown in AML patient samples compromised their ability to reconstitute AML upon transplantation into recipient mice (Wang et al., 2011; Rouault-Pierre et al., 2013). Although the subtype and molecular classification of AML samples used in these studies were not specified, the authors implied that HIF-1 and HIF-2 are independently required for the maintenance of AML leukemic stem cells (LSCs), suggesting that HIF-1 and HIF-2 are potential therapeutic targets for AML (Wang et al., 2011; Rouault-Pierre et al., 2013).
Considering the caveats of shRNA-mediated gene knockdown approaches, a recent study used a conditional Hif-1α knockout and reported that, surprisingly, conditional Hif-1α deletion does not compromise the development and maintenance of mouse LSCs generated by the MLL-ENL fusion, its downstream effectors Meis1 and Hoxa9, and Aml1-Eto9a (Velasco-Hernandez et al., 2014). In fact, loss of Hif-1α accelerated the development of Meis1/Hoxa9-induced AML or enhanced propagation of disease induced by Aml1-Eto9a (Velasco-Hernandez et al., 2014). This study concluded that Hif-1α can suppress LSC development or propagation and is dispensable for AML LSC maintenance, sparking a debate over the therapeutic benefit of targeting HIF-1 function (Vyas, 2014).
To date, the impact of conditional deletion of Hif-2α or loss of both Hif-1α and Hif-2α on leukemic transformation has not been examined. Therefore, in this study, we set out to investigate the requirement for Hif-2α or both Hif-1α and Hif-2α in the development and maintenance of AML LSCs.
RESULTS AND DISCUSSION
Hif-2α suppresses the development of LSCs but has no impact on AML propagation in a Meis1/Hoxa9-induced murine AML
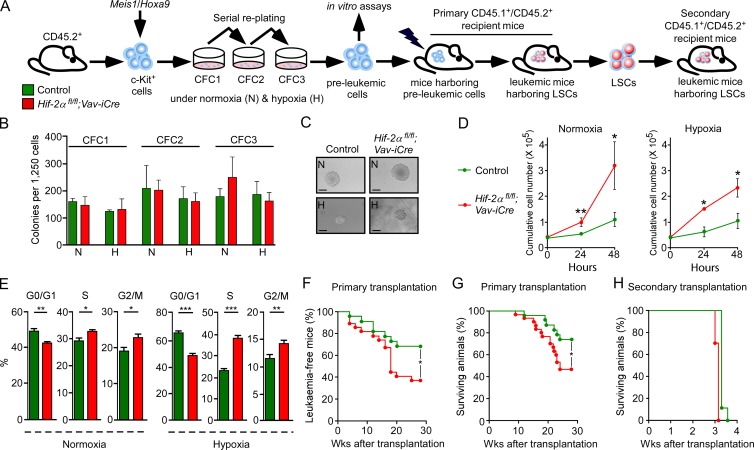
To investigate the requirement for Hif-2α in leukemogenesis, we used a well-characterized mouse model of AML in which the development and maintenance of LSCs is driven by coexpression of Meis1 and Hoxa9 oncogenes (Lessard and Sauvageau, 2003; Wang et al., 2010; Lehnertz et al., 2014). These two oncogenes are frequently overexpressed in several human AML subtypes (Lawrence et al., 1999; Drabkin et al., 2002) and their overexpression in mouse hematopoietic stem and progenitor cells (HSPCs) promotes self-renewal and perturbs their differentiation, resulting in the generation of self-renewing LSCs (Kroon et al., 1998). In the Meis1/Hoxa9 model used in this study, the BM c-Kit+ HSPC population, which contains all of the cellular targets for leukemic transformation, is transduced with retroviruses expressing Meis1 and Hoxa9. After three rounds of replating, it gives rise to the population of preleukemic cells that, upon transplantation to primary recipients, generates LSCs, thereby causing AML with a long latency (Fig. 1 A). LSCs are defined by their capacity to propagate AML with short latency in secondary recipients (Fig. 1 A). To establish the functional significance of Hif-2α in leukemogenesis, we used mice with Vav-iCre–mediated hematopoiesis-specific deletion of Hif-2α (Hif-2αfl/fl;Vav-iCre mice), which we previously demonstrated to have normal numbers of hematopoietic stem cells (HSCs) and progenitor cells and display no hematopoietic defects (Guitart et al., 2013). We transduced c-Kit+ cells from Hif-2αfl/fl;Vav-iCre and control (Hif-2αfl/fl without Vav-iCre) mice with Meis1/Hoxa9 retroviruses and serially replated them under normoxic and hypoxic conditions. Hif-2α–deficient and control cells displayed similar replating capacity and generated comparable numbers of compact colonies at each passage (Fig. 1, B and C). Operetta high-content automated microscope analyses revealed that the colonies of both genotypes had comparable sizes (unpublished data). Flow cytometry analyses of the CFC3 colonies using c-Kit, Mac-1, and Gr-1 markers revealed no differences in immunophenotypic composition of colonies of the two genotypes (unpublished data).
Figure 1.
Hif-2α deletion accelerates LSC development but has no impact on LSC maintenance. (A) CD45.2+c-Kit+ cells from Hif-2αfl/fl;Vav-iCre and control (Vav-iCre-negative) mice were cotransduced with Meis1 and Hoxa9 retroviruses and serially replated under normoxic (N) and hypoxic (H; 1% O2) conditions and were subjected to in vitro assays. 100,000 c-Kit+ preleukemic cells generated under normoxia were transplanted into lethally irradiated CD45.1+/CD45.2+ recipients. CD45.2+c-Kit+ LSCs from primary recipients were transplanted to secondary recipients. (B) Colony-forming cell (CFC) assay counts at each replating. Data are mean ± SEM (n = 3–5). (C) Representative colonies generated at CFC3. Bars, 70 µm. (D) Proliferation curves of preleukemic cells in liquid cultures under normoxia and hypoxia. Data are mean ± SEM (n = 3). (E) The cell cycle analysis of preleukemic cells under normoxia and hypoxia. Data are mean ± SEM (n = 3–4). (F) The percentage of leukemia-free mice defined as mice that have <1% of leukemic cells in the peripheral blood (n = 8–10 recipients per biological replicate [n = 3]). (G) Kaplan-Meier survival curve of recipients transplanted with 100,000 c-Kit+ preleukemic cells (n = 8–10 recipients per biological replicate [n = 3]). (H) Secondary transplantation. 10,000 CD45.2+c-Kit+ LSCs sorted from primary recipients were transplanted into secondary recipients. Data are mean ± SEM (n = 8–10 recipients per biological replicate [n = 2]). At least two independent experiments were performed for all analyses. Statistical analysis: Mann-Whitney test. *, P < 0.05; **, P < 0.005; ***, P < 0.001.
Proliferation assays and cell cycle analyses on Hif-2α–deficient preleukemic cells (i.e., cells obtained after the third round of replating) cultured under hypoxia and normoxia revealed that Hif-2α–deficient cells had increased proliferative capacity (Fig. 1 D) and that a larger proportion of Hif-2α–deficient cells were in S and G2/M phases of cell cycle compared with control cells (Fig. 1 E). Thus, Hif-2α is not required for the generation of preleukemic cells but restricts their proliferative capacity.
To investigate the impact of Hif-2α deletion on LSC generation, we transplanted Hif-2α–deficient and control preleukemic cells generated under normoxic conditions into primary recipient mice (Fig. 1 A). Blood sampling revealed that a significantly smaller proportion of recipients of Hif-2α–deficient cells remained leukemia-free compared with recipients of control preleukemic cells (Fig. 1 F). Consequently, recipients of Hif-2α–deficient cells had decreased survival compared with recipients of control preleukemic cells (Fig. 1 G). Analyses of BM from leukemic primary recipient mice revealed that Meis1/Hoxa9-transduced control and Hif-2αfl/fl;Vav-iCre preleukemic cells generate leukemia with the same immunophenotypic characteristics (unpublished data). In conclusion, Hif-2α suppresses the establishment of LSCs and delays the onset of Meis1/Hoxa9-induced AML.
To investigate the ability of Hif-2α–deficient LSCs to propagate leukemia, we sorted the CD45.2+ donor-derived c-Kit+ cell population (that contains LSCs; Somervaille et al., 2009) from the two cohorts of leukemic primary recipients and transplanted equal numbers of c-Kit+ cells into secondary recipients. LSCs of both genotypes generated aggressive AML with similar latency (Fig. 1 H). Therefore, Hif-2α is not required for the maintenance of LSCs and their ability to propagate Meis1/Hoxa9–induced AML.
Hif-2α deletion accelerates LSC development but does not affect LSC maintenance in a mouse model of Mll-AF9–driven AML
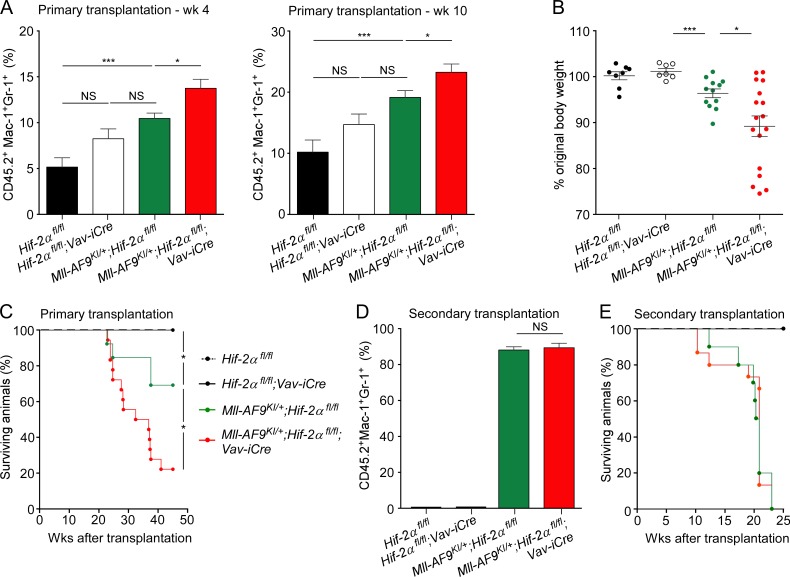
We next set out to confirm the ability of Hif-2α to suppress the development of AML in the Mll-AF9 knock-in (Mll-AF9KI/+) mouse model of human AML (Dobson et al., 1999; Chen et al., 2008; Kumar et al., 2009). This model is thought to closely mimic the human disease, as each cell contains a single copy of the Mll-AF9 fusion oncogene, physiologically expressed from the endogenous Mll promoter. In this model, Mll-AF9KI/+ Lin−Sca-1+c-Kit+ (LSK) stem and primitive progenitor cells are sufficient to initiate leukemia with long latency upon transplantation. We generated Mll-AF9KI/+;Hif-2αfl/fl;Vav-iCre mice and controls, i.e., Mll-AF9KI/+;Hif-2αfl/fl (without Vav-iCre), Hif-2αfl/fl;Vav-iCre, and Hif-2αfl/fl mice and transplanted LSK cells from these mice into primary recipients (together with 200,000 WT CD45.1+ unfractionated BM cells). We found that recipients of Mll-AF9KI/+;Hif-2αfl/fl;Vav-iCre and Mll-AF9KI/+;Hif-2αfl/fl LSK cells had similar percentages of CD45.2+ donor-derived cells indicating that the overall reconstitution capacity of LSK cells of both genotypes was not different. Furthermore, LSK cells of both Mll-AF9KI/+;Hif-2αfl/fl;Vav-iCre and Mll-AF9KI/+;Hif-2αfl/fl genotypes had multilineage differentiation potential that was sustained for at least 19 wk after transplantation (not depicted). However, recipients of Mll-AF9KI/+;Hif-2αfl/fl;Vav-iCre LSK cells had increased percentages of donor-derived myeloid cells in the peripheral blood compared with recipients of Mll-AF9KI/+;Hif-2αfl/fl LSK cells (Fig. 2 A). Consequently, recipients of Mll-AF9KI/+;Hif-2αfl/fl;Vav-iCre LSK cells became cachectic and succumbed to AML (confirmed by flow cytometric analyses of the BM; not depicted) faster compared with recipients of Mll-AF9KI/+;Hif-2αfl/fl LSK cells (Fig. 2, B and C). This was in contrast to recipients of Hif-2αfl/fl;Vav-iCre and Hif-2αfl/fl LSK cells which, as expected (Guitart et al., 2013), did not develop leukemia (Fig. 2, B and C). To investigate whether Hif-2α deletion compromises LSC maintenance in the Mll-AF9KI/+ model, we sorted the CD45.2+ donor-derived Lin−c-Kit+Sca-1− (LK) cells from primary recipients and transplanted them into secondary recipients (together with 200,000 WT CD45.1+ unfractionated BM cells). LK cells of both Mll-AF9KI/+;Hif-2αfl/fl and Mll-AF9KI/+;Hif-2αfl/fl;Vav-iCre genotypes equally efficiently engrafted (Fig. 2 D), and propagated full-blown disease (Fig. 2 E) in secondary recipients. We also tested the LSC frequency in primary recipients of Mll-AF9KI/+;Hif-2αfl/fl or Mll-AF9KI/+;Hif-2αfl/fl;Vav-iCre LSK cells that developed leukemia by performing a secondary transplantation of donor-derived LK cells sorted from primary leukemic recipients using an extreme limiting dilution assay (Hu and Smyth, 2009) and found no difference in LSC frequency between the genotypes (Table 1 and Table 2). We conclude that Hif-2α delays the development of LSCs in the Mll-AF9 model of AML, but once LSCs are established Hif-2α is dispensable for their maintenance.
Figure 2.
Hif-2α deletion in Mll-AF9KI/+ mice accelerates AML development but has no impact on its propagation. 2,000 CD45.2+Lin−Sca-1+c-Kit+ (LSK) cells were sorted from 18-wk-old Hif-2αfl/fl (Vav-iCre-negative), Hif-2αfl/fl;Vav-iCre, Mll-AF9KI/+;Hif-2αfl/fl, and Mll-AF9KI/+;Hif-2αfl/fl;Vav-iCre mice and transplanted into lethally irradiated CD45.1+/CD45.2+ recipients (together with 200,000 CD45.1+ BM cells per recipient). (A) Percentage of CD45.2+Mac-1+Gr-1+ cells in peripheral blood at week 4 and 10. Data are mean ± SEM (n = 3–8 recipients per biological replicate [n = 2]). (B) Ratio of weight of primary recipients at week 17 compared with week 4. (C) Kaplan-Meier survival curve of primary recipients transplanted with 2,000 LSK cells of the indicated genotypes. Data are mean ± SEM (n = 3–8 recipients per biological replicate [n = 2]). (D) 5,000 CD45.2+Lin−c-Kit+Sca-1− (LK) cells were sorted from primary recipients and transplanted into secondary recipients (together with 200,000 CD45.1+ BM cells per recipient). Percentage of CD45.2+Mac-1+Gr-1+ cells in peripheral blood at week 9. Data are mean ± SEM (n = 5–6 recipients per biological replicate [n = 2]). (E) Kaplan-Meier survival curve of secondary recipient mice transplanted with 5,000 LK cells of the genotypes indicated in Fig. 2 C (n = 5–6 recipients per biological replicate [n = 2]). At least two independent experiments were performed for all analyses. Statistical analysis: Mann-Whitney test. *, P < 0.05; ***, P < 0.001.
Table 1. Extreme limiting dilution assay (ELDA).
| Cell dose | Mll-AF9KI/+;Hif-2αfl/fl | Mll-AF9KI/+;Hif-2αfl/fl;Vav-iCre |
|---|---|---|
| 50 | 4/8 | 6/12 |
| 500 | 8/8 | 10/10 |
| 5,000 | 10/10 | 15/15 |
Secondary recipients were transplanted with 50, 500, or 5,000 LK cells of the indicated genotypes sorted from primary recipients. Number of mice (out of total injected) that showed engraftment of CD45.2+ cells in the peripheral blood at week 9 (n = 6–15 per group) are listed. All calculations and statistical analyses were done using the ELDA algorithm (Hu and Smyth, 2009). Three independent experiments were performed.
Table 2. ELDA confidence intervals.
| LSC frequency | Confidence intervals (95%) | |
|---|---|---|
| Mll-AF9KI/+;Hif-2αfl/fl | Mll-AF9KI/+;Hif-2αfl/fl;Vav-iCre | |
| Lower | 1/185 | 1/157 |
| Estimate | 1/71.7 | 1/71.9 |
| Upper | 1/27.9 | 1/33.1 |
Secondary recipients were transplanted with 50, 500, or 5,000 LK cells of the indicated genotypes sorted from primary recipients. Lower, estimate, and upper LSC frequency represented as 1/(indicated number of cells) at a 95% confidence interval are shown. All calculations and statistical analyses were done using the ELDA algorithm (Hu and Smyth, 2009). Three independent experiments were performed.
CRISPR-Cas9–mediated HIF-2α ablation has no impact on human AML cells
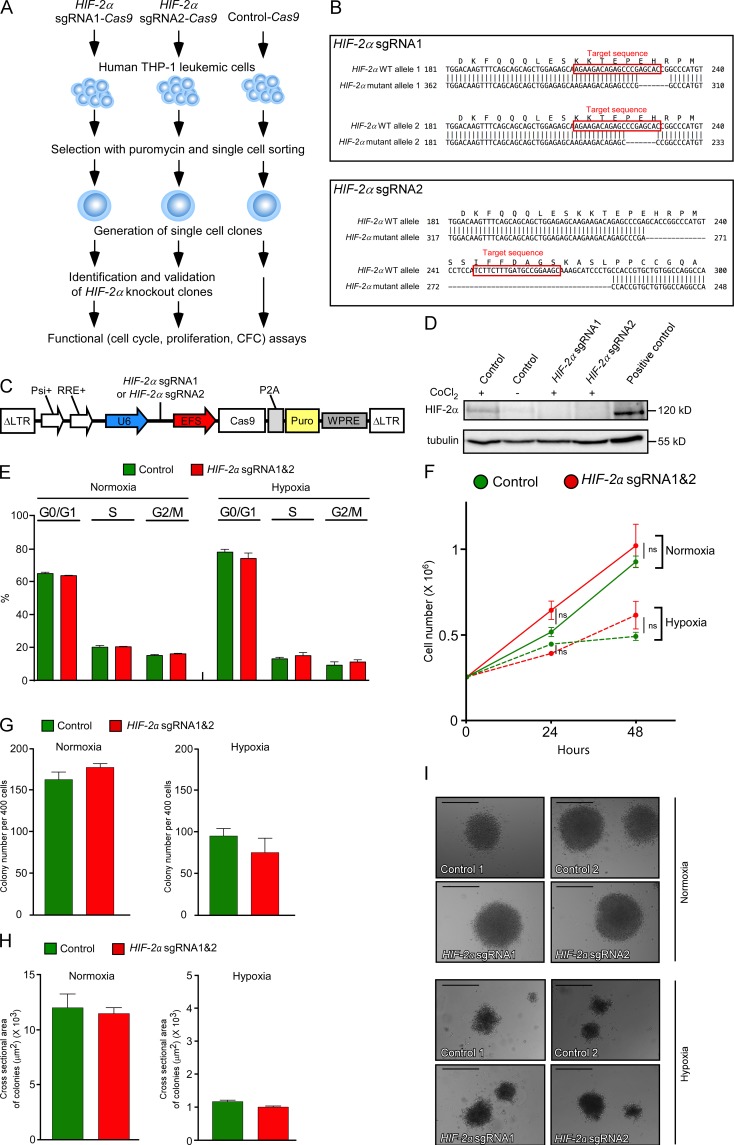
Given our surprising findings, which indicated that Hif-2α is not required for mouse AML propagation, we next determined the impact of HIF-2α ablation in human leukemic cells using the CRISPR-Cas9 genome editing approach. We transduced human AML (M5) THP-1 cells harboring MLL-AF9 translocation with lentiviruses expressing two independent single guide RNAs (sgRNAs) targeting exon 12 of the HIF-2α (EPAS1) gene (Fig. 3, A and B) and coexpressing a mammalian codon-optimized Cas9 nuclease (Sanjana et al., 2014). These lentiviruses will be referred to as HIF-2α sgRNA1-Cas9 and HIF-2α sgRNA2-Cas9. We isolated two independent clones, one harboring HIF-2α sgRNA1-Cas9 lentivirus and the second harboring sgRNA2-Cas9 lentivirus (Fig. 3 A). Both clones had frameshifting mutations (Fig. 3 B), resulting in the loss of HIF-2α protein (Fig. 3 D). Two clones of THP-1 cells transduced with Cas9 lentivirus lacking sgRNA were used as control. Our analyses performed under normoxic and hypoxic conditions revealed that the loss of HIF-2α expression had no impact on the cell cycle (Fig. 3 E), proliferation rate (Fig. 3 F), and colony formation capacity (Fig. 3, G–I) of THP-1 cells under normoxic and hypoxic conditions. Thus, consistent with our findings in the mouse models of AML, HIF-2α is not required for propagation of human leukemic cells under normoxic and hypoxic conditions.
Figure 3.
CRISPR-Cas9–mediated HIF-2α gene targeting in human THP-1 leukemic cells has no impact on their survival, proliferation, or colony formation. (A) Experimental design. Human THP-1 leukemic cells were transduced with bi-cistronic lentiviruses expressing one of two independent sgRNAs targeting exon 12 of human HIF-2α (these sgRNAs are referred to as HIF-2α sgRNA1 and HIF-2α sgRNA2) and a mammalian codon-optimized Cas9. Control-Cas9 constructs lack the 20-nt sequence within the sgRNA necessary for HIF-2α targeting. After single-cell cloning, the clones with the correctly targeted HIF-2α gene (and two control clones) were subjected to functional assays. (B) Design of sgRNAs to knock out human HIF-2α. Two independent sgRNAs (i.e., HIF-2α gRNA1 and HIF-2α gRNA2) were designed to target exon 12 of human HIF-2α gene. Target sequences are indicated in red. Top sequences indicate the WT allele of HIF-2α. Bottom sequences indicate the deletions in the HIF-2α gene, all of which resulted in frameshift mutations. (C) Schematic representation of a bi-cistronic lentiviral vector allowing for the expression of HIF-2α sgRNA1 or HIF-2α sgRNA2 from a U6 promoter (U6) and Cas9 from an EF1α short promoter (EFS). Psi+, Psi packaging signal; RRE, Rev response element; P2A, Picorna virus–derived 2A self-cleaving peptide; Puro, puromycin selection cassette; WPRE, Woodchuck hepatitis virus posttranscriptional regulatory element. (D) THP-1 cells transduced with Control-Cas9, HIF-2α sgRNA1-Cas9, and HIF-2α sgRNA2-Cas9 lentiviruses were incubated with CoCl2 to enhance HIF-2α stability. Western blot analysis was performed to determine the levels of HIF-2α. Hypoxic MCF-7 breast cancer cell line was used as a positive control for HIF-2α. (E) The cell cycle analysis of THP-1 cells transduced with Control-Cas9, HIF-2α sgRNA1-Cas9, and HIF-2α sgRNA2-Cas9 lentiviruses incubated under normoxic and hypoxic (1% O2) conditions. Data are mean ± SEM. (F) Proliferation assays under normoxia and hypoxia. Data are mean ± SEM, (G) Colony numbers automatically counted at day 11 after plating. Cells were plated in methylcellulose at a density of 400 cells per well in a 96-well plate and grown under either normoxic or hypoxic conditions. Plates were imaged at 37°C and 5% CO2 on an Operetta automated microscope on day 11. For each well, six fields of view were acquired at each of the five focal planes separated by 150 µm, ensuring no double counting of colonies. Data are mean ± SEM. (H) Colony size was calculated in an unbiased way by the Operetta microscope software at day 11 after plating using the Harmony texture-based image analysis software. Data are mean ± SEM. (I) Morphology of representative colonies cultured under normoxia and hypoxia and photographed at day 11 after plating. Bars, 120 µm. For A–I, the data with two independent control THP-1 cell clones and two independent HIF-2α–deficient clones (expressing HIF-2α sgRNA1-Cas9 and HIF-2α sgRNA2-Cas9, respectively) are shown. Technical triplicates for each clone were used. At least two independent experiments were performed for all analyses. Statistical analysis: Mann-Whitney test.
Hif-1α and Hif-2α synergize to suppress the development of AML but are dispensable for disease propagation
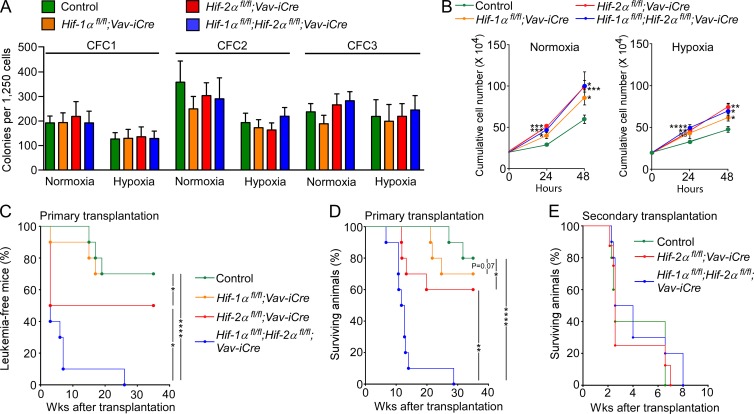
The tumor suppressor function of Hif-2α in AML development, taken together with the recent study indicating that Hif-1α can also suppress leukemic transformation (Velasco-Hernandez et al., 2014), suggested that Hif-1α and Hif-2α may collaborate to suppress leukemogenesis. To test this, we transduced BM-derived c-Kit+ cells from Hif-1αfl/fl;Vav-iCre, Hif-2αfl/fl;Vav-iCre, Hif-1αfl/fl;Hif-2αfl/fl;Vav-iCre, and control mice with Meis1/Hoxa9 retroviruses. Although cells of each genotype cultured under normoxia and hypoxia self-renewed equally in serial replating assays (Fig. 4 A), preleukemic cells lacking Hif-1α, Hif-2α, or both had increased proliferative capacity compared with control cells (Fig. 4 B). After transplantation, the cell cycle profile of Hif-1α–deficient cells was not different from that of control cells but a slightly larger proportion of Hif-2α–deficient and Hif-1α/Hif-2α–deficient cells were in S/G2/M phases of the cell cycle compared with Hif-1α–deficient cells (notably there was no difference in the cell cycle profile between Hif-2α–deficient and Hif-1α/Hif-2α–deficient cells; unpublished data). Remarkably, upon transplantation, preleukemic cells lacking both Hif-1α and Hif-2α generated dramatically accelerated AML compared with those lacking either Hif-1α or Hif-2α (Fig. 4, C and D). Therefore, Hif-1α and Hif-2α synergize to suppress the development of LSCs. Finally, control, Hif-2α- and Hif-1α/Hif-2α–deficient BM c-Kit+ cells sorted from primary leukemic recipients equally efficiently generated aggressive leukemia in secondary recipients (Fig. 4 E). Thus, despite the hypoxic nature of the BM, at least in AML involving Meis1/Hoxa9-mediated transformation the disease is propagated through Hif-independent mechanisms.
Figure 4.
Hif-1α and Hif-2α synergize to suppress LSC development in a murine Meis1/Hoxa9-mediated AML. (A) CFC assay counts at each replating. Data are mean ± SEM (n = 5). (B) Proliferation curves of preleukemic cells in liquid cultures under normoxia and hypoxia. Data are mean ± SEM (n = 3). (C) The percentage of leukemia-free recipient mice transplanted with control (Vav-iCre-negative), Hif-1α–deficient, Hif-2α–deficient, and Hif-1α/Hif-2α–deficient preleukemic cells (n = 4–5 recipients per biological replicate [n = 2]). (D) Kaplan-Meier survival curve of recipients transplanted with 100,000 c-Kit+ preleukemic cells of the indicated genotypes. n = 4–5 recipients per biological replicate (n = 2). (E) Secondary transplantation. 10,000 CD45.2+c-Kit+ LSCs sorted from primary recipients of control, Hif-2α–deficient and Hif-1α/Hif-2α–deficient preleukemic cells were retransplanted into secondary recipients. Data are mean ± SEM (n = 8–10 recipients). At least two independent experiments were performed for all analyses. Statistical analysis: Mann-Whitney test. *, P < 0.05; **, P < 0.005; ***, P < 0.001; ****, P < 0.0001.
Pharmacological inhibition of the HIF pathway does not affect human AML cell survival and proliferation
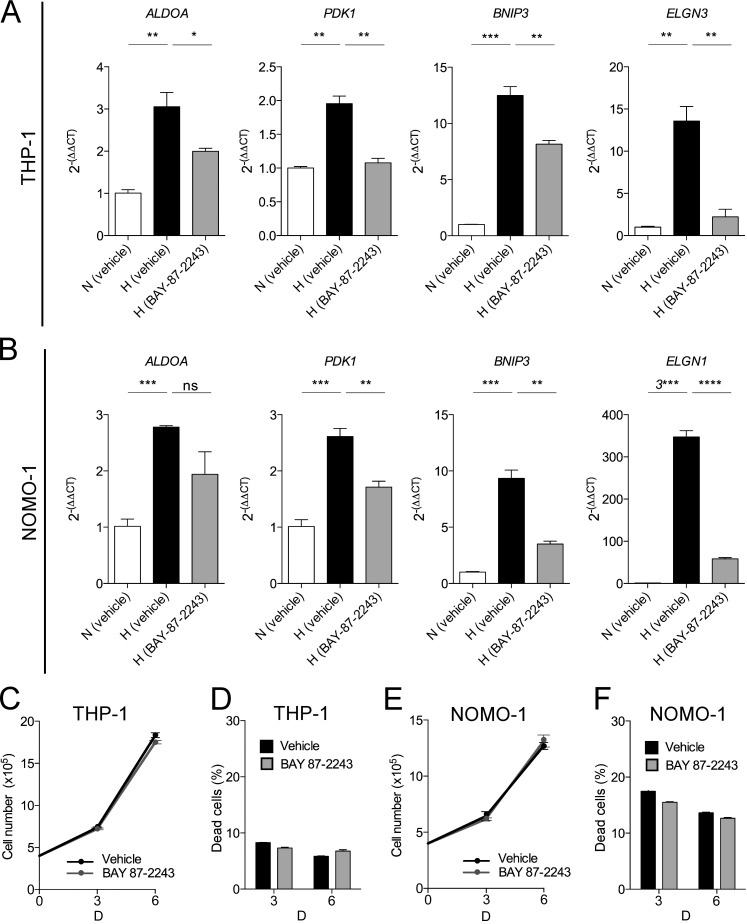
To test the impact of inhibiting the HIF pathway in human AML cells, we incubated (M5) THP-1 cells and NOMO-1 cells, both harboring the MLL-AF9 fusion, with BAY 87–2243, which inhibits HIF-1α and HIF-2α protein accumulation under hypoxia and decreases HIF target gene expression in tumor cells (Ellinghaus et al., 2013; Helbig et al., 2014; Chang et al., 2015). As expected, BAY 87–2243 treatment inhibited the expression of HIF target genes (ALDOA, PDK1, BNIP3, and EGLN3) in both THP-1 (Fig. 5 A) and NOMO-1 (Fig. 5 B) cells under hypoxia. Under these conditions, BAY 87–2243 had no effect on the proliferative capacity and survival of THP-1 (Fig. 5, C and D) and NOMO-1 (Fig. 5, E and F) cells. Thus, inhibition of the HIF pathway in two independent, established human AML cells with MLL-AF9 translocation has no impact on their survival and growth.
Figure 5.
Pharmacological targeting of the HIF pathways in human leukemic cells has no impact on their survival under hypoxic conditions. THP-1 and NOMO-1 cells were incubated for 6 d under hypoxic conditions (1% O2) with BAY 87–2243 (10nM) or vehicle control (0.001% EtOH [vol/vol]). Expression levels of HIF target genes ALDOA, PDK1, BNIP3, and EGLN3 in (A) THP-1 cells and in (B) NOMO-1 cells. Results are expressed as fold change (2−(ΔΔCT)) over vehicle control cultured in normoxia. HPRT was used as the reference gene and data are represented as mean ± SEM (n = 3 technical replicates per cell line). *, P < 0.05; **, P < 0.005; ***, P < 0.001; ****, P < 0.0001. THP-1 (C and D) and NOMO-1 (E and F) cells were plated at a density of 400,000 cells/ml and incubated with 10 nM BAY 87–2243 or vehicle control (0.001% EtOH [vol/vol]) under hypoxic conditions (1% O2). At day 3 and 6, the cells were incubated with 7-AAD and their proliferative capacity (number of 7-AAD− cells) and percentage of dead (7-AAD+) cells was measured by flow cytometry using the BD Accuri C6 Flow Cytometer. (C) Proliferation and (D) percentage of dead THP-1 cells treated with BAY 87–2243 or vehicle control under hypoxic conditions is shown. (E) Proliferation and (F) percentage of dead NOMO-1 cells treated with BAY 87–2243 or vehicle control under hypoxic conditions is shown. For A–F, data are mean ± SEM (n = 3 technical replicates). At least two independent experiments were performed for all analyses. Statistical analysis: Mann-Whitney test.
Hif-1α and Hif-2α deletion promotes a gene expression signature that facilitates survival and proliferation of preleukemic cells
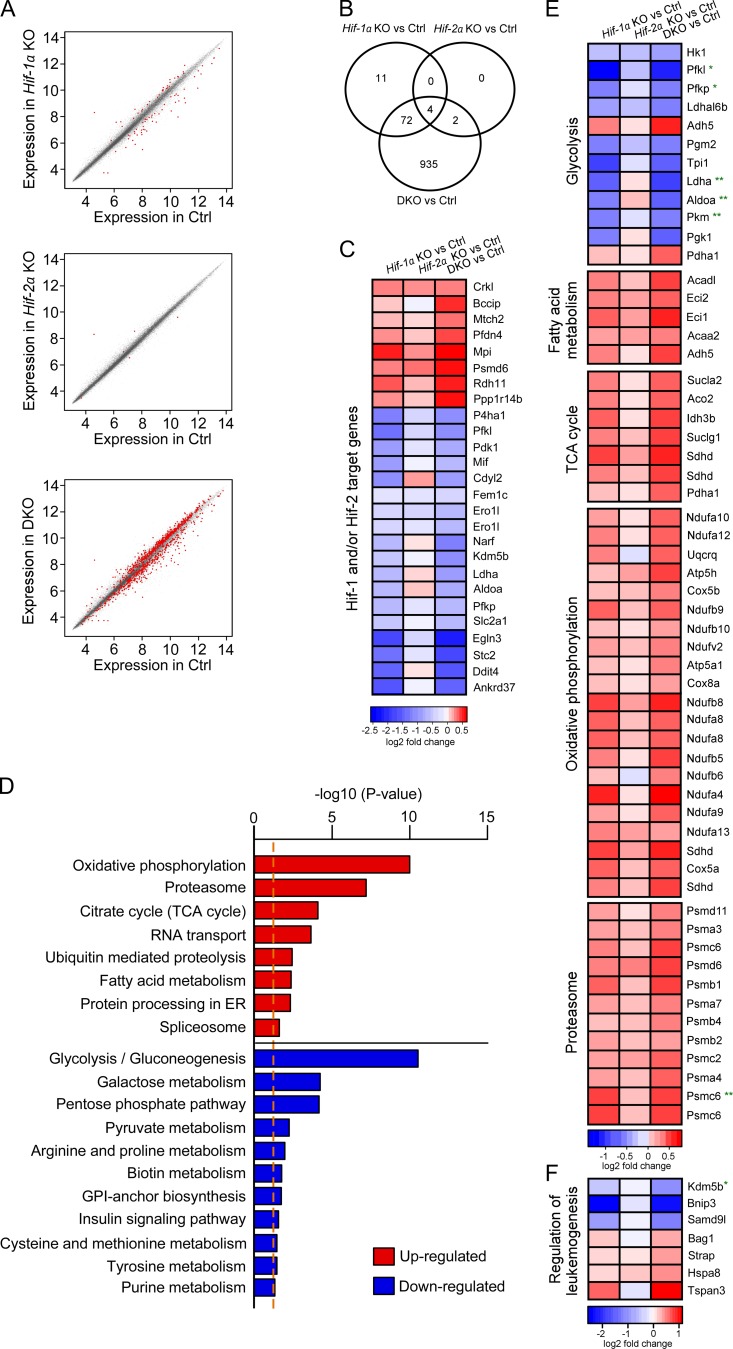
To understand the molecular signatures associated with accelerated leukemia development upon Hif-1α and Hif-2α deletion, we performed global gene expression profiling of Meis1/Hoxa9-transduced Hif-1αfl/fl;Vav-iCre, Hif-2αfl/fl;Vav-iCre, Hif-1αfl/fl;Hif-2αfl/fl;Vav-iCre, and control preleukemic cells incubated under hypoxic conditions (1% O2) for 8 d. Although deletion of Hif-1α or Hif-2α had a minor effect on gene expression (i.e., 87 and 6 genes were differentially expressed in Hif-1αfl/fl;Vav-iCre and Hif-2αfl/fl;Vav-iCre cells, respectively, compared with control cells), deletion of both Hif-1α and Hif-2α deregulated the expression of 1,013 genes (Fig. 6, A and B; FDR< 0.05), indicating the synergism between Hif-1α and Hif-2α in the control of gene expression. Indeed, multiple known Hif target genes identified previously in nonhematopoietic cells (Mole et al., 2009; Schödel et al., 2011) were deregulated in Hif-1αfl/fl;Hif-2αfl/fl;Vav-iCre preleukemic cells (Fig. 6 C). Strikingly, pathways analyses revealed that the most significantly affected group of genes deregulated upon Hif-1α and Hif-2α deletion were genes involved in energy metabolism. Whereas genes involved in glycolysis were significantly down-regulated in Hif-1αfl/fl;Hif-2αfl/fl;Vav-iCre cells, those controlling fatty acid degradation, the TCA cycle, and mitochondrial oxidative phosphorylation pathways were up-regulated in preleukemic cells lacking Hif-1α and Hif-2α (Fig. 6, D and E). Thus, the ability of Hif-1αfl/fl;Hif-2αfl/fl;Vav-iCre cells to generate accelerated leukemia is associated with the gene signature indicative of the metabolic switch from glycolysis to fatty acid oxidation and mitochondrial oxidative phosphorylation.
Figure 6.
Molecular signature of preleukemic cells lacking Hif-1α, Hif-2α or both. Gene expression profiling was performed in Hif-1αfl/fl;Vav-iCre, Hif-2αfl/fl;Vav-iCre, Hif-1αfl/fl;Hif-2αfl/fl;Vav-iCre, and control preleukemic cells (n = 4–5 biological replicates per genotype) cultured under hypoxic conditions (1% O2) for 8 d. (A) Expression scatterplots showing relative mean expression of Affymetrix probes between control (x-axis) and Hif-1αfl/fl;Vav-iCre, Hif-2αfl/fl;Vav-iCre or Hif-1αfl/fl;Hif-2αfl/fl;Vav-iCre (DKO) cells (y-axis). Significantly deregulated genes are shown in red (P < 0.05). (B) Venn diagram shows significantly deregulated (P < 0.05) genes in Hif-1αfl/fl;Vav-iCre, Hif-2αfl/fl;Vav-iCre, and Hif-1αfl/fl;Hif-2αfl/fl;Vav-iCre preleukemic cells. (C) Heat map showing a log2 fold change in the expression of HIF-1 and/or HIF-2 target genes (Schödel et al., 2011) in Hif-1α–, Hif-2α–, and Hif-1α/Hif-2α–deficient preleukemic cells relative to control preleukemic cells. The heat map shows all HIF-1 and/or HIF-2 target genes that are significantly deregulated in Hif-1αfl/fl;Hif-2αfl/fl;Vav-iCre cells (P < 0.05) compared with control cells. (D) KEGG pathway annotations in Hif-1αfl/fl;Hif-2αfl/fl;Vav-iCre (DKO) versus control cells. Pathway enrichment is presented as –log10 (p-value). The dotted orange line indicates P = 0.05. (E) Heat map showing a log2 fold change (vs. control preleukemic cells) in expression of genes belonging to the glycolysis, fatty acid metabolism, TCA cycle, oxidative phosphorylation, and proteasome pathways that are significantly (P < 0.05) deregulated in Hif-1αfl/fl;Hif-2αfl/fl;Vav-iCre cells. (F) Heat map showing a log2 fold change (vs. control preleukemic cells) in expression of genes involved in AML leukemogenesis. All genes are significantly deregulated in Hif-1αfl/fl;Hif-2αfl/fl;Vav-iCre cells (P < 0.05) compared with control cells. (E and F) *, HIF-1 target gene; **, HIF-1 and HIF-2 target gene (Schödel et al., 2011).
In addition to the pathways analyses, our data revealed that a subset of modulators of AML leukemogenesis was deregulated upon Hif-1α and Hif-2α deletion. The expression of several negative regulators of leukemogenesis was decreased, and the expression of distinct genes that promote AML was increased (Fig. 6 F). For example, the expression of a direct Hif target Kdm5b (Jarid1b) which functions as a H3K4-specific demethylase that negatively regulates leukemogenesis in murine and human MLL-rearranged AML cells (Wong et al., 2015), was decreased in Hif-1αfl/fl;Hif-2αfl/fl;Vav-iCre cells (notably, Kdm5b knockdown in MLL-rearranged cells decreases the latency of AML development; Wong et al., 2015). Levels of Tspan3 (encoding tetraspanin 3), which is required for the development and propagation of AML (Kwon et al., 2015), were increased (Fig. 6 F). A hypoxia-inducible gene Bnip3 (Sowter et al., 2001), encoding a proapoptotic protein that is silenced in hematopoietic malignancies, including 17% of AML cases (Murai et al., 2005), was down-regulated in Hif-1αfl/fl;Hif-2αfl/fl;Vav-iCre cells (Fig. 6 F). Furthermore, one of the most significantly up-regulated pathways in Hif-1αfl/fl;Hif-2αfl/fl;Vav-iCre cells was the proteasome pathway (Fig. 6, D and E), whose expression and activity is increased in malignancies, including leukemia (Kumatori et al., 1990; Ma et al., 2009), and inhibition of which reduces survival of human and mouse AML cells (Servida et al., 2005; Bernot et al., 2013; van der Helm et al., 2015). Finally, consistent with their increased proliferation, numerous genes involved in cell cycle progression or tumorigenesis were differentially expressed in Hif-1α/Hif-2α–deficient preleukemic cells compared with control (unpublished data). Therefore, Hif-1α and Hif-2α deletion promotes a molecular signature that facilitates survival and proliferation of preleukemic cells.
The role of Hif-1α and Hif-2α in leukemic transformation remains a subject of intense debate (Losman and Kaelin, 2013; Gezer et al., 2014; Vyas, 2014). Here, we used a conditional gene deletion approach to investigate the requirement for Hif-1α and Hif-2α in leukemogenesis. Neither Hif-1α nor Hif-2α was essential for the generation of preleukemic cells in vitro under normoxic and hypoxic conditions. Notably, however, loss of Hif-2α accelerated the conversion of preleukemic cells to LSCs and shortened AML latency, and this effect was potentiated by Hif-1α codeletion. Our data, taken together with the tumor suppressor functions of Hif-1α (Velasco-Hernandez et al., 2014), imply that Hif-1α and Hif-2α synergize within the hypoxic bone marrow to suppress LSC generation and AML development. Although deletions in HIF-1α or HIF-2α have not been thus far identified in any subtype of human AML (Ley et al., 2008; Cancer Genome Atlas Research Network, 2013; Andersson et al., 2015; Lavallée et al., 2015), it will be of interest to explore the possibility that HIF-1α or HIF-2α are mutated or silenced in leukemias with MLL translocations or in those expressing high levels of MEIS1 or HOXA9.
Gene knockdown studies suggested the requirement for HIF-1α or HIF-2α in human LSC functions, indicating that HIFs are potential therapeutic targets for AML LSC elimination (Wang et al., 2011; Rouault-Pierre et al., 2013). However, the interpretation of these knockdown studies is confounded by the lack of information regarding the subtype and cytogenetics of AML samples. Here, we report that, surprisingly, Hif-2α is dispensable for the ability of established LSCs to sustain murine AML induced by Mll-AF9 or Meis1/Hoxa9. Remarkably, established LSCs lacking both Hif-1α and Hif-2α efficiently propagate AML. Furthermore, CRISPR-Cas9–mediated knockout of HIF-2α or pharmacological inhibition of the HIF pathway in human AML cells with MLL-AF9 translocation had no impact on their survival and proliferation under hypoxic conditions. Thus, our study in mouse and human cells, taken together with the recent demonstration that Hif-1α is dispensable for disease propagation in MLL-ENL– and Meis1/Hoxa9-driven mouse AML (Velasco-Hernandez et al., 2014), indicates that HIF antagonists are unlikely to have a major beneficial therapeutic potential in AML subsets resulting from the activation of the Mll–AF9–Meis1/Hoxa9 pathway. Finally, given the heterogeneity of AML, we propose that the systematic dissection of the roles of HIF-1α and HIF-2α in different AML subsets is required to identify those that are sensitive to the inhibition of HIF-1 and/or HIF-2.
Collectively, we provide genetic evidence that Hif-2α acts as a tumor suppressor in the development of AML, but is dispensable for LSC maintenance, at least within Mll–AF9–Meis1/HoxA9-driven leukemia. In concordance, the HIF pathway is not essential in human AML cells harboring the MLL–AF9 translocation. Finally, at least in the Meis1/Hoxa9 AML model, Hif-1α and Hif-2α are not required for LSC maintenance and AML propagation, but they act synergistically to suppress LSC development. Considering that LSCs serve as a paradigm for other cancer stem cells, it will be interesting to investigate the synergistic tumor suppressor functions of Hif-1 and Hif-2 in other cancers.
MATERIALS AND METHODS
Mice
Hif-1αfl/fl;Vav-iCre, Hif-2αfl/fl;Vav-iCre, and Hif-1αfl/fl;Hif-2αfl/fl;Vav-iCre mice were described previously (Guitart et al., 2013). Mll-AF9KI/+ mice (Chen et al., 2008) were obtained from The Jackson Laboratory. 8–10-wk-old sex-matched mice were used as donors of c-Kit+ cells. Animal experiments were approved by the Animal Welfare and Ethical Review Body of the University of Edinburgh and authorized by the UK Home Office.
FACS
BM and blood samples were stained and analyzed as described previously (Kranc et al., 2009; Guitart et al., 2013).
RNA isolation and quantitative real-time polymerase chain reaction (qRT-PCR)
Total RNA was isolated using the RNeasy Mini kit (QIAGEN) and cDNA synthesis was performed with the High-Capacity cDNA Reverse Transcription kit (Applied Biosystems). qRT-PCR was performed in triplicate with the Light Cycler 480-II (Roche) using the human TaqMan Gene Expression Assay probe and primer sets for HPRT (Hs02800695_m1), BNIP3 (Hs00969291_m1), PDK1 (Hs01561850_m1), EGLN3 (Hs00222966_m1), and ALDOA (Hs00605108_g1; Life Technologies). Relative gene expression was analyzed by the ΔΔCt method using HPRT as reference control and an assigned calibrator.
Gene expression profiling and bioinformatics analyses
Preleukemic cells cultured in hypoxia (1% O2) were collected and resuspended in TRIzol (Life Technologies). RNA was isolated by standard phenol/chloroform extraction. RNA samples (500 ng) were processed and labeled for array hybridization using the Ambion WT Expression kit (Life Technologies). Labeled, fragmented cDNA (GeneChip WT Terminal Labeling and Controls kit; Affymetrix) was hybridized to Mouse Gene 2.0 arrays for 16 h at 45°C (at 60 rpm; GeneChip Hybridization, Wash, and Stain kit; Affymetrix). Arrays were washed and stained using the Affymetrix Fluidics Station 450, and scanned using the Hewlett-Packard GeneArray Scanner 3000 7G.
For bioinformatic analyses, a total of 18 arrays (n = 5 for control, n = 4 for Hif-1αfl/fl;Vav-iCre, n = 5 for Hif-2αfl/fl;Vav-iCre, n = 4 for Hif-1αfl/fl;Hif-2αfl/fl;Vav-iCre) were QC (quality control) analyzed using arrayQualityMetrics in Bioconductor. Raw data were quantile normalized across all arrays. Pairwise group comparisons were undertaken using linear modeling, using the limma package in Bioconductor. Subsequently, empirical Bayesian analysis was applied, including vertical (within a given comparison) p-value adjustment for multiple testing, which controls for false-discovery rate. Functional-enrichment hypergeometric test analyses were performed for KEGG pathways using the appropriate packages. Focused genes of interest lists were assembled from the literature and other publically available resources.
Immunoblotting
THP-1 cells were incubated with 150 µM CoCl2 (Sigma-Aldrich) for 16 h, washed in ice-cold phosphate-buffered saline (PBS), and prepared in lysis buffer containing 6.7 M urea, 10 mM Tris-Cl, pH 6.8, 10% glycerol, 1% SDS, 1 mM dithiothreitol, supplemented with Complete Protease Inhibitor and PhosStop (Roche). 20 µg of proteins were separated by electrophoresis on a 10% Tris-Glycine gel and transferred to Immun-Blot polyvinylidene difluoride membranes (Bio-Rad Laboratories). Membranes were blocked in 3% milk 3% BSA in Tris-buffered saline – 0.1% Tween-20 (TBST), and then incubated with HIF-2α primary antibody (NB100-122; Novus) in 3% milk 3% BSA TBST overnight, washed in TBST, and incubated with horseradish peroxidase (HRP)–labeled secondary antibodies (R&D Systems) in 3% milk TBST. Immunodetection was performed using the Supersignal West Dura Extended Duration Substrate (Thermo Fisher Scientific) and Odyssey Fc (Licor Biosciences).
Leukemic transformation assays
BM cells were extracted from 8–10-wk-old mice. After c-Kit enrichment using MACS LS columns (Miltenyi Biotec), cells were transduced with MSCV-Meis1a-puro and MSCV-Hoxa9-neo retroviruses and serially replated every 6 d in MethoCult M3231 (STEMCELL Technologies) supplemented with SCF, IL-3, IL-6, and GM-CSF in normoxia (20% O2) and hypoxia (1% O2).
Cell cycle and proliferation assays
Live cells were counted with Accuri C6 Flow Cytometer (BD) at the time points indicated in the figures. For cell cycle analyses, cells were resuspended in the DAPI/NP-40 solution and acquired on FACSFortessa V (BD).
CRISPR/Cas9 gene targeting
Guide RNAs to target the human HIF-2α gene were designed using the Zhang laboratory web resource (www.genome-engineering.org). Two individual sgRNAs were designed to target exon 12 of HIF-2α (sgRNA1, 5′-AGAAGACAGAGCCCGAGCAC-3′; sgRNA2, 5′-GCTTCCGGCATCAAAGAAGA-3′), which is the most 5′ exon common to all UCSC-annotated transcript variants. sgRNA-encoding oligonucleotides were cloned into pLentiCRISPRv2 vector (Sanjana et al., 2014) using standard procedures (www.genome-engineering.org). Lentiviral production was performed as described previously (Kranc et al., 2009). After lentiviral transduction and selection, THP-1 cells were subjected to single-cell sorting (FACSAria Fusion; BD), and individual clones were expanded and screened for frameshift mutations. In brief, a region spanning the target site was amplified from genomic DNA isolated from individual clones. PCR products were subsequently cloned into TOPO TA Cloning Sequencing vector (Invitrogen). 15 sequences were analyzed per clone by aligning them to the WT HIF-2α sequences using BLAST. Only the clones with frameshifting indel mutations and no detectable WT allele were used for subsequent validation and in vitro assays.
Cell culture for drug studies
THP-1 and NOMO-1 cells were cultured at 400,000 cells/ml in RPMI-1640 GlutaMAX containing 10% FBS, 25 mM HEPES, 100 U/ml penicillin, and 100 µg/ml streptomycin and incubated with 10 nM BAY 87–2243 (Selleckchem) or vehicle control (0.001% EtOH [vol/vol]) for 6 d under hypoxic conditions (1% O2). At days 3 and 6, the cells were incubated with 7-AAD, and their proliferative capacity (number of 7-AAD− cells) and percentage of dead (7-AAD+) cells was measured by flow cytometry using the Accuri C6 Flow Cytometer (BD).
Transplantation assays
Mll-AF9KI/+, Hif-1αfl/fl, Hif-2αfl/fl, and Vav-iCre mice were CD45.2+. For transplantations with Meis1/Hoxa9-expressing preleukemic cells, 100,000 c-Kit+ preleukemic cells were transplanted together with 200,000 WT CD45.1+ unfractionated BM cells into lethally irradiated (10 Gy) CD45.1+/CD45.2+ recipients. The mice were monitored for AML development. For secondary transplants with Meis1/Hoxa9-expressing LSCs, 10,000 CD45.2+c-Kit+ cells were sorted from primary recipients and transplanted into secondary recipients together with 200,000 WT CD45.1+ unfractionated BM cells. For Mll-AF9KI/+ cell transplantations, lethally irradiated CD45.1+/CD45.2+ syngeneic recipients were transplanted with 2,000 LSK or 5,000 LK cells (together with 200,000 CD45.1+ BM cells per recipient) as indicated in Fig. 2.
Statistical analysis
Kaplan-Meier survival curve statistics were determined using the Mantel-Cox test. LSC frequencies were determined using ELDA statistics software (Hu and Smyth, 2009). All other statistical significance was determined using the Mann-Whitney test.
Acknowledgments
We are grateful to Dr. Tim Somervaille for Meis1 and Hoxa9 constructs. We thank Dr. Vladimir Benes and Jelena Pistolic from the Genomics Core facility of the European Molecular Biology Laboratory (EMBL, Heidelberg) for performing the gene expression profiling using Affymetrix GeneChip arrays. We thank Fiona Rossi and Dr. Claire Cryer for their help with flow cytometry.
K.R. Kranc is a Cancer Research UK Senior Fellow. K.R. Kranc's laboratory is supported by grants from Cancer Research UK, Bloodwise (formerly called Leukaemia and Lymphoma Research), Edinburgh Cancer Research UK Centre Development Fund, The Wellcome Trust ISSF award, Medical Research Council, and the Kay Kendall Leukaemia Fund. T.L. Holyoake was supported by Cancer Research UK (C11074/A11008).
The authors declare no competing financial interests.
Footnotes
Abbreviations used:
- AML
- acute myeloid leukemia
- Hif-1α
- hypoxia-inducible factor-1α
- Hif-2α
- hypoxia-inducible factor-2α
- HSC
- hematopoietic stem cell
- LSC
- leukemic stem cell
References
- Andersson A.K., Ma J., Wang J., Chen X., Gedman A.L., Dang J., Nakitandwe J., Holmfeldt L., Parker M., Easton J., et al. St. Jude Children’s Research Hospital–Washington University Pediatric Cancer Genome Project . 2015. The landscape of somatic mutations in infant MLL-rearranged acute lymphoblastic leukemias. Nat. Genet. 47:330–337. 10.1038/ng.3230 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernot K.M., Nemer J.S., Santhanam R., Liu S., Zorko N.A., Whitman S.P., Dickerson K.E., Zhang M., Yang X., McConnell K.K., et al. 2013. Eradicating acute myeloid leukemia in a Mll(PTD/wt):Flt3(ITD/wt) murine model: a path to novel therapeutic approaches for human disease. Blood. 122:3778–3783. 10.1182/blood-2013-06-507426 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cancer Genome Atlas Research Network 2013. Genomic and epigenomic landscapes of adult de novo acute myeloid leukemia. N. Engl. J. Med. 368:2059–2074. 10.1056/NEJMoa1301689 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang E., Liu H., Unterschemmann K., Ellinghaus P., Liu S., Gekeler V., Cheng Z., Berndorff D., and Gambhir S.S.. 2015. 18F-FAZA PET imaging response tracks the reoxygenation of tumors in mice upon treatment with the mitochondrial complex I inhibitor BAY 87-2243. Clin. Cancer Res. 21:335–346. 10.1158/1078-0432.CCR-14-0217 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen W., Kumar A.R., Hudson W.A., Li Q., Wu B., Staggs R.A., Lund E.A., Sam T.N., and Kersey J.H.. 2008. Malignant transformation initiated by Mll-AF9: gene dosage and critical target cells. Cancer Cell. 13:432–440. 10.1016/j.ccr.2008.03.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dobson C.L., Warren A.J., Pannell R., Forster A., Lavenir I., Corral J., Smith A.J., and Rabbitts T.H.. 1999. The mll-AF9 gene fusion in mice controls myeloproliferation and specifies acute myeloid leukaemogenesis. EMBO J. 18:3564–3574. 10.1093/emboj/18.13.3564 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drabkin H.A., Parsy C., Ferguson K., Guilhot F., Lacotte L., Roy L., Zeng C., Baron A., Hunger S.P., Varella-Garcia M., et al. 2002. Quantitative HOX expression in chromosomally defined subsets of acute myelogenous leukemia. Leukemia. 16:186–195. 10.1038/sj.leu.2402354 [DOI] [PubMed] [Google Scholar]
- Ellinghaus P., Heisler I., Unterschemmann K., Haerter M., Beck H., Greschat S., Ehrmann A., Summer H., Flamme I., Oehme F., et al. 2013. BAY 87-2243, a highly potent and selective inhibitor of hypoxia-induced gene activation has antitumor activities by inhibition of mitochondrial complex I. Cancer Med. 2:611–624. 10.1002/cam4.112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gezer D., Vukovic M., Soga T., Pollard P.J., and Kranc K.R.. 2014. Concise review: genetic dissection of hypoxia signaling pathways in normal and leukemic stem cells. Stem Cells. 32:1390–1397. 10.1002/stem.1657 [DOI] [PubMed] [Google Scholar]
- Guitart A.V., Subramani C., Armesilla-Diaz A., Smith G., Sepulveda C., Gezer D., Vukovic M., Dunn K., Pollard P., Holyoake T.L., et al. 2013. Hif-2α is not essential for cell-autonomous hematopoietic stem cell maintenance. Blood. 122:1741–1745. 10.1182/blood-2013-02-484923 [DOI] [PubMed] [Google Scholar]
- Helbig L., Koi L., Brüchner K., Gurtner K., Hess-Stumpp H., Unterschemmann K., Baumann M., Zips D., and Yaromina A.. 2014. BAY 87-2243, a novel inhibitor of hypoxia-induced gene activation, improves local tumor control after fractionated irradiation in a schedule-dependent manner in head and neck human xenografts. Radiat. Oncol. 9:207 10.1186/1748-717X-9-207 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu Y., and Smyth G.K.. 2009. ELDA: extreme limiting dilution analysis for comparing depleted and enriched populations in stem cell and other assays. J. Immunol. Methods. 347:70–78. 10.1016/j.jim.2009.06.008 [DOI] [PubMed] [Google Scholar]
- Kranc K.R., Schepers H., Rodrigues N.P., Bamforth S., Villadsen E., Ferry H., Bouriez-Jones T., Sigvardsson M., Bhattacharya S., Jacobsen S.E., and Enver T.. 2009. Cited2 is an essential regulator of adult hematopoietic stem cells. Cell Stem Cell. 5:659–665. 10.1016/j.stem.2009.11.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kroon E., Krosl J., Thorsteinsdottir U., Baban S., Buchberg A.M., and Sauvageau G.. 1998. Hoxa9 transforms primary bone marrow cells through specific collaboration with Meis1a but not Pbx1b. EMBO J. 17:3714–3725. 10.1093/emboj/17.13.3714 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar A.R., Li Q., Hudson W.A., Chen W., Sam T., Yao Q., Lund E.A., Wu B., Kowal B.J., and Kersey J.H.. 2009. A role for MEIS1 in MLL-fusion gene leukemia. Blood. 113:1756–1758. 10.1182/blood-2008-06-163287 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumatori A., Tanaka K., Inamura N., Sone S., Ogura T., Matsumoto T., Tachikawa T., Shin S., and Ichihara A.. 1990. Abnormally high expression of proteasomes in human leukemic cells. Proc. Natl. Acad. Sci. USA. 87:7071–7075. 10.1073/pnas.87.18.7071 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwon H.Y., Bajaj J., Ito T., Blevins A., Konuma T., Weeks J., Lytle N.K., Koechlein C.S., Rizzieri D., Chuah C., et al. 2015. Tetraspanin 3 Is Required for the Development and Propagation of Acute Myelogenous Leukemia. Cell Stem Cell. 17:152–164. 10.1016/j.stem.2015.06.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lavallée V.P., Baccelli I., Krosl J., Wilhelm B., Barabé F., Gendron P., Boucher G., Lemieux S., Marinier A., Meloche S., et al. 2015. The transcriptomic landscape and directed chemical interrogation of MLL-rearranged acute myeloid leukemias. Nat. Genet. 47:1030–1037. 10.1038/ng.3371 [DOI] [PubMed] [Google Scholar]
- Lawrence H.J., Rozenfeld S., Cruz C., Matsukuma K., Kwong A., Kömüves L., Buchberg A.M., and Largman C.. 1999. Frequent co-expression of the HOXA9 and MEIS1 homeobox genes in human myeloid leukemias. Leukemia. 13:1993–1999. 10.1038/sj.leu.2401578 [DOI] [PubMed] [Google Scholar]
- Lehnertz B., Pabst C., Su L., Miller M., Liu F., Yi L., Zhang R., Krosl J., Yung E., Kirschner J., et al. 2014. The methyltransferase G9a regulates HoxA9-dependent transcription in AML. Genes Dev. 28:317–327. 10.1101/gad.236794.113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lessard J., and Sauvageau G.. 2003. Bmi-1 determines the proliferative capacity of normal and leukaemic stem cells. Nature. 423:255–260. 10.1038/nature01572 [DOI] [PubMed] [Google Scholar]
- Ley T.J., Mardis E.R., Ding L., Fulton B., McLellan M.D., Chen K., Dooling D., Dunford-Shore B.H., McGrath S., Hickenbotham M., et al. 2008. DNA sequencing of a cytogenetically normal acute myeloid leukaemia genome. Nature. 456:66–72. 10.1038/nature07485 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Losman J.A., and Kaelin W.G. Jr. 2013. What a difference a hydroxyl makes: mutant IDH, (R)-2-hydroxyglutarate, and cancer. Genes Dev. 27:836–852. 10.1101/gad.217406.113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma W., Kantarjian H., Bekele B., Donahue A.C., Zhang X., Zhang Z.J., O’Brien S., Estey E., Estrov Z., Cortes J., et al. 2009. Proteasome enzymatic activities in plasma as risk stratification of patients with acute myeloid leukemia and advanced-stage myelodysplastic syndrome. Clin. Cancer Res. 15:3820–3826. 10.1158/1078-0432.CCR-08-3034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mole D.R., Blancher C., Copley R.R., Pollard P.J., Gleadle J.M., Ragoussis J., and Ratcliffe P.J.. 2009. Genome-wide association of hypoxia-inducible factor (HIF)-1alpha and HIF-2alpha DNA binding with expression profiling of hypoxia-inducible transcripts. J. Biol. Chem. 284:16767–16775. 10.1074/jbc.M901790200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murai M., Toyota M., Satoh A., Suzuki H., Akino K., Mita H., Sasaki Y., Ishida T., Shen L., Garcia-Manero G., et al. 2005. Aberrant DNA methylation associated with silencing BNIP3 gene expression in haematopoietic tumours. Br. J. Cancer. 92:1165–1172. 10.1038/sj.bjc.6602422 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rouault-Pierre K., Lopez-Onieva L., Foster K., Anjos-Afonso F., Lamrissi-Garcia I., Serrano-Sanchez M., Mitter R., Ivanovic Z., de Verneuil H., Gribben J., et al. 2013. HIF-2α protects human hematopoietic stem/progenitors and acute myeloid leukemic cells from apoptosis induced by endoplasmic reticulum stress. Cell Stem Cell. 13:549–563. 10.1016/j.stem.2013.08.011 [DOI] [PubMed] [Google Scholar]
- Sanjana N.E., Shalem O., and Zhang F.. 2014. Improved vectors and genome-wide libraries for CRISPR screening. Nat. Methods. 11:783–784. 10.1038/nmeth.3047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schödel J., Oikonomopoulos S., Ragoussis J., Pugh C.W., Ratcliffe P.J., and Mole D.R.. 2011. High-resolution genome-wide mapping of HIF-binding sites by ChIP-seq. Blood. 117:e207–e217. 10.1182/blood-2010-10-314427 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Semenza G.L. 2014. Oxygen sensing, hypoxia-inducible factors, and disease pathophysiology. Annu. Rev. Pathol. 9:47–71. 10.1146/annurev-pathol-012513-104720 [DOI] [PubMed] [Google Scholar]
- Servida F., Soligo D., Delia D., Henderson C., Brancolini C., Lombardi L., and Deliliers G.L.. 2005. Sensitivity of human multiple myelomas and myeloid leukemias to the proteasome inhibitor I. Leukemia. 19:2324–2331. 10.1038/sj.leu.2403987 [DOI] [PubMed] [Google Scholar]
- Somervaille T.C., Matheny C.J., Spencer G.J., Iwasaki M., Rinn J.L., Witten D.M., Chang H.Y., Shurtleff S.A., Downing J.R., and Cleary M.L.. 2009. Hierarchical maintenance of MLL myeloid leukemia stem cells employs a transcriptional program shared with embryonic rather than adult stem cells. Cell Stem Cell. 4:129–140. 10.1016/j.stem.2008.11.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sowter H.M., Ratcliffe P.J., Watson P., Greenberg A.H., and Harris A.L.. 2001. HIF-1-dependent regulation of hypoxic induction of the cell death factors BNIP3 and NIX in human tumors. Cancer Res. 61:6669–6673. [PubMed] [Google Scholar]
- Spencer J.A., Ferraro F., Roussakis E., Klein A., Wu J., Runnels J.M., Zaher W., Mortensen L.J., Alt C., Turcotte R., et al. 2014. Direct measurement of local oxygen concentration in the bone marrow of live animals. Nature. 508:269–273. 10.1038/nature13034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Helm L.H., Bosman M.C., Schuringa J.J., and Vellenga E.. 2015. Effective targeting of primitive AML CD34(+) cells by the second-generation proteasome inhibitor carfilzomib. Br. J. Haematol. 171:652–655. 10.1111/bjh.13418 [DOI] [PubMed] [Google Scholar]
- Velasco-Hernandez T., Hyrenius-Wittsten A., Rehn M., Bryder D., and Cammenga J.. 2014. HIF-1α can act as a tumor suppressor gene in murine acute myeloid leukemia. Blood. 124:3597–3607. 10.1182/blood-2014-04-567065 [DOI] [PubMed] [Google Scholar]
- Vyas P. 2014. Targeting HIF function: the debate continues. Blood. 124:3510–3511. 10.1182/blood-2014-10-605055 [DOI] [PubMed] [Google Scholar]
- Wang Y., Krivtsov A.V., Sinha A.U., North T.E., Goessling W., Feng Z., Zon L.I., and Armstrong S.A.. 2010. The Wnt/beta-catenin pathway is required for the development of leukemia stem cells in AML. Science. 327:1650–1653. 10.1126/science.1186624 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y., Liu Y., Malek S.N., Zheng P., and Liu Y.. 2011. Targeting HIF1α eliminates cancer stem cells in hematological malignancies. Cell Stem Cell. 8:399–411. 10.1016/j.stem.2011.02.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong S.H., Goode D.L., Iwasaki M., Wei M.C., Kuo H.P., Zhu L., Schneidawind D., Duque-Afonso J., Weng Z., and Cleary M.L.. 2015. The H3K4-Methyl Epigenome Regulates Leukemia Stem Cell Oncogenic Potential. Cancer Cell. 28:198–209. 10.1016/j.ccell.2015.06.003 [DOI] [PMC free article] [PubMed] [Google Scholar]