Abstract
Nonribosomal peptide synthetases (NRPSs) are microbial enzymes that produce a wealth of important natural products by condensing substrates in an assembly line manner. The proper sequence of substrates is obtained by tethering them to phosphopantetheinyl arms of holo carrier proteins (CPs) via a thioester bond. CPs in holo and substrate-loaded forms visit NRPS catalytic domains in a series of transient interactions. A lack of structural information on substrate-loaded carrier proteins has hindered our understanding of NRPS synthesis. Here, we present the first structure of an NRPS aryl carrier protein loaded with its substrate via a native thioester bond, together with the structure of its holo form. We also present the first quantification of NRPS CP backbone dynamics. Our results indicate that prosthetic moieties in both holo and loaded forms are in contact with the protein core, but they also sample states in which they are disordered and extend in solution. We observe that substrate loading induces a large conformational change in the phosphopantetheinyl arm, thereby modulating surfaces accessible for binding to other domains. Our results are discussed in the context of NRPS domain interactions.
Keywords: Nonribosomal peptide synthetase, NMR solution structure, holo aryl carrier protein, loaded aryl carrier protein, ArCP, thiolation domain
Introduction
Nonribosomal peptide synthetases (NRPSs) are enzymatic systems found in bacteria and fungi responsible for the production of a myriad of secondary metabolites. These systems are capable of generating exceptionally complex and diverse natural products from simple starting materials such as amino and aryl acids by utilizing a modular architecture. Multiple modules are arranged in an assembly-line fashion to comprise the full synthetase, an organization also encountered in related fatty acid synthases (FASs) and modular polyketide synthases (PKSs).1–9 Each module within an NRPS is comprised of at least three core domains whose combined action leads to the selection, activation, and incorporation of a single small molecule into the growing peptide.10–12 NRPS modules select starting materials from a pool of hundreds of small molecules including the 20 standard L-amino acids, aryl acids, and D-amino acids. Assembling these small molecules in a combinatorial fashion creates the potential to generate enormous chemical and functional diversity. A central aspect of this successful strategy is the covalent tethering of chemical substrates to the assembly line, which occurs through so-called carrier proteins.
Each module within a NRPS is typically composed of at least an adenylation (A) domain, a condensation (C) domain, and a thiolation domain, also called a carrier protein (CP). Apo-CPs must first be activated to holo-CPs via attachment of a 4′-phosphopantetheine (PP) moiety to a conserved serine by a phosphopantetheinyl transferase.13 The PP provides a thiol by which activated monomers and intermediate products are covalently tethered to the synthetase through loaded-CPs. A domains load the substrates onto CPs by catalyzing two distinct reactions. First, they select the amino or aryl acid to be incorporated and activate it using ATP via formation of a high energy acyladenylate. Second, they load the activated monomer onto CPs via formation of a thioester with the PP. C domains then catalyze amide bond formation between products loaded on adjacent CPs, passing intermediates from an upstream donor CP to a downstream acceptor CP and extending the peptide by a single monomer. After the final monomer is incorporated, a thioesterase domain found in the final module releases the peptide via hydrolysis or macrocyclization. During synthesis, NRPS carrier proteins must interact with at least three different catalytic domains: a PPTase, an A domain, and one or more C domains. Crystallographic and nuclear magnetic resonance (NMR) studies have indicated that these interactions do not occur within the confines of a rigid assembly of NRPS domains but through a succession of transient interactions involving a dynamic quaternary structure.14–19 Understanding the role that chemical modifications of CPs play in orchestrating this series of transient protein-protein interactions is key to elucidating the molecular mechanism of NRPS synthesis.
Progress in understanding the molecular influence of PP and substrates on the function of carrier proteins has been impeded by the lack of structural information on substrate-loaded NRPS carrier proteins. There are several NMR and crystal structures of apo- and holo-CPs, either isolated or together with other NRPS domains.12,14,15,20–28 Of these, the only two solution structures of isolated holo carrier proteins have produced contradictory results with respect to the influence of the PP moiety. In the first system, PP was found not only to bind to its CP but also to dramatically influence conformational fluctuations.22 In the second study, the protein core of CP was thought to interact only weakly, if at all, with the PP, leading to the conclusion that phosphopantetheinylation does not affect the carrier protein in a relevant manner.27 Perhaps more importantly, there are currently no structures available of substrate-loaded CPs from an NRPS and it remains unclear whether the substrate is simply tethered on an unstructured PP arm or if it interacts with its carrier protein. Lack of success in studying structures of loaded carrier proteins results in part from rapid hydrolysis of the thioester bond, so we recently proposed a means to bypass this limitation for monomeric substrates.29 Determining if the loaded substrate directly interacts with the protein core of a CP or remains unbound will shape our understanding of the role it plays in directing protein-protein interactions.
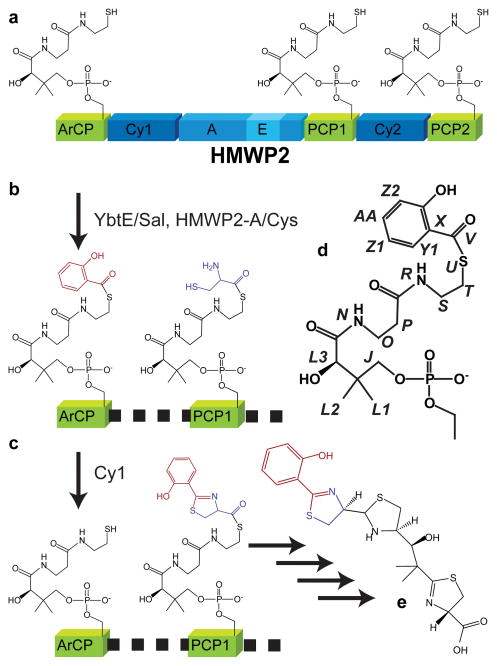
In order to study the influence of substrate loading on the structure of a CP, we determined the solution structures of the aryl carrier protein (ArCP) from yersiniabactin synthetase in its holo and substrate-loaded forms. Yersiniabactin (Figure 1e) is an iron chelator and virulence factor for Yersinia pestis, the causative agent of the bubonic plague, and its biosynthesis has been extensively characterized.30–36 The yersiniabactin synthetase system is comprised of the stand-alone A domain YbtE, the multidomain proteins HMPW2 (Figure 1a) and HMWP1, and the reductase YbtU. ArCP is the first carrier protein involved in yersiniabactin synthesis. It composes the N-terminal 100 residues of HMPW2, which also contains two cyclization domains (Cy1 and Cy2), an adenylation domain (A), two peptidyl carrier proteins (PCP1 and PCP2), and an epimerization domain (E) (Figure 1a). Cyclization domains are related to condensation domains yet catalyze a cyclodehydration in addition to condensation. As a starter carrier protein, ArCP has a relatively simple life-cycle. Following activation of carrier proteins to holo forms, YbtE loads ArCP with salicylic acid while the A domain of HMPW2 loads PCP1 with cysteine (Figure 1b). Cy1 then catalyzes peptide bond formation and cyclization between the substrates loaded on ArCP and PCP1, regenerating holo-ArCP and producing PCP1 now loaded with a 2-hydroxyphenylthiazoline moiety (Figure 1c). Two forms of ArCP communicate with catalytic domains during synthesis: holo-ArCP is a substrate for YbtE and a product of Cy1, whereas ArCP loaded with salicylate (hereafter referred to as ‘loaded-ArCP’) is a product of YbtE and a substrate for Cy1.
Figure 1.
Role of ArCP in yersiniabactin synthesis. a) ArCP is the first carrier protein domain of HMWP2, which also comprises two cyclization domains (Cy1 and Cy2), two peptidyl carrier proteins (PCP1 and PCP2), an adenylation domain for cysteine (A), and an epimerization domain embedded in the A-domain (E). b) The A-domain loads cysteine onto PCP1 and PCP2, and the stand alone A-domain YbtE loads salicylate onto ArCP. c) Cy1 catalyzes the condensation and cyclodehydration of Sal and Cys, forming 2-hydroxyphenylthiazoline on PCP1 and returning ArCP to its holo form. Not shown: Cy2, HMWP1 (a mixed PKS NRPS protein), and YbtU complete the synthesis of yersiniabactin (e). d) Nomenclature used to assign phosphopantetheine (J-U) and salicylate (V-AA) is shown in italics.
We have previously established a method for analysis of ArCP loaded with salicylate in its native thioester form by nuclear magnetic resonance spectroscopy29. The method exploits the non-invasive nature of NMR and isotope editing to allow for sustained NMR measurements on loaded-ArCP. Loaded-ArCP is generated in situ and the appropriate concentration of substrates and adenylation domain, ATP, Sal and YbtE, ensure that the regeneration of loaded-ArCP outperforms hydrolysis without influencing the chemical shift perturbations (CSPs) observed upon substrate loading. We found that these CSPs occur along multiple distinct secondary structural elements of the core of the protein. However, CSPs cannot differentiate among a direct interaction with the substrate, structural changes in the ArCP, a modulation of protein dynamics, or some combination of these effects. Here, we present the solution structures of ArCP in the holo and salicylate-loaded forms together with a characterization of the dynamics of holo and loaded-ArCP, including the PP. We show that holo-ArCP has a transient yet well-defined interaction with the PP and that the protein core of ArCP has a direct interaction with loaded salicylate, and we discuss the role these observations may play in influencing protein-protein interactions in NRPS systems.
Methods
Cloning strategies are described in the supporting information (SI).
Purification of holo-ArCP made from coexpression with Sfp
The purification of holo-ArCP resulting from coexpression with Sfp is identical to that reported for apo-ArCP29 except for the following modifications. pET-DUET-SfpHis6-GB1-TEV-ArCP14-93 was transformed into ΔEntD cells (courtesy Drs. Chalut and Guilhot, CNRS, Toulouse, France). Ampicillin was used instead of kanamycin. Following overnight cell growth at 37°C, the temperature was lowered to 18 °C when reaching an optical density of 0.4. At an optical density of 0.6, IPTG was added to 0.5 mM, and growths continued at 18 °C. Cells were harvested 4–4.5 hours after induction at an optical density of 1.1–1.2. Following digestion by TEV protease and HisTrap purification, dithiothreitol (DTT) was added to a final concentration of 10 mM.
In vitro phosphopantetheinylation of apo-ArCP was performed as described previously.29 Completion of the phosphopantetheinylation reaction was confirmed by HN-HSQC.
NMR data for assignment and structure determination
All spectra were collected at 25 °C on a 600 MHz Bruker Avance III spectrometer equipped with a QCI cryoprobe. All NMR spectra were processed using NMRPipe37 and analyzed using CARA.38 See SI for details of NMR acquisitions.
To ensure long-term stability of NMR samples, samples were buffer exchanged (>125-fold) into freshly prepared NMR buffer containing 0.05% (w/v) sodium azide by repeated concentration and dilution immediately prior to use. D2O was added to all samples to a final concentration of 10% and DSS was used for internal referencing. For samples in D2O, NMR buffer was prepared as described above in 99.8% D2O (Aldrich Chemistry) and the pD adjusted using sodium deuteroxide (40% in D2O, 99.5% D, Cambridge Isotope Laboratories) to a pH of 6.40 (pD=6.80).
15N-Holo-ArCP used for the holo and loaded samples was generated in vitro using apo-ArCP, purified Sfp, and unlabeled coenzyme A. The NOESY-HN-HSQC of holo-ArCP was run on a 520 μM sample in the standard NMR buffer containing 1 mM DTT instead of 500 μM TCEP. To prepare the loaded sample, salicylic acid and ATP were added to 2 mM and YbtE added to 100 nM to 400 μM ArCP and the loading reaction monitored by HN-HSQC. The NOESY-HN-HSQC was begun immediately upon completion of loading.
13C,15N apo-ArCP has previously been assigned29 and its assignment was used as a starting point for assigning holo- and loaded-ArCP resonances.
13C,15N-Holo-ArCP was generated by coexpression with Sfp such that the phosphopantetheine cofactor is also labeled. A first sample was prepared in buffered H2O to complete resonance assignments and collect dihedral angle constraints (SI).
A 390 μM sample was prepared in D2O by repeated concentration and dilution in NMR buffer prepared in D2O until a 900-fold dilution had been achieved. This sample was used for aromatic side-chain resonance assignment and to collect distance constraints involving aliphatic and aromatic protons.
13C,15N-Loaded-ArCP was prepared by adding salicylic acid and ATP to 2 mM (final concentration) and YbtE to 100 nM (final concentration) to 413 μM holo-ArCP and monitoring the loading reaction by HN-HSQC. After completion of the loading reaction, the sample was diluted 15-fold in identical buffer to lower the concentration of adenosine monophosphate (AMP) and pyrophosphate (PPi) produced by the loading reaction and additional YbtE added to increase the final concentration to 250 nM. This was concentrated to a final ArCP concentration of 360 μM. This sample was used to complete resonance assignments and collect dihedral angle constraints (SI).
A sample in D2O was used for aromatic side-chain resonance assignment and to collect distance constraints involving aliphatic and aromatic protons. Loading was performed with 50 μM holo-ArCP, 50 nM YbtE, 2 mM ATP, and 500 μM 13C-salicylate in a total volume of 3.95 ml in 90% H2O/10% D2O and loading monitored by HN-HSQC. After loading was complete, the sample was buffer exchanged 560-fold into NMR buffer prepared in D2O containing 2 mM ATP and 500 μM 13C-salicylate and concentrated to a final loaded-ArCP concentration of 360 μM.
Relaxation experiments
15N-Holo-ArCP was generated by coexpression with Sfp such that the phosphopantetheine cofactor is also labeled. 15N longitudinal and transverse relaxation rates, R1 and R2, and {HN}-heteronuclear-NOESY were measured as described in the supporting information (SI S2).
Salicylate-loaded ArCP was generated by incubating 50 μM 15N-holo-ArCP with 2 mM unlabeled salicylic acid, 2 mM ATP, and either 100 nM (R1, {HN}-Heteronuclear-NOESY) YbtE in 3.5 ml total volume or 20 nM (R2 measurement) YbtE in 3.75 ml total volume and monitoring loading by HN-HSQC. Upon completion of loading, the sample was concentrated to 1 ml, diluted 15-fold in NMR buffer with 2 mM unlabeled salicylic acid and 2 mM ATP, and concentrated to a final protein concentration of 300 μM (R1, {HN}-Heteronuclear NOESY) or 390 μM (R2).
Relaxation parameters were fit using the program nlinLS, part of the NMRPipe software package.37 In all experiments, line shapes in the 1H dimension were fit using a Gaussian function and line shapes in the 15N dimension were fit using a Fourier-transformed, apodized, exponentially decaying sinusoid. Residues 20, 29, 34, 40, 88 and 91 were excluded from fitting in holo-ArCP due to severe overlap. In loaded-ArCP, residues 20, 29, 32, 34, 75, 88, and 91 were excluded for the same reason.
Lipari-Szabo model-free analysis was performed with the program ROTDIF.39,40 ROTDIF fits both the overall rotational diffusion tensor as well as the model-free parameters at each residue, including Rex. Here, a first pass was performed to identify residues with order parameters of 0.75 or below. These residues were then excluded from analysis when fitting the global rotational diffusion tensor but were included when fitting the model-free parameters. In holo-ArCP the excluded residues were 14, 15, 51, 69, 90, and 93. In loaded-ArCP residues 14, 15, 51, 69 and 93 were excluded.
The “sausage” representations of Figure 3 were created using the PyMOL41 “putty” feature after replacing each residue’s b-factor with its corresponding value of 1-S2.
Structure calculation
Assignment of NOESY cross-peaks was performed manually using CARA.38 1636 unambiguous restraints were assigned for holo-ArCP and 1314 for loaded-ArCP. In addition, 131 and 142 angle constraints were obtained with TALOS-N.42 Structure calculations were performed using CYANA version 2.1.43 For the final structure calculation, 100 structures were calculated using 50,000 steps. The final CYANA target functions were 2.65 (for holo) and 2.99 (for loaded). There were no distance violations bigger than 0.5 Å and no angle violations larger than 3.5° in either NMR ensemble. The average rmsd to mean for these conformers were 0.37 Å (backbone) and 0.88 Å (heavy) for holo and 0.36 Å (backbone) and 0.85 Å (heavy) for loaded. Other rmsd’s are described in the remainder of the text. The 20 structures with the lowest target function were chosen for water refinement in explicit solvent using CNS.44 Modified parameter and topology files were generated using the ACPYPE45 web application and refinement run using modified RECOORD46 scripts. The NMR ensembles were analyzed with the protein structure validation suite, PSVS,47 that includes PROCHECK_NMR48 and MolProbity.49,50 Ramachandran statistics (PROCHECK) are: 92.5% in most favored region, 7.5% in additionally allowed, 0.0% in generously allowed, and 0.0% in disallowed region for holo-ArCP and 89.4%, 10.6%, 0.0% and 0.0%, respectively, for loaded-ArCP. See also Supporting Information (SI), Tables S1 and S2.
Surface potentials were generated using the APBS Tools2.1 plugin51 for PyMOL using the default parameters. Input files were generated from pdb files of holo- and loaded-ArCP in which the phosphopantetheinylated serine was replaced with a standard serine using the pdb2pqr52,53 online server.
Structures (PDB 2N6Y and 2N6Z) were analyzed with Py-MOL41 and MOLMOL.54
Results and Discussion
Acyl, aryl, and peptidyl carrier proteins play a central role in PKSs, FASs, and NRPSs as they shuttle substrates between various catalytic sites during synthesis. In the past few years, various structural and functional studies have suggested that NRPSs are not rigid assemblies but are subject to inter-domain dynamics. Notably, rather than simply swinging the phosphopantetheinyl arm between active sites, the entire carrier protein is expected to visit partner domains in a series of transient domain interactions14–18. Understanding the biosynthesis of all associated products thus requires an understanding of the molecular parameters that modulate these successive transient domain interactions. Here, we will describe how both the phosphopantetheinyl arm and its tethered substrate interact with the core of an NRPS aryl carrier protein in its holo and substrate-loaded form. We found that although these interactions are well-defined they are nevertheless transient, as a substantial population of both holo- and loaded-ArCP possesses prosthetic groups subject to large amplitude motions. Our results provide novel insights into interactions between carrier proteins and PP and tethered substrates, and our observations will be discussed within the framework of domain interactions.
Structures of holo- and loaded-ArCP
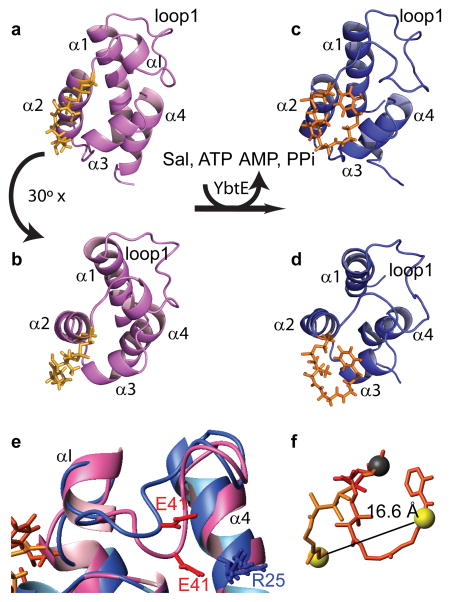
Holo-ArCP and loaded-ArCP both display the right-handed helical bundle fold typical of carrier proteins20,55,56 (Figure 2a–d). This fold consists of three long helices with an up, down, down arrangement (α1, A19-E31, α2, S52-K64, and α4, L80-M87) and an additional shorter helix (α3, L71-A76). Here, secondary structure boundaries are defined as observed in the mean structure of holo-ArCP. The post-translational modification site, S52, is located at the N-terminal end of α2. In ArCP, the four helices are packed predominantly by a hydrophobic core substantiated with aromatic interactions. α1 and α2 are linked by loop1, which is well-defined (Figure S1) and features a single-turn helix, αI (L45-A48). Loop1 is held in place by a number of hydrophobic interactions with α1, α3, and α4 and likely with an ionic interaction between E41 and R25 in the holo form. α3 lies in between loop2 (G66-T70) and the very short loop3 (A77-T79). Overall, ArCP in both holo and loaded forms adopts a well-defined and compact protein fold.
Figure 2.
Solution structures of holo- (a,b) and loaded-ArCP (c,d). The lowest energy conformer of the NMR ensemble is shown for each form of the protein under two different views. e) Detail of loop1 shown for a superposition of holo- (pink) and loaded-ArCP (blue). Structures were aligned with each other using helices α1 through α4. f) Mean structures of Ser-PP from holo-ArCP (orange) and Ser-PP-Sal from loaded-ArCP (orange-red). The moieties were translated to overlay Ser 52 Cα of each form.
A large number of NOESY cross-peaks revealed contacts between PP and ArCP in holo-ArCP and between PP, Sal and ArCP in loaded-ArCP (Figures S3 – S6). nOe’s within the PP arm have the same sign as the source peaks (so-called diagonal signals in NOESY experiments) which demonstrates that the PP arm is subject to the same molecular tumbling as the carrier protein. nOe’s between the protein core and PP denote a contact between PP and ArCP. Similarly, in loaded-ArCP, NOESY cross-peaks indicate a stable, well-defined interaction between loaded-ArCP and its prosthetic group.
Substrate loading alters the conformation of the PP arm in a dramatic manner and modifies the surface of the protein sustained by α2, α3 and nearby regions. In holo-ArCP, the phosphopantetheinyl arm is extended and docks on ArCP along α2 on one side and α3 on the other side (Figure 2a, b). Such a conformation masks a large area of the solvent-exposed surface of α2 and α3. Upon substrate loading, the phosphopantetheinyl arm adopts a curled conformation that accommodates salicylate binding (Figure 2c, d, f). Sal binds at the surface of ArCP in a region defined by the N-terminal end of α2, the C-terminal end of loop1, and the C-terminal end of α3. The change of conformation induced by tethering of salicylate exposes a region previously covered by PP in holo-ArCP and masks a new region of ArCP. Both regions are involved in domain communication and the significance of this alteration in surface access will be discussed within the framework of domain interactions, below.
Comparison of the protein cores of holo- and loaded-ArCP reveals that subtle changes occur upon substrate loading. Three helices, α1, α2, and α4, are slightly shorter after substrate loading leading to a slight variation in the relative distances between helices 1, 2, and 3. The N-terminal end of α2 moves towards α1 and away from α3, in a manner that accommodates substrate docking in loaded-ArCP (Figure 2b and d). While doing so, α1 rotates slightly and the phosphate group on S52 is moved towards the outside of the protein (perhaps best seen in Figures S6 and S9), and the helices in the bundle become more parallel to one another. A major difference between the NMR ensembles of holo-ArCP and loaded-ArCP lies in a change of conformation in loop1, between L39 and N44 (Figure 2e). This region packs against the helical bundle with hydrophobic interactions towards the core, but also with interactions involving side-chains of α1 and α4 that are more peripheral. In holo the latter provide more NMR constraints while in the loaded form constraints with the core dominate. This change in conformation shifts the mean position of the amide proton of E41 by 4 Å. As a consequence, the ionic interaction previously mentioned between E41 and R25 can only occur in the holo form. Loop1 has been shown to be involved in a number of interactions with NRPS catalytic domains19,21–23,57–59 and its change in conformation is discussed further below.
Dynamics in the ArCP core
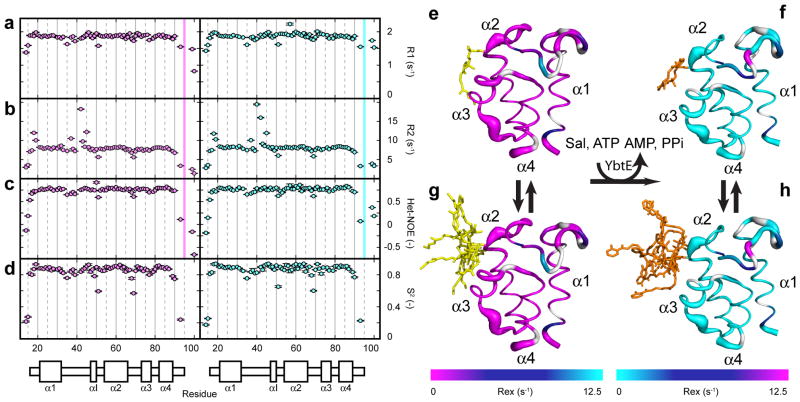
The protein cores of holo- and loaded-ArCP are mainly rigid on fast time-scales but undergo local conformational fluctuations at slower time-scales. To assess the dynamics of holo- and loaded-ArCP, we measured the nitrogen longitudinal (Figure 3a) and transverse (Figure 3b) relaxation rates (R1 and R2, respectively) as well as the heteronuclear nOe between amide protons and nitrogens (HN-NOE, Figure 3c). R2 is sensitive to both motions in ps-ns time-scale and in μs or slower time-scales. R1 is sensitive exclusively to ps-ns motions. HN-NOE is a direct reporter of ps-ns fluctuations in bond orientations. We applied the Lipari-Szabo formalism60 as developed by Palmer and co-workers61 and as implemented by Fushman and co-workers39,40 to obtain order parameters, S2, that provide a measure of the amplitude of ps-ns motions (a value of 1 indicates rigidity and 0 denotes complete disorder). Figures 3d–h reveal that all helices and a large part of the connecting loops are relatively rigid for both forms, and only a few selected residues display increased flexibility. They are L34 and T35 in the beginning of loop1, D51, the residue preceding the phosphopantetheinylation site and the last residue of loop1, and R68 and L69, both in loop2. No less than nine studies reported flexibility in the N-terminal half of loop1 in FAS and PKS acyl carrier proteins (ACPs).62–70 We speculate that such flexibility may be required to perform different interactions with different NRPS partner domains. The very position of L34 (i-18 with respect to S52) is used to stabilize binding with an adenylation domain in a related aryl carrier protein (EntB).19
Figure 3.
NMR dynamics of holo- and loaded-ArCP. (a–d) Residue-specific NMR relaxation parameters for holo (left, magenta) and loaded (right, cyan). The secondary structure of ArCP is illustrated below the plots. In (a–c) the relaxation parameters for the NN and NR positions of the PP are displayed at residues 99 and 100 respectively. A thick, colored line distinguishes them from the remaining residues. (a) R1 relaxation rates. (b) R2 relaxation rates. (c) Heteronuclear NOE parameterized by Isat/Iref where Isat and Iref are the amplitudes of signals in the proton-saturated and reference experiment, respectively. (d) Order parameter S2. (e–h) Dynamics visualization of holo- (magenta) and loaded-ArCP (cyan), residues 16–90. A thicker ribbon corresponds to a reduced order parameter and increased flexibility. Colors represent the Rex parameters fit during Lipari-Szabo analysis. Data are not available for residues in white due to overlap (e) holo-ArCP with PP in its bound state (f) loaded-ArCP with the PP in its bound state (g) holo-ArCP with PP in its unbound state (h) loaded-ArCP with PP in its unbound state
D51 stands out as the single residue with marked fast dynamics at the C-terminal end of loop1 in both holo and loaded forms (Figure 3d, e and f). This is a rather critical observation as D51 is the predecessor of the PP site, a position that has been shown to be actively involved in many domain interactions14,15,19,23,25,28,57,58 and even hypothesized to be participating in enzymatic activity,57 so this position must have access to many conformations to satisfy its role in these protein interactions. Upon re-inspecting previously reported relaxation rates and order parameters, we found that this position is flexible in five ACPs.64–70 This conserved flexibility likely reflects the versatile role that the position preceding the PP site plays during synthesis.
Fast internal motions have also been detected in the region encompassing R68 and L69 (loop2) in FAS and PKS ACPs. Flexibility has been consistently probed in loop2,62–64,66,67,70 sometimes extending to the adjacent α3.68,69 α3 has often been found to be subject to conformational exchange, and its conformation, as well as its relative orientation to α2 are modulated upon interactions with substrates and partner domains.22,62,63,67 The conserved flexibility of loop2 likely permits modulation of the relative orientation between helices α2 and α3.
Overall, substrate loading does not affect the flexibility of ArCP in a dramatic manner. Nevertheless, comparison of order parameters indicates a trend for a rigidification at ps-ns time-scales for a few residues. The small amplitude of these effects, however, prevents us from discussing their significance.
Slower conformational fluctuations (μs) increase the magnitudes of R2 for two groups of residues (Figure 3b, color coded in Figures 3e and f). A18, A19, and D20, are located at the N-terminal end of α1, and Q37, H40, E42, S43 and L45, are all in the center of loop1, up to the single turn helix, αI. All residues display Rex in the Model Free analysis (Figure S7). These two groups are remote in the structures of ArCP and these fluctuations likely reflect separate events.
The conformational fluctuations observed in loop1 are likely relevant to ArCP’s activity. Conformational exchange has been detected in corresponding regions of FAS and PKS ACPs64,70 and in nearby regions of loop1.68,69,71 Most strikingly, the two residues that display the largest R2 rates are H40 and E42 that flank E41 (Figures 3e and f). E41 could not be detected, presumably because its NMR resonances have been broadened by exchange beyond detection. We have already mentioned that E41 can form a salt bridge in holo-ArCP but not in loaded-ArCP because the surrounding region of loop1 changes conformation upon substrate loading. We observe an overall reduction of Rex upon substrate loading, indicating that the substrate affects related conformational fluctuations (Figures 3b and S7). Indeed, salicylate docks in the vicinity of αI, which signals the end of the malleable region, and the docking of Sal may modulate the dynamics of loop1. In summary, the change on conformation that we observed when comparing holo- and loaded-ArCP structures results from a change in conformational equilibria and is not a static effect. In this context, the flexibility of T35 and L34 discussed above may reflect a hinge in this region, used to provide malleability to the region encompassing Q37-L45. Various residues in loop1 have been shown to interact with NRPS catalytic domains19,21–23,57–59 and the significance of the malleability of loop1 will be discussed below.
Transient interactions between ArCP and its prosthetic groups
Our data reveal that, although they are well-defined, the interactions we see between ArCP and its prosthetic groups are transient in nature. Our relaxation data indicate that the NMR signals of PP reflect an extreme amount of motional averaging, characteristic of disorder (Figures 3a–c). However, nOe’s between ArCP and its prosthetic moieties as well as nOe’s within PP with the same sign as those of the protein indicate that PP is bound to ArCP. Together, these observations indicate an averaging of NMR parameters72 due to an equilibrium between a bound form of holo-ArCP, b-holo-ArCP, and a form in which PP does not bind and is disordered, u-holo-ArCP (unbound). Likewise, loaded-ArCP exists both in bound and unbound forms.
The detection of positive nOe’s with signals that seemingly display high motional averaging is reminiscent of the well-known transferred nOe’s that occur for small molecules binding to a large protein.73–78 The detection of nOe’s between PP and ArCP through signals of the unstructured form of PP is a special case of population averaged nOe’s, in which the nOe of u-holo-ArCP is zero. Similar effects have been discussed for nOe’s in protein cores and even using E. coli ACP as a model system.65,79 In the end, our results unequivocally indicate that holo-ArCP and loaded-ArCP are subject to equilibria between bound and unbound forms (Figures 3e and g). Because the bound forms of holo- and loaded-ArCP display well-defined interactions between ArCP and its prosthetic groups and given the dramatic alterations in domain binding surfaces that occur upon substrate loading, future characterization of the populations of the bound forms and of the time-scale of the equilibrium are warranted.
In addition to our investigations of ArCP’s flexibility and conformational fluctuations, we have observed signals indicative of a minor conformer of ArCP both in holo and loaded forms. Similar observations have been made for the tyrocidin A peptidyl carrier protein in apo and holo form and solution structures were determined by NMR using nOe constraints.22 Unfortunately, very low signal amplitudes provided limited NOESY cross-peaks in our spectra which would result in conformers that artificially appear partially unfolded. Similar signs of slow exchange have been ubiquitously observed for PKS and FAS ACPs.62,63,66–69,71,79,80 In one study, the second conformer was shown to involve a second binding site for the PP arm accompanied with alterations in α3.63 We do see a second set of signals for PP in holo-ArCP and loaded-ArCP, and a similar scenario cannot be excluded for ArCP. A limited chemical shift perturbation (Figure S8) indicates that these conformers are likely subject to subtle changes in conformations and do not belong to an unfolded or unfolded-like state. Further studies are needed to characterize these minor conformers.
Implications for domain communication
All our observations can be revisited within the context of domain interactions and catalytic steps that occur during biosynthesis. Following post-translational modification, ArCP interacts with the adenylation domain YbtE to harvest salicylate as well as with the cyclization domain Cy1 to catalyze the condensation of salicylate with cysteine.31,81 In this chain of events, holo-ArCP is the substrate of YbtE and loaded-ArCP is its product. Conversely, loaded-ArCP is the substrate for Cy1 and holo-ArCP is a product of Cy1.
As noted above, interactions between the core of the protein and the tethered salicylate alter the conformation of the PP between holo- and loaded-ArCP. This change in PP conformation in turn alters the electrostatic surface presented by loop1 and helices 2 and 3. Loop1, α2, and α3 are all involved in binding with other domains, and altering the solvent exposed surfaces as described necessarily modulates domain affinities. Indeed, studies of related PKS ACPs have shown that changes in surface potentials could explain the success or failure in ACP domain swaps82. Inspection of the surface potential of ArCP reveals a positively charged region defined by the second half of α2, loop2, and R72 in α3, as well as a negatively charged region delimited by the C-terminal end of loop1, the beginning of α2, and part of α3 and loop2 (SI Figure S9). In holo-ArCP much of the positively charged area is covered by the PP arm, while the negatively charged region is accessible. Upon substrate loading, part of the positively charged area becomes accessible while the negatively charged surface is obfuscated by Sal and the end of the PP arm. We hypothesize that such a dramatic modification of the surface potential in a region consistently involved in domain binding19,22,23,58,59 likely participates in modulating the binding affinity of ArCP towards its partner domains.
The equilibrium between unbound and bound forms of holo-ArCP may be modulated as it interacts with its adenylation domain. Like all adenylation domains, YbtE comprises a large N-terminal sub-domain, A(N), and a smaller C-terminal sub-domain, A(C). YbtE catalyzes two distinct steps. First, salicylate is selected and adenylated to produce an activated mixed anhydride Sal-AMP. Second, the thiol group of holo-ArCP reacts with the activated carbonyl to form a thioester bond and, hence, to load ArCP with salicylate. It has been shown that the relative orientation of A(N) and A(C) changes by about 140° during the two-step reaction, with an adenylation conformer AA active for adenylation and a thioester conformer AT responsible for thioesterification.16,17 The proposed mechanism invokes a transition from AA to AT upon completion of the adenylation reaction and prior to binding of holo-ArCP. Thioesterification requires PP to be extended towards the activated adenylate, e-holo-ArCP, and thus the relevant complex consists of e-holo-ArCP bound to AT (see Figure 4b). Our observations of equilibria between bound and unbound forms of holo-ArCP raise questions as to whether the bound form is used during A/CP recognition, and which mechanisms are compatible with such an equilibrium. An active participation of b-holo-ArCP would mean that the complex competent for thioesterification, e-holo-ArCP/AT, is obtained following formation of an encounter complex involving b-holo-ArCP in a manner reminiscent of that described by Burkart and coworkers for type II FAS ACP and its partner, FabA.83 Alternatively, AT may select e-holo-ArCP through u-holo-ArCP.
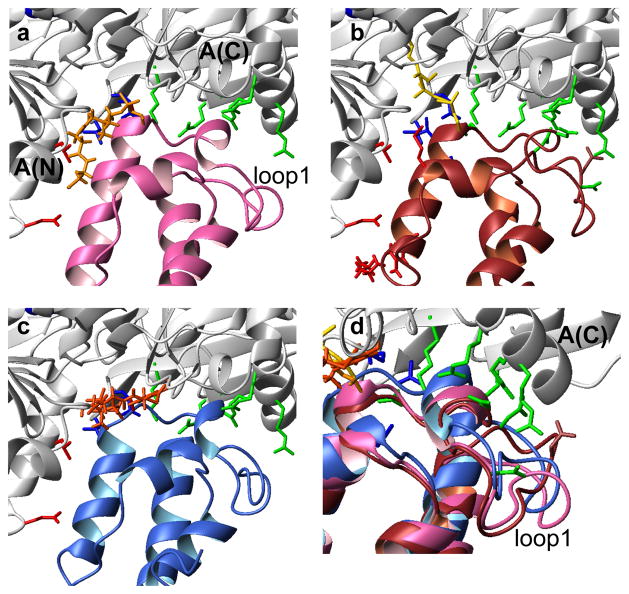
Figure 4.
Comparison of holo- (a, in pink) and loaded-ArCP (c, in blue) in complex with an adenylation domain in AT conformation (in white). The original structure of EntB-ArCP (brown) in complex with EntE is shown in b) (2ROG). In b), EntB-ArCP sidechains colored in red highlight interactions between A(N) and α2, those in blue show interactions between A(C) and ArCP, and those in green denote interactions between A(C) and loop1. The same color scheme was used for side-chains of EntE that are displayed in a–c). d) Detail showcasing changes in the conformation of loop1. The orientation in d is obtained by rotation of 30° around the Y vertical axis.
Combined insight from existing structures and the structures described here suggest how the positioning of PP may influence interactions with A domains. Complexes of e-holo-ArCP bound to AT have been crystallized, notably with an aryl carrier protein closely related to ArCP, EntB-ArCP.19 Loop1 in EntB-ArCP interacts with A(C) (Figure 4b), whereas α2 interacts both with A(C) and with A(N). Structural alignment of b-holo-ArCP onto EntB-ArCP reveals that b-holo-ArCP could maintain interactions with A(C) but not A(N) (Figure 4a). Thus, an encounter complex between AT and b-holo-ArCP would necessarily occur with a different domain organization than that seen in Figure 4b.
In either unbound or bound form, ArCP can interact with A(C). This is an important observation given that the domain reorientation in A domains was proposed to provide a means of shuttling carrier proteins between binding sites.15,19 If A(C) is used to shuttle holo-ArCP towards A(N), an interaction between A(C) and ArCP such as that shown in Figure 4 would likely shift the equilibrium towards b-holo-ArCP as many conformations of u-holo-ArCP would be incompatible with binding. Upon reaching a conformation AT, the adenylation domain must then select for e-holo-ArCP. This model would correspond to a sequence of conformational selection events and the equilibrium between b-holo-ArCP and u-holo-ArCP may then be a means to ensure simultaneously an interaction between A(C) and ArCP (with b-holo-ArCP paying an entropic cost and opening an interaction surface) while maintaining access to an extended form that must be selected through the unbound form for catalysis. The many mechanisms that are compatible with our observations beg for further investigations.
The loaded form of ArCP in bound form prevents interaction between ArCP and A(N) but allows for a stable interaction with A(C). Loaded-ArCP is the product of the thiolation reaction, and hence loaded-ArCP must dissociate from the A-domain. Inspection of Figure 4c demonstrates that the curled conformation of PP in the bound form would prevent any functional interaction between b-loaded-ArCP and A(N). Thus, b-loaded-ArCP likely helps ensure that NRPS synthesis moves productively to the next step. Like b-holo-ArCP, b-loaded-ArCP can interact with A(C). Again, this observation is compatible with A(C) shuttling loaded-ArCP towards the next catalytic partner, here Cy1.
Inspection of the conformation of loop1 within the context of an A/CP complex suggests that substrate loading may help ArCP interact with the C-terminal subdomain of YbtE. Figure 4d emphasizes that loop1 moves towards A(C) when comparing holo-ArCP, loaded-ArCP, and the EntB-ArCP. Loop1 simultaneously adopts a conformation that is increasingly open with respect to the ArCP core protein. With EntB-ArCP representing an optimal interaction, this observation suggests that loaded-ArCP adopts a conformation more suitable for an interaction with A(C). Thus, the bound form of loaded-ArCP may affect the affinity towards A domains in two manners, first by disrupting interactions with A(N) and adopting a conformation incompatible with a competent complex, and second by promoting an interaction between ArCP and A(C) by stabilizing loop1 in an open form. Substrate loading would then actively contribute to release from A(N) and shuttling of ArCP by A(C). Further studies are needed to probe this hypothesis.
The bound form of loaded-ArCP likely modulates interactions with partner cyclization or condensation domains, while access to the extended form needed for catalysis is provided by the unbound form. ArCP is the first carrier protein of the yersinibactin synthetase and, as such, Sal-loaded-ArCP is exclusively a substrate-donor carrier protein. There are no structures of condensation domains in complex with substrate-donor carrier proteins and we cannot make detailed mechanistic predictions for loaded-ArCP. Fortunately, combinatorial mutagenesis and selection studies, conducted specifically with ArCP or EntB-ArCP, have identified residues necessary for communication with condensation domains.59,84 They span the solvent accessible surface of N-terminal α2 (I53 and M56), C-terminal α3 (Y75 and A76) and one residue in loop1 (N44). Strikingly, all these residues interact with PP or Sal in b-loaded-ArCP. It may well be that selection was achieved by optimizing interactions with the tethered substrate. If so, this suggests that bound loaded-ArCP is used as a substrate for an encounter complex. Access to e-loaded-ArCP through its unbound form would then be used to reach the Cy (or C) domain active site, which is located far from the surface of these domains. Clearly, our observations must be revisited once the structure of a donor ArCP in complex with a cyclization or condensation domain is available.
In summary, the structures of b-holo-ArCP and b-loaded-ArCP that we have determined, together with our observations of equilibria between bound and free forms set a framework to elucidate mechanisms for domain recognition and domain rearrangements during NRPS synthesis.
Conclusions
We have presented the first solution structure of a nonribosomal peptide synthetase carrier protein loaded with its substrate, the backbone dynamics analysis of an NRPS carrier protein, and the first solution structure of an aryl carrier protein, both in holo and loaded forms.
The solution structures of ArCP indicate that the phosphopantetheinyl arm interacts with the protein core of ArCP both in holo and in loaded forms. Substrate loading induces a large conformational change in the phosphopantetheinyl arm that alters the nature of the protein surface in surrounding regions, which are involved in domain recognition. In holo-ArCP, PP lies in an extended conformation between helices α2 and α3, whereas in loaded-ArCP PP curls back to allow for substrate binding in a region near the phosphopantetheinylation site. The repositioning of the PP arm modulates access to regions of ArCP with distinct electrostatic potentials, providing a rationale for altering the binding affinity of ArCP to its partner domains.
Joint analysis of NOESY spectra and NMR spin relaxation indicate that ArCP interacts with its prosthetic groups in a transient yet well-defined manner. Our findings indicate that both holo- and loaded-ArCP undergo conformational equilibria between unbound and bound forms. Interactions with PP have been occasionally observed in NMR studies of NRPS, PKS and FAS carrier proteins and the (well-established) motional averaging of NMR parameters we discussed can be applied to revisit observations made for these systems.
Many molecular properties have been discussed within the context of NRPS, PKS, and FAS domain communication and our results suggest that the dynamics of the prosthetic group must be considered as well. That is, binding to carrier proteins must occur through a conformational selection of unbound or bound forms, potentially with subsequent induced fits or conformational selection events. In addition, we have observed dynamics in the protein cores of holo- and loaded-ArCP that cover all time-scales. Notably, we have found that substrate loading affects conformational fluctuations occurring in loop1, a region involved in domain communication. Together, our results substantiate a mechanism for NRPS synthesis that relies on the interplay between chemical modification and modulation of molecular properties, such as dynamics and surface potentials, to direct domain communication.
Supplementary Material
Acknowledgments
We thank Drs. Mike Burkart and Craig Townsend for stimulating discussions, and the Burkart lab for coordinating publication following acceptance of their manuscript. This research was supported by the National Institutes of Health (grants GM 104257). A.C.G. is supported by an American Heart Association Mid-Atlantic Affiliate Predoctoral Fellowship 14PRE20460253.
Footnotes
ASSOCIATED CONTENT
Details of cloning and NMR acquisitions, NMR ensembles, NOESY strips for prosthetic moieties, Model-Free Analysis, Chemical Shift Perturbations of minor and major forms, Surface potentials, NMR assignments, PDB coordinates.
References
- 1.Khosla C. Chem Rev. 1997;97:2577. doi: 10.1021/cr960027u. [DOI] [PubMed] [Google Scholar]
- 2.Jenni S, Leibundgut M, Maier T, Ban N. Science. 2006;311:1263. doi: 10.1126/science.1123251. [DOI] [PubMed] [Google Scholar]
- 3.Jenni S, Leibundgut M, Boehringer D, Frick C, Mikolasek B, Ban N. Science. 2007;316:254. doi: 10.1126/science.1138248. [DOI] [PubMed] [Google Scholar]
- 4.Leibundgut M, Jenni S, Frick C, Ban N. Science. 2007;316:288. doi: 10.1126/science.1138249. [DOI] [PubMed] [Google Scholar]
- 5.Khosla C. J Org Chem. 2009;74:6416. doi: 10.1021/jo9012089. [DOI] [PubMed] [Google Scholar]
- 6.Khosla C, Kapur S, Cane DE. Curr Opin Chem Biol. 2009;13:135. doi: 10.1016/j.cbpa.2008.12.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dutta S, Whicher JR, Hansen DA, Hale WA, Chemler JA, Congdon GR, Narayan AR, Hakansson K, Sherman DH, Smith JL, Skiniotis G. Nature. 2014;510:512. doi: 10.1038/nature13423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Whicher JR, Dutta S, Hansen DA, Hale WA, Chemler JA, Dosey AM, Narayan AR, Hakansson K, Sherman DH, Smith JL, Skiniotis G. Nature. 2014;510:560. doi: 10.1038/nature13409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Smith JL, Skiniotis G, Sherman DH. Curr Opin Struct Biol. 2015;31:9. doi: 10.1016/j.sbi.2015.02.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mootz HD, Schwarzer D, Marahiel MA. Chembiochem: a European journal of chemical biology. 2002;3:490. doi: 10.1002/1439-7633(20020603)3:6<490::AID-CBIC490>3.0.CO;2-N. [DOI] [PubMed] [Google Scholar]
- 11.Finking R, Marahiel MA. Annu Rev Microbiol. 2004;58:453. doi: 10.1146/annurev.micro.58.030603.123615. [DOI] [PubMed] [Google Scholar]
- 12.Samel SA, Schoenafinger G, Knappe TA, Marahiel MA, Essen LO. Structure. 2007;15:781. doi: 10.1016/j.str.2007.05.008. [DOI] [PubMed] [Google Scholar]
- 13.Lambalot RH, Gehring AM, Flugel RS, Zuber P, LaCelle M, Marahiel MA, Reid R, Khosla C, Walsh CT. Chem Biol. 1996;3:923. doi: 10.1016/s1074-5521(96)90181-7. [DOI] [PubMed] [Google Scholar]
- 14.Frueh DP, Arthanari H, Koglin A, Vosburg DA, Bennett AE, Walsh CT, Wagner G. Nature. 2008;454:903. doi: 10.1038/nature07162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tanovic A, Samel SA, Essen LO, Marahiel MA. Science. 2008;321:659. doi: 10.1126/science.1159850. [DOI] [PubMed] [Google Scholar]
- 16.Yonus H, Neumann P, Zimmermann S, May JJ, Marahiel MA, Stubbs MT. J Biol Chem. 2008;283:32484. doi: 10.1074/jbc.M800557200. [DOI] [PubMed] [Google Scholar]
- 17.Gulick AM. ACS chemical biology. 2009;4:811. doi: 10.1021/cb900156h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Strieker M, Tanovic A, Marahiel MA. Curr Opin Struct Biol. 2010;20:234. doi: 10.1016/j.sbi.2010.01.009. [DOI] [PubMed] [Google Scholar]
- 19.Sundlov JA, Shi C, Wilson DJ, Aldrich CC, Gulick AM. Chem Biol. 2012;19:188. doi: 10.1016/j.chembiol.2011.11.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Weber T, Baumgartner R, Renner C, Marahiel MA, Holak TA. Structure. 2000;8:407. doi: 10.1016/s0969-2126(00)00120-9. [DOI] [PubMed] [Google Scholar]
- 21.Drake EJ, Nicolai DA, Gulick AM. Chem Biol. 2006;13:409. doi: 10.1016/j.chembiol.2006.02.005. [DOI] [PubMed] [Google Scholar]
- 22.Koglin A, Mofid MR, Lohr F, Schafer B, Rogov VV, Blum MM, Mittag T, Marahiel MA, Bernhard F, Dötsch V. Science. 2006;312:273. doi: 10.1126/science.1122928. [DOI] [PubMed] [Google Scholar]
- 23.Mitchell CA, Shi C, Aldrich CC, Gulick AM. Biochemistry. 2012;51:3252. doi: 10.1021/bi300112e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Allen CL, Gulick AM. Acta Crystallogr D Biol Crystallogr. 2014;70:1718. doi: 10.1107/S1399004714008311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Haslinger K, Brieke C, Uhlmann S, Sieverling L, Sussmuth RD, Cryle MJ. Angew Chem. 2014;53:8518. doi: 10.1002/anie.201404977. [DOI] [PubMed] [Google Scholar]
- 26.Lohman JR, Ma M, Cuff ME, Bigelow L, Bearden J, Babnigg G, Joachimiak A, Phillips GN, Jr, Shen B. Proteins. 2014;82:1210. doi: 10.1002/prot.24485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Haslinger K, Redfield C, Cryle MJ. Proteins. 2015;83:711. doi: 10.1002/prot.24758. [DOI] [PubMed] [Google Scholar]
- 28.Liu Y, Zheng T, Bruner SD. Chem Biol. 2011;18:1482. doi: 10.1016/j.chembiol.2011.09.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Goodrich AC, Frueh DP. Biochemistry. 2015;54:1154. doi: 10.1021/bi501433r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gehring AM, DeMoll E, Fetherston JD, Mori I, Mayhew GF, Blattner FR, Walsh CT, Perry RD. Chem Biol. 1998;5:573. doi: 10.1016/s1074-5521(98)90115-6. [DOI] [PubMed] [Google Scholar]
- 31.Gehring AM, Mori I, Perry RD, Walsh CT. Biochemistry. 1998;37:11637. doi: 10.1021/bi9812571. [DOI] [PubMed] [Google Scholar]
- 32.Pelludat C, Rakin A, Jacobi CA, Schubert S, Heesemann J. J Bacteriol. 1998;180:538. doi: 10.1128/jb.180.3.538-546.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Keating TA, Miller DA, Walsh CT. Biochemistry. 2000;39:4729. doi: 10.1021/bi992923g. [DOI] [PubMed] [Google Scholar]
- 34.Keating TA, Suo Z, Ehmann DE, Walsh CT. Biochemistry. 2000;39:2297. doi: 10.1021/bi992341z. [DOI] [PubMed] [Google Scholar]
- 35.Suo Z, Tseng CC, Walsh CT. Proc Natl Acad Sci U S A. 2001;98:99. doi: 10.1073/pnas.021537498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Miller DA, Luo L, Hillson N, Keating TA, Walsh CT. Chem Biol. 2002;9:333. doi: 10.1016/s1074-5521(02)00115-1. [DOI] [PubMed] [Google Scholar]
- 37.Delaglio F, Grzesiek S, Vuister GW, Zhu G, Pfeifer J, Bax A. JBNMR. 1995;6:277. doi: 10.1007/BF00197809. [DOI] [PubMed] [Google Scholar]
- 38.Keller RLJ. The Computer Aided Resonance Assignment Tutorial. Cantina Verlag; Goldau: 2004. [Google Scholar]
- 39.Berlin K, Longhini A, Dayie TK, Fushman D. J Biomol NMR. 2013;57:333. doi: 10.1007/s10858-013-9791-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Walker O, Varadan R, Fushman D. J Magn Reson. 2004;168:336. doi: 10.1016/j.jmr.2004.03.019. [DOI] [PubMed] [Google Scholar]
- 41.Schrodinger, LLC 2010.
- 42.Shen Y, Bax A. J Biomol NMR. 2013;56:227. doi: 10.1007/s10858-013-9741-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Guntert P. Methods Mol Biol. 2004;278:353. doi: 10.1385/1-59259-809-9:353. [DOI] [PubMed] [Google Scholar]
- 44.Brunger AT, Adams PD, Clore GM, DeLano WL, Gros P, Grosse-Kunstleve RW, Jiang JS, Kuszewski J, Nilges M, Pannu NS, Read RJ, Rice LM, Simonson T, Warren GL. Acta Crystallogr D Biol Crystallogr. 1998;54:905. doi: 10.1107/s0907444998003254. [DOI] [PubMed] [Google Scholar]
- 45.Sousa da Silva AW, Vranken W, Laue ED. Manuscript to be submitted. [Google Scholar]
- 46.Nederveen AJ, Doreleijers JF, Vranken W, Miller Z, Spronk CA, Nabuurs SB, Guntert P, Livny M, Markley JL, Nilges M, Ulrich EL, Kaptein R, Bonvin AM. Proteins. 2005;59:662. doi: 10.1002/prot.20408. [DOI] [PubMed] [Google Scholar]
- 47.Bhattacharya A, Tejero R, Montelione GT. Proteins. 2007;66:778. doi: 10.1002/prot.21165. [DOI] [PubMed] [Google Scholar]
- 48.Laskowski R, Rullmann JA, MacArthur M, Kaptein R, Thornton J. J Biomol NMR. 1996;8:477. doi: 10.1007/BF00228148. [DOI] [PubMed] [Google Scholar]
- 49.Davis IW, Leaver-Fay A, Chen VB, Block JN, Kapral GJ, Wang X, Murray LW, Arendall WB, 3rd, Snoeyink J, Richardson JS, Richardson DC. Nucleic Acids Res. 2007;35:W375. doi: 10.1093/nar/gkm216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Chen VB, Arendall WB, 3rd, Headd JJ, Keedy DA, Immormino RM, Kapral GJ, Murray LW, Richardson JS, Richardson DC. Acta Crystallogr D Biol Crystallogr. 2010;66:12. doi: 10.1107/S0907444909042073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Baker NA, Sept D, Joseph S, Holst MJ, McCammon JA. Proc Natl Acad Sci USA. 2001;98:10037. doi: 10.1073/pnas.181342398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Dolinsky TJ, Nielsen JE, McCammon JA, Baker NA. Nucleic Acids Res. 2004;32:W665. doi: 10.1093/nar/gkh381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Dolinsky TJ, Czodrowski P, Li H, Nielsen JE, Jensen JH, Klebe G, Baker NA. Nucleic Acids Res. 2007;35:W522. doi: 10.1093/nar/gkm276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Koradi R, Billeter M, Wuthrich K. J Mol Graphics. 1996;14:51. doi: 10.1016/0263-7855(96)00009-4. [DOI] [PubMed] [Google Scholar]
- 55.Holak TA, Kearsley SK, Kim Y, Prestegard JH. Biochemistry. 1988;27:6135. doi: 10.1021/bi00416a046. [DOI] [PubMed] [Google Scholar]
- 56.Crump MP, Crosby J, Dempsey CE, Parkinson JA, Murray M, Hopwood DA, Simpson TJ. Biochemistry. 1997;36:6000. doi: 10.1021/bi970006+. [DOI] [PubMed] [Google Scholar]
- 57.Parris KD, Lin L, Tam A, Mathew R, Hixon J, Stahl M, Fritz CC, Seehra J, Somers WS. Structure. 2000;8:883. doi: 10.1016/s0969-2126(00)00178-7. [DOI] [PubMed] [Google Scholar]
- 58.Tufar P, Rahighi S, Kraas FI, Kirchner DK, Lohr F, Henrich E, Kopke J, Dikic I, Guntert P, Marahiel MA, Dotsch V. Chem Biol. 2014;21:552. doi: 10.1016/j.chembiol.2014.02.014. [DOI] [PubMed] [Google Scholar]
- 59.Zhou Z, Lai JR, Walsh CT. Proc Natl Acad Sci USA. 2007;104:11621. doi: 10.1073/pnas.0705122104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Lipari G, Szabo A. J Am Chem Soc. 1982;104:4546. [Google Scholar]
- 61.Mandel AM, Akke M, Palmer AG., III J Mol Biol. 1995;246:144. doi: 10.1006/jmbi.1994.0073. [DOI] [PubMed] [Google Scholar]
- 62.Li Q, Khosla C, Puglisi JD, Liu CW. Biochemistry. 2003;42:4648. doi: 10.1021/bi0274120. [DOI] [PubMed] [Google Scholar]
- 63.Sharma AK, Sharma SK, Surolia A, Surolia N, Sarma SP. Biochemistry. 2006;45:6904. doi: 10.1021/bi060368u. [DOI] [PubMed] [Google Scholar]
- 64.Kim Y, Kovrigin EL, Eletr Z. Biochem Biophys Res Commun. 2006;341:776. doi: 10.1016/j.bbrc.2006.01.025. [DOI] [PubMed] [Google Scholar]
- 65.Kim Y, Prestegard JH. Biochemistry. 1989;28:8792. doi: 10.1021/bi00448a017. [DOI] [PubMed] [Google Scholar]
- 66.Chan DI, Chu BC, Lau CK, Hunter HN, Byers DM, Vogel HJ. J Biol Chem. 2010;285:30558. doi: 10.1074/jbc.M110.128298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Zornetzer GA, Tanem J, Fox BG, Markley JL. Biochemistry. 2010;49:470. doi: 10.1021/bi9014659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Lim J, Kong R, Murugan E, Ho CL, Liang ZX, Yang D. PloS one. 2011;6:e20549. doi: 10.1371/journal.pone.0020549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Lim J, Xiao T, Fan J, Yang D. Angew Chem Int Ed. 2014;53:2358. doi: 10.1002/anie.201308512. [DOI] [PubMed] [Google Scholar]
- 70.Findlow SC, Winsor C, Simpson TJ, Crosby J, Crump MP. Biochemistry. 2003;42:8423. doi: 10.1021/bi0342259. [DOI] [PubMed] [Google Scholar]
- 71.Lim J, Sun H, Fan J-S, Hameed Iman F, Lescar J, Liang Z-X, Yang D. Biophys J. 2012;103:1037. doi: 10.1016/j.bpj.2012.08.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Wennerström H. Mol Phys. 1972;24:69. [Google Scholar]
- 73.Balaram P, Bothnerb Aa, Breslow E. J Am Chem Soc. 1972;94:4017. [Google Scholar]
- 74.Balaram P, Bothner-By AA, Dadok J. J Am Chem Soc. 1972;94:4015. doi: 10.1021/ja00766a063. [DOI] [PubMed] [Google Scholar]
- 75.Clore GM, Gronenborn AM. J Magn Reson. 1982;48:402. [Google Scholar]
- 76.Lippens GM, Cerf C, Hallenga K. J Magn Reson. 1992;99:268. [Google Scholar]
- 77.Nageswara Rao BD. In: Methods Enzymol. Norman JO, Thomas LJ, editors. Vol. 176. Academic Press; 1989. p. 279. [DOI] [PubMed] [Google Scholar]
- 78.Nirmala NR, Lippens GM, Hallenga K. J Magn Reson. 1992;100:25. [Google Scholar]
- 79.Kim Y, Ohlrogge JB, Prestegard JH. Biochem Pharmacol. 1990;40:7. doi: 10.1016/0006-2952(90)90171-g. [DOI] [PubMed] [Google Scholar]
- 80.Oswood MC, Kim Y, Ohlrogge JB, Prestegard JH. Proteins. 1997;27:131. [PubMed] [Google Scholar]
- 81.Suo Z, Walsh CT, Miller DA. Biochemistry. 1999;38:14023. doi: 10.1021/bi991574n. [DOI] [PubMed] [Google Scholar]
- 82.Alekseyev VY, Liu CW, Cane DE, Puglisi JD, Khosla C. Protein Sci. 2007;16:2093. doi: 10.1110/ps.073011407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Nguyen C, Haushalter RW, Lee DJ, Markwick PR, Bruegger J, Caldara-Festin G, Finzel K, Jackson DR, Ishikawa F, O’Dowd B, McCammon JA, Opella SJ, Tsai SC, Burkart MD. Nature. 2014;505:427. doi: 10.1038/nature12810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Lai JR, Fischbach MA, Liu DR, Walsh CT. Proc Natl Acad Sci U S A. 2006;103:5314. doi: 10.1073/pnas.0601038103. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.