Abstract
The Drosophila melanogaster histone lysine methyltransferase (HKMT) Eggless (Egg/dSETDB1) catalyzes methylation of Histone H3 lysine 9 (H3K9), a signature of repressive heterochromatin. Our previous studies showed that H3K9 methylation by Egg is required for oogenesis. Here we analyze a set of EMS-induced mutations in the egg gene, identify the molecular lesions of these mutations, and compare the effects on oogenesis of both strong loss-of-function and weak hypomorphic alleles. These studies show that H3K9 methylation by Egg is required for multiple stages of oogenesis. Mosaic expression experiments show that the egg gene is not required intrinsically in the germ cells for their early differentiation, but is required in the germ cells for their survival past stage 5 of oogenesis. egg is also required in germ stem cells for their maintenance, since egg− germ stem cells initially survive but are not maintained as females age. Mosaic analysis also reveals that the early egg chamber budding defects in egg− ovaries are due to an intrinsic requirement for egg in follicle stem cells and their descendents, and that egg plays a non-autonomous role in somatic cells in the germarium to influence the differentiation of early germ cells.
Keywords: heterochromatin, methylation, epigenetics, oogenesis, stem cells
Introduction
Histone tails are subject to extensive post-translational modifications (Peterson and Laniel, 2004). The complexity of these modifications are at the core of the histone code hypothesis, which predicts that combinatorial histone signatures are bound by proteins that recognize these signatures, with specific outcomes for chromatin structure and function (Strahl and Allis, 2000). In fact histone modifications impact many phenomena, including gene expression, chromosome architecture, DNA replication, and nuclear architecture (Greer and Shi, 2012). One type of histone modification, methylation of histone lysine residues, illustrates the diverse consequences of any given type of histone modification.
Histone lysine methylation has distinct outcomes for chromatin structure and gene expression depending on the particular histone and lysine residue that is modified, as well as the degree of its methylation (Lachner and Jenuwein, 2002). The enzymes responsible for this modification, histone lysine methyltransferases (HKMTs), are targeted by associated proteins and non-coding RNAs, and their methylated lysine marks are recognized and bound by proteins that regulate chromatin structure and gene expression (Black et al., 2012). The consequences of a particular histone lysine mark can vary depending on its location within the genome. For instance, Histone H3 methylated at lysine 9 (H3K9me) is bound by Heterochromatin Protein 1a (HP1a), an essential step in the establishment of heterochromatin (Bannister et al., 2001; Lachner et al. 2001). This chromatin state is usually associated with gene repression (Grewal and Jia, 2007). However, in some instances H3K9me and HP1a are associated with active genes, illustrating that the biological consequences of this specific histone mark are themselves complex and context-dependent (Yasuhara and Wakimoto, 2006; Lundberg et al., 2013). HKMTs have profound effects on development (Dambacher, 2010), and altered expression of these enzymes has been recognized in an increasing number of human diseases (Moss and Wallrath, 2007; Decarlo and Hadden, 2012).
To address the importance of HKMTs in vivo, we are studying the developmental roles of Eggless/dSETDB1, the Drosophila ortholog of human SETDB1 (mouse ESET). We identified the gene in a mutagenesis screen and named the gene eggless (egg), based on the oogenesis defects of mutant females (Clough et al., 2007), but it is also called dSETDB1 (Seum et al., 2007a; Tzeng et al., 2007) and dEset (Stabell et al., 2006). The Egg protein, like its mammalian ortholog, contains a SET domain (the catalytic methyltransferase domain), two tandem Tudor domains, and a methyl-CpG binding (MBD) domain (Schultz et al., 2002; Yang et al., 2002; Stabell et al., 2006; Clough et al., 2007; Seum et al., 2007a).
Our previous studies showed that Egg is required for trimethylation of H3K9 (H3K9me3) in the germarium of the ovary, and that oogenesis arrests at very early stages in females carrying strong loss-of-function egg mutations (Clough et al., 2007). In this study we characterize our entire set of EMS-induced egg mutations and report the nature of their molecular lesions. A more in-depth analysis of the oogenesis defects caused by strong loss-of-function alleles demonstrates that egg is required very early in oogenesis for the differentiation of germ stem cell daughter cells, while an analysis of weak, hypomorphic alleles reveals that egg is also required for H3K9 methylation at late stages of oogenesis, where it is essential for terminal stages of oocyte development. Using methods of mosaic gene expression to address which cells in the ovary require egg activity, we show that egg has both cell-autonomous and non-autonomous effects on germ cell development at early stages of oogenesis. In germ cells, egg is required intrinsically for the maintenance of germ stem cells, and for the survival of developing germline cells past stage 5 of oogenesis. egg is required in follicle stem cells and their daughter cells for the formation and budding of egg chambers, and has a non-autonomous role in somatic cells in the germarium that influences the early differentiation of the germ cells.
Materials and Methods
Fly Stocks and genetics
All flies were maintained at 25°C or RT (22° – 25°C) on cornmeal-molasses medium supplemented with live yeast. The genotypes of strains containing the egg mutations were: w; cn bw egg*/GFPCyO, where * denotes any of the following allele numbers: A97, A510, A711, 178, 235, 238, 391, 1020, 1024, 1473, 1372, 1670, 2138. The GFPCyO chromosome expresses EGFP in the kruppel pattern (Casso et al., 2000). Oogenesis was analyzed in egg*/Df(2R)Dll-Mp females. Df(2R)Dll-Mp/Cyo was obtained from the Bloomington Drosophila Stock Center. Clonal analysis utilized the following recombinant lines: w1118; FRTG13, bw egg2138/SM1 and w1118; FRTG13, bw egg1473/SM1. The dSETDB1null allele and the UAST-dSETDB1 transgenic line were obtained from C-K.J. Shen’s lab (Tzeng et al., 2007). tj-Gal4 (P{GawB}NP1624-5-1) was obtained from the Drosophila Genome Resource Centre in Kyoto. The genotype of the Gal4-rescued flies was w1118; tj-Gal4,dSETDB1null/Df(2R)Dll-Mp;UAST-dSETDB1/+. A stock carrying the w1118 mutation on the X chromosome was used as a wildtype control.
Sequencing of alleles
Genomic DNA was extracted from hemizygous egg flies (egg*/Df(2R)Dll-MP) by homogenizing single flies in 50 µl of extraction buffer (10 mM Tris, pH 8.2, 1 mM EDTA, 25 mM NaCl, 200 µg/ml proteinase K). The homogenate was incubated at 37°C for 30 minutes followed by a three-minute incubation at 94°C to inactivate the proteinase K. Amplified DNA from four to six independent PCR reactions using egg-specific primers was pooled, purified (Qiagen), and used as template for sequencing with primers designed at intervals of 200–300 bp encompassing the entire egg coding sequence. Identification of each egg mutation was confirmed by repeat sequencing runs of independent DNA samples, and by sequencing the opposite strand at the location of the mutation. Sequencing was performed at the Columbia University Herbert Irving Comprehensive Cancer Center’s DNA Analysis and Sequencing Facility or by Genewiz.
Clonal analysis
The genotypes of female flies used in these experiments included the following (GFP indicates Ubi-GFP): (1) y w hsFLP12/w; FRTG13, egg1473/FRTG13, Ubi-GFP, (2) y w hsFLP12/w; FRTG13, egg2138/FRTG13, Ubi-GFP, and (3) y w hsFLP12/w; FRT G13/FRT G13, Ubi-GFP. egg− clones were identified by the absence of the Ubi-GFP marker.
Clones were induced by heat shock in either 3rd instar larvae or 1–2 day old adults. Larvae were heat shocked once for 2 hours, while adults were heat shocked 1 hour twice a day (with a 7 hour recovery period) for 3 days. Heat shocks were performed by placing vials with flies in a water bath at 37°C for the indicated lengths of time.
Antibody labeling and microscopy
For antibody labeling, the ovaries were dissected in PBS, fixed for 9 minutes in 5.9% formaldehyde in 1X Ephrussi buffer (16.7 mM KH2PO4, 75 mM KCl, 25 mM NaCl, 3.3 mM MgCl2), rinsed twice briefly, and then washed 3 X 5 minutes in PBS with 0.1% Triton-X100 (PBTx) and blocked for several hours in 1% BSA in PBTx. Primary and secondary antibodies were diluted to the appropriate concentration in PBTx with 0.1% BSA. The ovaries were incubated with the primary antibody overnight at 4°C, incubated with the appropriate secondary antibody for two hours at room temperature, and washed 3 × 10 minute in PBTx. DNA was stained with with propidium iodide (10 µg/ml) or sytox green (1:10, Molecular Probes) following a one-hour incubation in RNAseA (1 mg/ml) at 37 °C and three five-minute washes in PBTx. The ovaries were mounted on microscope slides in Fluoromount G (Southern Biotech). Primary antibodies included rabbit anti-H3K9me3 (1:1000, Upstate), mouse anti-Hts1B1 (1:25, Developmental Studies Hybridoma Bank, DSHB), rabbit anti-Vasa (1:500, R. Lehmann), rabbit anti-Cleaved Caspase-3 (1:500; Cell Signaling), mouse anti-Fas3 (1:10, DSHB), rabbit anti-Anillin (1:1000, Christine Field), rat anti-BamC (1:10; D. McKearin), and mouse anti-Sxl (1:10, DSHB). Secondary antibodies included FITC, rhodamine, or Cy5-conjugated goat or donkey anti-mouse, anti-rabbit or anti-rat F(ab’)2 fragments (Jackson ImmunoResearch), and were used at 1:500. Microscopy was performed on an inverted Olympus1X71 confocal microscope, using Fluoview and Image J software.
Results
Molecular characterization of a set of EMS-induced egg mutations
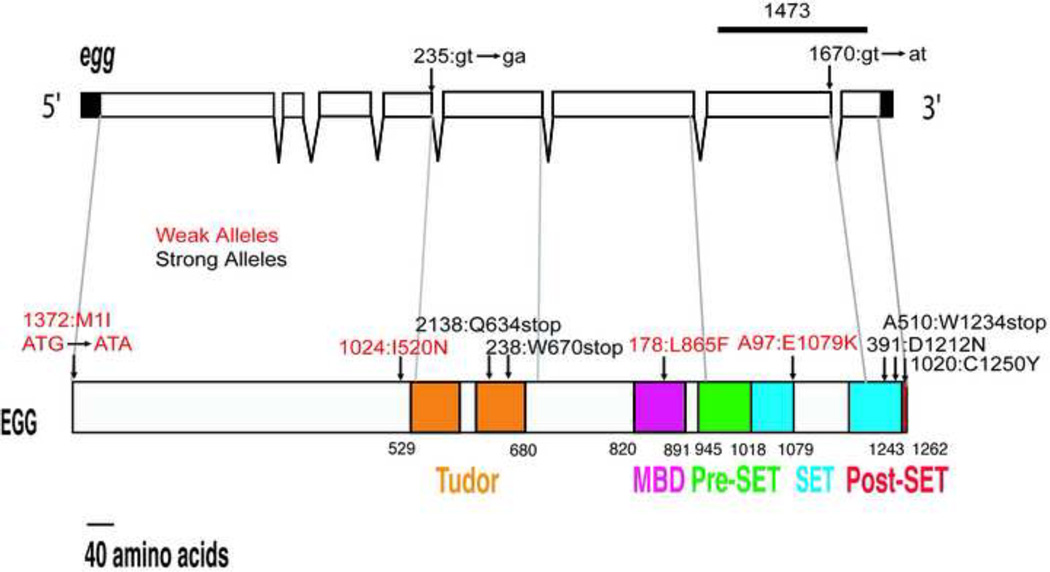
The Drosophila Egg protein contains the following domains: a catalytic SET domain, which is interrupted by a long intervening (SET-I) region; conserved Pre- and Post-SET domains; two tandem Tudor domains; and a MBD domain (Fig. 1). Previously we reported the isolation of 13 female-sterile EMS-induced mutations in egg (Clough et al., 2007). These mutations were categorized as strong or weak, based on the phenotypes of mutant females: strong alleles were associated with very early arrest of oogenesis and reduced viability, whereas weak alleles allowed oogenesis to proceed to later stages and did not affect viability. One allele in this original collection was subsequently lost; the remaining 12 mutations were analyzed in this study.
Figure 1. EMS-induced egg mutations.
The position and nature of the egg mutations characterized in this study are shown mapped to the egg transcript (top) and protein (bottom). The bar above the transcript indicates the region deleted in the egg1473 allele. The strong alleles are shown in black and the weak alleles in red.
Two strong alleles, egg235 and egg1473 were sequenced earlier: egg235, a mutation at the donor site of the 4th intron, introduces a premature stop codon in the mRNA if this intron is not removed, and egg1437 contains an in frame deletion of the entire SET domain (Clough et al., 2007). The molecular lesions of the remaining 10 mutations were determined by sequencing, and all 12 mutations are shown in Fig. 1. The eight strong alleles include nonsense mutations (egg2138, egg238 and eggA510), splice site mutations (egg235 and egg1670), missense mutations (egg391, and egg1020) and an internal deletion that removes the entire SET domain (egg1473). The four weak alleles are all missense mutations (egg1372, egg1024, egg178, and eggA97).
Effects of strong egg/dSETDB1 mutations on viability
All of the strong egg mutations affect viability. To quantify these effects, the survival rates of several strong alleles were determined. The mutations chosen for this analysis included mutations that introduced early premature stop codons (egg235, egg2138, egg238), a deletion that removes the SET domain (egg1473), and two alleles isolated by others (Seum et al., 2007a; Tzeng et al., 2007) that are deleted for most or all of the coding sequence (dmSetdb110.1a and dSETDB1null). Each allele was tested in heterozygotes carrying a deficiency for the region, Df(2R)Dll-Mp (breakpoints 60E1-2; 60E6). Since Seum et al (2007a) reported that their allele, dmSetdb110.1a, was lethal using a different deficiency for their analysis, Df(2R)ED4065 (breakpoints 60C8;60E8), we compared survival over both deficiencies. In heterozygous combination with Df(2R)Dll-Mp, all of the strong alleles reduced viability by 50% or more, and the effects on viability were more pronounced in males than in females (Table 1). Viability was reduced further in Df(2R)ED4065 heterozygotes (table 1), suggesting that this larger deficiency removes a gene (or genes) that enhances the lethal effects of the egg/dsetdb1 mutations.
Table 1.
Viability of strong egg mutations
| egg235 | egg2138 | egg238 | egg1473 | dSetDB1null | dSetdb110.1a | |
|---|---|---|---|---|---|---|
| Dll-MP | 40:217 [37%] [0.08] n=257 |
86:344 [50%] [0.02] n=430 |
70:322 [43%] [0.17] n=392 |
54:359 [30%] [0.17] n=413 |
39:181 [43%] [0.50] n=220 |
43:294 [29%] [0.13] n=337 |
| ED4065 | 5:191 [5%] [n.a.] n=196 |
24:266 [18%] [0.09] n=290 |
18:331 [11%] [0.06] n=349 |
5:155 [6%] [n.a.] n=160 |
3:249 [2%] [n.a.] n=252 |
4:80 [10%] [n.a.] n=84 |
Shown are the numbers of progeny obtained from crosses of egg/CyO males with Df(2R)Dll-Mp or Df(2R)ED4065 females. Each entry includes: the number of egg/Df: CyO adult progeny (top line); the percentage of obtained/expected egg/Df progeny [in brackets]; the egg/Df male : female ratio (in parentheses; n.a. indicates no male progeny had eclosed), and n = total number of progeny scored.
The egg adults that eclosed had held out wings that were incompletely expanded, and the flies moved with shaky, uncoordinated movements. The wing and locomoter defects were more pronounced in males, and the few surviving males died within 2–3 days of eclosion. Thus, egg mutations are more detrimental to the development and survival of males than females. The lethal phase of egg mutants was determined, using the egg1473 mutation as a representative strong allele. Using a GFP-marked CyO balancer, the number of egg1473/ egg1473 and egg1473/GFPCyO progeny was scored at 3rd instar larval, pupal and adult stages after a cross between egg1473/GFPCyO parents (CyO homozygotes die as embryos or early first instar larvae). Whereas the expected 33% of pupae were egg1473 homozygotes (N=176), this percentage dropped to 20% in the adult sample (N=148). Pharate egg1473 adults that had initiated eclosion but remained stuck in their pupal cases were frequently observed. Thus, egg is first required for survival late in pupation, and its loss leads to defects in morphogenesis and late pupal death.
egg is required at multiple stages of oogenesis
The ovaries of Drosophila contain 15–20 ovarioles with egg chambers at progressive stages of development. Oogenesis is initiated in the germarium, at the anterior tip of each ovariole, where 2–3 germ stem cells (GSCs) are maintained in a stem cell niche (Xie and Spradling, 2000; Eliazer and Buszczak, 2011). Differentiation of a germ stem cell daughter, the cystoblast (CB), occurs when the CB moves away from this niche, removing it from signals from the cap and terminal filament cells that repress differentiation. The CB undergoes four rounds of incomplete mitotic divisions to form a cyst of 16 interconnected germ cells. Associated somatic cells, the escort cells, send signals to the germ cells that influence their early differentiation (Kirilly et al., 2011). The germline cysts are eventually encapsulated by prefollicular cells, produced by follicle stem cells (FSCs), forming stage 1 egg chambers that bud off from the germarium and progress through several distinct stages of oogenesis to form mature stage 14 oocytes. We showed previously that Egg is expressed in both germ cells and somatic cells in the germarium, where it mediates trimethylation of histone H3 at its lysine 9 residue (H3K9), and that its loss leads to very early arrest of oogenesis (Clough et al., 2007). To learn more about the possible roles of Egg at all stages of oogenesis, we compared the effects of both weak and strong alleles in our collection.
H3K9 methylation by Egg is required for late stages of oogenesis
Four missense alleles in our collection (egg1372, egg1024, egg178, and eggA97) were classified as weak alleles because they did not affect viability in tests with Df(2R)Dll-Mp, but were female-sterile. In contrast to the very early arrest of oogenesis caused by strong alleles, oogenesis proceeds to later stages in the ovaries of females bearing any of the weak alleles. Analysis of these weak alleles, in particular egg1372, has shown that Egg-dependent H3K9 methylation occurs in late stage egg chambers, and is required for the successful completion of oogenesis.
egg1372 is a point mutation in the initiator methionine (ATG to ATA). egg1372/Df (2R)Dll-Mp females are fertile at 25°C, but sterile at 29°C. Although egg1372 females lay eggs at 25°C, some eggs have fragile chorions and abnormally short dorsal appendages. Surprisingly, since the mutation lies in the AUG start codon, egg1372 is the weakest allele of the set, allowing oogenesis to proceed to very late stages, and even at the higher temperature egg1372 adult flies are fully viable.
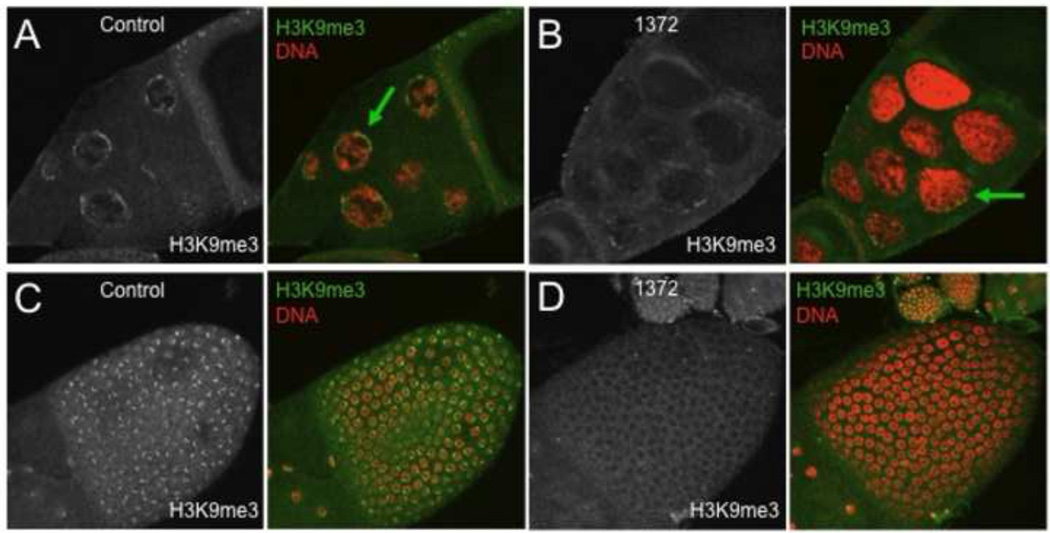
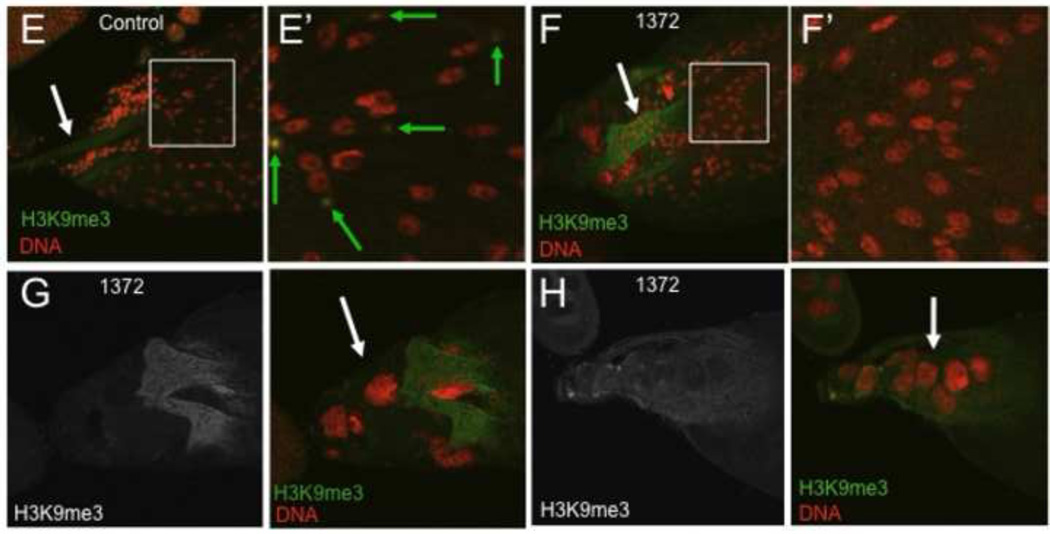
To determine if egg is required for H3K9 methylation in late oogenesis, control and egg1372 ovaries (from females grown at 29 °C) were labeled with an antibody to H3K9me3 (Fig. 2). Wildtype egg chambers had high H3K9me3 levels in the nurse cell nuclei and the follicle cells of stage 10B and 11 egg chambers (Fig. 2A,C), but H3K9me3 was strongly reduced in both the germ cells and the follicle cells of egg1372 egg chambers (Fig. 2 B, D). In the final stages of oogenesis, H3K9me3 was detected in some follicle cells overlying stage 14 oocytes in control ovaries, but not all of the follicle cells expressed this mark, suggesting that it may define a subpopulation of follicle cells at this late stage (Fig. 2E–E’). H3K9me3 was undetectable in all egg1372 stage 14 follicle cells (Fig. 2F–F’). The ovaries of egg1372 females contained many late stage egg chambers that had not undergone or completed nurse cell dumping (Fig. 2G). These oocytes were noticeably smaller than wildtype oocytes at the same stage. Also present were relatively normal-sized stage 14 oocytes with persistent nurse cells (Fig. 2H). The dorsal appendages of late stage egg1372 oocytes were usually short and malformed (e.g. Fig 2F,G). Another weak allele, eggA97, exhibited similar phenotypes. Thus H3K9 methylation, mediated by Egg, is required for successful completion of major events in the late stages of oogenesis, including nurse cell dumping and the production by the follicle cells of the eggshell structures, and may also play a role in the programmed death of late stage nurse cells.
Figure 2. Weak egg alleles affect H3K9 methylation at late stages of oogenesis.
egg1372/Df(2R)Dll-Mp (1372) and control ovaries from flies raised at 29 °C were labeled with antibodies to H3K9me3 and stained with the DNA dye propidium iodide. (A) H3K9me3 is present at the periphery (green arrow) of nurse cell nuclei of control stage 10b egg chambers. (B) H3K9me3 is strongly reduced in stage 10B 1372 nurse cell nuclei (green arrow). (C) H3K9me3 is present in distinct nuclear domains in follicle cells overlying the oocyte in control stage 10b egg chambers. (D) H3K9me3 is not detected in the follicle cell nuclei of 1372 stage 10B egg chambers. (E – E’) Stage 14 egg chambers from control flies have slender paired dorsal appendages (white arrow) and the nurse cells have been removed by programmed cell death. High levels of H3K9me3 are present in a subpopulation of stage 14 follicle cells (green arrows in the expanded boxed region sown in E’). (F – F’) Stage 14 egg chambers from 1372 flies have thick, club-shaped dorsal appendages (white arrow), and H3K9me3 is not detected in the follicle cells (expanded boxed region shown in F’). G. 1372 ovaries contain frequent small oocytes (the short dorsal appendages of a oocyte are visible here) associated with nurse cells that have not transferred their cytoplasm to the oocyte (white arrow). H. Persistent nurse cell nuclei (white arrow) are often associated with mature 1372 oocytes.
Strong egg alleles affect the early differentiation of germ cells
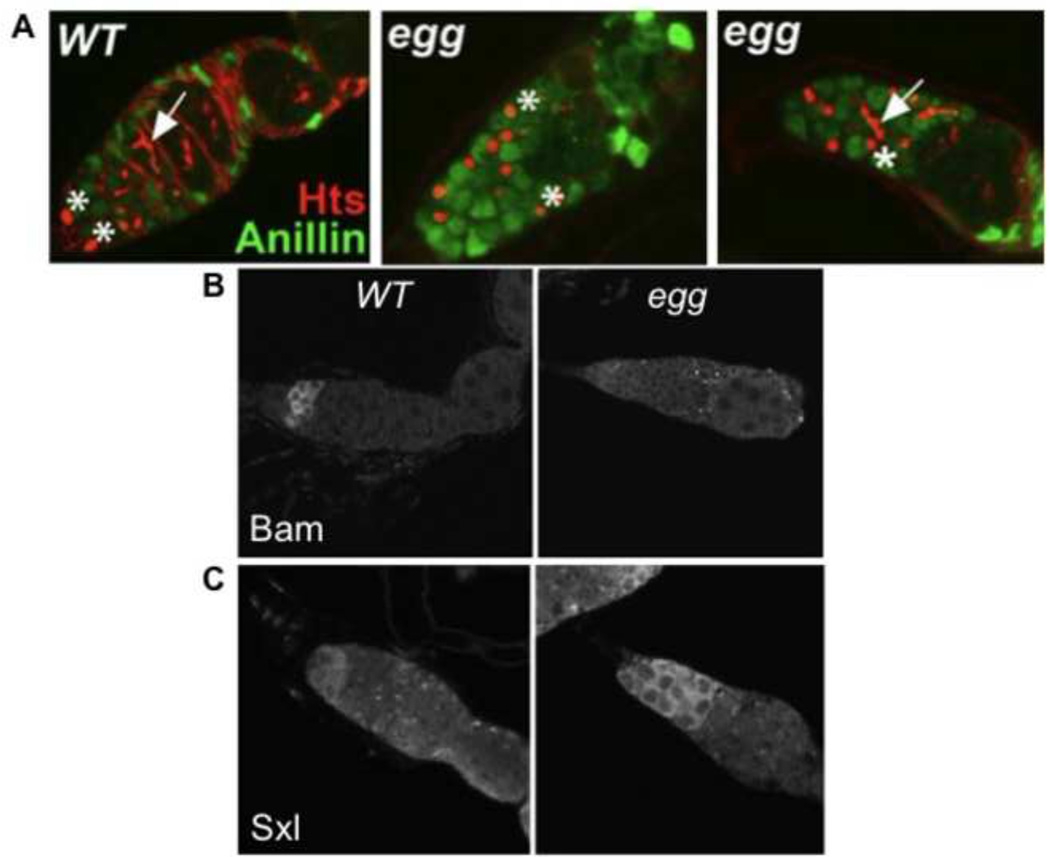
The ovaries of females carrying strong egg mutations contain germaria with arresting germ cells, as well as some germaria that have become agametic, and very rarely are budded egg chambers observed in these females (Clough et al., 2007). To better characterize when and how the germ cells arrest, the expression of early germ cell markers was examined in mutant females (Fig. 3). Hts (hui li tai shao) antibody labels the spectrosome, a spherical membranous structure present in germ stem cells and cystoblasts (Lin et al., 1994; Petrella et al., 2007). During germline cyst divisions the spectrosome morphs into a branched structure called the fusome (de Cuevas and Spradling, 1998). The cell cycle status of dividing germ cells can be assessed by the localization of Anillin, an Actin-binding protein that accumulates in the nuclei of interphase cells, but is released to the cytoplasm during mitosis (Field and Alberts, 1995). Spectrosomes and some branched fusomes were both present in egg mutant germaria. However, most germaria contained an excess number of spectrosome-bearing germ cells (Fig. 3A), suggesting that early germ cells were arresting as GSCs or CBs, or possibly as undifferentiated GSC daughter cells. The excess spectrosome-bearing cells had large nuclei typical of GSCs and CBs, and most contained nuclear Anillin, suggesting that the germ cells were arresting in interphase.
Figure 3. Strong egg alleles cause early germ cell arrest.
Shown are germaria from the ovaries of w1118 control (WT) or egg1473/Df(2R)Dll-Mp (egg) females. (A) Germaria labeled for Hts (red) and Anillin (green). Two GSCs with spectrosomes (*s) are located at the anterior tips of the control germarium. The egg germaria contain an excess number of germ cells with spectrosomes, some of which have moved away from the niche (the most posterior are indicated with *s). Germline cysts with branched fusomes (arrows) are also present. (B) Germaria labeled with an antibody to Bam. Bam is expressed in CBs and early germline cysts in the control germarium (left panel), but is strongly reduced or absent in the egg germarium (right panel). (C) Germaria labeled with an antibody to Sxl. Sxl levels are highest in the GSCs and CBs of the control germarium (left panel). The egg germarium (right panel) contains an expanded domain of Sxl-expressing germ cells.
In wildtype ovaries, Bam (Bag-of-Marbles) expression is repressed in the GSCs, and is first detected in the CB daughters of GSCs, where it promotes their differentiation (McKearin and Spradling, 1990; Song et al., 2004). Bam expression was undetectable or strongly reduced in all early germ cells in egg− germaria (Fig. 3B). Sxl (Sex-lethal) is normally expressed most strongly in a restricted area of the germarium, in both GSCs and CBs (Chau et al., 2009). In egg− germaria, Sxl was expressed strongly in an expanded domain of germ cells (Fig. 3C). Since Sxl and Bam are both required to coordinately regulate the transition of GSC daughters to the CB cell fate (Chau et al., 2012), their combined expression pattern in egg− germaria (lack of Bam and an expanded domain of Sxl) suggests that the germ cells in egg germaria are arresting as either GSCs or as undifferentiated GSC daughters. Since the cells are no longer anchored, like GSCs, to the cap cells in the stem cell niche, but instead move away from this niche, we favor the idea that they are behaving as undifferentiated GSC daughters.
egg has both cell-autonomous and non-autonomous effects on early germ cell development in the ovary
Since egg is expressed in germ cells and somatic cells in the ovary (Clough et al., 2007), the early oogenesis arrest and germ cell differentiation defects in mutant females could be due to a requirement for egg activity in the germ cells, the somatic cells, or both. To distinguish these possibilities we used the FLP-FRT system (Golic, 1991; Chou and Perrimon, 1996) to produce egg germline and somatic cell clones in larval and adult ovaries – induction at both time points had similar effects. Two independent strong alleles were tested, egg1473 (a deletion of the SET domain) and egg2138 (a premature stop codon), and egg− cells were identified by loss of a GFP marker (see materials and methods for protocols and genotypes).
egg is required intrinsically in germ cells for survival past stage 5 of oogenesis
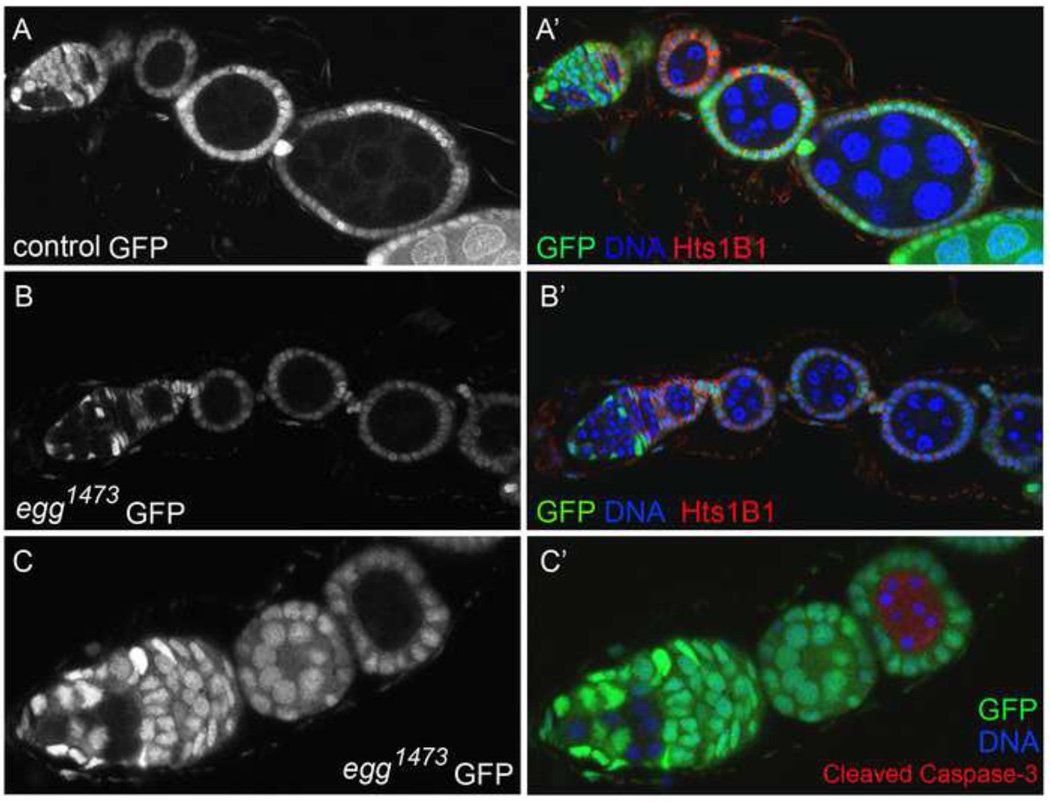
When clones were induced in adult females and the ovaries were examined 8 days later, egg− germline clones were present and differentiated normally at the earliest stages of oogenesis (Fig. 4B). Bam was expressed normally in CB daughters and early germline cysts (not shown), and no excess spectrosome-bearing germ cells were observed in the germline clones. The egg chambers bearing egg− germ cells budded off the germarium and developed normally until stage 5 of oogenesis, but most arrested at this stage with pycnotic nuclei, typical of apoptotic cells. Labeling with an antibody to activated Caspase-3 confirmed that the germ cells were dying by apoptosis (Fig. 4C). Thus egg is not required intrinsically in germ cells for the differentiation of GSC daughters, or for the formation of egg chambers, but is required in germ cells for their continued survival past stage 5 of oogenesis.
Figure 4. egg germline clones differentiate normally but arrest at stage 5 of oogenesis.
Ovaries containing control and egg1473 germline clones were dissected and fixed 8 (A and B) or 12 days (C) after clone induction, and labeled with an antibody to Hts 1B1 or activated (cleaved) Caspase-3. Marked germ cell clones were identified in the ovaries by their lack of GFP. The left panels show the GFP signal alone, and the right panels are merged images of all 3 labels. (A–A’) A control ovariole containing a marked wildtype GSC that has produced several germline cysts and developing egg chambers. (B–B’) An ovariole containing two egg1473 GSCs. The mutant GSCs have given rise to developing germline cysts and egg chambers that have budded off the germarium normally, but the egg chambers do not progress beyond stage 5 of oogenesis. (C–C’) A germarium with a single egg1473 GSC. The stage 5 egg chamber on the right contains egg germ cells that are apoptotic, with pycnotic nuclei and activated Caspase-3.
egg is required intrinsically in germ stem cells for their maintenance
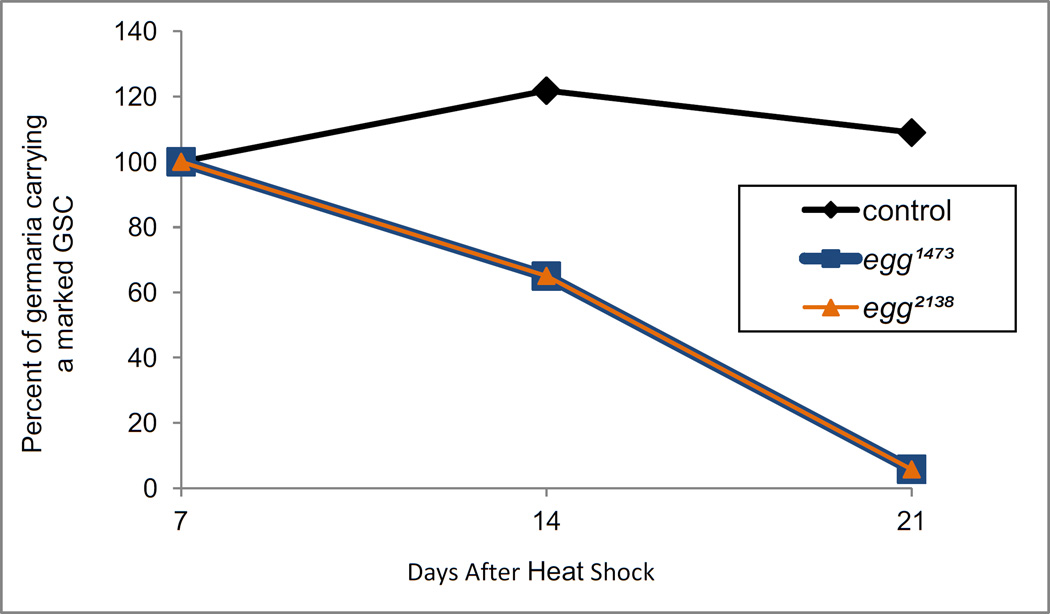
We next asked if egg is required in GSCs for their maintenance over time. This possibility was tested by comparing the number of egg− germ stem cell clones in adult ovaries at 1, 2 and 3 weeks after the induction of clones in 3rd instar larvae (Fig. 5). More than 100 germaria for each genotype were analyzed (Table S1). The ovaries were labeled with an antibody to Hts (which labels the spectrosome and fusome, and also the cell membranes of follicle cells), and GSC clones were identified by their lack of GFP, the presence of a spectrosome, and their location at the apical end of each germarium. One week after clone induction, egg mutant GSCs were present at frequencies equal to or greater than wildtype control clones, but by 2 weeks the percentage of germaria containing egg mutant GSCs had decreased dramatically. At 3 weeks, egg mutant GSCs were extremely rare. The same pattern of germ stem cell loss was observed when clones were induced in adult females (not shown). The striking loss of egg mutant GSCs was in contrast to control wildtype GSC clones, which were maintained over the time course of this experiment. Thus, egg is required intrinsically in ovary GSCs for their maintenance over time.
Figure 5. egg is required for germ stem cell maintenance.
The normalized percentage of germaria containing at least one marked control or egg (egg1473 or egg2138) germ stem cell is shown 7, 14, and 21 days after clone induction. (See table S1 for the actual numbers scored.) Clones were induced in third instar larvae with one two-hour heat shock at 37 °C, and the ovaries were labeled with Hts1B1 antibody and propidium iodide. The clones were identified by the absence of GFP, and GSCs were defined as the spectrosome-bearing cells at the apical end of the germarium adjacent to the cap cells.
egg has both cell-autonomous and inductive effects in somatic cells
The fact that egg− germline clones initially survived to produce egg chambers that bud off of the germarium, and developed up to stage 5 of oogenesis, suggested that both the early germ cell differentiation and the egg chamber budding defects of egg− females were due to a requirement for egg in somatic cells. Follicle stem cells (FSCs) reside in the Drosophila ovary at the junction between regions 2a and 2b of the germarium (Margolis and Spradling, 1995). Division of FSCs gives rise to the follicular cell epithelium that encapsulates the germline cyst in region 2b of the germarium, prior to budding of the egg chamber. Follicle stem cell clones can be identified in the ovary 8 days and later after clone induction, when all marked follicle cells are the products of FSC clones – transient clones (those induced in dividing follicle cells) have exited the ovariole and persistent marked follicle cells are the products of FSC clones (Zhang and Kalderon, 2001).
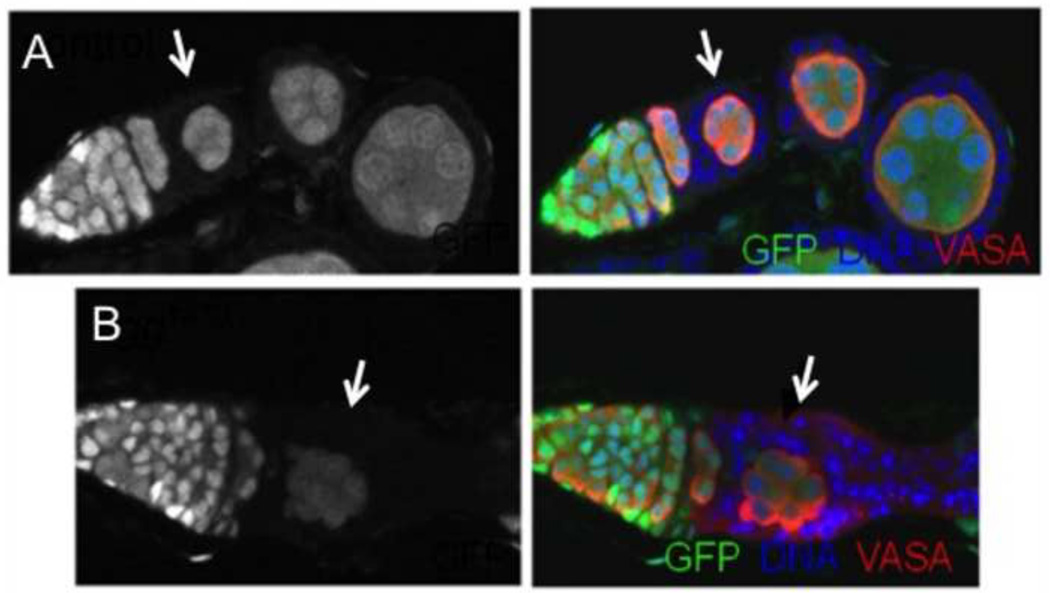
When clones were induced in young adult females and fixed ovaries, labeled with Hts antibody and PI, were examined at 8, 15 and 22 days after clone induction, egg− FSC clones were less frequent than control FSC clones at the earliest time point, and were not maintained over time. In one experiment, wildtype FSC clones were present in 16% of the germaria (N = 229) when measured at 15 days after clone induction, and this frequency was maintained at 22 days (Table S2). In contrast, only 2% of germaria contained egg1473 FSC clones at 15 days (N = 194), and the number of clones was further reduced at 22 days. Similar low frequencies were observed when clones were induced with the egg2138 allele. In cases where an egg− FSC clone was present, its egg− prefollicular daughter cells often formed disorganized aggregates around germline cysts, and dying cells with pycnotic nuclei were present among the mutant somatic cells. This phenotype was confirmed in FSC clones in ovaries labeled with an antibody to Vasa, to mark the germ cells (Fig 6).
Figure 6. egg− prefollicular cells fail to properly encapsulate germline cysts.
Shown are follicle stem cell (FSC) clones from ovaries that were dissected and fixed 8 days after clone induction, and labeled with an antibody to Vasa to mark the germ cells. (A) A wildtype control FSC clone has given rise to follicle cells that envelope the germ cells of a stage 1 egg chamber (arrow), and several budded egg chambers. (B) The prefollicular descendents of an egg1473 FSC have formed multilayer aggregates (arrow) around a germline cyst and, judging by the appearance of their pycnotic nuclei, many appear to be dying.
Since egg− FSCs are not maintained, and egg− prefollicular cells are defective, we used a complementary approach employing the Gal4 system (Brand and Perrimon, 1993) to determine if selective expression of the egg gene in somatic cells could rescue any of the early oogenesis defects of egg mutants. For these experiments, egg expression was driven in somatic cells using a UAST-dSETDB1 transgene constructed by C.-K. J. Shen’s lab (Tzeng et al, 2007), and the tj-Gal4 driver. Like the traffic jam gene (Li et al., 2003), the tj-Gal4 driver is expressed very weakly in the cap cells, and strongly in the escort cells that associate with developing germline cysts and the FSCs, prefollicular cells, and follicle cells throughout oogenesis (Fig. S1). Rescue was tested in egg− females carrying the tj-Gal4 driver along with UAST-dSETDB1 (see materials and methods for the exact genotype), referred to here as “tj-Gal4-rescued” females. Ovaries from 2–3 day old females were fixed and labeled with antibodies to Vasa, to identify germ cells, and Hts, to assess the presence of GSCs, CBs and germline cysts (by their characteristic spectrosomes and fusomes). Hts also provided a convenient label for the somatic prefollicular cells and follicle cells, where it marks the cell membranes.
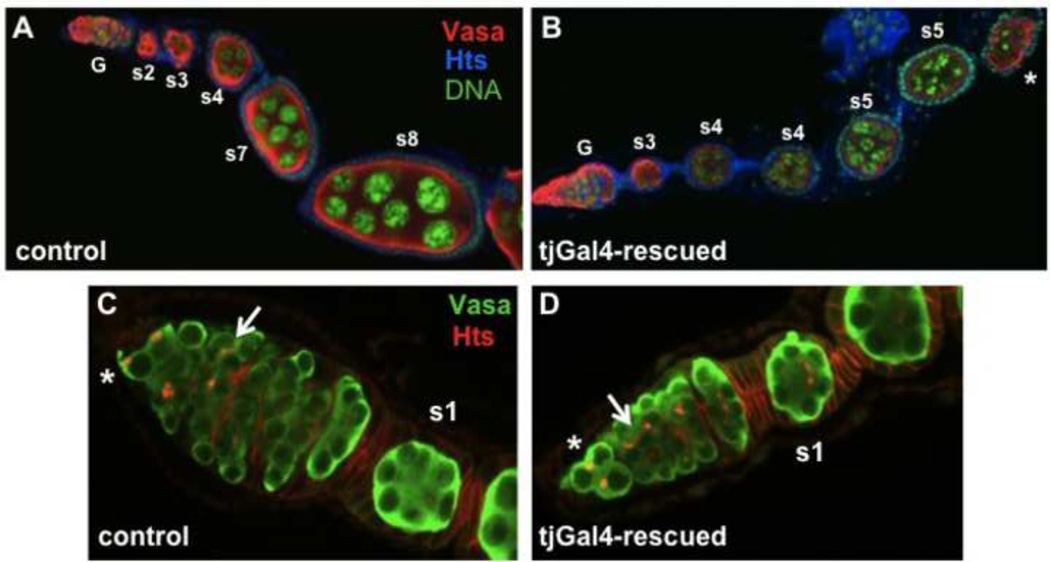
tj-Gal4-driven UAST-Setdb1 substantially rescued both the early germ cell differentiation defects, and the encapsulation and budding defects (Fig. 7). The ovarioles of tj-Gal4-rescued females contained budded egg chambers that arrested at stage 5 of oogenesis (Fig. 7B). Although the tj-Gal4 driver is expressed in follicle cells at later stages, the arrest of egg chambers at stage 5 is consistent with our analysis of germline clones (above), which showed that egg is required in the germ cells themselves for continued development past this stage. Rescue of the encapsulation and budding defects by tjGal4-driven expression in the somatic cells is strong positive evidence that egg is required in the prefollicular and early follicle cells for these events in early oogenesis.
Figure 7. Gal4-driven expression of egg/dSETDB1 in somatic cells rescues the early oogenesis defects of egg mutations.
Shown are ovarioles from 2–3 day old wildtype control females (A and C) and egg− females carrying the tj-Gal4 driver and a UAST-dSETDB1 transgene (“tj-Gal4-rescued” females; B and D), labeled with antibodies to Vasa and Hts. (A–B) The tjGal4 driver rescues both the early germ cell differentiation defects and egg chamber budding defects of the strong egg mutation (compare to Fig 3A). The germaria (G) and the stages of successive egg chambers are indicated. (C–D) Higher magnification views of control and tjGal4-rescued germaria. The tj-Gal4-rescued germarium, like the wildtype control, contains germ cells at all stages of development, including germ stem cells (*), developing germline cysts (arrow), and fully formed egg chambers budding off the germarium (s1). No excess spectrosome-bearing cells are present in the tjGal4-rescued germarium.
In many of the tj-Gal4-rescued ovaries, early germ cell differentiation was normal. All germ cell stages were present in these germaria, including spectrosome-containing GSCs and CBs and dividing germline cysts with branched fusomes in region 1, fully formed cysts in region 2a, disc-shaped cysts in region 2b, and newly formed egg chambers in region 3 (Fig. 7D). Excess-spectrosome bearing germ cells were not present in these germaria. Rescue of the early germ cell differentiation defects is strong evidence that differentiation of germ cell daughters is affected by expression of Egg in somatic cells of the germarium.
Despite the substantial rescue observed in tj-Gal4 rescued females, the germaria were not all normal – some had a reduced number of germ cells or were agametic. Thus, in one set of tj-Gal4-rescued germaria (N = 37) from 2–3 day old females, 49% (18/37) appeared normal, 35% (13/37) had a reduced number of germ cells, and 16% (6/37) were agametic. The observed germ cell loss is most likely due to the absence of egg expression in the germ cells, since clonal analysis (above) showed that egg is required in the GSCs for their maintenance. GSCs may be lost more rapidly in tj-Gal4 rescued females, than in egg− clones, because their germ cells have been without Egg for all of development, whereas in the GSC clones egg gene function was removed by recombination late in development and some Egg, or its H3K9me mark, may perdure over time in germ cell clones, leading to more gradual loss.
Discussion
Three Histone Lysine Methyltransferases (HKMTs), Su(Var)3-9, dG9a, and Egg/dSETDB1, methylate histone H3K9 in Drosophila. Su(Var)3-9, discovered for its role in position effect variegation, plays a central role in establishing centromeric heterochromatin, but is not an essential gene (Tschiersch et al., 1994; Schotta et al., 2002). dG9a affects position effect variegation and in addition targets euchromatic sites in the genome, and is also not essential (Mis et al., 2006; Seum et al., 2007b; Kato et al., 2008). Of these three only Egg, the Drosophila ortholog of human SetDB1, plays an essential role in development. Strong loss-of-function egg mutations affect survival and are female-sterile, whereas dG9a and Su(Var)3-9 mutants are both fertile and viable. Functional redundancy between these HKMTs may mask vital functions at some stages of development, but Egg alone is essential in late pupae and in adult tissues (Seum et al., 2007a; Brower-Toland et al., 2009; this report). To learn more about the role of H3K9 methylation in an adult tissue, the ovary, we analyzed a collection of EMS-induced egg mutations that includes both strong loss-of-function and weak hypomorphic alleles and determined their effects on oogenesis.
A previous report (Yoon et al., 2008) concluded that Egg and Su(Var)3-9 play sequential roles in oogenesis, with Egg being required only at the very earliest stages of oogenesis, in regions 1 and 2a of the germarium, and Su(Var)3-9 functioning later. We, however, find that Egg is an essential HKMT at several stages of oogenesis. While strong loss-of-function mutations (e.g. egg2138 and egg1473) cause very early oogenesis arrest and demonstrate Egg’s role in early oogenesis (Fig 3), the weak alleles are all associated with late oogenesis arrest. The H3K9me patterns in females carrying weak alleles (e.g. egg1372 and eggA97) demonstrate that Egg promotes H3K9 methylation in both the germ cells and somatic cells at late stages of oogenesis and may be the sole HKMT acting at these stages (Fig 2). Furthermore, the oogenesis defects produced by the weak egg alleles show that Egg is required for at least two late events in oogenesis: nurse cell dumping and formation of the eggshell and its structures. The presence of persistent nurse cells associated with mature stage 14 oocytes may also indicate a unique role of H3K9 methylation in the programmed death of nurse cells in late oogenesis (Pritchett et al., 2009), or nurse cell death could be delayed as a consequence of defects in nurse cell dumping (Foley and Cooley, 1998; Riparbelli et al., 2007). Future studies in our lab will address whether H3K9me promotes specific changes in chromatin structure that accompany these terminal events in oogenesis, or if its primary function is to regulate the expression of genes required for these events.
Four egg alleles are missense mutations that lie in identified domains of the protein (Fig 1): three are in the SET or POST-SET domains (eggA97, egg391, and egg1020) and one is in the MBD domain (egg178). The weak eggA97 mutation (E1079K) lies at the border of the SET-I domain, a region that may affect substrate specificity (Zhang et al., 2002; Wu et al., 2010). The strong egg1020 allele is a missense mutation that changes one of the Post-SET conserved cysteines (C1250Y). The equivalent mutation in human SETDB1 (C1279Y) eliminated its catalytic activity (Schultz et al., 2002). The strong egg391 (D1212N) mutation alters an amino acid in the SET domain that is conserved in other H3K9 targeting HKMTs, but not in HKMTs that target other lysines (Qian and Zhou, 2006). The weak egg178 allele is a missense mutation (L865F) in a highly conserved residue of the MBD domain (Dhasarathy and Wade, 2008). The presence of methylated DNA in Drosophila has been controversial (Krauss and Reuter, 2011), but recent evidence argues against its existence (Raddatz et al., 2013). If so, then the function of this domain in the Egg protein remains to be elucidated. Nevertheless, the presence of all four mutations point to the importance of these domains for Egg function. Alternatively, any or all of these mutations may affect the stability of the protein, rather the function of particular domains.
The egg1372 allele is one of the more interesting mutations in our collection since it is a point mutation in the start codon. One might have expected it to be a strong mutation, but it is the weakest allele in our collection. The weak phenotype may indicate that an internal methionine codon is used as a start codon in egg1372, and that a polypeptide missing the N-terminus contributes to development but is insufficient to sustain the late stages of oogenesis. The production of antibodies designed to detect specific N- and C-terminal segments of the protein may help to distinguish these possibilities, and lead to a better understanding of how the egg gene is regulated during development.
Our analysis of egg− germline clones has shown that egg is not required intrinsically in germ cells for their early differentiation and the formation of egg chambers, but is required in the germ cells for survival past stage 5 of oogenesis (Fig 4). Why would stage 5 egg chambers be particularly sensitive to a change in H3K9 methylation? At this stage, the nurse cells undergo striking structural changes in their endoreplicated chromosomes, which switch from a polytene state to a dispersed polyploid state (Dej and Spradling, 1999). In egg chambers containing egg germline clones the mutant nurse cells develop the aggregated chromosome arms typical of early stage 5 egg chambers, but most became apoptotic before progressing to a more dispersed chromatin state, suggesting that H3K9 methylation could play a role in these chromatin transitions. Also, at this stage of oogenesis replication of heterochromatin is normally blocked in nurse cells (Royzman et al., 2002). Since H3K9me is required for the establishment of heterochromatin, this block may not occur in the egg mutant nurse cells, and inappropriate replication of repetitive DNA could contribute to the observed germ cell death.
Our analysis of germ cell clones also demonstrates that egg is required for germ stem cell maintenance (Fig 5 and Table S1), in agreement with a previous study (Wang et al., 2011). Mechanisms that guarantee self-renewal are fundamental to stem cell biology. In Drosophila ovaries, each time a GSC divides one daughter chooses the germ stem cell fate, and the other differentiates and enters the oogenesis pathway. While signaling from somatic cap and terminal filament cells in the niche plays a major role in GSC maintenance, less is known about the role of intrinsic factors in GSC maintenance (Eliazer and Buszczak, 2011). One possible intrinsic role for Egg in this self-renewal process is that H3K9 methylation by Egg represses the expression of differentiation genes in GSCs. However, ectopic expression of at least one critical differentiation factor, Bam, was not observed in mutant GSCs (Fig. 3). A second possibility is that egg− GSCs are lost by cell death, which could be difficult to detect by traditional methods, such as TUNEL labeling, if dying GSCs are rapidly phagocytosed by surrounding somatic cells. Peng and Karpen (2009) have shown that loss of Su(Var)3-9, which itself is not an essential HKMT, nevertheless causes destabilization of heterochromatin that can lead to DNA damage in repetitive DNA and cell death, and one would expect these effects to be more extreme in the case of an essential HKMT like Egg. Another interesting scenario is suggested by work by Rangan et al. (2011), who found reduced expression of piRNAs (Piwi-interacting small RNAs) in egg mutants, and increased levels of several retrotransposon transcripts, the targets of piRNAs (Aravin et al., 2009). Since retrotransposons move by way of an RNA intermediate, excessive mobilization could occur in egg mutants and would likely result in widespread DNA damage and cell death. Alternatively, Egg, like its mammalian counterpart Setdb1/Eset (Schultz et al., 2002), is also likely to target a large number of genes, including euchromatic genes (Brower-Toland et al., 2009; Stabell et al., 2006; Tzeng et al., 2007), and derepression of these genes, including pro-apoptotic genes, could lead to cell death.
Two lines of evidence in this study provide strong evidence that Egg plays both intrinsic and non-autonomous roles in somatic cells at early stages of oogenesis. First, the analysis of somatic cell clones showed that Egg is required intrinsically in FSCs for their maintenance and in the prefollicular and early follicle cells for the formation and budding of egg chambers from the germarium (Fig 6 and Table S2). Second, expression of Egg exclusively in somatic cells using a Gal4 driver independently confirmed that Egg is required in the FSCs and the prefollicular cells for their survival and normal functions, and also showed that Egg expression in somatic cells influences the differentiation of early germ stem cell daughters (Fig 7). This last result complements RNAi studies demonstrating a similar requirement in somatic cells in the ovary for early germ cell differentiation (Rangan et al., 2011; Wang et al., 2011). We suspect that the escort cells are the critical somatic cells in the germarium providing this instructive role for germ cell differentiation. The driver used in our experiments to express Egg in somatic cells, tjGal4, is expressed strongly in the escort cells and only weakly and intermittently in the cap cells. The cap cells are unlikely candidates for this rescue effect since Egg expression has not been detected in the cap cells (Clough et al., 2007). Escort cells are known to play an instructive role in the early differentiation of the germ cells (Decotto and Spradling, 2005; Guo and Wang, 2009; Eliazar et al., 2011; Morris and Spradling, 2011). This action occurs, at least in part, by restricting access of BMP signaling from the niche, and involves repression of Dally (an enhancer of Dpp ligand activity) and Dpp expression in the escort cells (Kirilly et al., 2011). Egg could directly repress dpp and/or Dally by targeting these genes for H3K9 methylation. Alternatively, Egg may be required for the survival of the escort cells. In support of the latter possibility, we have observed extensive apoptotic cell death in egg− germaria, including dying somatic cells (Clough et al., 2007).
A full understanding of Egg’s role in oogenesis will require future studies to determine its full repertoire of gene targets. Several studies have looked at the effects of egg mutations on gene expression throughout the genome, and H3K9me by Egg has been implicated in both gene activation and gene repression (Seum et al., 2007; Tzeng et al., 2007; Brower-Toland et al. 2009; Rangan et al., 2011; Lundberg et al., 2013). This complexity may in part reflect the fact that heterochromatin, which requires H3K9me for its formation, is not only a repressive chromatin state: in some cases genes that reside in heterochromatin require this environment for their optimal expression (Yasuhara and Wakimoto, 2006). Egg may also, like its mammalian counterparts, target euchromatic genes, and its effects on gene expression may depend on gene context, other epigenetic marks, and associated transcription factors (Lundberg et al, 2013). And finally, in addition to its known histone target, Histone H3K9, Egg could have non-histone targets, as has been reported for human SetDB1 and other HKMTs (Van Duyne et al., 2008; Zhang et al., 2012). In support of this possibility, we have observed high levels of Egg protein in the cytoplasm of germ stem cells (Clough et al., 2007).
Supplementary Material
Figure S1 tjGal4 expression pattern. Shown are fixed ovarioles from 2-3 day old females carrying the tj-Gal4 driver along with a UAST-EGFP reporter. (A) The tj-Gal4 driver is expressed strongly in most somatic cells in the germarium, and in follicle cells until late stages of oogenesis. (B) In the germarium, expression of tj-Gal4 is strong in the escort cells (triangles) and also the FSCs (*), and prefollicular and follicle cells. tj-Gal4 is also expressed weakly, and variably, in the cap cells.
Table S1 egg− ovarian germ stem cells are not maintained. Larvae (see materials and methods for the complete genotypes) were given one two-hour heat shock on the first day of the third larval instar. 7, 14, and 21 days after clone induction the ovaries of adult flies were scored for the presence of GSC clones.
Table S2 egg− ovarian follicle stem cells are not maintained. 1–2 day old adult females (see materials and methods for the complete genotypes) were given two one-hour heat shocks for 3 days, and scored for persistent follicle stem cell clones 8, 15 and 22 days after the final heat shock.
Highlights.
Eggless is an essential Drosophila Histone H3K9 methyltransferase.
Eggless is required for the maintenance of germ and somatic stem cells.
H3K9 methylation is required for both early and late stages of Drosophila oogenesis.
Acknowledgements
We thank Ying Wang for excellent technical support in the final stages of this work. We are grateful to C-K.J Shen, C. Seum and S. Bontron, the Bloomington Drosophila stock center, and the Drosophila Genome Resource Centre in Kyoto for providing fly stocks, and to D. McKearin, R. Lehmann, C. Field, and the Iowa Developmental Studies Hybridoma Bank for providing antibodies. This work was supported by grants to T.H. from the National Science Foundation, The New York State Department of Health Stem Cell Program (NYSTEM) and the March of Dimes Organization. E.C. was supported by an NIH training grant to the Columbia University’s Department of Biological Sciences.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Literature Cited
- Aravin AA, Hannon GJ, Brenneke J. The Piwi-piRNA pathway provides an adaptive defense in the transposon arms race. Sciences. 2009;318:761–764. doi: 10.1126/science.1146484. [DOI] [PubMed] [Google Scholar]
- Bannister AJ, Zegerman P, Partridge JF, Miska EA, Thomas JO, Allshire RC, Kouzarides T. Selective recognition of methylated lysine 9 on histone H3 by the HP1 chromo domain. Nature. 2001;410:120–124. doi: 10.1038/35065138. [DOI] [PubMed] [Google Scholar]
- Black JC, Van Rechem C, Whetstine JR. Histone lysine methylation dynamics: establishment, regulation, and biological impact. Mol Cell. 2012;48(4):491–507. doi: 10.1016/j.molcel.2012.11.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brand AH, Perrimon N. Targeted gene expression as a means of altering cell fates and generating dominant phenotypes. Development. 1993;118(2):401–415. doi: 10.1242/dev.118.2.401. [DOI] [PubMed] [Google Scholar]
- Brower-Toland B, Riddle NC, Jiang H, Huisinga KL, Elgin SC. Multiple SET methyltransferases are required to maintain normal heterochromatin domains in the genome of Drosophila melanogaster . Genetics. 2009;181:1303–1319. doi: 10.1534/genetics.108.100271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casso D, Ramírez-Weber F, Kornberg TB. GFP-tagged balancer chromosomes for Drosophila melanogaster . Mech Dev. 2000;91:451–454. doi: 10.1016/s0925-4773(00)00248-3. [DOI] [PubMed] [Google Scholar]
- Chau J, Kulnane LS, Salz HK. Sex-lethal facilitates the transition from germline stem cell to committed daughter cell in the Drosophila ovary. Genetics. 2009;182(1):121–132. doi: 10.1534/genetics.109.100693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chau J, Kulnane LS, Salz HK. Sex-lethal enables germline stem cell differentiation by down-regulating Nanos protein levels during Drosophila oogenesis. Proc Natl Acad Sci U S A. 2012;109(24):9465–9470. doi: 10.1073/pnas.1120473109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chou T, Perrimon N. The autosomal FLP-DFS technique for generating germline mosaics in Drosophila melanogaster . Genetics. 1996;144:1673–1679. doi: 10.1093/genetics/144.4.1673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clough E, Moon W, Wang S, Smith K, Hazelrigg T. Histone methylation is required for oogenesis in Drosophila . Development. 2007;134:157–165. doi: 10.1242/dev.02698. [DOI] [PubMed] [Google Scholar]
- Dambacher S, Hahn M, Schotta G. Epigenetic regulation of development by histone lysine methylation. Heredity. 2010;105(1):24–37. doi: 10.1038/hdy.2010.49. [DOI] [PubMed] [Google Scholar]
- Dhasarathy A, Wade PA. The MBD protein family-reading an epigenetic mark? Mutat Res. 2008;647(1–2):39–43. doi: 10.1016/j.mrfmmm.2008.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Decarlo D, Hadden MK. Oncoepigenomics: making histone lysine methylation count. Eur J Med Chem. 2012;56:179–194. doi: 10.1016/j.ejmech.2012.08.010. [DOI] [PubMed] [Google Scholar]
- DeCotto E, Spradling AC. The Drosophila ovarian and testis stem cell niches: similar somatic stem cells and signals. Dev. Cell. 2005;9:501–510. doi: 10.1016/j.devcel.2005.08.012. [DOI] [PubMed] [Google Scholar]
- de Cuevas M, Spradling AC. Morphogenesis of the Drosophila fusome and its implications for oocyte specification. Development. 1998;125:2781–2789. doi: 10.1242/dev.125.15.2781. [DOI] [PubMed] [Google Scholar]
- Dej KJ, Spradling AC. The endocycle controls nurse cell polytene chromosome structure during Drosophila oogenesis. Development. 1999;126:293–303. doi: 10.1242/dev.126.2.293. [DOI] [PubMed] [Google Scholar]
- Eliazer S, Buszczak M. Finding a niche: studies from the Drosophila ovary. Stem Cell Res Ther. 2011;2(6):45. doi: 10.1186/scrt86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Field CM, Alberts BM. Anillin, a contractile ring protein that cycles from the nucleus to the cell cortex. J. Cell Biol. 1995;131(1):165–178. doi: 10.1083/jcb.131.1.165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foley K, Cooley L. Apoptosis in late stage nurse cells does not require genes within the H99 deficiency. Devt. 1998;125:1075–1082. doi: 10.1242/dev.125.6.1075. [DOI] [PubMed] [Google Scholar]
- Golic KG. Site-specific recombination between homologous chromosomes in Drosophila . Science. 1991;252:958–961. doi: 10.1126/science.2035025. [DOI] [PubMed] [Google Scholar]
- Greer EL, Shi Y. Histone methylation: a dynamic mark in health, disease and inheritance. Nat Rev Genet. 2012;13(5):343–357. doi: 10.1038/nrg3173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grewal SI, Jia S. Heterochromatin revisited. Nat. Rev. Genet. 2007;8(1):35–46. doi: 10.1038/nrg2008. [DOI] [PubMed] [Google Scholar]
- Guo Z, Wang Z. The glypican Dally is required in the niche for the maintenance of germline stem cells and short-range BMP signaling in the Drosophila ovary. Development. 2009;136(21):3627–3635. doi: 10.1242/dev.036939. [DOI] [PubMed] [Google Scholar]
- Kato Y, Kato M, Tachibana M, Shinkai Y, Yamaguchi M. Characterization of Drosophila G9a in vivo and identification of genetic interactants. Genes Cells. 2008;13(7):703–722. doi: 10.1111/j.1365-2443.2008.01199.x. [DOI] [PubMed] [Google Scholar]
- Kirilly D, Wang S, Xie T. Self-maintained escort cells form a germline stem cell differentiation niche. Development. 2011;138(23):5087–5097. doi: 10.1242/dev.067850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krauss V, Reuter G. DNA methylation in Drosophila--a critical evaluation. Prog Mol Biol Transl Sci. 2011;101:177–191. doi: 10.1016/B978-0-12-387685-0.00003-2. [DOI] [PubMed] [Google Scholar]
- Lachner M, Jenuwein T. The many faces of histone methylation. 2002. Curr. Opin. Cell Biol. 2002;14(3):286–298. doi: 10.1016/s0955-0674(02)00335-6. [DOI] [PubMed] [Google Scholar]
- Lachner M, O’Carroll D, Rea S, Mechtler K, Jenuwein T. Methylation of histone H3 lysine 9 creates a binding site for HP1 proteins. Nature. 2001;410:116–120. doi: 10.1038/35065132. [DOI] [PubMed] [Google Scholar]
- Li MA, Alls JD, Avancini RM, Koo K, Godt D. The large Maf factor Traffic Jam controls gonad morphogenesis in Drosophila . Nat. Cell. Biol. 2003;5(11):994–1000. doi: 10.1038/ncb1058. [DOI] [PubMed] [Google Scholar]
- Lin H, Yue L, Spradling AC. The Drosophila fusome, a germline-specific organelle, contains membrane skeletal proteins and functions in cyst formation. Development. 1994;120:947–956. doi: 10.1242/dev.120.4.947. [DOI] [PubMed] [Google Scholar]
- Lundberg LE, Stenberg P, Larsson J. HP1a, Su(Var)3-9, SETDB1 and POF stimulate or repress gene expression depending on genomic position, gene length and expression pattern in Drosophila melanogaster . Nucleics Acids Research. 2013;41(8):4481–4494. doi: 10.1093/nar/gkt158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Margolis J, Spradling AC. Identification and behavior of epithelial stem cells in the Drosophila ovary. Development. 1995;121:3797–3807. doi: 10.1242/dev.121.11.3797. [DOI] [PubMed] [Google Scholar]
- McKearin DM, Spradling AC. bag-of-marbles: a Drosophila gene required to initiate both male and female gametogenesis. Genes Dev. 1990;4:2242–2251. doi: 10.1101/gad.4.12b.2242. [DOI] [PubMed] [Google Scholar]
- Mis JM, Ner SS, Grigliatti TA. Identification of three histone methyltransferases in Drosophila: dG9a is a suppressor of PEV and is required for gene silencing. Mol. Gen. Gen. 2006;275:513–526. doi: 10.1007/s00438-006-0116-x. [DOI] [PubMed] [Google Scholar]
- Morris LX, Spradling AC. Long-term live imaging provides new insight into stem cell regulation and germline-soma coordination in the Drosophila ovary. Development. 2011;138(11):2207–2215. doi: 10.1242/dev.065508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moss TJ, Wallrath LL. Connections between epigenetic gene silencing and human disease. Mutat. Res. 2007;618(1–2):163–174. doi: 10.1016/j.mrfmmm.2006.05.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng JC, Karpen GH. Heterochromatic genome stability requires regulators of histone H3 K9 methylation. PLoS Genet. 2009;5(3):e1000435. doi: 10.1371/journal.pgen.1000435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peterson CL, Laniel MA. Histones and histone modifications. Curr. Biol. 2004;14:R546–R551. doi: 10.1016/j.cub.2004.07.007. [DOI] [PubMed] [Google Scholar]
- Petrella LN, Smith-Leiker T, Cooley L. The Ovhts polyprotein is cleaved to produce fusome and ring canal proteins required for Drosophila oogenesis. Development. 2007;134(4):703–712. doi: 10.1242/dev.02766. [DOI] [PubMed] [Google Scholar]
- Pritchett TL, Tanner EA, McCall K. Cracking open cell death in the Drosophila ovary. Apoptosis. 2009;14:969–979. doi: 10.1007/s10495-009-0369-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qian C, Zhou MM. SET domain protein lysine methyltransferases: Structure, specificity and catalysis. Cell. Mol. Life Sci. 2006;63:2755–2763. doi: 10.1007/s00018-006-6274-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raddatz G, Guzzardo PM, Olova N, Fantappié MR, Rampp M, Schaefer M, Reik W, Hannon GJ, Lyko F. Dnmt2-dependent methylomes lack defined DNA methylation patterns. Proc Natl Acad Sci U S A. 2103;110(21):8627–8631. doi: 10.1073/pnas.1306723110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rangan P, Malone CD, Navarro C, Newbold SP, Hayes PS, Sachidanandam R, Hannon GJ, Lehmann R. piRNA production requires heterochromatin formation in Drosophila . Curr Biol. 2011;21(16):1373–1379. doi: 10.1016/j.cub.2011.06.057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riparbelli MG, Gigliotti S, Callaini G. The Drosophila Nucleoporin gene nup154 is required for correct microfilament dynamics and cell death during oogenesis. Cell Motility and the Cytoskeleton. 2007;64:590–604. doi: 10.1002/cm.20206. [DOI] [PubMed] [Google Scholar]
- Royzman I, Hayashi-Hagihara A, Dej KJ, Bosco G, Lee JY, Orr-Weaver TL. The E2F cell cycle regulator is required for Drosophila nurse cell DNA replication and apoptosis. Mech Dev. 2002;119(2):225–237. doi: 10.1016/s0925-4773(02)00388-x. [DOI] [PubMed] [Google Scholar]
- Schotta G, Ebert A, Krauss V, Fischer A, Hoffmann J, Rea S, Jenuwein T, Dorn R, Reuter G. Central role of Drosophila Su(Var)3-9 in histone H3-K9 methylation and heterochromatic gene silencing. EMBO J. 2002;21(5):1121–1131. doi: 10.1093/emboj/21.5.1121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schultz DC, Ayyanathan K, Negorev D, Maul GG, Rauscher FJ., III SETDB1: a novel KAP-1 associated histone H3, lysine 9-specific methyltransferase that contributes to HP1-mediated silencing of euchromatic genes by KRAB zinc-finger proteins. Genes Dev. 2002;16:919–932. doi: 10.1101/gad.973302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seum C, Reo E, Peng H, Rauscher FJ, III, Spierer P, Bontron S. Drosophila SETDB1 is required for chromosome 4 silencing. PLoS Genetics. 2007a;3:e76. doi: 10.1371/journal.pgen.0030076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seum C, Bontron S, Reo E, Delattre M, Spierer P. Drosophila G9a is a nonessential gene. Genetics. 2007b;177:1955–1957. doi: 10.1534/genetics.107.078220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song X, Wong MD, Kawase E, Xi R, Ding BC, McCarthy JJ, Xie T. Bmp signals from niche cells directly repress transcription of a differentiation-promoting gene, bag of marbles, in germline stem cells in the Drosophila ovary. Development. 2004;131(6):1353–1364. doi: 10.1242/dev.01026. [DOI] [PubMed] [Google Scholar]
- Stabell M, Bjorkmo M, Aalen RB, Lambertsson A. The Drosophila SET domain encoding gene dEset is essential for proper development. Hereditas. 2006;143:177–188. doi: 10.1111/j.2006.0018-0661.01970.x. [DOI] [PubMed] [Google Scholar]
- Strahl BD, Allis CD. The language of covalent histone modifications. Nature. 2000;403:41–45. doi: 10.1038/47412. [DOI] [PubMed] [Google Scholar]
- Tschiersch B, Hoffman A, Krauss V, Dorn R, Korge G, Reuter G. The protein encoded by the Drosophila position-effect variegation suppressor gene Su(Var)3-9 combines domains of antagonistic regulators of homeotic gene complexes. EMBO J. 1994;13:3822–3831. doi: 10.1002/j.1460-2075.1994.tb06693.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tzeng TY, Lee CH, Chan LW, Shen CK. Epigenetic regulation of the Drosophila chromosome 4 by the histone H3K9 methyltransferase dSETDB1. Proc. Nat. Acad. Sci. 2007;104:12691–12696. doi: 10.1073/pnas.0705534104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Duyne R, Easley R, Wu W, Berro R, Pedati C, Klase Z, Kehn-Hall K, Flynn EK, Symer DE, Kashanchi F. Lysine methylation of HIV-1 Tat regulates transcriptional activity of the viral LTR. Retrovirology. 2008;22(5):40. doi: 10.1186/1742-4690-5-40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Pan L, Wang S, Zhou J, McDowell W, Park J, Haug J, Staehling K, Tang H, Xie T. Histone H3K9 trimethylase Eggless controls germline stem cell maintenance and differentiation. PLoS Genet. 2011;7(12):e1002426. doi: 10.1371/journal.pgen.1002426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu H, Min J, Lunin VV, Antoshenko T, Dombrovski L, Zeng H, Allali-Hassani A, Campagna-Slater V, Vedadi M, Arrowsmith CH, Plotnikov AN, Schapira M. Structural biology of human H3K9 methyltransferases. PLoS One. 2010;5(1):e8570. doi: 10.1371/journal.pone.0008570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie T, Spradling AC. A niche maintaining germ line stem cells in the Drosophila ovary. Science. 2000;290(5490):328–330. doi: 10.1126/science.290.5490.328. [DOI] [PubMed] [Google Scholar]
- Yang L, Xia L, Wu DY, Wang H, Chansky HA, Schubach WH, Hickstein DD, Zhang Y. Molecular cloning of ESET, a novel histone H3-specific methyltransferase that interacts with ERG transcription factor. 2002;21:148–152. doi: 10.1038/sj.onc.1204998. [DOI] [PubMed] [Google Scholar]
- Yasuhara JC, Wakimoto BT. Oxymoron no more: the expanding world of heterochromatic genes. TIG. 2006;22(6):330–338. doi: 10.1016/j.tig.2006.04.008. [DOI] [PubMed] [Google Scholar]
- Yoon J, Lee KS, Park JS, Yu K, Paik SG, Kang YK. dSETDB1 and Su(Var)3-9 sequentially function during germline-stem cell differentiation in Drosophila melanogaster . PLoS ONE. 2008;3(5):e2234. doi: 10.1371/journal.pone.0002234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Kalderon D. Hedgehog acts as a somatic stem cell factor in the Drosophila ovary. Nature. 2001;410:599–604. doi: 10.1038/35069099. [DOI] [PubMed] [Google Scholar]
- Zhang X, Tamaru J, Khan SI, Horton JR, Keefe LJ, Selker EU, Cheng X. Structure of the Neurospora SET domain protein DIM-5, a histone H3 lysine methyltransferase. Cell. 2002;111:117–127. doi: 10.1016/s0092-8674(02)00999-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X, Wen H, Shi X. Lysine methylation: beyond histones. Acta Biochim Biophys Sin (Shanghai) 2012;44(1):14–27. doi: 10.1093/abbs/gmr100. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1 tjGal4 expression pattern. Shown are fixed ovarioles from 2-3 day old females carrying the tj-Gal4 driver along with a UAST-EGFP reporter. (A) The tj-Gal4 driver is expressed strongly in most somatic cells in the germarium, and in follicle cells until late stages of oogenesis. (B) In the germarium, expression of tj-Gal4 is strong in the escort cells (triangles) and also the FSCs (*), and prefollicular and follicle cells. tj-Gal4 is also expressed weakly, and variably, in the cap cells.
Table S1 egg− ovarian germ stem cells are not maintained. Larvae (see materials and methods for the complete genotypes) were given one two-hour heat shock on the first day of the third larval instar. 7, 14, and 21 days after clone induction the ovaries of adult flies were scored for the presence of GSC clones.
Table S2 egg− ovarian follicle stem cells are not maintained. 1–2 day old adult females (see materials and methods for the complete genotypes) were given two one-hour heat shocks for 3 days, and scored for persistent follicle stem cell clones 8, 15 and 22 days after the final heat shock.