Summary
Many plant viruses encode proteins that suppress the antiviral RNA silencing response mounted by the host. The suppressors p19 from tombusvirus and p21 from Beet yellows virus appear to block silencing by directly binding siRNA, a critical mediator in the process. Here, we report the crystal structure of p21, which reveals an octameric ring architecture with a large central cavity of 90 Å diameter. The all α-helical p21 monomer consists of N- and C-terminal domains that associate with their neighboring counterparts through symmetric head-to-head and tail-to-tail interactions. A putative RNA binding surface is identified in the conserved, positive-charged inner surface of the ring. In contrast to the specific p19-siRNA duplex interaction, p21 is a general nucleic acid binding protein, interacting with 21 nt or longer single- and double-stranded RNAs in vitro. This study reveals an RNA binding structure adopted by the p21 silencing suppressor.
Introduction
Small RNAs mediate many gene regulatory events, including posttranscriptional gene silencing (PTGS) in plants, RNA interference (RNAi) in animals, as well as microRNA (miRNA) pathways and related events at the genome level (see reviews: Baulcombe, 2004; Bartel, 2004; Meister and Tuschl, 2004; Matzke and Birchler, 2005). These processes are collectively known as RNA silencing. In plants, one of the major functions of RNA silencing is to act as a defense against invading viruses by targeting their RNAs (Baulcombe, 2004; Wang and Metzlaff, 2005). The defense response is triggered by double-stranded (ds) RNA molecules generated in replicating RNA viruses or synthesized from viral mRNAs by the host RNA-dependant RNA polymerases (RDPs) or simply from long helical regions in viral RNAs (Voinnet, 2005). Dicer-like enzymes recognize and process these dsRNAs into small interfering RNAs (siRNAs) (Bernstein et al., 2001). siRNAs are characterized by ~19 base pairs (bp), two nucleotide (nt) overhangs, 5′-phosphates, and 3′-hydroxyls (Hamilton and Baulcombe, 1999; Elbashir et al., 2001). siRNAs mediate the silencing by guiding a nuclease complex, RNA-induced silencing complex (RISC), to recognize and cleave target RNAs (Hammond et al., 2000). The specificity of targeting is dictated by the base paring interaction between guide and target RNA strands. In addition to cell-autonomous silencing, plants also generate a mobile signal at the site of silencing initiation, after which the signal spreads out and induces specific gene suppression in distant parts of the plant (Voinnet and Baulcombe, 1997). The local and systemic silencing phenomena create an immune-like system by which to protect plants from virus infection.
Viruses have developed ways to suppress the host silencing response (Baulcombe, 2004; Ding et al., 2004; Roth et al., 2004; Silhavy and Burgyan, 2004; Voinnet, 2005). Proteins that suppress silencing were first identified in potyvirus and cucumovirus (Anandalakshmi et al., 1998; Brigneti et al., 1998; Kasschau and Carrington, 1998). The number of viral suppressors has grown to about 20 at last count (Voinnet, 2005). Some suppressors were even found in animal viruses (Li et al., 2002, 2004; Bucher et al., 2004; Delgadillo et al., 2004), raising an intriguing question as to whether RNA silencing functions as an antiviral mechanism in animals. The expression of these suppressors is believed to be a means for viruses to overcome the host antiviral silencing defense. On the other hand, other types of RNA silencing may also be affected by certain suppressors because of the shared mechanism among related silencing processes. Disruption of the miRNA pathway by silencing suppressors has been shown to cause defects in plant development processes under miRNA control (Kasschau et al., 2003; Chapman et al., 2004; Dunoyer et al., 2004).
These viral suppressors vary widely in sequence, structure, effect, and resulting phenotype (Voinnet, 2005). Their diversity suggests that they have evolved independently and may function by different mechanisms. However, for most viral suppressors, little is known about how they block silencing events.
Recent studies have pointed out that siRNA is an attractive target for viral intervention. This was first shown for the tombusviral p19 suppressor that physically binds to siRNA duplexes in vitro (Silhavy et al., 2002) and in vivo (Lakatos et al., 2004; Dunoyer et al., 2004; Chapman et al., 2004). p19 was proposed to block RNA silencing by sequestering siRNAs, thereby preventing them from mediating RNA silencing. Biochemical analysis showed that p19 exhibits high binding specificity for 19–21 bp RNA duplexes, but not for ssRNA and long dsRNA (Silhavy et al., 2002; Vargason et al., 2003; Ye et al., 2003). The molecular basis for such size-selective binding has been revealed in recent cocrystal structures of p19 bound to 19 bp siRNA duplexes (Vargason et al., 2003; Ye et al., 2003). In the structure of the complex, p19 forms a homodimer that binds one face of the RNA duplex through interactions with backbone phosphate and 2′-hydroxyl groups in a non-sequence-specific manner. In addition, critical tryptophan residues projecting from symmetrically positioned reading head helices were found to cap both ends of the 19 bp duplex, thereby providing a caliper-like mechanism for measuring duplex length.
Beet yellows virus (BYV), the type member of the genus Closterovirus, is a positive strand RNA virus that features a large 15.5 kb genome and long filamentous virions (Dolja, 2003). The p21 protein, one of nine proteins encoded in the BYV genome, was the only one that suppressed the silencing of green fluorescence protein (GFP) induced by GFP dsRNA in a transitive agrobacterium infiltration experiment (Reed et al., 2003). Interestingly, a related Citrus tristeza virus (CTV), also of genus Closterovirus, encodes at least three suppressors (Lu et al., 2004): one suppressor is a homolog of BYV p21, and two others seem to have no counterparts in BYV. Additional biochemical studies showed that p21 binds siRNA duplexes both in vitro and in vivo, but not single-stranded miRNA (Chapman et al., 2004). Therefore, it has been suggested that p21, like p19, functions by inactivating siRNA duplexes.
The molecular mechanism of the p21-siRNA interaction has not been elucidated, particularly regarding the question as to whether the two suppressors, p19 and p21, are similar in terms of structure and RNA binding. As a first step, we have determined the crystal structure of p21 in the free state. The structure shows that p21 forms an octameric ring of previously unknown topology. Conservation and charge analysis suggest that the inner surface of the ring might be involved in RNA binding. We also show that p21 has no strict binding specificity for siRNA duplexes. Conversely, it appears to be a general nucleic acid binding protein, implicating additional p21-mediated pathways for suppression of RNA silencing.
Results and Discussion
p21 Forms Higher-Order RNA Binding Oligomers
We overexpressed the BYV p21 in E. coli and purified the protein though three chromatography steps. In the final gel filtration step, we observed that p21 elutes as two overlapping peaks, corresponding to species of apparent molecular weights of 250 and 500 kDa, respectively (Figure 1A). The exact elution profile varied between samples, reflecting a change in composition. In denatured SDS-PAGE (Figure 1B), all eluates migrate as a single 22 kDa species, as expected for a p21 monomer. The gel filtration analysis indicates that p21 forms higher-order oligomers in solution.
Figure 1. p21 Forms Large siRNA Binding Oligomers.
(A) Elution profile of p21 in a Hiload 26/60 Superdex 200 column in a buffer of 0.1 M KCl and 10 mM HEPES (pH 7.6). Also indicated are molecular standards: Aldolase (158k), Ferritin (440k), and Thy-roglobulin (669k). Two main peaks appear at the 250 kDa and 500 kDa regions.
(B) SDS-PAGE of fractions 11–19 showing protein purity.
(C and D) EMSA showing complex formation between a 21 nt siRNA (50 pmol) and 10 μl p21 from fractions 11–19 in 0.1 M KCl, 1 mM DTT, 1 mM MgCl2, and 10 mM HEPES (pH 7.6). The total amounts of protein used are indicated for each lane. Roughly, more than four p21 monomers are required to bind one siRNA duplex. The experiment was done immediately after the protein was eluted from the column; if the experiment is not performed immediately, the RNP pattern will change. Lane 12 compares the migration of the p19-siRNA complex. The gel was stained by ethidium bromide to show RNA in (C) and by Coomassie to reveal protein in (D).
A previous study has shown that p21 is capable of binding siRNA duplexes in vitro and in vivo (Chapman et al., 2004). We tested whether our preparation of p21 binds siRNAs in a electrophoretic mobility shift assay (EMSA). Proteins in fractions 11–19 (Figure 1A) were mixed with a 21 nt siRNA duplex, and the reaction was resolved in a 5% native gel. Under binding conditions of excess RNA, all of the added protein forms complexes with RNA, as evident by the presence of free RNA bands and the absence of free protein bands in lanes 2–10. Multiple and distinct RNA-protein complexes (RNP) were visible in the gel, with two species, labeled RNP1 and RNP2, predominating under these conditions. The distribution of RNP1 and RNP2 in the gel correlates with that of the 250 kDa and 500 kDa peaks, respectively, in the gel filtration profile. This suggests that the faster-migrating RNP1 is made up of the 250 kDa species, while the slower-migrating RNP2 is derived from the 500 kDa species. Figure 1 also compares the migration of p21 RNPs with that of the p19-siRNA complex, a well-characterized RNA silencing suppressor-siRNA complex consisting of the p19 (monomeric size: 20 kDa) dimer and an siRNA duplex (Vargason et al., 2003; Ye et al., 2003). RNP1 migrates only one-third of the distance that the p19-siRNA complex does, supporting the large size of RNP1.
Complexes of RNP3 and RNP4, though present in smaller amounts, are also clearly observed in early eluates in lanes 2–4. In samples stored at 4°C, the protein tends to form complexes of higher order over time. These results establish that p21 exists as a mixture of oligomers in solution and that each oligomer is capable of forming distinct complexes with siRNA.
Structure Determination
We have successfully crystallized p21 in the free state despite its existence as a multiple oligomeric species in solution. The structure was solved by the method of Multiple Isomorphous Replacement and Anomalous Scattering (MIRAS) based on mercury and osmium derivatives. The experimental map calculated at 3.8 Å was readily interpretable and has permitted the tracing of the majority of the polypeptide chain. The N-terminal engineered His tag (20 resides) and residues 65–76 were not modeled due to missing density. The final model was refined at 3.3 Å resolution and has an R factor of 21.0%, an Rfree of 24.4%, and excellent stereochemistry (Table 1).
Table 1.
Crystallographic Statistics
| Crystal | Native | EMTS (Hg) | K2OsO4 |
|---|---|---|---|
| Space group | P3221 | P3221 | P3221 |
| Cell parameters a = b/c (Å) | 199.6/56.1 | 198.9/55.9 | 199.3/56.2 |
| X-ray source | Cu-Kα | BNL-X14 | Cu-Kα |
| Wavelength (Å) | 1.5418 | 1.009 | 1.5418 |
| Resolution (Å) | 20–3.3 | 20–3.8 | 20–4.0 |
| Last shell (Å) | (3.42–3.3) | (3.93–3.8) | (4.14–4.0) |
| Number of total reflections | 220,293 | 284,136 | 167,300 |
| Number of unique reflections | 19,491 | 12,653 | 10,916 |
| Redundancy | 11.3 | 22.5 | 15.3 |
| Completeness as I > 0 (%) | 98.8 (95.8) | 97.4 (93.8) | 97.5 (94.4) |
| I/σ | 21.6 (3.0) | 31.6 (7.7) | 25.7 (4.6) |
| Rsym (%) | 0.11 (0.66) | 0.13 (0.49) | 0.12 (0.61) |
|
| |||
| Phasing Statistics | |||
|
| |||
| Number of sites | 4 | 8 | |
| Phasing power (iso/ano) | 1.36/3.46 | 0.70/2.68 | |
| MIRAS figure of merit | 0.45 | 0.45 | |
|
| |||
| Refinement | |||
|
| |||
| Rcryst (%) | 21.0 (30.8) | ||
| Rfree (%) | 24.4 (39.5) | ||
| Rmsd bond length (Å) | 0.019 | ||
| Rmsd bond angles (°) | 1.812 | ||
| Atom number | 5,388 | ||
| Average B factor of each molecule (Å2) | 96, 140, 151, 147 | ||
| Ramachandran plot (favorable/allowed, %) | 93.5/5.8 | ||
Numbers in parentheses represent statistics for the highest-resolution shell of data.
Structure Description
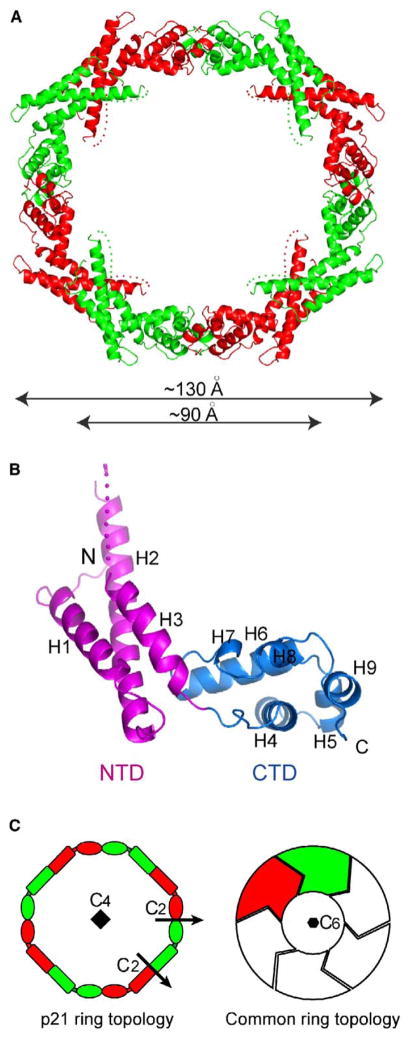
The crystal structure reveals that p21 is an oligomer of eight monomer subunits that assemble into a closed ring (Figure 2A). The large octameric ring measures ~130 Å in the outer diameter, ~90 Å in the inner diameter, and ~20 Å in thickness.
Figure 2. Structure of the p21 Octameric Ring.
(A) Ribbon representation of the entire p21 octamer. Neighboring monomers are alternatively colored with green and red. The dots stand for disordered regions.
(B) Structure of the p21 monomer with the secondary structure elements labeled. The NTD and CTD domains are colored magenta and blue, respectively.
(C) Topology diagrams of the p21 octameric ring structure (left panel). Each p21 monomer, colored the same as in (A), is represented by a rectangle (NTD) and an ellipse (CTD). Symmetry elements consist of a 4-fold axis perpendicular to the plane and two types of dyad axes in the plane. A diagram of other common ring structures with a hexameric ring as an example is shown (right panel).
Individual p21 monomers adopt an all α-helical structure (Figure 2B). The structure is built upon nine α helices and can be divided into amino (NTD)- and carboxy (CTD)-terminal domains. The NTD (residues 1–93) is mainly a left-handed three-helical bundle plus an ordered N-terminal tail. The bundle consists of the H1, H2, and H3 helices arranged in an up-and-down fashion. The H2 and H3 helices are connected by a disordered 10 residue loop. In the CTD (residues 94–174), six contiguous α helices, including the short 310 helix H7, fold into a two-layered array. The first layer includes helices H4, H5, and H9, whereas helices H6–H8 constitute the second layer. Notably, all adjacent helices in the CTD are approximately orthogonal to each other.
We attempted to find structural homologs of p21 in Dali-based searches by using the NTD, CTD, or the entire monomer structure as a search model (Holm and Sander, 1998). Related structures can only be identified for the NTD part of the structure. The left-handed three-helical NTD bundle is a common structural motif belonging to the spectrin repeat-like fold in SCOP, found in proteins of diverse function (Murzin et al., 1995). Dali hits for the CTD are not significant (Z score < 2.5), suggesting that the CTD adopts a new fold. Based on these data, together with the ring topology discussed below, we conclude that the p21 octamer represents a novel, to our knowledge, structure.
Oligomer Organization
Forming a ring is a common way to build high-order oligomeric scaffolds. Examples of ring structure can be found for GroEL/GroES chaperones (Sigler et al., 1998), Sm or Sm-like spliceosomal proteins (Toro et al., 2002), the Trp RNA binding attenuation protein (Antson et al., 1999), the 20S proteasome (Bochtler et al., 1999), replicative hexameric DNA helicases (Patel and Picha, 2000), and the recently described RNA binding protein HutP, which consists of two stacked trimeric rings (Kumarevel et al., 2005). In these known ring structures, subunits assemble through a single type of interface, referred to here as head-to-tail association. Such an association involves nonequivalent segments of adjacent subunits and lacks 2-fold symmetry in the interface. Subunits are related to each other by n-fold rotation symmetry along the ring axis perpendicular to the ring plane; n stands for the order of the ring (see the diagram of common head-to-tail ring topology in Figure 2C, right panel).
By contrast, the eight p21 monomer subunits associate into the closed ring structure through two types of head-to-head and tail-to-tail arrangements (Figure 2C). The NTD joins with the NTD of an adjacent monomer on one side (head-to-head), while the CTD associates with the CTD of a neighboring monomer on the other side (tail-to-tail). The NTD and CTD association interfaces for an individual monomer subunit are roughly opposite of each other, with one facing inward and the other facing outward (Figure 2A). Both types of the molecular interface contain a dyad symmetry in the ring plane. There is no 8-fold symmetry in the structure; rather, alternative subunits (same colored subunits in Figure 2C, left panel) are related to each other by a 4-fold rotation symmetry normal to the ring. To the best of our knowledge, this is the first example of such a ring topology in a protein structure.
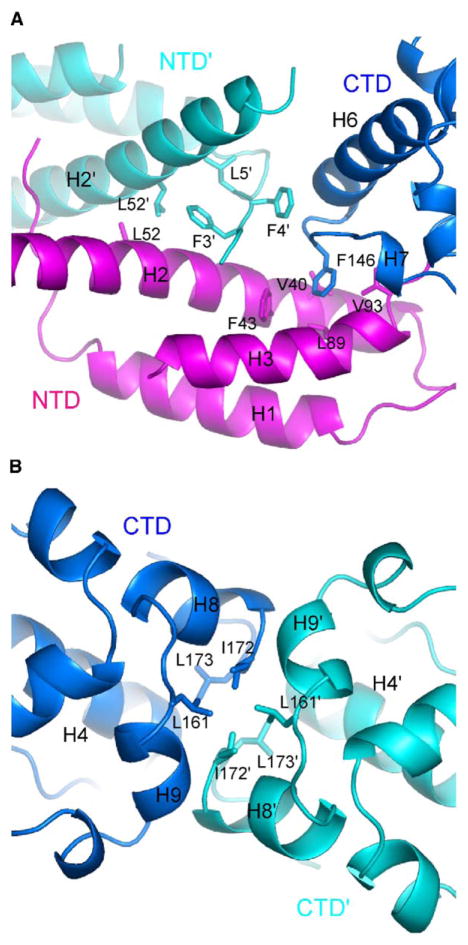
The association between adjacent NTDs involves the H2 helices and the N-terminal tails (Figure 3A) and buries a solvent-accessible area of 1142 Å2 per subunit. The H2 helix and its counterpart intersect at near right angles (80°) with a single contact involving Leu52. In addition, the N-terminal tail plays an important role in intermolecular association. It inserts into the space enveloped by its own NTD and by the NTD and the CTD of the neighboring monomer subunit. Conserved hydrophobic residues 3–5 in the tail serve as a nucleation center in the anchoring of the three domains. Each of residues 3–5 anchor one of three domains: residue Leu5 interacts with H2 of its own NTD; residue Phe3 contacts H2 of the neighboring NTD; residue Phe4 interacts with the C-terminal region of H6 and its adjacent loop in the neighboring CTD.
Figure 3. Interactions that Build up the p21 Octameric Ring.
(A) The NTDs interface with key interacting residues shown in stick representation. The N-terminal tail plays a critical role in the association of neighboring NTDs. Also shown are the conserved interactions that stabilize the orientation between NTD and CTD within one monomer subunit. The NTD, CTD, and the neighboring NTD′ are colored magenta, blue, and cyan, respectively. The prime symbol in the labels denotes the other molecule.
(B) The hydrophobic interface between the blue-colored CTD and its neighboring cyan-colored CTD′.
The CTDs associate along a flat surface joined by the N terminus of helix H4 and by helices H8 and H9 (Figure 3B). The central segment of the association interface is lined by residues Ile172, Leu173, and Leu161, which form hydrophobic patches holding adjacent CTDs together. The association of CTDs buries a solvent-accessible area of 500 Å2 per subunit.
The structure can be alternatively viewed as an equal-angled octagon with two alternating sides. One side is represented by two joining CTDs, and the other is represented by two joining NTDs. The two sides meet at the junction of the NTD and CTD within individual monomer subunits related by 145°. This angle is realized by maintaining a fixed orientation of the NTD and CTD in a monomer. The interdomain orientation is maintained by a loop connecting helices H3 and H4 and by additional constraints provided by interdomain interactions. Most notably, the benzene ring of Phe146 in the CTD projects out and reaches into a hydrophobic depression, converged by residues Phe43, Val40, Leu89, and Val93, on the surface of the NTD of the same monomer subunit (Figure 3A). The five residues forming the hydrophobic cluster are highly conserved among p21 homologs, highlighting their importance for maintenance of a stable orientation between the two domains to generate the octagonal arrangement. Among the four monomer subunits in the crystallographic asymmetric unit, the variation in the interdomain orientation is minimal, with an average of 5.4° and a maximum of 8.3°. It should be noted that no rational constraint was imposed during refinement between the two domains, which belong to independent noncrystallographic symmetry (NCS) groups.
p21 as an Octamer
The crystal structure shows p21 to be an integral octamer. However, the protein exists as multiple oligomeric species in solution. What is the relationship between the crystallographic octamer and these oligomers in solution? By taking advantage of the power of native gel electrophoresis, the oligomer composition can be estimated within the RNP. Comparison of the gel filtration elution profile and the RNP distribution pattern in the native gel implies that RNP1 and RNP2 are derived from the 250 kDa and 500 kDa species, respectively. Given that p21 is arranged into an octameric alignment in the crystal structure, it appears likely that RNP1, which represents the smallest complex, corresponds to a single p21 octamer bound to siRNA. The p21 octamer has a total molecular weight of 170 kDa and, considering its nonglobular shape, could give rise to the observed 250 kDa peak in the gel filtration profile. RNP2, derived from the 500 kDa species, most likely corresponds to dimerization of the p21 octamer-siRNA complex. The larger RNPs, which migrate in a ladder-like manner, may result from further association of octameric species. Oligomers smaller than an octamer are likely to be unstable, since p21 was not observed in the gel filtration fractions following the 250 kDa peak, nor have complexes smaller than RNP1 been visible in the native gel. Therefore, we conclude that the octamer is the minimal stable structure of p21.
The formation of the higher-order oligomers is not due to intermolecular disulfide bond formation since the reducing agent DTT was included in the binding reaction. Moreover, inspection of the structure indicates that neither Cys39 nor Cys114, the only two cysteine residues in the p21 sequence, is accessible for disulfide bond formation: Cys39, which forms the mercury derivative, is located within the inner circumference of the ring, while Cys114 is buried. The formation of higher-order oligomers is not caused by the presence of a His tag either, because a nontagged p21 displayed the same elution profile during gel filtration (data not shown). It is not known whether self-association of the p21 octamer occurs in vivo, but a large portion of p21 expressed in plants was found in the inclusion body fraction, suggesting its associative nature (Reed et al., 2003). It remains to be determined whether the self-association of the p21 octamer has any biological role.
A Putative RNA Binding Surface inside the Ring
With the structure in hand, we next analyzed the conserved features of the structure for their likely involvement in RNA binding. p21 homologs can be found in six different virus species, all of which belong to the genus Closterovirus. The multiple sequence alignment generated by CLUSTALW (Thompson et al., 1994) shows a high divergence of sequence, except between GLRaV-2 and GLRSaV (Figure 4). These six sequences exhibit 20% pairwise identity and 40% pairwise similarity on average (16% and 37% respectively, if the comparison between the GLRaV-2 and GLRSaV sequences is excluded). The high sequence diversity is helpful for identification of those functionally important residues that are less prone to change during evolution. Mapping conserved residues into the three-dimensional structure shows that the majority of them participate in the formation of the hydrophobic core structure and in oligomerization. Some of them are assigned to different functional groups, which can be seen in Figure 4. The invariance of residue Ser141 most likely reflects the requirement of its side chain hydroxyl for hydrogen bonding with the backbone atoms of F146-O and V148-N in the structure; this binding thereby stabilizes a local turn. However, four absolutely conserved residues located along the inner surface of the ring cannot be assigned to any structural role (Figures 5A and 5B). They consist of basic residues Lys2 (from the neighboring monomer), Arg90, and Arg130 and polar reside Thr149 and could play important functional roles, such as RNA binding.
Figure 4. Sequence Alignment of p21 Homologs from the Closterovirus Genus.
Aligned are the sequences of AAC55667 (GenBank ID) of Beet yellows stunt virus (BYSV), NP_041877 of Beet yellows virus (BYV), NP_042870 of Citrus tristeza virus (CTV), AAC40863 of Grapevine leafroll-associated virus 2 (GLRaV-2), NP_835252 of Grapevine rootstock stem lesion-associated virus (GRSLaV), and YP_224098 of Mint virus 1 (MV-1). The secondary structure observed in the crystal structure is displayed on the top. Residues are shaded according to their degree of conservation. Functional groups are defined.
Figure 5. A Putative RNA Binding Surface.
(A) Conserved residues located at the inner surface of the ring.
(B) Close-up view of (A). Conserved residues are shown in stick representation.
(C) Electrostatic potential shows a contrasting distribution of positive charge (blue) in the inside of the ring and negative charge (red) on the outside.
The rings in (A) and (C) have the same orientation, which is related to the ring in Figure 2A by a horizontal 60° rotation.
In the electrostatic potential view (Figure 5C), the ring surface displays a highly polarized distribution of charge: negative charge (red) dominates the outer surface of the ring, while positive charge (blue) dominates the inner surface of the ring. The positively charged inner surface might be able to bind RNA via electrostatic interactions. Importantly, the four conserved basic and polar resides are also located at this positively charged surface. The conservation and charge analysis together define a putative RNA binding surface within the inner circumference of the ring.
To test whether conserved residues Lys2, Arg90, Arg130, and Thr149 mediate RNA binding, we have generated alanine mutations one at a time at each of these positions. In addition, we generated mutations at positions Arg16, Arg73, Arg83, Arg92, Arg121, and Arg126, which serve as controls, and studied their binding to siRNAs by using a filter binding assay. These mutational experiments were inconclusive, since large uncertainties were observed in the measured apparent disassociation constants (Kd). For example, the Kd values for wild-type protein fluctuates between 2 nM and 130 nM (as monomer concentration) in different measurements. We found that factors that affect the measurement may include the freshness of the sample, oligomer composition in the sample, storage conditions, and possibly other as yet undefined contributors. Similar problems were also observed for all of the other mutant proteins. Given the large uncertainty in the measured Kd values, we were unable to conclusively detect the effect of a single mutation involving as much as a ~10-fold change of Kd. Nevertheless, all mutant proteins still bind RNA with Kd values less than 1 μM, suggesting that none of the tested residues play a single decisive role in the binding process. Other experimental methods are needed to definitively pinpoint the RNA binding site.
The binding stoichiometry between p21 and siRNA can be roughly estimated from the EMSA experiment shown in Figure 1, though it is conceivable that different RNPs may exhibit different protein:RNA ratios. In lane 16 of Figure 1C, where the amount of protein (216 pmol) is four times more than that of siRNA (50 pmol), RNA is still in excess; thus, the p21 monomer binds siRNA with an average molar ratio of at least 4:1. Titration experiments with individual fractions enriched with RNP1 or RNP2 (Figure S1; see the Supplemental Data available with this article online) indicate that the protein:RNA ratio is about 6:1 in RNP1 and 8:1 in RNP2. It seems that one p21 octamer ring is likely to bind 1–2 siRNA duplexes under our experimental conditions. A standard 19 bp A-form RNA duplex has an approximate length of 60 Å and a diameter of 22 Å. The large central cavity (~90 Å diameter) of the p21 ring could readily accommodate one or two siRNA duplexes.
p21 Is a General Nucleic Acid Binding Protein
Two proteins, p19 and p21, have been reported to date to suppress RNA silencing by interacting with siRNA. The structure of the p21 octamer reported here bears no similarity with that of dimeric p19, suggesting that they have evolved independently. Moreover, the caliper-like apparatus in the p19 structure, which provides the duplex size selectivity, is apparently absent in the p21 ring structure, suggesting that the two proteins are likely to bind RNA in different ways. To define the RNA binding specificity of p21, we studied the interaction between p21 and various RNAs by using EMSA (Figure 6). Surprisingly, p21 binds both the 21 nt double-stranded siRNA duplex and ssRNA (lanes 1–4), as well as their DNA counterparts (lanes 5–8). The protein also efficiently binds longer RNAs of 56, 110, and 401 nt, both in single-stranded and duplex forms (lanes 9–20). In contrast to p19, which exhibits a strict binding requirement for a RNA duplex of defined length, p21 appears to be a promiscuous RNA binder in terms of specificity for helix structure and length. In a more quantitative characterization (data not shown), the binding affinities are generally 4–9 times higher for duplex than for single-stranded RNA, are about 2–3 times worse for DNA than for RNA, and increased with the length of RNA. These characteristics indicate that p21 is a general nucleic acid binding protein.
Figure 6. EMSA Showing that p21 Forms Complexes with Various Nucleic Acid Species.
The protein (1 μM) was assembled with 5′-end 32P-labeled oligonucleotides of different lengths and structures (~0.1 nM) in 10 μl binding buffer (see Experimental Procedures). Single-stranded nucleic acids normally migrate faster than double-stranded nucleic acids of the same length, except for the RNAs of 110 and 401 nt length, in which the reverse is true. For 110 nt dsRNA, and 401 nt ssRNAs and dsRNAs, some RNPs are stuck in the wells, probably as a result of multiple protein association. In lane 13, the 110 nt ssRNA is partially degraded; nevertheless, all degraded fragments still form RNPs in lane 14. In contrast to what is seen in Figure 1, in which multiple RNPs are obvious, only one form of RNP species predominates in this figure. The reason for this difference is due to the different experimental conditions used. EMSA in Figure 1D is done with the RNA in excess, and literally all oligomers in solution, irrespective of their different RNA binding affinities, form the RNPs shown in the gel. Thus, the composition of RNP is a faithful representation of that of oligomers in solution. While experiments in this figure were done with negligible amounts of 32P-labeled RNA, p21 oligomers actually are competing for limited RNA, and only the oligomer with the highest binding affinity will first form RNP. The RNP picture does not reflect the true composition of the oligomer in this case. It is for this reason that only one band of RNP is visible in some lanes.
Other Suppression Mechanisms of p21
The promiscuous RNA binding by p21 also raises an interesting question regarding its mechanism of silencing suppression. Could other RNAs in addition to siRNA duplexes be targeted by p21 in silencing suppression? This is possible because different types of RNA involved in the RNA silencing process are theoretical targets of suppression. In the initiation stage, trigger molecules are long dsRNAs derived from viral RNAs, which are then transformed into ~21 nt siRNA duplexes during dicing. Upon duplex unwinding and strand selection, siRNAs become single-stranded guide RNAs bound in effector complexes. Long dsRNAs and siRNA duplexes, as well as single-stranded guide RNAs, are all potential targets of p21, though their accessibility might vary in vivo. Single-stranded guide RNAs bound in effector complexes are likely to have less accessibility than other RNAs. Among these RNAs, siRNA duplexes are most likely targeted by p21, because it has been shown that transgenically expressed p21 can coimmnuoprecipitate siRNAs in vivo (Chapman et al., 2004). Nevertheless, the question of whether the function of other RNAs is affected by p21 needs further study.
Experimental Procedures
Cloning, Protein Expression, and Purification
The full-length p21 gene of 177 amino acid residues (GenBank ID NP_041877) of Beet yellows virus was synthesized by a PCR-based approach (Casimiro et al., 1997) and was cloned into the His tag vector pET28a by using the NdeI and EcoRI restriction sites. The coding sequence was optimized for expression in E. coli (sequence available upon request). The expression vector with the correct gene sequence was transformed into BL21(DE3)-Gold E. coli. Protein expression was induced with 0.1 mM IPTG in mid-log cells, and the cultures were further grown overnight at a reduced temperature of 20°C. Harvested cells were resuspended in buffer A (0.1 M KCl and 50 mM phosphate [pH 7.6]) and broken by sonication. Clarified cell lysate was loaded onto a 5 ml Histrap column (Amersham Biosciences). After a wash by 50 mM imidazole in buffer A, the His-tagged protein was eluted with 500 mM imidazole in buffer A. The eluate was supplemented with 5 mM DTT and directly applied onto a 5 ml Hitrap Heparin column equilibrated with buffer B (20 mM HEPES [pH 7.6]). The bound p21 was eluted at ~0.5 M KCl in a linear gradient of buffer C (1 M KCl, 20 mM HEPES [pH 7.6]). The fractions containing p21 were pooled, concentrated, and further purified with gel filtration (Superdex 200) in buffer D (0.1 M KCl, 5 mM HEPES [pH 7.6]). For crystallization, the protein was supplemented with 10 mM DTT, concentrated to ~20 mg/ml, and used immediately or stored at −80°C. The His tag was refractory to thrombin cleavage and was left intact. The protein concentration was determined by UV absorbance at 280 nM by using a calculated extinction coefficient of 8375 M−1 cm−1. The p21 concentration is described in monomer units.
Crystallization and Data Collection
The protein was crystallized at 20°C by the vapor diffusion hanging drop method by mixing 1 μl of 20 mg/ml protein solution with 1 μl of a well solution containing 0.2 M K/Na Tartrate, 100 mM HEPES (pH 7.0). For cryoprotection, crystals were transferred to a 2 μl drop of well solution plus 5% glycerol at 4°C, and the glycerol concentration was gradually raised to 33% by stepwise addition of an equal volume of well solution plus 60% glycerol.
The crystals belong to space group P3221 and have unit cell dimensions of a = b = 199.63 Å, c = 56.12 Å and a solvent content of 65%. The asymmetric unit of the crystal contains four p21 monomer subunits.
For heavy atom derivatives, crystals were soaked in the well solution containing 5% glycerol and either 5 mM ethyl mercury thiosalicylate (EMTS) for 2 hr, or 5 mM K2OsO4 for 7 days. The data on the mercury derivative were collected at the peak wavelength of mercury (1.009 Å) at beamline X14 at Brookhaven National Laboratory. The native and osmium derivative data sets were collected at a wavelength of 1.5418 Å on our in-house Rigaku diffractometer.
Structure Determination and Refinement
Data were reduced by using HKL2000 (Otwinowski and Minor, 1997). SHELXD found four mercury sites (Uson and Sheldrick, 1999), which were used to calculate initial phases by the method of single isomorphous replacement with anomalous scattering (SIRAS) in CNS (Brunger et al., 1998). The map based on the SIRAS phases was already interpretable. Crossphasing identified eight sites in the osmium derivative, which was used in combination to calculate MIRAS phases and a map with improved quality. The experimental map at 3.8 Å showed clear features of protein secondary structures. Many bulky side chains were recognizable, allowing us to register the correct amino acid sequence into the structure. One of the four monomers displayed electron density of the best quality, which was then primarily used in model building. Density averaging was not used due to its adverse effect on the best part of the map. The model was built with the program O (Jones and Kjeldgaard, 1997).
The structure was refined in CNS until Rfree reached 33%, then refined by Refmac in CCP4 with the option of TLS parameters, which define group anisotropy (Murshudov et al., 1999; CCP4, 1994). Two noncrystallographic symmetry (NCS) groups, tightly constrained, were defined for four NTDs and for four CTDs. A total of eight TLS groups were defined for each NTD and CTD of the four molecules. The final Rfree dropped to 24.4%, and the R factor to 21.0%, after the Refmac refinement, largely due to the introduction of TLS parameters. The final model contains residues 1–64 and 77–177 for each of four p21 molecules. The four molecules in the asymmetric unit have average B factors of 96, 140, 151, and 147 Å2 respectively. The B factor was converted from the TLS parameters and the residual B factor by the program tlsanl in CCP4 (CCP4, 1994). Variation in the interdomain orientation was calculated as the additional rotation, expressed as Kappa angle in the spherical mode, that is required to superimpose the NTDs of each pair of molecules that have been superimposed by their CTDs. Structure figures were prepared by PyMol (DeLano, 2002) and Grasp (Nicholls et al., 1991).
Electrophoretic Mobility Shift Assay
We preformed two kinds of EMSA assays with either nonlabeled RNAs in μM concentrations (Figure 1D) or 5′-end 32P-labeled RNAs in sub-nM concentrations (Figure 6). The p21 protein was assembled with RNAs in the binding buffer of 0.1 M KCl, 1 mM DTT, 1 mM MgCl2, and 10 mM HEPES (pH 7.6). The reaction proceeded for 15 min before the addition of 2 μl loading buffer of 50% glycerol and dyes. The reactions were resolved in a 5% native polyacrylamide gel for 1 hr at constant power of 5 W at room temperature and in a buffer of 25 mM Tris, 192 mM glycine (pH 8.3). In case of nonlabeled RNA, the gel is stained by ethidium bromide and subsequently by Commassie blue. In case of labeled RNA, the autoradiograph was read by phosphor imaging.
RNA Preparation
The 21 nt siRNA duplex was formed by annealing ssRNAs of si1 (5′-CGUACGCGGAAUACUUCGAUU-3′) and si2 (5′-UCGAAGUAUUCCGCGUACGUU-3′). si2 was also used as the 21 nt ssRNA in experiments. The corresponding DNA versions have the same sequence. Short nucleic acids were chemically synthesized without 5′-phosphate.
Long RNAs were derived from the GST gene sequence and were prepared by in vitro transcription on the dsDNA templates amplified from pGEX-4T-2 plasmid by PCR (Amersham Biosciences). The PCR primers were (from 5′ to 3′):
GCGTAATACGACTCACTATAGGTTATTGGAAAATTAAGGG
GGTTATTGGAAAATTAAGGG
GCGTAATACGACTCACTATAGGATATTCCAAAAGAAGTCGA
GGATATTCCAAAAGAAGTCGA
GCGTAATACGACTCACTATAGGCCATTTATCACCTTCATCG
GGCCATTTATCACCTTCATCG
GCGTAATACGACTCACTATAGGACATAAACGATCTTCGAAC
GGACATAAACGATCTTCGAAC.
Primers 1 and 4 were used to amplify the transcription template for the 56 nt sense RNA; primers 2 and 3 were used for the 56 nt antisense RNA; primers 1 and 6 were used for the 110 nt sense RNA; primers 2 and 5 were used for the 110 nt antisense RNA; primers 1 and 8 were used for the 401 nt sense RNA; and primers 2 and 7 were used for the 402 nt antisense RNA. Duplexes were formed by annealing complementary sense and antisense RNAs of corresponding length. Antisense RNAs were used as ssRNAs in Figure 6.
Duplexes were annealed in 0.1 M KCl, 2 mM Mg, and 10 mM HEPES (pH 7.6) by a 1 min heating treatment at 95°C and a 1 hr incubation at 37°C. RNAs prepared by in vitro transcription were dephosporylated by CIP (Roche) at 50°C, followed by phenol extraction. Nucleic acids were 5′-end labeled with 32P-γ-ATP by T4 kinase (NEB) under standard conditions and were separated from unincorporated nucleotide by a mini-quick-spin-oligo column (Roche).
Supplementary Material
Acknowledgments
We thank Yu-Ren Yuan for help in synchrotron diffraction data collection and structure calculation. We are grateful to the scientists of beamline X14 at National Synchrotron Light Source, Brookhaven National Laboratory for assistance with data collection. This research is supported by the Abby Rockefeller Mauze Trust and the Dewitt Wallace and Maloris Foundations.
Footnotes
Supplemental Data including one figure showing stoichiometry of p21-siRNA binding are available at http://www.structure.org/cgi/content/full/13/9/1375/DC1/.
Accession Numbers
The structural coordinates have been deposited in the Protein Data Bank with the accession code 2CWO.
References
- Anandalakshmi R, Pruss GJ, Ge X, Marathe R, Mallory AC, Smith TH, Vance VB. A viral suppressor of gene silencing in plants. Proc Natl Acad Sci USA. 1998;95:13079–13084. doi: 10.1073/pnas.95.22.13079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Antson AA, Dodson EJ, Dodson G, Greaves RB, Chen X, Gollnick P. Structure of the trp RNA-binding attenuation protein, TRAP, bound to RNA. Nature. 1999;401:235–242. doi: 10.1038/45730. [DOI] [PubMed] [Google Scholar]
- Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–297. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- Baulcombe DC. RNA silencing in plants. Nature. 2004;431:356–363. doi: 10.1038/nature02874. [DOI] [PubMed] [Google Scholar]
- Bernstein E, Caudy AA, Hammond SM, Hannon GJ. Role for a bidentate ribonuclease in the initiation step of RNA interference. Nature. 2001;409:363–366. doi: 10.1038/35053110. [DOI] [PubMed] [Google Scholar]
- Bochtler M, Ditzel L, Groll M, Hartmann C, Huber R. The proteasome. Annu Rev Biophys Biomol Struct. 1999;28:295–317. doi: 10.1146/annurev.biophys.28.1.295. [DOI] [PubMed] [Google Scholar]
- Brigneti G, Voinnet O, Li WX, Ji LH, Ding SW, Baulcombe DC. Viral pathogenicity determinants are suppressors of transgene silencing in Nicotiana benthamiana. EMBO J. 1998;17:6739–6746. doi: 10.1093/emboj/17.22.6739. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Brunger AT, Adams PD, Clore GM, DeLano WL, Gros P, Grosse-Kunstleve RW, Jiang JS, Kuszewski J, Nilges M, Pannu NS, et al. Crystallography & NMR system: a new software suite for macromolecular structure determination. Acta Crystallogr D Biol Crystallogr. 1998;54:905–921. doi: 10.1107/s0907444998003254. [DOI] [PubMed] [Google Scholar]
- Bucher E, Hemmes H, de Haan P, Goldbach R, Prins M. The influenza A virus NS1 protein binds small interfering RNAs and suppresses RNA silencing in plants. J Gen Virol. 2004;85:983–991. doi: 10.1099/vir.0.19734-0. [DOI] [PubMed] [Google Scholar]
- Casimiro DR, Wright PE, Dyson HJ. PCR-based gene synthesis and protein NMR spectroscopy. Structure. 1997;5:1407–1412. doi: 10.1016/s0969-2126(97)00291-8. [DOI] [PubMed] [Google Scholar]
- Chapman EJ, Prokhnevsky AI, Gopinath K, Dolja VV, Carrington JC. Viral RNA silencing suppressors inhibit the microRNA pathway at an intermediate step. Genes Dev. 2004;18:1179–1186. doi: 10.1101/gad.1201204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- CCP4 (Collaborative Computational Project Number 4) The CCP4 suite: programs for protein crystallography. Acta Crystallogr D Biol Crystallogr. 1994;50:760–763. doi: 10.1107/S0907444994003112. [DOI] [PubMed] [Google Scholar]
- DeLano WL. The PyMOL User’s Manual. San Carlos, CA: Delano Scientific; 2002. [Google Scholar]
- Delgadillo MO, Saenz P, Salvador B, Garcia JA, Simon-Mateo C. Human influenza virus NS1 protein enhances viral pathogenicity and acts as an RNA silencing suppressor in plants. J Gen Virol. 2004;85:993–999. doi: 10.1099/vir.0.19735-0. [DOI] [PubMed] [Google Scholar]
- Ding SW, Li H, Lu R, Li F, Li WX. RNA silencing: a conserved antiviral immunity of plants and animals. Virus Res. 2004;102:109–115. doi: 10.1016/j.virusres.2004.01.021. [DOI] [PubMed] [Google Scholar]
- Dolja VV. Beet yellows virus: the importance of being different. Mol Plant Pathol. 2003;4:91–98. doi: 10.1046/j.1364-3703.2003.00154.x. [DOI] [PubMed] [Google Scholar]
- Dunoyer P, Lecellier CH, Parizotto EA, Himber C, Voinnet O. Probing the microRNA and small interfering RNA pathways with virus-encoded suppressors of RNA silencing. Plant Cell. 2004;16:1235–1250. doi: 10.1105/tpc.020719. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Elbashir SM, Lendeckel W, Tuschl T. RNA interference is mediated by 21- and 22-nucleotide RNAs. Genes Dev. 2001;15:188–200. doi: 10.1101/gad.862301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamilton AJ, Baulcombe DC. A species of small anti-sense RNA in posttranscriptional gene silencing in plants. Science. 1999;286:950–952. doi: 10.1126/science.286.5441.950. [DOI] [PubMed] [Google Scholar]
- Hammond SM, Bernstein E, Beach D, Hannon GJ. An RNA-directed nuclease mediates post-transcriptional gene silencing in Drosophila cells. Nature. 2000;404:293–296. doi: 10.1038/35005107. [DOI] [PubMed] [Google Scholar]
- Holm L, Sander C. Touring protein fold space with Dali/FSSP. Nucleic Acids Res. 1998;26:316–319. doi: 10.1093/nar/26.1.316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones TA, Kjeldgaard M. Electron-density map interpretation. Methods Enzymol. 1997;227:174–208. doi: 10.1016/s0076-6879(97)77012-5. [DOI] [PubMed] [Google Scholar]
- Kasschau KD, Carrington JC. A counterdefensive strategy of plant viruses: suppression of posttranscriptional gene silencing. Cell. 1998;95:461–470. doi: 10.1016/s0092-8674(00)81614-1. [DOI] [PubMed] [Google Scholar]
- Kasschau KD, Xie Z, Allen E, Llave C, Chapman EJ, Krizan KA, Carrington JC. P1/HC-Pro, a viral suppressor of RNA silencing, interferes with Arabidopsis development and miRNA unction. Dev Cell. 2003;4:205–217. doi: 10.1016/s1534-5807(03)00025-x. [DOI] [PubMed] [Google Scholar]
- Kumarevel T, Mizuno H, Kumar PK. Structural basis of HutP-mediated anti-termination and roles of the Mg2+ ion and L-histidine ligand. Nature. 2005;434:183–191. doi: 10.1038/nature03355. [DOI] [PubMed] [Google Scholar]
- Lakatos L, Szittya G, Silhavy D, Burgyan J. Molecular mechanism of RNA silencing suppression mediated by p19 protein of tombusviruses. EMBO J. 2004;23:876–884. doi: 10.1038/sj.emboj.7600096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Li WX, Ding SW. Induction and suppression of RNA silencing by an animal virus. Science. 2002;296:1319–1321. doi: 10.1126/science.1070948. [DOI] [PubMed] [Google Scholar]
- Li WX, Li H, Lu R, Li F, Dus M, Atkinson P, Brydon EW, Johnson KL, Garcia-Sastre A, Ball LA, et al. Interferon antagonist proteins of influenza and vaccinia viruses are suppressors of RNA silencing. Proc Natl Acad Sci USA. 2004;101:1350–1355. doi: 10.1073/pnas.0308308100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu R, Folimonov A, Shintaku M, Li WX, Falk BW, Dawson WO, Ding SW. Three distinct suppressors of RNA silencing encoded by a 20-kb viral RNA genome. Proc Natl Acad Sci USA. 2004;101:15742–15747. doi: 10.1073/pnas.0404940101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matzke MA, Birchler JA. RNAi-mediated pathways in the nucleus. Nat Rev Genet. 2005;6:24–35. doi: 10.1038/nrg1500. [DOI] [PubMed] [Google Scholar]
- Meister G, Tuschl T. Mechanisms of gene silencing by double-stranded RNA. Nature. 2004;431:343–349. doi: 10.1038/nature02873. [DOI] [PubMed] [Google Scholar]
- Murshudov GN, Vagin AA, Lebedev A, Wilson KS, Dodson EJ. Efficient anisotropic refinement of macromolecular structures using FFT. Acta Crystallogr D Biol Crystallogr. 1999;55:247–255. doi: 10.1107/S090744499801405X. [DOI] [PubMed] [Google Scholar]
- Murzin AG, Brenner SE, Hubbard T, Chothia C. SCOP: a structural classification of proteins database for the investigation of sequences and structures. J Mol Biol. 1995;247:536–540. doi: 10.1006/jmbi.1995.0159. [DOI] [PubMed] [Google Scholar]
- Nicholls A, Sharp KA, Honig B. Protein folding and association: insights from the interfacial and thermodynamic properties of hydrocarbons. Proteins. 1991;11:281–296. doi: 10.1002/prot.340110407. [DOI] [PubMed] [Google Scholar]
- Otwinowski Z, Minor W. Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol. 1997;276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- Patel SS, Picha KM. Structure and function of hexameric helicases. Annu Rev Biochem. 2000;69:651–697. doi: 10.1146/annurev.biochem.69.1.651. [DOI] [PubMed] [Google Scholar]
- Reed JC, Kasschau KD, Prokhnevsky AI, Gopinath K, Pogue GP, Carrington JC, Dolja VV. Suppressor of RNA silencing encoded by Beet yellows virus. Virology. 2003;306:203–209. doi: 10.1016/s0042-6822(02)00051-x. [DOI] [PubMed] [Google Scholar]
- Roth BM, Pruss GJ, Vance VB. Plant viral suppressors of RNA silencing. Virus Res. 2004;102:97–108. doi: 10.1016/j.virusres.2004.01.020. [DOI] [PubMed] [Google Scholar]
- Sigler PB, Xu Z, Rye HS, Burston SG, Fenton WA, Horwich AL. Structure and function in GroEL-mediated protein folding. Annu Rev Biochem. 1998;67:581–608. doi: 10.1146/annurev.biochem.67.1.581. [DOI] [PubMed] [Google Scholar]
- Silhavy D, Burgyan J. Effects and side-effects of viral RNA silencing suppressors on short RNAs. Trends Plant Sci. 2004;9:76–83. doi: 10.1016/j.tplants.2003.12.010. [DOI] [PubMed] [Google Scholar]
- Silhavy D, Molnar A, Lucioli A, Szittya G, Hornyik C, Tavazza M, Burgyan J. A viral protein suppresses RNA silencing and binds silencing-generated, 21- to 25-nucleotide double-stranded RNAs. EMBO J. 2002;21:3070–3080. doi: 10.1093/emboj/cdf312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson JD, Higgins DG, Gibson TJ. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toro I, Basquin J, Teo-Dreher H, Suck D. Archaeal Sm proteins form heptameric and hexameric complexes: crystal structures of the Sm1 and Sm2 proteins from the hyperthermophile Archaeoglobus fulgidus. J Mol Biol. 2002;320:129–142. doi: 10.1016/S0022-2836(02)00406-0. [DOI] [PubMed] [Google Scholar]
- Uson I, Sheldrick GM. Advances in direct methods for protein crystallography. Curr Opin Struct Biol. 1999;9:643–648. doi: 10.1016/s0959-440x(99)00020-2. [DOI] [PubMed] [Google Scholar]
- Vargason JM, Szittya G, Burgyan J, Tanaka Hall TM. Size selective recognition of siRNA by an RNA silencing suppressor. Cell. 2003;115:799–811. doi: 10.1016/s0092-8674(03)00984-x. [DOI] [PubMed] [Google Scholar]
- Voinnet O. Induction and suppression of RNA silencing: insights from viral infections. Nat Rev Genet. 2005;6:206–220. doi: 10.1038/nrg1555. [DOI] [PubMed] [Google Scholar]
- Voinnet O, Baulcombe DC. Systemic signaling in gene silencing. Nature. 1997;389:553. doi: 10.1038/39215. [DOI] [PubMed] [Google Scholar]
- Wang MB, Metzlaff M. RNA silencing and antiviral defense in plants. Curr Opin Plant Biol. 2005;8:216–222. doi: 10.1016/j.pbi.2005.01.006. [DOI] [PubMed] [Google Scholar]
- Ye K, Malinina L, Patel DJ. Recognition of small interfering RNA by a viral suppressor of RNA silencing. Nature. 2003;426:874–878. doi: 10.1038/nature02213. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.