Abstract
G-quadruplexes and Z-DNA are two important non-B forms of DNA architecture. Results on novel structural elements, folding and unfolding kinetics, and interactions with small molecules and proteins have been reported recently for these forms. These results will enhance our understanding of the biology of these structures and provide a platform for drug design.
Introduction
Besides the canonical right-handed double helix (B-DNA) first proposed by Watson and Crick, DNA can adopt other biologically relevant structures. Four-stranded structures (G-quadruplexes) and left-handed duplexes (Z-DNA) are two important examples. Although both these structures were first discovered several decades ago [1•,2•], they have recently received particular attention, as their biological relevance has become more clear. Here, we review the latest studies on G-quadruplex and Z-DNA structures published between 2003 and 2005.
G-quadruplex structures
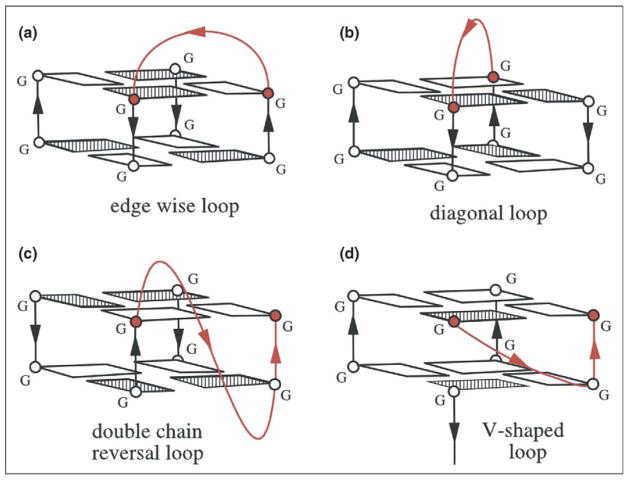
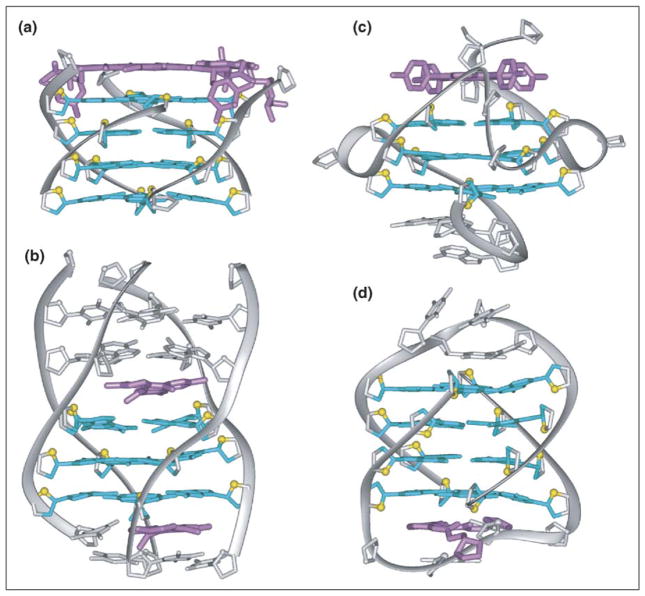
Guanine-rich oligonucleotides can form four-stranded G-quadruplex structures through the stacking of planar G-tetrads [1•]. Cations, such as K+ or Na+, stabilize G-quadruplexes by coordinating electronegative carbonyl groups of guanines, which are directed towards the interior of G-tetrads. Because of the size difference, Na+ ions are positioned mainly in the plane of the G-tetrads, whereas K+ ions are positioned between G-tetrad planes. G-quadruplex architecture is dependent on the nature of cations and, generally, K+ stabilizes G-quadruplexes better than Na+. G-quadruplexes are highly polymorphic, as regards three mutually related factors: the orientation of the strands, the syn/anti glycosidic conformation of guanines and the loop connectivities. Oligonucleotides containing one, two or four G-stretches can form tetrameric, dimeric or monomeric G-quadruplexes, respectively [1•,3,4]. In dimeric and monomeric G-quadruplexes, loops connect G-stretches. These loops can be classified into four major families: edgewise loops connecting two adjacent antiparallel strands (Figure 1a), diagonal loops connecting two opposing antiparallel strands (Figure 1b), double-chain-reversal loops connecting adjacent parallel strands (Figure 1c) and V-shaped loops connecting two corners of a G-tetrad core in which one supporting column is lacking (Figure 1d). These various loop motifs, in combination with different strand orientations and syn/anti distributions, have been found in many different monomeric (intramolecular) and dimeric G-quadruplexes [1•,3,4].
Figure 1.
Polymorphism of the loops connecting individual strands of the G-quadruplex: (a) edgewise loop, (b) diagonal loop, (c) double-chain-reversal loop and (d) V-shaped loop. Adapted with permission from [69].
New topologies
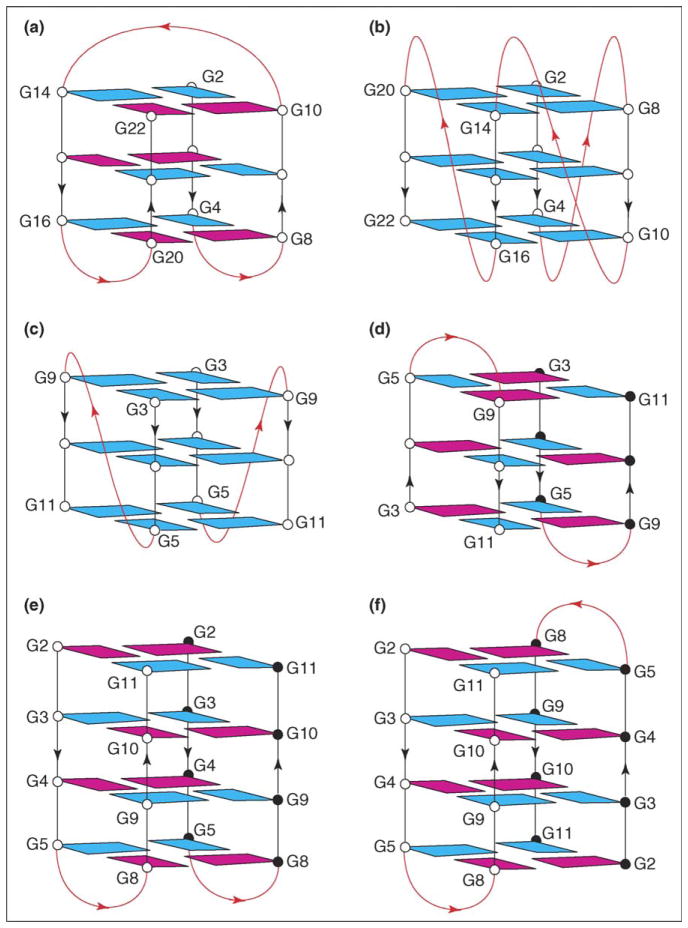
Telomeric DNA has probably attracted the greatest attention of G-quadruplex researchers because of the natural existence of a G-rich single-strand overhang at telomere 3′ ends and its relevance to a potential anticancer strategy. The first solution structure of a four-repeat human telomeric sequence, d[AGGG(TTAGGG)3], was characterized in 1993 by NMR in Na+ solution [5]. This sequence forms an intramolecular G-quadruplex involving three stacked G-tetrads with anti•anti•syn•syn alignments around each tetrad. Three connecting TTA loops adopt successive edgewise, diagonal and edgewise alignments, such that each strand has both parallel and anti-parallel adjacent strands (Figure 2a). A decade later, Parkinson et al. [6] reported the crystal structure of a completely different G-quadruplex formed by the same sequence in the presence of K+. In this structure, known as ‘propeller type’, all four strands are parallel, the connecting TTA loops are double-chain-reversal motifs and all guanines adopt anti glycosidic conformations (Figure 2b). This latter structure was very different from all G-quadruplexes reported previously, and could readily facilitate higher order telomere folding and unfolding [6]. However, data on the same human telomeric sequence in K+ solution, derived using different physical and chemical techniques, and reported in a large number of subsequent papers, indicated the presence of a mixture of several G-quadruplex forms [7••,8–12,13•,14•].
Figure 2.
Telomeric G-quadruplexes. (a,b) Structures formed by the human telomeric sequence d[AGGG(TTAGGG)3] (a) in Na+ solution [5] and (b) in K+ crystal [6]. (c,d) Structures formed by the human telomeric sequence d(TAGGGTTAGGGT) in K+ solution [7••]: (c) parallel form and (d) antiparallel form. (e,f) Structures formed by the Tetrahymena telomeric sequence d(TGGGGTTGGGGT) in Na+ solution [15]: (e) head-to-head form and (f) head-to-tail form. Loops are colored red; anti and syn guanines are colored cyan and magenta, respectively.
For the two-repeat human telomeric sequence d(TAGGGTTAGGGT), both parallel (Figure 2c) and antiparallel G-quadruplexes (Figure 2d) were found to co-exist and interconvert in K+ solution [7••]. The parallel-stranded structure is symmetrical and similar to the ‘propeller-type’ G-quadruplex crystal structure [6], with double-chain-reversal loops and all anti guanines (Figure 2c). The antiparallel-stranded structure is asymmetrical, with all adjacent strands antiparallel, two edgewise loops and syn•anti•syn•anti G-tetrad alignments (Figure 2d).
The two-repeat Tetrahymena telomeric sequence d(TGGGGTTGGGGT), which differs from the human sequence by only one G-for-A replacement in each repeat, interconverts between two asymmetric dimeric G-quadruplex structures in Na+ solution (Figure 2e,f) [15]. Both structures include a core of four stacked G-tetrads and two edgewise loops. The adjacent strands of the G-tetrad core are alternately parallel and antiparallel. All G-tetrads adopt syn•syn•anti•anti alignments, which occur with 5′-syn-anti-syn-anti-3′ alternations along G-tracts. In the first structure (head-to-head), two loops are at one end of the G-tetrad core (Figure 2e); in the second structure (head-to-tail), two loops are located on opposite ends of the G-tetrad core (Figure 2f).
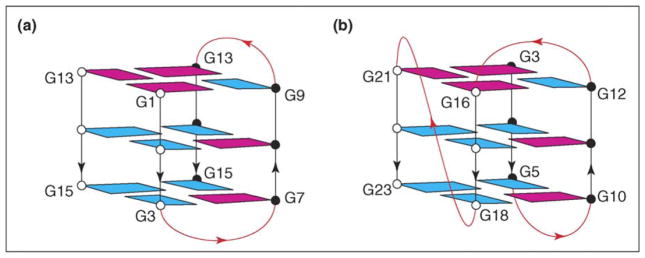
The three-repeat human telomeric sequence d(GGGTTAGGGTTAGGGT) forms an asymmetric dimeric quadruplex in Na+ solution, in which the G-tetrad core involves all three G-tracts of one strand and only the last 3′-end G-tract of the other strand (Figure 3a) [16••]. A three-repeat human telomeric sequence can also associate with a single-repeat human telomeric sequence, forming a structure with the same topology called a (3+1) quadruplex assembly. In this G-quadruplex assembly, there are one syn•syn•syn•anti and two anti•anti•anti•syn syn G-tetrads, two edgewise loops, and three G-tracts oriented in one direction and the fourth in the opposite direction. This is the first G-quadruplex structure shown to involve the heterodimeric association of two strands of such different lengths. There is striking similarity between this (3+1) G-tetrad core and the core of both the four-repeat Tetrahymena telomeric d(T2G4)4 sequence reported in 1994 (Figure 3b) [17] and a variant of the four-G-tract segment of the human bcl-2 promoter reported in 2006 [18•].
Figure 3.
G-quadruplex topologies containing the (3+1) G-tetrad core. (a) (3+1) G-quadruplex formed through dimerization of a three-repeat human telomeric sequence [16••]. (b) Intramolecular G-quadruplex formed by the Tetrahymena d(T2G4)4 sequence [17]. Color coded as in Figure 2.
Recently reported G-quadruplex structures formed by non-telomeric G-rich sequences also revealed a number of new structural features. Double-chain-reversal or propeller-type parallel-stranded G-quadruplexes were observed in K+ solution for G-rich sequences containing four G-tracts from the c-myc promoter [19•,20•,21]. Besides loops containing two or six residues, single-residue (A or T) double-chain-reversal loops were observed systematically to bridge three layers of G-tetrads [19•]. This study suggested that a one-residue loop forms the most stable double-chain-reversal loop bridging three G-tetrad layers and a six-residue loop is less stable than a two-residue loop [19•], in agreement with the results of a simulation study [22•]. More recent studies [23••,24••] showed that the structure of single-residue double-chain-reversal loops bridging three G-tetrad layers does not depend much on the nature of the single base, either T or A.
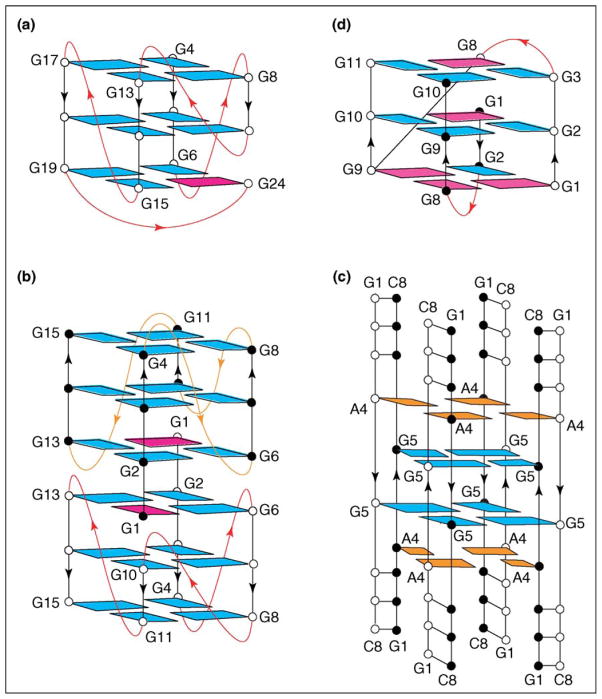
A five-G-tract sequence from the c-myc promoter was found to adopt a distinct novel parallel-stranded fold-back G-quadruplex topology in K+ solution (Figure 4a) [23••]. The study revealed several new structural motifs not observed previously. In particular, a guanine (G24) from the 3′ end is plugged back into the G-tetrad core by participating in G-tetrad formation and displacing another guanine (G10) of a continuous G-tract into a loop. This configuration is stabilized by formation of a diagonal loop, which contains a G•(A-G) triad that stacks on and caps the G-tetrad core. These new folding features are direct consequences of the presence of five G-tracts in the sequence [23••], in contrast to the four G-tracts in the sequences studied previously [19•,20•,21].
Figure 4.
Novel structural elements in G-quadruplexes. (a) Fold-back G-quadruplex of the five-G-tract c-myc promoter sequence [23••], (b) interlocked dimeric G-quadruplex of 93del [24••], (c) i-motif of G-quartets [27] and (d) dimeric G-quadruplex of d(GGGTTTTGGGG) [28•]. Color coded as in Figure 2.
The 93del d(GGGGTGGGAGGAGGGT) oligonucleotide, an inhibitor of HIV-1 integrase, was shown to form a very stable twofold-symmetric dimeric quadruplex in K+ solution [24] (Figure 4b). Each monomer subunit contains two G•G•G•G tetrads and one A•(G•G•G•G) pentad, in which all the G-stretches are parallel and linked by three single-nucleotide double-chain-reversal loops. Dimer formation is achieved through the mutual pairing of G1 of one monomer with G2, G6 and G13 of the other monomer, to complete G•G•G•G tetrad formation. The compact interlocking of symmetry-related subunits through (3+1) G2•G6•G1•G13 tetrad formation across the dimeric interface (Figure 4b) constitutes an interesting feature of the 93del architecture and highlights a new principle for robust dimeric quadruplex folding. Another example of the interlocking of two G-quadruplex subunits, in which one G-tetrad at the interface involves two guanines from each subunit, has also been reported [25].
The d(GCGAGAGC) sequence, which does not even contain a GG-tract, nevertheless adopts a novel eight-stranded topology in the crystalline state under low K+ conditions (Figure 4c), in which G5 participates in G•G•G•G tetrad formation and A4 in A•A•A•A tetrad formation. In this multistranded topology, a given G-tetrad is intercalated between flanking G- and A-tetrads, as a result of extension at the G4-A5 step [26•]. This intercalation feature has also been observed in the eight-stranded architecture formed by the r(U)d(BrG)r(GUGU) sequence, which forms a mixture of U•U•U•U and G•G•G•G tetrads [27]. Intercalation of guanine residues as part of a G-tetrad by guanine residues of flanking G-tetrads has also been observed for the dimeric G-quadruplex formed by the d(GGGTTTTGGGG) sequence in solution (Figure 4d), whereby G1 is intercalated between an extended G8-G9 step [28•].
Folding and unfolding kinetics
Functions of nucleic acids depend not only on their structure and stability, but also on their dynamics. The folding and unfolding kinetics of G-quadruplexes have been studied for quite some time [29,30], but only recently have more quantitative data been obtained systematically for various sequences using different techniques [7••,8,31–33,34•,35,36]. In order to monitor and measure the unfolding rates of G-quadruplexes, Raghuraman and Cech, in their pioneering work [29], trapped the unfolded strand with the complementary C-rich strand. Adopting the same approach, Balasubramanian and coworkers [8,31] used fluorescence resonance energy transfer (FRET), Halder and Chowdhury [32], and Zhao et al. [33] used surface plasmon resonance (SPR), and Phan and Patel [7••] used NMR spectroscopy. The advantage of the last technique is that it enables the monitoring, at high resolution, of the simultaneous unfolding of different G-quadruplexes that coexist in solution, thereby obtaining the unfolding rates by single-exponential fits of signal decays [7••]. The advantages of the two former techniques are simplicity and small sample requirements; furthermore, FRET can allow measurements at single-molecule levels [8]. G-quadruplex unfolding can also be characterized using concentration-jump or temperature-jump approaches [7••].
Because the dissociation of G-quadruplexes is very slow, Mergny et al. [34•] used non-equilibrium denaturation curves obtained from UV spectroscopy to systematically determine the dissociation rates of a number of tetrameric G-quadruplexes. The association rates were also systematically measured. A study by Merkina and Fox [35] characterized the dissociation rates of tetrameric G-quadruplexes using fluorescence spectroscopy techniques. Petraccone et al. [36] used CD to characterize the association kinetics of quadruplexes formed by PNA/DNA chimeras. Recently, Lee et al. [14•] used single-molecule FRET spectroscopy to characterize the structure and dynamics of intramolecular human telomeric G-quadruplexes, and found that a four-repeat human telomeric sequence can exist in short-lived and long-lived states.
The general conclusions of these studies are the very slow folding (association) and unfolding (dissociation) kinetics of G-quadruplexes. The folding and unfolding times of a G-quadruplex depend strongly on the number of G-tetrads in the structure and on the presence of cations. They can range from few seconds to days or years at room temperature.
Interaction with small molecules
Because G-quadruplexes in both telomeres and oncogenic promoters have been established as promising anticancer targets [37,38], development of small molecules that interact with and stabilize G-quadruplexes remains of great interest to a number of academic laboratories and pharmaceutical companies. Structures of some ligand–quadruplex complexes have recently been reported [23••,39•,40,41••] and should help the structure-based design of new drugs.
Clark et al. [39•] reported the crystal structure of a parallel G-quadruplex–daunomycin complex. In this complex, three daunomycin molecules stack on the 5′ end of the G-tetrad core, with daunosamine sugar moieties forming hydrogen-bonding interactions and/or van der Waals contacts with the quadruplex grooves (Figure 5a). Gavathiotis et al. [40] reported the NMR solution structure of the parallel G-quadruplex formed by the human telomeric sequence d(TTAGGGT) bound to two molecules of a fluorinated polycyclic methyl-acridinium salt. The structure shows that two ligand molecules stack on the two ends of the G-tetrad core; the partial positive charge of the ligands is positioned above and below the ion channel at the center of the G-tetrad core (Figure 5b).
Figure 5.
Interaction of G-quadruplexes with small molecules, as reported by (a) Clark et al. [39•] for the parallel quadruplex–daunomycin complex, (b) Gavathiotis et al. [40] for the parallel G-quadruplex bound to two molecules of a fluorinated polycyclic methyl-acridinium salt, (c) Phan et al. [23••] for the G-quadruplex–TMPyP4 complex and (d) Haider et al. [41••] for the complex between an antiparallel G-quadruplex and a disubstituted aminoalkylamido acridine. Guanine bases are colored cyan, O4′ are colored yellow and ligands are colored magenta.
The structure of a quinobenzoxazine–G-quadruplex complex has been characterized by solid-state NMR [42]. The study showed that the ligand stacks on the 3′ end of the G-tetrad core of the parallel-stranded G-quadruplex formed by the human telomeric sequence d(TAGGGTTA). Cocco et al. [43] used solution NMR to show that distamycin, a DNA duplex minor groove ligand, stacks on both ends of the G-quadruplex formed by the same sequence in K+ solution.
Recently, Phan et al. [23••] reported the NMR solution structure of the complex between a G-quadruplex formed by a five-G-tract sequence from the c-myc promoter and the cationic porphyrin TMPyP4. In this structure, TMPyP4 is well defined and stacked on the top G-tetrad (5′ end of the parallel-stranded core), somewhat shifted towards one corner of this G-tetrad (Figure 5c). The positive charges of the ligand are in close contact with several negatively charged phosphates. The structure of this G-quadruplex–TMPyP4 complex revealed how stacking and electrostatic interactions contribute to the stability of the complex.
Haider et al. [41••] reported the crystal structure of the complex between a dimeric antiparallel G-quadruplex formed by the Oxytricha telomeric sequence d(GGGGTTTTGGGG) and a di-substituted aminoalkylamido acridine compound (Figure 5d). In this structure, the ligand is bound at one end of the G-tetrad core, within one of the T4 diagonal loops. It is held in place by a combination of stacking interactions and specific hydrogen bonds with thymine bases.
The emphasis to date has been on aromatic-ring-containing ligands that have the potential to stack over terminal G-tetrads positioned towards either end of the quadruplex. Nevertheless, opportunities exist for targeting the four grooves of the quadruplex, whose width, depth and accessibility depend on strand alignment and connecting loop topologies. Clearly, double-chain-reversal loops occlude access to the grooves, but no such constraints exist for edgewise and diagonal loops. The Watson–Crick and major groove edges of a guanine are involved in G-tetrad formation, leaving the minor groove edge available for further recognition. Indeed, it has been shown that an adenine can align with the minor groove edge of a G-tetrad guanine, to form a sheared G•A non-canonical pair, resulting in formation of an A•(G•G•G•G) pentad [24••]. The structure-based design of ligands tailor-made to target individual grooves of defined G-quadruplexes represents an ongoing challenge in the field.
Biological existence and functions
In nature, G-rich sequences are found in a number of important DNA regions, such as telomeres and centromeres, immunoglobulin switch regions and mutational hot spots, and act as regulatory elements within oncogenic promoters and as repeat elements implicated in triplet repeat expansion diseases. Recently, the prevalence of putative G-quadruplexes in the human genome has been systematically examined [44,45]. It has been found that as many as 376,000 potential quadruplexes could coexist. The possible existence and roles of G-quadruplexes in vivo have been corroborated by the detection of proteins that bind specifically to G-quadruplexes [46,47] and proteins that have biological activities, such as helicases [48] and nucleases [49], and are specific for G-quadruplexes.
Duquette et al. [50•] used EM to detect a novel DNA structure called the G-loop, which is formed during transcription when the non-template strand is guanine rich. G-loops contain G-quadruplex structures on one strand and a stable DNA–RNA hybrid on the other.
Among the most convincing evidence for the biological existence of G-quadruplexes is the generation of a G-quadruplex-specific antibody [51]. Using this antibody, Paeschke et al. [52••] have demonstrated G-quadruplex formation in the telomeres of nanochromosomes of Stylonychia. The authors showed that the telomere-end-binding proteins TEBPα and TEBPβ cooperate to control the formation of G-quadruplex structure in telomeres in vivo. Furthermore, such G-quadruplex formation is regulated by the cell-cycle-dependent phosphorylation of TEBPβ.
It has been shown that formation of intramolecular G-quadruplexes by the telomeric G-rich strand inhibits the activity of telomerase [53–55] and that the POT1 (protection of telomere 1) protein, which binds the overhang end single strand, plays a role in disrupting G-quadruplex structures in telomeric DNA, thereby allowing proper elongation by telomerase [54]. Stabilizing intramolecular telomeric G-quadruplexes and the G-quadruplexes formed by sequences in oncogenic promoters is an attractive strategy for the development of anticancer drugs [37,38]. Indeed, some G-quadruplex ligands exhibit anticancer activities [38,56–58].
Future challenges and prospects
The G-rich tracts found in telomeres and oncogenic promoter sequences adopt a diversity of G-quadruplex scaffolds in vitro that appear to be dependent on the length of individual G-tracts, loop composition and size, and type of cation. Indeed, the same four-repeat human telomeric sequence adopts distinct folds in Na+ [5] and K+ (unpublished) solution, and in K+-containing crystals [6]. It is likely that some of these G-quadruplex conformations are isoenergetic, enabling interconversion between two or more distinct topologies, as shown for human [7••] and Tetrahymena [15] two-repeat telomeric sequences.
Currently, no definitive information has emerged that addresses the critical issue as to which G-quadruplex structure(s) are most relevant in vivo. Nevertheless, increasing attention is being paid to the (3+1) G-quadruplex scaffold containing a single double-chain-reversal loop, initially identified in 1994 for the four-repeat Tetrahymena telomere sequence [17] and thought to be an outlier at that time. The (3+1) G-quadruplex scaffold has now been observed for a range of sequences including both telomeres [16••,17] and oncogenic promoters [18•] in physiologically relevant K+ solution. Indeed, the Tetrahymena telomere [17] and the human bcl-2 promoter sequence [18•] adopt the same (3+1) quadruplex scaffold despite different loop compositions and size.
New approaches are needed to clarify the extent and type of G-quadruplex formation in vivo. The probes themselves have to avoid induction of quadruplex architecture and/or bias towards certain scaffolds. To date, these probes have been limited to antibodies that recognize quadruplexes in macronuclei [51].
Z-DNA
Z-DNA, the left-handed form of the double helix, can be formed by certain sequences containing alternating purine and pyrimidine bases. The history of more than two decades of Z-DNA research was excellently summarized in a recent review [2•]. The discovery of proteins that bind to Z-DNA with high affinity and specificity [2•] indicated a biological role for this structure and stimulated much research in this area. In the past couple of years, two more families of proteins that specifically recognize Z-DNA have been reported: poxvirus protein E3L [59] and an ortholog of the protein kinase PKR [60]. Furthermore, there are data on the binding activity and the function of the Z-DNA-binding domain of E3L in vivo [61].
B-Z junction
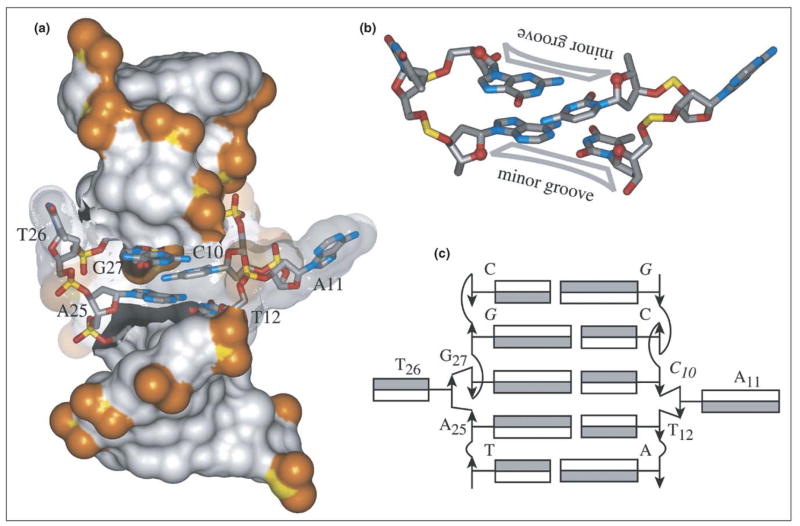
One of the most important advances in recent Z-DNA research is the crystal structure of a junction between the right-handed B-DNA and the left-handed Z-DNA forms of the double helix [62••]. Z-DNA is less stable than B-DNA at physiological salt concentrations, but can be stabilized by negative supercoiling or by protein binding [2•]. The structure of the B-Z junction is important because such a junction should be formed each time a double-helical DNA segment turns into Z-DNA. Ha et al. [62••] used Z-DNA-binding proteins to keep one end of a DNA duplex fragment in the Z-form, while the other end remained in the B-form (Figure 6a). The handedness of the DNA duplex is completely reversed at the junction by breaking only one base pair and projecting the bases out of the duplex. At the junction, the minor grooves of two adjacent steps are on two opposite sides of the duplex (Figure 6b). Base pairs of the B-DNA and Z-DNA segments are continuously stacked across the junction. Note that, if one considers ‘black’ and ‘white’ faces of the bases (in the nomenclature of Lavery et al. [63]), stacking contacts are black-black and white-white across the junction, as opposed to black-white within the B-DNA or Z-DNA segments (Figure 6c). The structure of the B-Z junction maximizes base pairing and stacking, thereby minimizing the energetic cost of the junction and most probably facilitating the use of Z-DNA more widely in nature [62••].
Figure 6.
Structure of the B-Z DNA junction [62••]. Views highlighting (a) flipped-out bases of the junctional pair, (b) opposing minor grooves flanking looped-out bases of the junctional pair, and (c) stacking arrangement of base faces at junctional and flanking sites.
Conclusions
The polymorphism of DNA G-quadruplex structures depends strongly on sequence and experimental conditions. Several new folding topologies and structural elements have been discovered, such as the (3+1) interlocking structure [24••] and fold-back motifs [23••]. Some general rules for G-quadruplex folding have emerged, such as the robustness of the single-residue double-chain-reversal loop in K+ solution. Methods for studying the folding/unfolding kinetics of G-quadruplexes have been proposed and applied to various structures. More systematic studies are required to understand these important properties. The interaction of some G-quadruplexes with small molecules has been reported, providing a platform for the design of new specific ligands. Reported evidence for the proposed biological existence and function of G-quadruplexes should stimulate more research into this non-canonical DNA architecture. Research on Z-DNA has expanded available insights into its functional role [2•] and the new structure of the B-Z junction [62••] should open fresh avenues of research.
Update
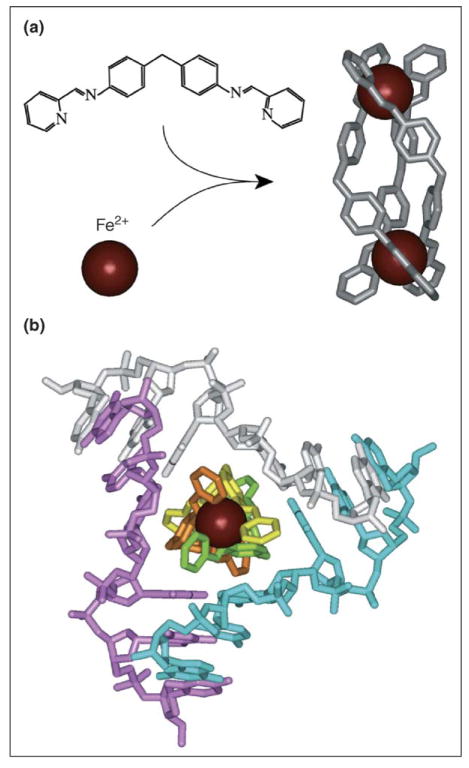
Oleksi et al. [64••] have just published a seminal structural paper on a novel supramolecular ligand that targets higher order DNA architecture with high affinity and structural specificity, thus opening new avenues for the development of anti-DNA therapeutic agents. Specifically, a synthetic tetracationic metallosupramolecular helicate, generated from three bis-pyrimidine organic strands wrapped around two Fe2+ ions (Figure 7a), was found to match the size and shape of the central trigonal hydrophobic cavity of a three-way DNA junction formed by the palindromic sequence d(CGTACG) (Figure 7b). Molecular recognition within this triangular hydrophobic binding site is associated with a combination of face-to-face π-stacking intercalation and minor groove sandwiching interactions, supplemented by electrostatic and C-H•••X type hydrogen-bonding interactions. The three-way DNA junction observed in this complex adopts the same Y-shaped topology, with stable Watson–Crick-aligned junctional pairs, reported previously for three-way DNA junctions observed in complexes with Cre recombinase [65] and Ku [66]. The design and implementation of ligand-based recognition and stabilization of junctional folds could impact the emerging field of DNA-based nanotechnology research [67]. The research on the metallosupramolecular helicate targeted to a three-way DNA junction [64••] nicely complements earlier research highlighting shape complementarity between a chiral bicyclic Diels–Alder product bound within the central hydrophobic cavity of a three-way RNA helical junction [68].
Figure 7.
Recognition of a three-way DNA junction by a metallosupramolecular helicate. (a) [Fe2L3]4+ (L = C25H20N4) tetracationic supramolecular helicate, with the Fe2+ ions represented as red spheres. (b) Structure of a three-way DNA junction bound to [Fe2L3]4+. Three DNA strands are colored cyan, magenta and gray, respectively.
Acknowledgments
This research was supported by National Institutes of Health grant GM034504. Phan, Anh Tuân dedicates this work to his late beloved wife, Luu, Kim Ngoc.
References and recommended reading
Papers of particular interest, published within the annual period of review, have been highlighted as:
• of special interest
•• of outstanding interest
- 1•.Davis JT. G-quartets 40 years later: from 5′-GMP to molecular biology and supramolecular chemistry. Angew Chem Int Ed Engl. 2004;43:668–698. doi: 10.1002/anie.200300589. This recent review of G-tetrads (or G-quartets) discusses areas ranging from structural biology and medicinal chemistry to supramolecular chemistry and nanotechnology. [DOI] [PubMed] [Google Scholar]
- 2•.Rich A, Zhang S. Timeline: Z-DNA: the long road to biological function. Nat Rev Genet. 2003;4:566–572. doi: 10.1038/nrg1115. A recent review of 24 years of Z-DNA research. [DOI] [PubMed] [Google Scholar]
- 3.Patel DJ, Bouaziz S, Kettani A, Wang Y. Structures of guanine-rich and cytosine-rich quadruplexes formed in vitro by telomeric, centromeric, and triplet repeat disease DNA sequences. In: Neidle S, editor. Oxford Handbook of Nucleic Acid Structures. Oxford University Press; 1999. pp. 389–453. [Google Scholar]
- 4.Simonsson T. G-quadruplex DNA structures - variations on a theme. Biol Chem. 2001;382:621–628. doi: 10.1515/BC.2001.073. [DOI] [PubMed] [Google Scholar]
- 5.Wang Y, Patel DJ. Solution structure of the human telomeric repeat d[AG3(T2AG3)3] G-tetraplex. Structure. 1993;1:263–282. doi: 10.1016/0969-2126(93)90015-9. [DOI] [PubMed] [Google Scholar]
- 6.Parkinson GN, Lee MP, Neidle S. Crystal structure of parallel quadruplexes from human telomeric DNA. Nature. 2002;417:876–880. doi: 10.1038/nature755. [DOI] [PubMed] [Google Scholar]
- 7••.Phan AT, Patel DJ. Two-repeat human telomeric d(TAGGGTTAGGGT) sequence forms interconverting parallel and antiparallel G-quadruplexes in solution: distinct topologies, thermodynamic properties, and folding/unfolding kinetics. J Am Chem Soc. 2003;125:15021–15027. doi: 10.1021/ja037616j. This NMR study of G-rich human telomeric sequences showed that parallel and antiparallel G-quadruplexes coexist and interconvert in K+ solution. The thermodynamic properties and folding/unfolding kinetics of each form were determined. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ying L, Green JJ, Li H, Klenerman D, Balasubramanian S. Studies on the structure and dynamics of the human telomeric G quadruplex by single-molecule fluorescence resonance energy transfer. Proc Natl Acad Sci USA. 2003;100:14629–14634. doi: 10.1073/pnas.2433350100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ourliac-Garnier I, Elizondo-Riojas MA, Redon S, Farrell NP, Bombard S. Cross-links of quadruplex structures from human telomeric DNA by dinuclear platinum complexes show the flexibility of both structures. Biochemistry. 2005;44:10620–10634. doi: 10.1021/bi050144w. [DOI] [PubMed] [Google Scholar]
- 10.He Y, Neumann RD, Panyutin IG. Intramolecular quadruplex conformation of human telomeric DNA assessed with 125I-radioprobing. Nucleic Acids Res. 2004;32:5359–5367. doi: 10.1093/nar/gkh875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Rujan IN, Meleney JC, Bolton PH. Vertebrate telomere repeat DNAs favor external loop propeller quadruplex structures in the presence of high concentrations of potassium. Nucleic Acids Res. 2005;33:2022–2031. doi: 10.1093/nar/gki345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Qi J, Shafer RH. Covalent ligation studies on the human telomere quadruplex. Nucleic Acids Res. 2005;33:3185–3192. doi: 10.1093/nar/gki632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13•.Li J, Correia JJ, Wang L, Trent JO, Chaires JB. Not so crystal clear: the structure of the human telomere G-quadruplex in solution differs from that present in a crystal. Nucleic Acids Res. 2005;33:4649–4659. doi: 10.1093/nar/gki782. The results of biophysical experiments in solution and computational studies suggest that the propeller-type G-quadruplex structure observed in the K+-containing crystal [6] appears unlikely to be the major form in K+-containing solution. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14•.Lee JY, Okumus B, Kim DS, Ha T. Extreme conformational diversity in human telomeric DNA. Proc Natl Acad Sci USA. 2005;102:18938–18943. doi: 10.1073/pnas.0506144102. Single-molecule spectroscopy was used to probe the structure and dynamics of human telomeric G-quadruplexes. Long-lived and short-lived forms were detected. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Phan AT, Modi YS, Patel DJ. Two-repeat Tetrahymena telomeric d(TGGGGTTGGGGT) sequence interconverts between asymmetric dimeric G-quadruplexes in solution. J Mol Biol. 2004;338:93–102. doi: 10.1016/j.jmb.2004.02.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16••.Zhang N, Phan AT, Patel DJ. (3 + 1) Assembly of three human telomeric repeats into an asymmetric dimeric G-quadruplex. J Am Chem Soc. 2005;127:17277–17285. doi: 10.1021/ja0543090. A three-repeat human telomeric sequence is shown to associate with a single-repeat human telomeric sequence, forming a structure called the (3+1) G-quadruplex assembly. In this assembly, three G-tracts are oriented in one direction and the fourth in the opposite direction. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wang Y, Patel DJ. Solution structure of the Tetrahymena telomeric repeat d(T2G4)4 G-tetraplex. Structure. 1994;2:1141–1156. doi: 10.1016/s0969-2126(94)00117-0. [DOI] [PubMed] [Google Scholar]
- 18•.Dai J, Dexheimer TS, Chen D, Carver M, Ambrus A, Jones RA, Yang D. An intramolecular G-quadruplex structure with mixed parallel/antiparallel G-strands formed in the human bcl-2 promoter region in solution. J Am Chem Soc. 2006;128:1096–1098. doi: 10.1021/ja055636a. This paper shows that a four-G-tract segment of the bcl-2 promoter sequence adopts the same G-quadruplex topology as the four-repeat Tetrahymena telomere sequence [17], despite different loop compositions and size. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19•.Phan AT, Modi YS, Patel DJ. Propeller-type parallel-stranded G-quadruplexes in the human c-myc promoter. J Am Chem Soc. 2004;126:8710–8716. doi: 10.1021/ja048805k. Two different G-rich sequences containing four G-tracts from the c-myc promoter form a propeller-type intramolecular G-quadruplex. Besides loops containing two or six residues, single-residue (A or T) double-chain-reversal loops were observed systematically to bridge three layers of G-tetrads. This study suggested that a single residue forms the most stable double-chain-reversal loop bridging three G-tetrad layers, and a six-residue loop is less stable than a two-residue loop. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20•.Seenisamy J, Rezler EM, Powell TJ, Tye D, Gokhale V, Joshi CS, Siddiqui-Jain A, Hurley LH. The dynamic character of the G-quadruplex element in the c-MYC promoter and modification by TMPyP4. J Am Chem Soc. 2004;126:8702–8709. doi: 10.1021/ja040022b. This paper shows that a four-G-tract fragment from the human c-myc promoter can form a mixture of four different parallel-stranded propeller-type G-quadruplexes. See also [19•] [DOI] [PubMed] [Google Scholar]
- 21.Ambrus A, Chen D, Dai J, Jones RA, Yang D. Solution structure of the biologically relevant G-quadruplex element in the human c-MYC promoter. Implications for G-quadruplex stabilization. Biochemistry. 2005;44:2048–2058. doi: 10.1021/bi048242p. [DOI] [PubMed] [Google Scholar]
- 22•.Hazel P, Huppert J, Balasubramanian S, Neidle S. Loop-length- dependent folding of G-quadruplexes. J Am Chem Soc. 2004;126:16405–16415. doi: 10.1021/ja045154j. This study combined spectroscopic and simulation techniques to suggest that a single residue forms the most stable double-chain-reversal loop bridging three G-tetrad layers. Longer linkers form less stable double-chain-reversal loops and prefer to form different types of loop connections. [DOI] [PubMed] [Google Scholar]
- 23••.Phan AT, Kuryavyi V, Gaw HY, Patel DJ. Small-molecule interaction with a five-guanine-tract G-quadruplex structure from the human MYC promoter. Nat Chem Biol. 2005;1:167–173. doi: 10.1038/nchembio723. A DNA fragment containing five G-tracts from the c-myc promoter forms a novel G-quadruplex fold, which comprises a core of three stacked G-tetrads formed by four parallel G-tracts with all anti guanines and a snapback 3′-end syn guanine. Study of the interaction of this G-quadruplex with four different ligands indicated that they all stack on the top of the G-tetrad core. In particular, TMPyP4 binds to this G-quadruplex in slow exchange; the structure of the G-quadruplex–TMPyP4 complex revealed how stacking and electrostatic interactions contribute to the stability of the complex. This structural information provides a platform for the design of anticancer drugs that target multi-G-tract sequences found in c-myc and other oncogenic promoters, as well as in telomeres. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24••.Phan AT, Kuryavyi V, Ma JB, Faure A, Andreola ML, Patel DJ. An interlocked dimeric parallel-stranded DNA quadruplex: a potent inhibitor of HIV-1 integrase. Proc Natl Acad Sci USA. 2005;102:634–639. doi: 10.1073/pnas.0406278102. The 93del d(GGGGTGGGAGGAGGGT) oligonucleotide, a potent inhibitor of HIV-1 integrase, adopts a very stable dimeric quadruplex architecture in K+ solution. Within each 16-nucleotide monomeric subunit, all G-stretches are parallel and all guanines are anti with the exception of G1, which is syn. Dimer formation is achieved through the mutual pairing of G1 of one monomer with G2, G6 and G13 of the other monomer, to complete G•G•G•G tetrad formation. Assays on loop-modified sequences suggested a new strategy for the potential design of improved HIV-1 integrase inhibitors. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Krishnan-Ghosh Y, Liu D, Balasubramanian S. Formation of an interlocked quadruplex dimer by d(GGGT) J Am Chem Soc. 2004;126:11009–11016. doi: 10.1021/ja049259y. [DOI] [PubMed] [Google Scholar]
- 26•.Kondo J, Adachi W, Umeda S, Sunami T, Takenaka A. Crystal structures of a DNA octaplex with i-motif of G-quartets and its splitting into two quadruplexes suggest a folding mechanism of eight tandem repeats. Nucleic Acids Res. 2004;32:2541–2549. doi: 10.1093/nar/gkh575. Intercalation of G-tetrads was observed in the crystal structure of d(GCGAGAGC) at low K+ concentration, producing a structure termed ‘i-motif of G-quartets’. In the structure, four base-intercalated duplexes are assembled to form an octaplex, with eight G5 bases forming a pair of stacked G-tetrads in the central part of the structure. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pan B, Shi K, Sundaralingam M. Base-tetrad swapping results in dimerization of RNA quadruplexes: implications for formation of the i-motif RNA octaplex. Proc Natl Acad Sci USA. 2006;103:3130–3134. doi: 10.1073/pnas.0507730103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28•.Crnugelj M, Sket P, Plavec J. Small change in a G-rich sequence, a dramatic change in topology: new dimeric G-quadruplex folding motif with unique loop orientations. J Am Chem Soc. 2003;125:7866–7871. doi: 10.1021/ja0348694. The sequence d(GGGTTTTGGGG) forms an asymmetric dimeric G-quad-ruplex that contains interesting structural motifs. [DOI] [PubMed] [Google Scholar]
- 29.Raghuraman MK, Cech TR. Effect of monovalent cation-induced telomeric DNA structure on the binding of Oxytricha telomeric protein. Nucleic Acids Res. 1990;18:4543–4552. doi: 10.1093/nar/18.15.4543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wyatt JR, Davis PW, Freier SM. Kinetics of G-quartet-mediated tetramer formation. Biochemistry. 1996;35:8002–8008. doi: 10.1021/bi960124h. [DOI] [PubMed] [Google Scholar]
- 31.Green JJ, Ying L, Klenerman D, Balasubramanian S. Kinetics of unfolding the human telomeric DNA quadruplex using a PNA trap. J Am Chem Soc. 2003;125:3763–3767. doi: 10.1021/ja029149w. [DOI] [PubMed] [Google Scholar]
- 32.Halder K, Chowdhury S. Kinetic resolution of bimolecular hybridization versus intramolecular folding in nucleic acids by surface plasmon resonance: application to G-quadruplex/duplex competition in human c-myc promoter. Nucleic Acids Res. 2005;33:4466–4474. doi: 10.1093/nar/gki750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zhao Y, Kan ZY, Zeng ZX, Hao YH, Chen H, Tan Z. Determining the folding and unfolding rate constants of nucleic acids by biosensor. Application to telomere G-quadruplex. J Am Chem Soc. 2004;126:13255–13264. doi: 10.1021/ja048398c. [DOI] [PubMed] [Google Scholar]
- 34•.Mergny JL, De Cian A, Ghelab A, Sacca B, Lacroix L. Kinetics of tetramolecular quadruplexes. Nucleic Acids Res. 2005;33:81–94. doi: 10.1093/nar/gki148. A systematic study of the association/dissociation kinetics of tetrameric G-quadruplexes is presented. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Merkina EE, Fox KR. Kinetic stability of intermolecular DNA quadruplexes. Biophys J. 2005;89:365–373. doi: 10.1529/biophysj.105.061259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Petraccone L, Pagano B, Esposito V, Randazzo A, Piccialli G, Barone G, Mattia CA, Giancola C. Thermodynamics and kinetics of PNA-DNA quadruplex-forming chimeras. J Am Chem Soc. 2005;127:16215–16223. doi: 10.1021/ja0545923. [DOI] [PubMed] [Google Scholar]
- 37.Neidle S, Parkinson G. Telomere maintenance as a target for anticancer drug discovery. Nat Rev Drug Discov. 2002;1:383–393. doi: 10.1038/nrd793. [DOI] [PubMed] [Google Scholar]
- 38.Siddiqui-Jain A, Grand CL, Bearss DJ, Hurley LH. Direct evidence for a G-quadruplex in a promoter region and its targeting with a small molecule to repress c-MYC transcription. Proc Natl Acad Sci USA. 2002;99:11593–11598. doi: 10.1073/pnas.182256799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39•.Clark GR, Pytel PD, Squire CJ, Neidle S. Structure of the first parallel DNA quadruplex-drug complex. J Am Chem Soc. 2003;125:4066–4067. doi: 10.1021/ja0297988. The crystal structure of the parallel quadruplex–daunomycin complex shows three molecules of daunomycin stacked on the 5′ end of the G-tetrad core. [DOI] [PubMed] [Google Scholar]
- 40.Gavathiotis E, Heald RA, Stevens MF, Searle MS. Drug recognition and stabilisation of the parallel-stranded DNA quadruplex d(TTAGGGT)4 containing the human telomeric repeat. J Mol Biol. 2003;334:25–36. doi: 10.1016/j.jmb.2003.09.018. [DOI] [PubMed] [Google Scholar]
- 41••.Haider SM, Parkinson GN, Neidle S. Structure of a G-quadruplex-ligand complex. J Mol Biol. 2003;326:117–125. doi: 10.1016/s0022-2836(02)01354-2. The crystal structure of a ligand bound to an antiparallel G-quadruplex. The ligand is bound at one end of the G-tetrad core and interacts with loop bases. [DOI] [PubMed] [Google Scholar]
- 42.Mehta AK, Shayo Y, Vankayalapati H, Hurley LH, Schaefer J. Structure of a quinobenzoxazine-G-quadruplex complex by REDOR NMR. Biochemistry. 2004;43:11953–11958. doi: 10.1021/bi049697h. [DOI] [PubMed] [Google Scholar]
- 43.Cocco MJ, Hanakahi LA, Huber MD, Maizels N. Specific interactions of distamycin with G-quadruplex DNA. Nucleic Acids Res. 2003;31:2944–2951. doi: 10.1093/nar/gkg392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Todd AK, Johnston M, Neidle S. Highly prevalent putative quadruplex sequence motifs in human DNA. Nucleic Acids Res. 2005;33:2901–2907. doi: 10.1093/nar/gki553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Huppert JL, Balasubramanian S. Prevalence of quadruplexes in the human genome. Nucleic Acids Res. 2005;33:2908–2916. doi: 10.1093/nar/gki609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wen JD, Gray CW, Gray DM. SELEX selection of high-affinity oligonucleotides for bacteriophage Ff gene 5 protein. Biochemistry. 2001;40:9300–9310. doi: 10.1021/bi010109z. [DOI] [PubMed] [Google Scholar]
- 47.Muniyappa K, Anuradha S, Byers B. Yeast meiosis-specific protein Hop1 binds to G4 DNA and promotes its formation. Mol Cell Biol. 2000;20:1361–1369. doi: 10.1128/mcb.20.4.1361-1369.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Mohaghegh P, Karow JK, Brosh RM, Jr, Bohr VA, Hickson ID. The Bloom’s and Werner’s syndrome proteins are DNA structure-specific helicases. Nucleic Acids Res. 2001;29:2843–2849. doi: 10.1093/nar/29.13.2843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Sun H, Yabuki A, Maizels N. A human nuclease specific for G4 DNA. Proc Natl Acad Sci USA. 2001;98:12444–12449. doi: 10.1073/pnas.231479198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50•.Duquette ML, Handa P, Vincent JA, Taylor AF, Maizels N. Intracellular transcription of G-rich DNAs induces formation of G-loops, novel structures containing G4 DNA. Genes Dev. 2004;18:1618–1629. doi: 10.1101/gad.1200804. EM was used to detect a DNA structure called the G-loop, which is formed during transcription when the non-template strand is guanine rich. G-loops contain G-quadruplex structures on one strand and a stable DNA–RNA hybrid on the other. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Schaffitzel C, Berger I, Postberg J, Hanes J, Lipps HJ, Pluckthun A. In vitro generated antibodies specific for telomeric guanine-quadruplex DNA react with Stylonychia lemnae macronuclei. Proc Natl Acad Sci USA. 2001;98:8572–8577. doi: 10.1073/pnas.141229498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52••.Paeschke K, Simonsson T, Postberg J, Rhodes D, Lipps HJ. Telomere end-binding proteins control the formation of G-quadruplex DNA structures in vivo. Nat Struct Mol Biol. 2005;12:847–854. doi: 10.1038/nsmb982. Using a G-quadruplex-specific antibody, the authors have demonstrated G-quadruplex formation in the telomeres of nanochromosomes of Stylonychia. They also showed that the telomere-end-binding proteins TEBPα and TEBPβ cooperate to control the formation of G-quadruplex structure at telomeres in vivo. Furthermore, such G-quadruplex formation is regulated by the cell-cycle-dependent phosphorylation of TEBPβ. [DOI] [PubMed] [Google Scholar]
- 53.Zahler AM, Williamson JR, Cech TR, Prescott DM. Inhibition of telomerase by G-quartet DNA structures. Nature. 1991;350:718–720. doi: 10.1038/350718a0. [DOI] [PubMed] [Google Scholar]
- 54.Zaug AJ, Podell ER, Cech TR. Human POT1 disrupts telomeric G-quadruplexes allowing telomerase extension in vitro. Proc Natl Acad Sci USA. 2005;102:10864–10869. doi: 10.1073/pnas.0504744102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Oganesian L, Moon IK, Bryan TM, Jarstfer MB. Extension of G-quadruplex DNA by ciliate telomerase. EMBO J. 2006;25:1148–1159. doi: 10.1038/sj.emboj.7601006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Pennarun G, Granotier C, Gauthier LR, Gomez D, Hoffschir F, Mandine E, Riou JF, Mergny JL, Mailliet P, Boussin FD. Apoptosis related to telomere instability and cell cycle alterations in human glioma cells treated by new highly selective G-quadruplex ligands. Oncogene. 2005;24:2917–2928. doi: 10.1038/sj.onc.1208468. [DOI] [PubMed] [Google Scholar]
- 57.Burger AM, Dai F, Schultes CM, Reszka AP, Moore MJ, Double JA, Neidle S. The G-quadruplex-interactive molecule BRACO-19 inhibits tumor growth, consistent with telomere targeting and interference with telomerase function. Cancer Res. 2005;65:1489–1496. doi: 10.1158/0008-5472.CAN-04-2910. [DOI] [PubMed] [Google Scholar]
- 58.Binz N, Shalaby T, Rivera P, Shin-ya K, Grotzer MA. Telomerase inhibition, telomere shortening, cell growth suppression and induction of apoptosis by telomestatin in childhood neuroblastoma cells. Eur J Cancer. 2005;41:2873–2881. doi: 10.1016/j.ejca.2005.08.025. [DOI] [PubMed] [Google Scholar]
- 59.Ha SC, Lokanath NK, Van Quyen D, Wu CA, Lowenhaupt K, Rich A, Kim YG, Kim KK. A poxvirus protein forms a complex with left-handed Z-DNA: crystal structure of a yatapoxvirus Zα bound to DNA. Proc Natl Acad Sci USA. 2004;101:14367–14372. doi: 10.1073/pnas.0405586101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Rothenburg S, Deigendesch N, Dittmar K, Koch-Nolte F, Haag F, Lowenhaupt K, Rich A. A PKR-like eukaryotic initiation factor 2α kinase from zebrafish contains Z-DNA binding domains instead of dsRNA binding domains. Proc Natl Acad Sci USA. 2005;102:1602–1607. doi: 10.1073/pnas.0408714102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Kwon JA, Rich A. Biological function of the vaccinia virus Z-DNA-binding protein E3L: gene transactivation and antiapoptotic activity in HeLa cells. Proc Natl Acad Sci USA. 2005;102:12759–12764. doi: 10.1073/pnas.0506011102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62••.Ha SC, Lowenhaupt K, Rich A, Kim YG, Kim KK. Crystal structure of a junction between B-DNA and Z-DNA reveals two extruded bases. Nature. 2005;437:1183–1186. doi: 10.1038/nature04088. This paper describes the crystal structure of a junction between the right-handed B-DNA and the left-handed Z-DNA forms of the double helix. At the junction, only one base pair is broken and the bases are flipped out of the duplex. Base pairs of the B-DNA and Z-DNA segments are continuously stacked across the junction. The structure of the B-Z junction maximizes base pairing and stacking, thereby minimizing the energetic cost of the junction and facilitating the use of Z-DNA more widely in nature. [DOI] [PubMed] [Google Scholar]
- 63.Lavery R, Zakrzewska K, Sun JS, Harvey SC. A comprehensive classification of nucleic acid structural families based on strand direction and base pairing. Nucleic Acids Res. 1992;20:5011–5016. doi: 10.1093/nar/20.19.5011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64••.Oleksi A, Blanco AG, Boer R, Uson I, Aymani J, Rodger A, Hannon MJ, Coll M. Molecular recognition of a three-way DNA junction by a metallosupramolecular helicate. Angew Chem Int Edn Engl. 2006;45:1227–1231. doi: 10.1002/anie.200503822. This paper describes the role of shape complementarity in the recognition of the central hydrophobic cavity of a Y-shaped three-way DNA junction by a tetracationic metallosupramolecular helicate. This research has broad implications for ligand-based design targeted to structural scaffolds and their potential stabilization as part of DNA-based nanotechnology arrays. [DOI] [PubMed] [Google Scholar]
- 65.Woods KC, Martin SS, Chu VC, Balwin EP. Quasi-equivalence in site-specific recombinase structure and function: crystal structure and activity of trimeric Cre recombinase bound to a three-way LOX junction. J Mol Biol. 2001;313:49–69. doi: 10.1006/jmbi.2001.5012. [DOI] [PubMed] [Google Scholar]
- 66.Walker JR, Corpina RA, Goldberg J. Structure of Ku heterodimer bound to DNA and its implications for double-strand break repair. Nature. 2001;412:607–614. doi: 10.1038/35088000. [DOI] [PubMed] [Google Scholar]
- 67.Seeman N. Structural DNA nanotechnology: an overview. Methods Mol Biol. 2005;303:143–166. doi: 10.1385/1-59259-901-X:143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Serganov A, Keiper S, Malinina L, Tereshko V, Skripkin E, Hobartner C, Polonskaia A, Phan AT, Wombacher R, Micura R, et al. Structural basis for Diels-Alder ribozyme-catalyzed carbon-carbon bond formation. Nat Struct Mol Biol. 2005;12:218–224. doi: 10.1038/nsmb906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Zhang N, Gorin A, Majumdar A, Kettani A, Chernichenko N, Skripkin E, Patel DJ. V-shaped scaffold: a new architectural motif identified in an A•(G•G•G•G) pentad-containing dimeric DNA quadruplex involving stacked G(anti)•G(anti)• G(anti)•G(syn) tetrads. J Mol Biol. 2001;311:1063–1079. doi: 10.1006/jmbi.2001.4916. [DOI] [PubMed] [Google Scholar]