Abstract
Background
Cytochrome P450 sterol 14α-demethylase (CYP51) is an essential enzyme for sterol biosynthesis and a target for anti-parasitic drug design. However, the design of parasite-specific drugs that inhibit parasitic CYP51 without severe side effects remains challenging. The active site of CYP51 is situated in the interior of the protein. Here, we characterize the potential ligand egress routes and mechanisms in Trypanosoma brucei and human CYP51 enzymes.
Methods
We performed Random Acceleration Molecular Dynamics simulations of the egress of four different ligands from the active site of models of soluble and membrane-bound T. brucei CYP51 and of soluble human CYP51.
Results
In the simulations, tunnel 2 f, which leads to the membrane, was found to be the predominant ligand egress tunnel for all the ligands studied. Tunnels S, 1 and W, which lead to the cytosol, were also used in T. brucei CYP51, whereas tunnel 1 was the only other tunnel used significantly in human CYP51. The common tunnels found previously in other CYPs were barely used. The ligand egress times were shorter for human than T. brucei CYP51, suggesting lower barriers to ligand passage. Two gating residues, F105 and M460, in T. brucei CYP51 that modulate the opening of tunnels 2 f and S were identified.
Conclusions
Although the main egress tunnel was the same, differences in the tunnel-lining residues, ligand passage and tunnel usage were found between T. brucei and human CYP51s.
General Significance
The results provide a basis for the design of selective anti-parasitic agents targeting the ligand tunnels.
Keywords: Sterol 14α-demethylase, CYP51, Cytochrome P450, Random acceleration molecular dynamics, Membrane-bound protein, Ligand access and egress
1. Introduction
The sterol 14-α demethylase (CYP51) enzyme plays an important role in sterol biosynthesis [1]. The inhibition of this enzyme has been used as a strategy to develop anti-fungal drugs and, more recently, anti-parasitic drugs [1–7]. As a member of the cytochrome P450 (CYP) superfamily, eukaryotic CYP51 has features that are found in many CYPs, including: (1) embedding in the membrane via immersion of part of its globular domain and a single transmembrane helix, (2) a buried active site which requires the substrates and products to enter and exit from the active site through ligand tunnels, and (3) the need for electrons to be transferred from the redox partners for the catalytic cycle. However, distinct features of CYP51 compared to other CYPs have been observed in experiments and in molecular dynamics (MD) simulations. These features include high substrate specificity, binding site rigidity, different open tunnels between the active site and the protein surface, and plasticity of the two heme propionate groups [1,8,9]. Because of CYP51's potential as a target for anti-parasitic drug design, it is important to study these features of CYP51.
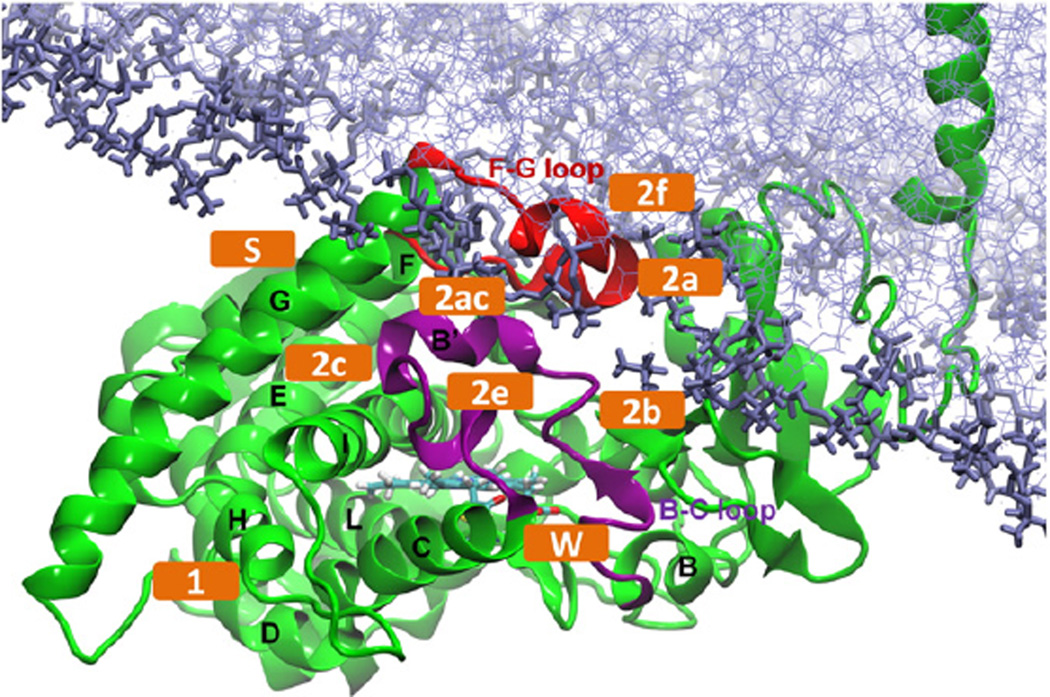
We have previously built and simulated a model of membrane-bound T. brucei CYP51 in a 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphocholine (POPC) bilayer based on a crystal structure of the globular domain of T. brucei CYP51 [8]. The mapping of ligand tunnels that have been found in other CYPs to the model of the membrane-bound T. brucei CYP51 are, according to Wade's nomenclature [10]: tunnel 2a (between the F-G loop, B-B’ loop and β1-1 sheet) and tunnel 2 f (between the A’ and F” helices and the tip of the β4 hairpin) which lead to the membrane, and tunnels 1 (between the C, D, H and L helices), 2b (between the B-B’ loop and the β1-2 and β1-4 sheets), 2c (between the G and I helices and the B’-C loop), 2 ac (between the B’ helix and G helix), 2e (through the B-C loop), S (the solvent tunnel, between the F and I helices and the β4 hairpin) and W (the water tunnel, between the C helix, the B-B’ loop and helix K” of the heme bulge segment) which lead to the solvent (Figs. 1, S1 and S2).
Fig. 1.
Model of membrane-bound ligand-free T. brucei CYP51 [8]. The important tunnels defined in Wade's nomenclature are labeled (orange). The protein is shown in cartoon representation with the B-C loop colored purple, the F-G loop red and the helices labelled. The heme (cyan) and the POPC bilayer (purple) are shown in stick representation.
Tunnels 2 f, S and W have been shown to be open to a probe with a radius of 1.4 Å, corresponding to the size of a water molecule, in standard MD simulations of T. brucei CYP51 [8]. However, it is challenging to simulate ligand egress in a feasible time in standard MD simulations. In order to make the simulation of ligand egress computationally possible, the random acceleration molecular dynamics (RAMD) simulation method was developed [11,12]. In RAMD simulations, an external, randomly oriented force is applied to the center of mass of the ligand to accelerate the ligand exit. As a result, the ligand is able to exit the active site on the time scale of a few hundreds of ps to a few ns and a large number of trajectories can be generated to explore alternative egress routes and mechanisms. RAMD has been applied to a wide range of proteins (e.g. [13–15]), including haloalkane dehalogenases for which it served to guide the engineering of tunnels to the active site to improve enzymatic activity [16]. RAMD has been successfully used to study the ligand egress pathways of many soluble forms of CYPs, including P450cam, CYP2B1, CYP2C5, CYP3A4, CYP2E1, CYP2A6 and CYP2C9 and membrane-bound forms of CYP19 [11,12,17–28]. The preferred ligand tunnels in these simulations are mostly the family 2 tunnels, including 2a, 2b, 2c, 2e and 2 ac. These are all lined by the B-C loop. Because of the distinct features of CYP51 compared to the other CYPs, it is interesting to study the ligand egress routes in CYP51, especially in the membrane-bound form. Thus, we here perform RAMD simulations to study how different ligands enter and egress from the active site of T. brucei CYP51, considering both soluble and membrane-bound forms of the protein. We also perform comparative simulations for the soluble form of human CYP51,which has 34% overall sequence identity and 52% sequence similarity to T. brucei CYP51,with 50% sequence identity in the binding site cavity (residues within 5 Å of a ligand bound in the active site).
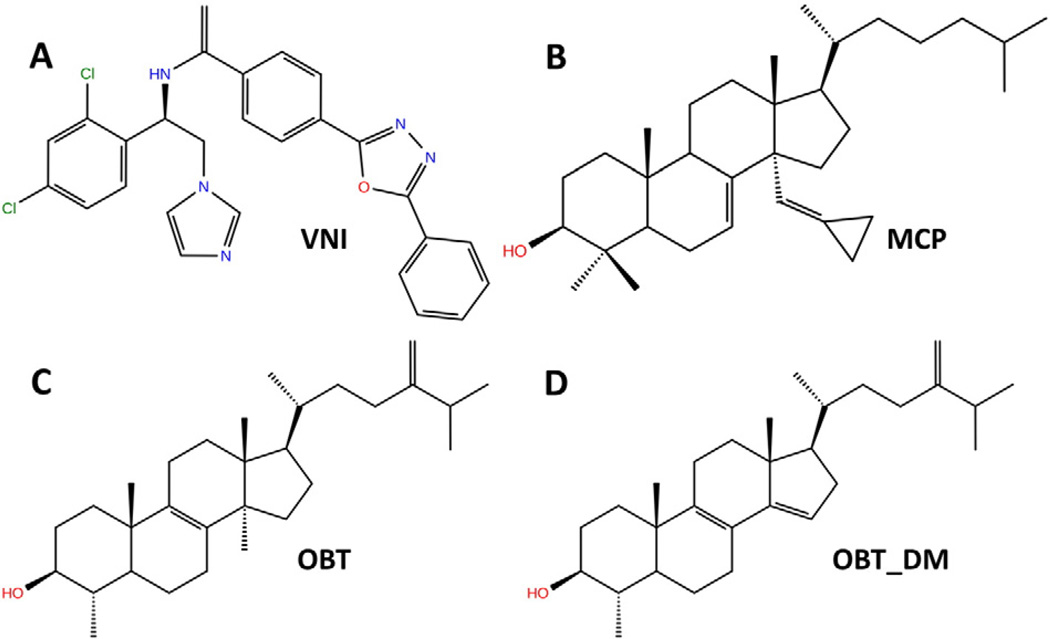
We consider the binding of four different compounds, representing different types of ligand: two inhibitors, one substrate and its product. The two inhibitors studied have had the structures of their complexes with T. brucei CYP51 determined crystallographically. One is a well studied azole compound, N-[(1R)-1-(2,4-dichlorophenyl)-2-(1 H-imidazol-1-yl)ethyl]-4-(5-phenyl-1,3,4-oxadiazol-2-yl)benzamide (VNI), which is potent against trypanosomal CYP51 while having low cytotoxicity and not inhibiting human CYP51 [1]. The other is the sterol derivative, 14α-methylenecyclopropyl-Δ7-24,25-dihydrolanosterol (MCP), which shows inhibition against all protozoan CYP51s [5]. The substrate studied, obtusifoliol (OBT) (4α,14-dimethyl-5α-ergosta-8,24(28)-dien-3β-ol), is an established substrate of both T. brucei and human CYP51s [29], and the product, with the 14α-methyl group removed, is δ4α-methyl-5α-ergosta-8,14,24 [28]-trien-3β-ol (OBT_DM). The structures of these four compounds are shown in Fig. 2. We carried out RAMD simulations of ligand egress for models of membrane-bound and soluble T. brucei CYP51 for all four compounds. For comparison, simulations of ligand egress were performed for soluble human CYP51 for the substrate, OBT, and product, OBT_DM.
Fig. 2.
Chemical structures of the four ligands studied colored according to atom type. VNI: N-[(1R)-1-(2,4-dichlorophenyl)-2-(1 H-imidazol-1-yl)ethyl]-4-(5-phenyl-1,3,4-oxadiazol-2-yl)benzamide; MCP: 14α-methylenecyclopropyl-Δ7-24,25-dihydrolanosterol, OBT: obtusifoliol, OBT_DM: 14α-demethylated obtusifoliol.
The simulations allow us to specifically address the following questions: Can the tunnels in CYP51 that are open in standard MD simulations be used by inhibitors, substrates and products to enter and exit the active site? Are different ligand tunnels used in the soluble and membrane-bound forms? Which residues gate and modulate the opening and closing of the ligand tunnels? How do ligand egress and the ligand tunnels differ between T. brucei CYP51 and human CYP51?
2. Materials and methods
2.1. Preparation of structures of the protein-ligand complexes
The models of soluble and membrane-bound T. brucei CYP51 were based on the crystal structure of ligand-free T. brucei CYP51 (PDB id: 3G1Q [1]). The model of membrane-bound ligand-free CYP51 in a lipid bilayer of 602 POPC molecules was built from this crystal structure using a protocol combining coarse-grained and all-atom simulations as described in [8,30]. The coarse-grained simulations were performed to find the predominant orientation of CYP51 above the membrane and the all-atom simulations were then performed to refine the membrane-bound model and investigate the dynamics of the system with this predominant orientation. The models of soluble human CYP51 were based on the crystal structure of human CYP51 (PDB id: 3LD6, [31]) from which the ligand was removed.
To enable comparison of simulation results for different ligands, all the complexes of soluble T. brucei CYP51 with ligands were built using the structure of ligand-free T. brucei CYP51. This could be done because of the similarity of the binding site shape in the crystal structures of T. brucei CYP51 in unliganded and liganded form (For example, the Cα RMSD between the crystal structures of ligand-free and VNI-bound T. brucei CYP51 is ca. 0.75 Å [1]). This procedure meant that possible artifacts in the simulations due to small differences in the protein structures in the crystal structures could be avoided. The coordinates of the inhibitors VNI and MCP were generated by superimposing the binding site of the ligand-bound crystal structures (PDB id: 3GW9 [1] and 3P99, [5]), respectively, on to the binding site of the ligand-free CYP51. Structures (sdf files) of the substrate (OBT) and product (OBT_DM) were downloaded from PubChem (CID 65252 and 443,237), respectively. These compounds were positioned in the active site of T. brucei CYP51 so as to adopt a similar orientation to MCP, which is a sterol-like inhibitor, and so that their site of metabolism was positioned next to the heme center.
For the models of membrane-bound T. brucei CYP51, ligand coordinates were generated by superimposing the last snapshot from equilibration (see below) of the corresponding soluble ligand-bound models onto the binding site of the refined model of ligand-free membrane-bound T. brucei CYP51.
For the human CYP51, the coordinates of the substrate (OBT) and product (OBT_DM) were generated by superimposing the OBT-bound and OBT_DM-bound structures of T. brucei CYP51 onto the crystal structure of human CYP51 with the ligand removed.
2.2. Parameterization of the systems for simulation
The ff99SB force field for the protein and the general Amber force field (GAFF) for the ligands and POPC phospholipids [32] were used [33], along with the TIP3P model [34] for water. The parameters for the ferric heme center were assigned as in [30,35]. The partial atomic charges of the four ligands were derived from Hartree-Fock 6-31G* ab initio calculations and restrained electrostatic potential fit (RESP) on the R.E.D server [36–38]. The force field parameters of VNI were derived without considering its coordination through a nitrogen atom with the heme iron as we wanted to explore the binding and unbinding of this compound, see also section 3.4. Other parameters were derived with the Antechamber program of AmberTools12 [39]. The AMBER parameter files are given in the Supplementary Information.
2.3. Standard molecular dynamics simulations
Simulations of soluble models were performed with the Amber11 and Amber 12 software [39,40]. Those of the membrane-bound models were performed with NAMD2.9 due to its better computational performance for these larger systems [41]. The same protonation, solvation, minimization and equilibration procedures as described in ref. [8] were used. These resulted in protein-ligand complexes modeled at pH 7 solvated in an octahedral box of water molecules and neutralized with counterions (soluble models) or embedded in a bilayer and solvated in a rectangular box of water molecules neutralized in 150 mM NaCl (membrane-bound models).
Short production runs of 2–15 ns (see Table 1) were performed before the RAMD simulations in order to generate starting coordinates for the RAMD simulations. The parameters of the simulations were the same as those validated and used previously [8,30] unless otherwise stated and the reader is referred to these references for details. In all simulations, all bonds to hydrogen atoms were constrained with the SHAKE algorithm [42]. For the soluble models, the non-bonded cutoff was set to 12 Å. Production runs were performed with a time step of 1 fs. Langevin dynamics was used to maintain the temperature at 300 K and isotropic position scaling kept the pressure at 1 bar. For the membrane-bound models, the non-bonded cutoff was set to 10 Å. Production runs were performed with a time step of 1.5 fs. In the production run, Langevin dynamics and the Nosé-Hoover Langevin Piston method with a surface tension of 60 dyn/cm applied to the lipid bilayer plane were used to maintain the temperature at 310 K and the pressure at 1 bar.
Table 1.
Simulations performed of T. brucei and human CYP51 enzymes.
| Systemi | Environmentii | Methodiii | No. of Trajectoriesiv |
rMin (Å)v |
Duration (ns)v |
|---|---|---|---|---|---|
| CYP51 + VNI (T. brucei) | SOL | MD | 1 | —– | 2 |
| RAMD | 30 | 0.025 | —– | ||
| 30 | 0.050 | —– | |||
| MEM | MD | 1 | —– | 15 | |
| RAMD | 30 | 0.025 | —– | ||
| 30 | 0.050 | —– | |||
| CYP51 + MCP (T. brucei) | SOL | MD | 1 | —– | 2 |
| RAMD | 30 | 0.025 | —– | ||
| 30 | 0.050 | —– | |||
| MEM | MD | 1 | —– | 15 | |
| RAMD | 30 | 0.025 | —– | ||
| 30 | 0.050 | —– | |||
| CYP51 + OBT (T. brucei) | SOL | MD | 1 | —– | 2 |
| RAMD | 30 | 0.025 | —– | ||
| 30 | 0.050 | —– | |||
| MEM | MD | 1 | —– | 3.75 | |
| RAMD | 30 | 0.025 | —– | ||
| 30 | 0.050 | —– | |||
| CYP51 + OBT_DM (T. brucei) | SOL | MD | 1 | —– | 2 |
| RAMD | 30 | 0.025 | —– | ||
| 30 | 0.050 | —– | |||
| MEM | MD | 1 | —– | 3 | |
| RAMD | 30 | 0.025 | —– | ||
| 30 | 0.050 | —– | |||
| CYP51 + OBT (human) | SOL | MD | 1 | —– | 2 |
| RAMD | 30 | 0.025 | —– | ||
| 30 | 0.050 | —– | |||
| CYP51 + OBT_DM (human) | SOL | MD | 1 | —– | 2 |
| RAMD | 30 | 0.025 | —– | ||
| 30 | 0.050 | —– |
Note:
The CYP51-ligand complexes studied. The full names of the ligands are given in the Introduction section and their structures are shown in Fig. 2.
The simulations were of models of soluble CYP51 (SOL) and membrane-bound CYP51 (MEM).
Simulation method: standard molecular dynamics (MD); random acceleration molecular dynamics (RAMD).
“1” indicates that the standard MD simulation was performed once to produce the input snapshots for RAMD. The last snapshot of the equilibration and the last snapshot of the production run were used for RAMD.
The length of the standard MD production simulation is listed. The exit time of different ligands via tunnel 2 f in RAMD simulations varies, see Fig. 5, but the maximum length of one RAMD simulation was set to 2 ns.
An additional 12 ns standard MD simulation of OBT_DM in the membrane in the presence of T. brucei CYP51 was performed directly after one RAMD simulation, in which the ligand exited via tunnel 2 f and entered the membrane at the end of the simulation. The last snapshot of the RAMD simulation was taken as the starting structure with all the velocities and periodic box information from that RAMD simulation. The production run was then performed directly with the same parameters as standard MD simulations of membrane-bound models of T. brucei CYP51. The new simulation of only OBT_DM in the membrane was started from the snapshot at 7.5 ns of the 12 ns simulation after the RAMD simulation and a 7.15 ns production run was directly performed with CYP51 removed from the system.
2.4. RAMD simulations
Random acceleration molecular dynamics (RAMD) as implemented in NAMD2.9 was used to study the ligand egress pathways from the buried active site of CYP51 [11,12,17,18,20]. The systems simulated and number of trajectories generated are given in Table 1. The same parameters as in the standard MD simulations were used for the RAMD simulations except that the pressure in the simulations of soluble CYP51 was controlled by a Berendsen barostat at 1 bar, for consistency with previous applications of RAMD, and a time step of 2 fs was used. An external acceleration of magnitude 0.035 kcal/Å·g was applied in a random direction on the ligand. The distance traveled by the ligand was monitored at 100 fs (50 steps with a time step of 2 fs) time intervals. If the distance traveled was greater than a pre-defined minimum threshold, the direction of the force was maintained. If not, the direction of the force was changed to another randomly chosen direction. For each system studied, 30 RAMD trajectories were run with a threshold of 0.025 Å and 30 RAMD trajectories with a threshold of 0.050 Å. When the accumulated distance traveled by the ligand was greater than the distance between the active site and the exterior of the protein, defined as 30 Å for soluble models and 40 Å for membrane-bound models, the RAMD simulation was terminated. If the ligand did not exit within 2 ns, the trajectory was stopped. For each model studied, the last snapshot from the equilibration and one snapshot from the production run (at 500 ps for the soluble models, 15 ns for the membrane-bound VNI- and MCP-bound models, 3.75 ns for the membrane-bound OBT-bound model and 3 ns for the membrane-bound OBT_DM-bound models) were used as initial structures for performing RAMD simulations. Two different snapshots were used for RAMD simulations for each system so that any sensitivity of the RAMD results to starting structure could be checked.
2.5. Analysis
The concatenation and analysis of the trajectories were performed with the cpptraj tool of AmberTools12 [39]. The trajectories were visualized in VMD1.9 [43]. The figures were made with VMD, PyMOL [44] and Maestro [45]. To plot the ligand 2D structures, the SMILES identifiers of VNI and MCP were taken from the PDB structures (PDB id: 3GW9 and 3P99) and those of OBT and OBT_DM from the PubChem database.
3. Results and discussion
3.1. Tunnel 2f is the predominant ligand egress route observed in RAMD simulations
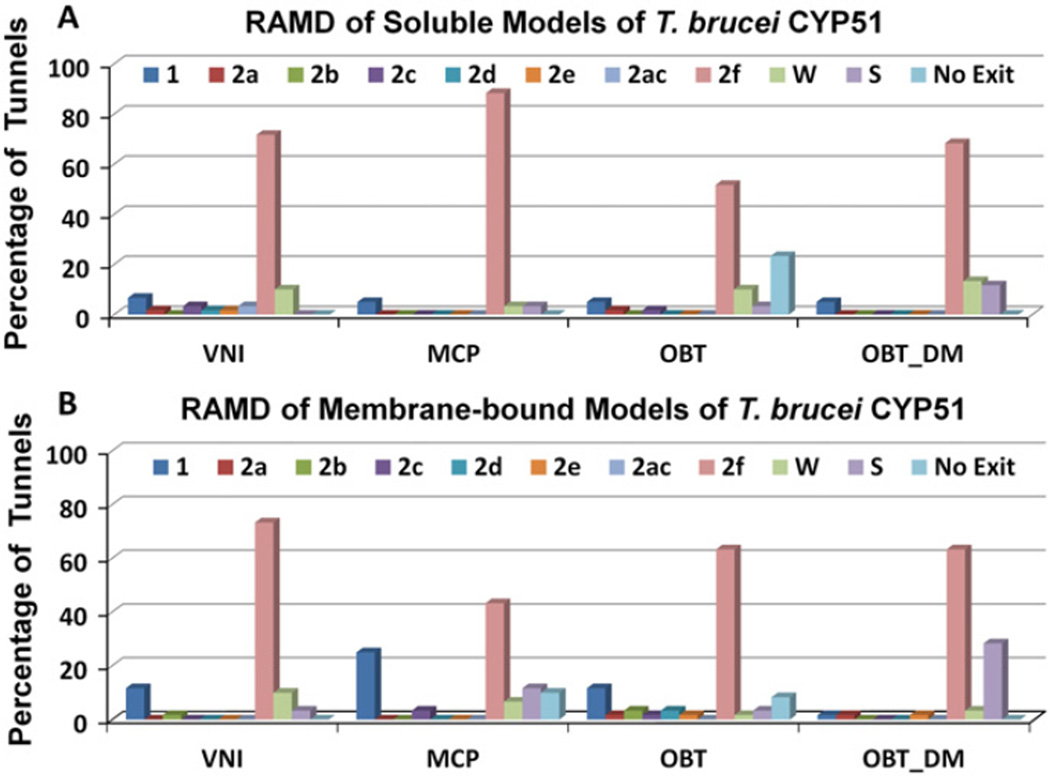
For all four ligands (VNI, MCP, OBT and OBT_DM) in both soluble and membrane-bound forms of T. brucei CYP51, the 2 f tunnel was found to be the predominant ligand exit tunnel in the RAMD simulations (Fig. 3). The 2 f tunnel is open in the crystal structures of parasitic CYP51s and was proposed to be the major ligand tunnel of the CYP51 family [1,46]. We found that the 2 f tunnel widened transiently to allow the ligands to exit from this tunnel and that this widening was facilitated by the flexible movements of the residues at the 2 f tunnel entrance.
Fig. 3.
The tunnels by which the four ligands are observed to exit from T. brucei CYP51 in RAMD simulations. The usage of the tunnels is given as a percentage of the 60 RAMD simulations performed for each system. Simulations were run for models of (A) soluble and (B) membrane-bound T. brucei CYP51.
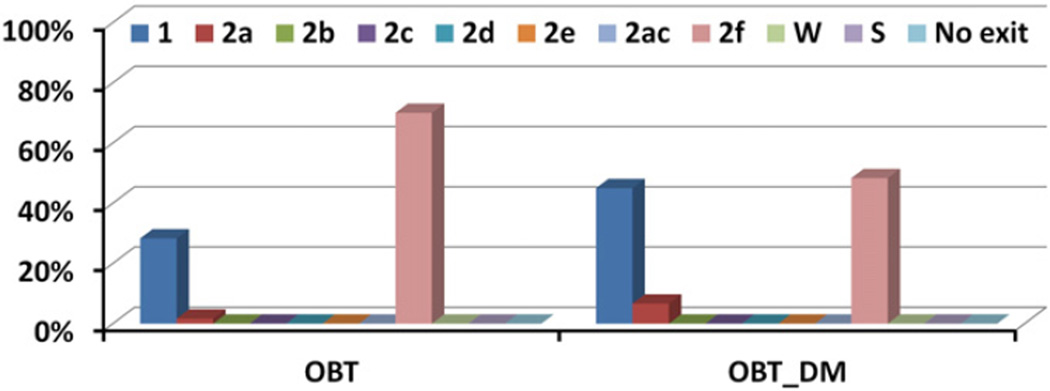
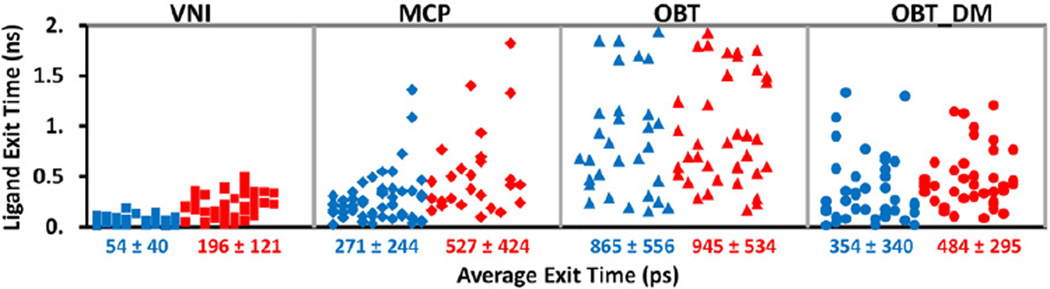
Tunnel 2 f was also found to be the predominant ligand tunnel in simulations of soluble models of human CYP51 (Fig. 4). However, differences in the passage of the ligands through tunnel 2 f in T. brucei CYP51 and human CYP51 are apparent from the much shorter exit times of the ligands in the RAMD simulations of human CYP51 with OBT and OBT_DM, than for T. brucei CYP51 (Figs. 5 and S3). For the substrate (OBT), average egress times were lower by a factor of about 8, for the product (OBT_DM), they were lower by a factor of about 3. The quick exit of ligands from the active site of human CYP51 by tunnel 2 f indicates less hindrance to the passage of ligands into and out of the human CYP51 active site in comparison to T. brucei CYP51. The catalytic rate for OBT is ca. 5 fold quicker for human CYP51 than T. brucei CYP51 in vitro (Fig. 6 of ref. [4]). Fast ligand access and egress in human CYP51 may contribute to the fast catalytic rate.
Fig. 4.
The tunnels by which the ligands, OBT and OBT_DM, are observed to exit from human CYP51 in RAMD simulations. The usage of the tunnels is given as a percentage of the 60 RAMD simulations performed for each system. The simulations were run for a model of the soluble human CYP51.
Fig. 5.
The times required for the four ligands to egress through tunnel 2 f in T. brucei CYP51 (blue: soluble model, red: membrane-bound model) in each RAMD trajectory. Each symbol represents the result of one RAMD trajectory and the average exit times of all simulations with exit through tunnel 2 f are given below the x-axis in ps. Times obtained in RAMD simulations are expected to be much shorter than the real exit times but their relative values indicate the extent to which ligand egress from the active site is hindered; longer average times correspond to higher barriers to ligand egress.
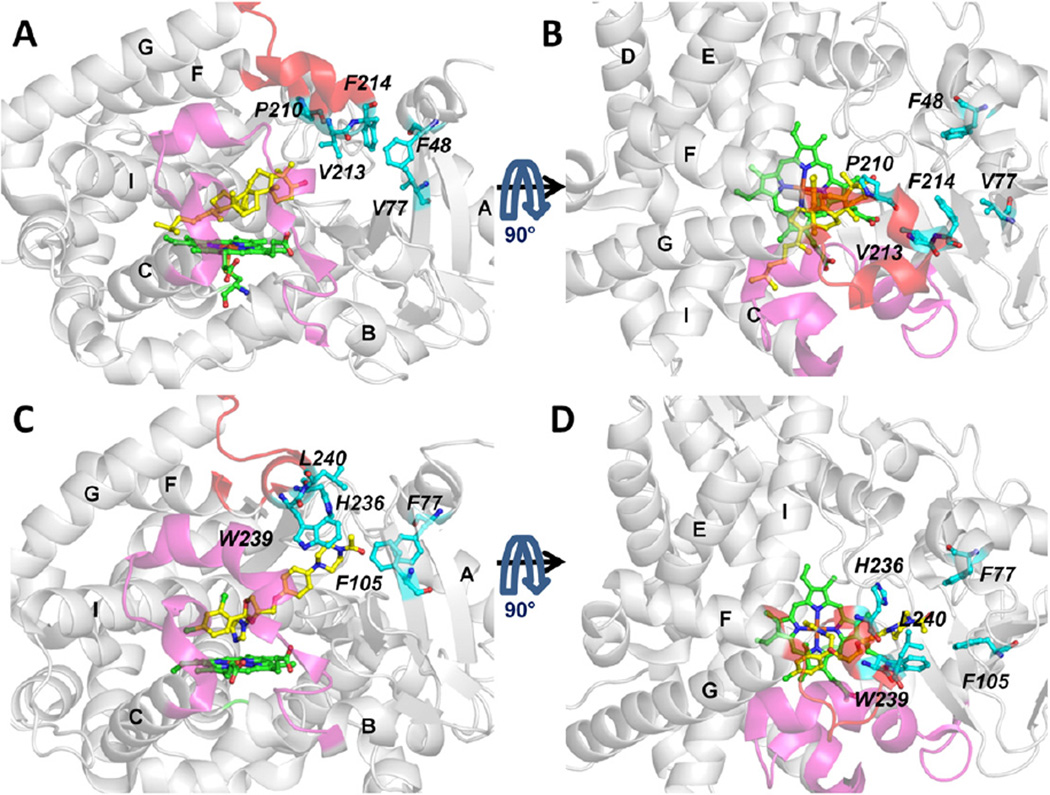
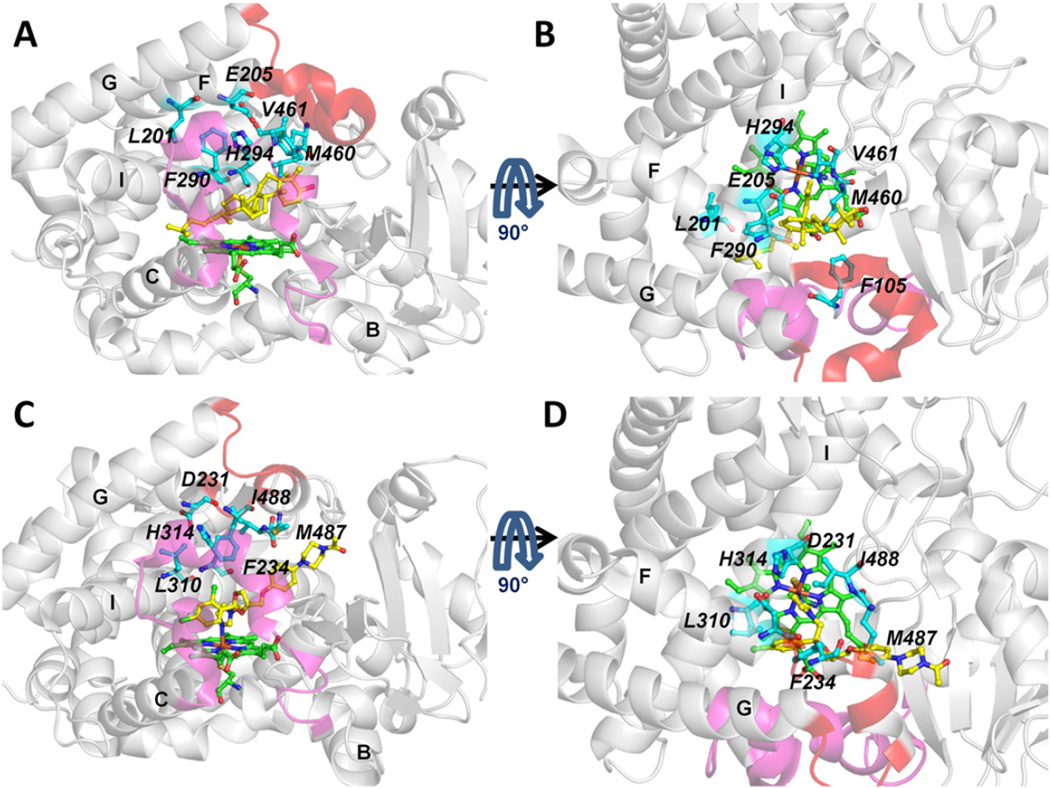
The tunnel entrance residues of tunnel 2 f are shown in human and T. brucei CYP51 in Fig. 6, The conformation of the F-G loop region differs which results in the 2 f tunnel in T. brucei CYP51 being more closed than in human CYP51, see Fig. S4. In the F-G loop region, the crystal structures show that both proteins have an F” helix whereas T. brucei CYP51 has a G’ helix but human CYP51 does not. This difference could contribute to the quicker exit via the 2 f tunnel of human CYP51. Furthermore, there are differences in the residues lining the tunnel which may affect ligand gating of tunnel 2 f (see section 3.4).
Fig. 6.
Comparison of the entrance residues of tunnel 2 f of T. brucei CYP51 (in complex with MCP, PDB ID: 3P99 [5]) (A, B) and human CYP51 (in complex with the inhibitor, ketoconazole, PDB ID: 3LD6 [31]) (C,D). Ligands and heme cofactor are shown in stick representation in yellow and green, respectively. The B-C loop is in magenta and the F-G loop in red. The tunnel entrance residues are labeled in italic and important secondary structures are labeled in bold. Each protein is shown in two orientations for clarity.
3.2. Tunnels 1, S and W In T. brucei CYP51, and tunnel 1 in human CYP51 are additional egress routes in RAMD simulations
Tunnel 1, tunnel S and tunnel W were observed to be ligand egress routes in the simulations of T. brucei CYP51 but were used much less often than tunnel 2 f (Fig. 3). The other tunnels (2a, 2b, 2c, 2d, 2e, 2 ac) were only rarely observed to be ligand egress routes. All the ligands showed exits through tunnel W in T. brucei CYP51 but the likelihood of this tunnel serving as a ligand tunnel in vivo is very low because the entrance of this tunnel is on the proximal side which is proposed to be the binding interface of the cytochrome P450 reductase [47]. Binding of the reductase would affect the dynamics of the proximal side of CYP51 and would block or hinder ligand egress through tunnel W, though the tunnel may still be sufficiently open to permit passage of water molecules. In the simulations of soluble human CYP51 however, tunnel W was not used at all and neither was tunnel S (Fig. 4). Tunnel 1 was often used. Of the other tunnels, only tunnel 2a was occasionally used.
The differences in the use of tunnels S and W for ligand egress is consistent with the tunnel opening observed in standard MD simulations of T. brucei and human CYP51 as revealed by analysis with CAVER in Fig. 5 of ref. [8]. In the CAVER analysis, the percentage of the simulation in which each of the tunnels was open to a water-sized probe sphere was monitored. Tunnel 2 f was overall the most open tunnel in the standard MD simulations of the two CYP51s. Tunnel W and tunnel S were mostly open in the simulations of T. brucei CYP51 whereas Tunnel W was found to be closed and tunnel S to be barely open (< 10%) in the simulation of human CYP51. Thus, both the RAMD and the standard MD simulations show different tunnel usage in human CYP51 in comparison to T. brucei CYP51.
Tunnel S has been proposed to be used for the passage of water molecules and was found to be open in standard MD simulations of other CYPs, including CYP2C9, CYP3A4 and CYP2D6 [30,48,49] and in RAMD simulations of human aromatase (CYP19) [25]. Tunnel 1, on the other hand, has only rarely been observed as a ligand exit tunnel in standard MD simulations [10] and RAMD simulations [11]. However, the fact that tunnel 1 was used in the RAMD simulations of both CYP51s, and rather often in the soluble model of human CYP51 shows the potential of tunnel 1 to serve as a ligand tunnel. Interestingly, in the crystal structure of Mycobacterium tuberculosis H37Rv CYP51 (PDB id: 2VKU), the bis(4-hydroxyphenyl) methanone ligand (with coordinates for two alternative positions) is trapped in the middle of tunnel 1 (Fig. S1) [50]. This suggests that tunnel 1 can potentially serve as a ligand exit tunnel although in this crystal structure, the opening of tunnel 1 results from the I helix being kinked towards the distal side (away from heme). In the RAMD simulations, this large conformational change of the I helix was not needed to allow the ligands to exit from tunnel 1. The three residues of the kinked region are relatively well conserved with “AGQ” in T. brucei and human CYP51 and “AGH” in M. tuberculosis CYP51 (Fig. S2). However, the Cα atoms of the I helix residues lining tunnel 1 (residue numbers 278–283) showed similar dynamics in standard simulations [8] and in RAMD simulations in which a ligand egressed by pathway 1 with root mean squared deviations (RMSD) of less than 1.5 Å from initial positions.
3.3. Ligand passage from the active site to the membrane is observed through the 2f tunnel
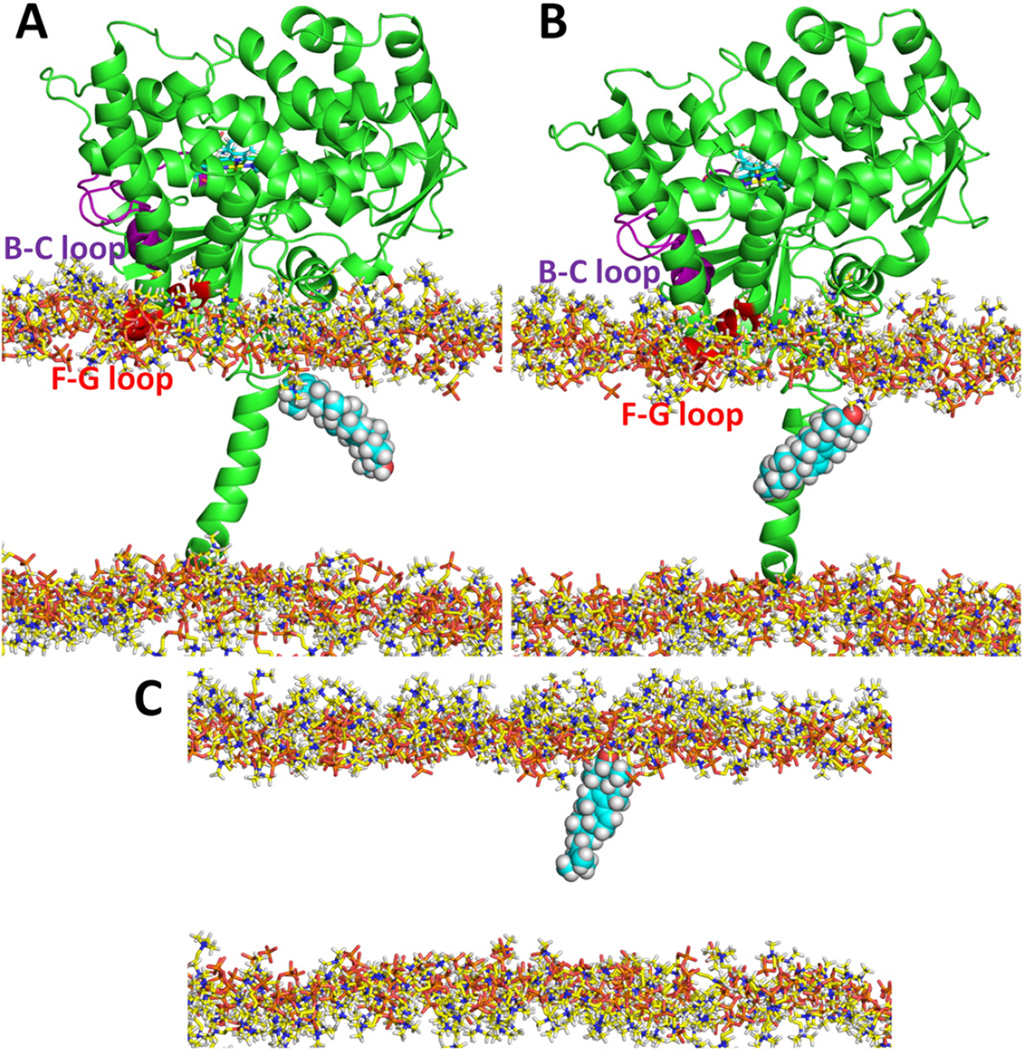
In the model of membrane-bound T. brucei CYP51, tunnel 2 f leads to the membrane bilayer. Of the tunnels used in the RAMD simulations, it is the only one leading to the membrane bilayer. Tunnel 2 f is open in the standard simulations of all the systems simulated but is notably wider open in the model of membrane-bound CYP51 (see Fig. S4B showing a ca. 4 Å greater distance between the F” and G’ helices and the A’ helix). Surprisingly, the association of T. brucei CYP51with the membrane resulted in longer average exit times of all four ligands through tunnel 2 f than for the soluble form of the protein in the RAMD simulations (Fig. 5). Thus, it is harder for the ligands to egress through tunnel 2 f into the membrane in the membrane-bound protein than into aqueous solvent in the soluble form of the protein. However, in almost 95% of the egress trajectories via tunnel 2 f in the membrane-bound form, the ligands entered the membrane after leaving the protein. For the other egress events via tunnel 2 f, the tunnel entrance widening was accompanied by large conformational changes enabling the ligands to enter the solvent next to the membrane. Thus, for the membrane-bound form, ligand egress via tunnel 2 f is predominantly into the membrane.
The crystal structure shows that the hydrophilic head of the sterol-derivative MCP in the active site faces towards the membrane. This means that when the sterol-derivative ligands enter the membrane from the protein, the ligands have the hydrophilic head pointing towards the middle of the bilayer. In the RAMD simulations, the hydrophilic head first interacts with the lipid head groups and then penetrates into the middle of the bilayer so that the ligand is in the opposite orientation to the phospholipids with their polar head groups on the bilayer surface (Fig. 7A). Ligand reorientation would be necessary for the ligand to exist in the membrane. This could potentially occur in the protein or in the membrane although it is not excluded that sterols may exit the CYP51 active site directly into the active site of the next enzyme in the sterol biosynthetic pathway, particularly since no detectable intermediates are present in the membranes if the pathway is not blocked. We did not observe any large reorientation of the ligand in the protein despite the enhanced sampling of ligand positions due to the RAMD force. To investigate whether such reorientation in the membrane was possible, an additional standard MD simulation with the natural product OBT_DM in the membrane in the presence of the protein was performed starting from the end snapshot of a RAMD simulation that had resulted in ligand egress to the middle of the membrane through the 2 f tunnel. In this simulation, after ca. 6.5 ns, OBT_DM rotated roughly 100° and changed its orientation from hydrophilic head down in the middle of the bilayer to up towards the head-group layer. This orientation is similar to that of the POPC phospholipids (Fig. 7B). A further simulation (of 7.15 ns length) without the protein allowed the OBT_DM to orient in the same way as the phospholipids. After 150 ps, it had positioned its hydrophilic head in the head group region of the bilayer (Fig. 7C) and it kept this orientation for the rest of the simulation. These results suggest that the substrates of CYP51 can come from the membrane before the catalytic reaction and, as products, re-enter the membrane afterwards (via tunnel 2 f), and reorient for the next step in their enzymatic processing.
Fig. 7.
The change in orientation of OBT_DM in the membrane upon exit from T. brucei CYP51. (A) At the end of a RAMD simulation of T. brucei CYP51, the OBT_DM was orientated with its hydrophilic head down in the middle of the bilayer. (B) After a subsequent standard MD simulation of 6.5 ns, OBT_DM had changed its orientation in the membrane by about 100° so that its hydrophilic head pointed up towards the lipid head groups. (C) A further subsequent simulation without the protein starting with the snapshot of (B) allowed the hydrophilic part of OBT_DM to orient in the head group regions of the phospholipid bilayer. Only the head groups of the POPC bilayer are shown for clarity. OBT_DM is shown in CPK representation and T. brucei CYP51 is shown in cartoon representation.
3.4. Tunnel entrance residues may play important role in ligand specificity
T. brucei and human CYP51 differ in their substrate and inhibitor specificity. For example, VNI is a potent inhibitor of T. brucei CYP51 but has little inhibitory effect on human CYP51 [3]. Figs. 5, S3 and S5 show that the four ligands studied differ in their egress times and in their usage of the different tunnels.
VNI on average exits notably faster than the other three ligands through tunnel 2 f of T. brucei CYP51 (Fig. 5). It also exits faster through most of the other tunnels (Fig. S5), These observations may initially seem surprising as the azole ring of VNI coordinates the heme iron atom at the sixth position. However, this strong N-Fe bonding was intentionally omitted in our model so that we could explore how the ligand could pass from the active site to the protein exterior and from this perhaps also gain insights into the (reverse) binding process. VNI's faster egress may be because of its greater flexibility as it is the only ligand studied that does not have a steroid ring system. It may also be due to its different interactions with the protein as it is more polar than the other ligands studied, which are very hydrophobic. In T. brucei CYP51, the product OBT_DM tends to exit quicker along tunnel 2 f than the substrate, OBT (Fig. 5). The lack of the 14-methyl group in OBT_DM may reduce steric hindrance to ligand egress through tunnel 2 f in T. brucei CYP51. On the other hand, the egress times for OBT_DM in human CYP51 tend to be slightly longer than for OBT. The wider opening of tunnel 2 f in human CYP51 than T. brucei CYP51 (Fig. S4) may mean, not only that ligand egress generally proceeds much faster, but also that ligand egress is less sensitive to the removal of a methyl group in the product.
The differences in ligand specificity between T. brucei CYP51 and human CYP51 may in part result from differences in the entrance residues of tunnel 2 f which are not conserved in the two proteins, as shown in Fig. 6. The secondary structure of the F-G loop region differs in the two proteins and the F-G loop region of T. brucei CYP51 is closer to the A’ helix region than the human CYP51 (Figs S4E and F). The entrance residues of tunnel 2 f are F48 (A’ helix), V77 (β1-2), P210, V213 and F214 (F” helix) in T. brucei CYP51 and F77 (A’ helix), F105 (β1-2), H236, W239 and L240 (F” helix) in human CYP51.
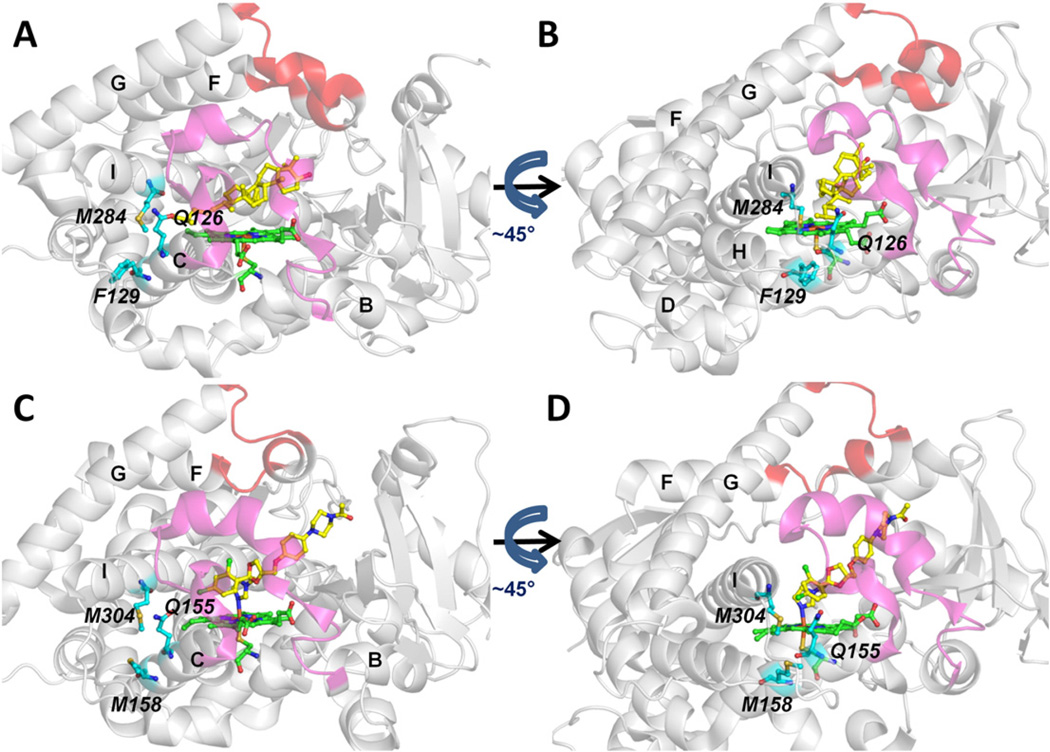
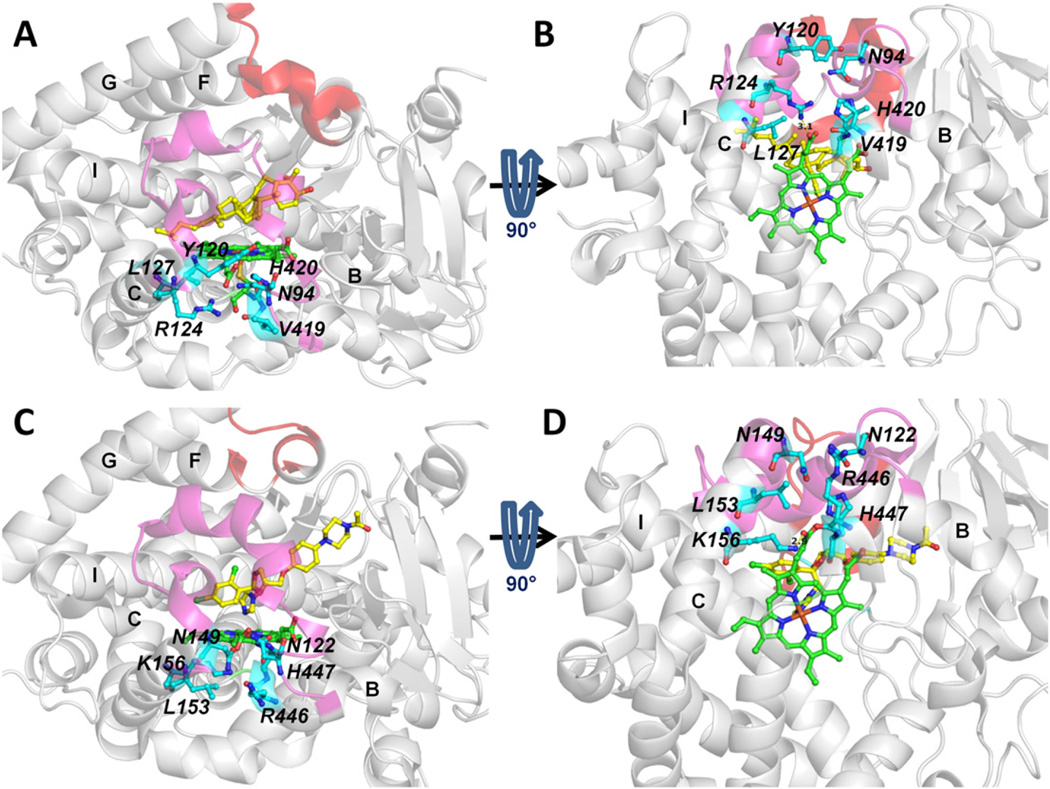
The entrance residues of tunnel 1 (Fig. 8) are better conserved than those of tunnel 2 f (Fig. 6). These residues are Q126, F129 and M284 in T. brucei CYP51 and Q155, M158 and M304 in human CYP51.When ligands exit from tunnel 1, both F129 in T. brucei CYP51 and M158 in human CYP51 need to undergo large side chain conformational changes to allow ligand passage via tunnel 1. However, conformational change of a flexible aliphatic side chain, such as M158 in human CYP51, can be expected to be easier than that of an aromatic sidechain, such as F129 in T. brucei CYP51. These differences in side-chain dynamics may result in ligand exit more often and with shorter egress times from tunnel 1 in human CYP51 than in T. brucei CYP51 (Figs. 3,4, S3 and S5). A clear difference between the four ligands studied in terms of ligand egress times or frequencies via tunnel 1 was not detected in either CYP51.
Fig. 8.
Comparison of the entrance residues of tunnel 1 of T. brucei CYP51 (PDB ID: 3P99) (A,B) and human CYP51 (PDB ID: 3LD6) (C,D). Representation as in Fig. 6.
The entrance residues of tunnel S are shown in Fig. 9. These residues are well conserved in T. brucei and human CYP51. However, tunnel S is used in T. brucei CYP51 but not in human CYP51. This difference is not due to the tunnel entrance residues but the relative openness of the tunnel. Tunnel S was found to be mostly closed in human CYP51 but to be open in T. brucei CYP51 in standard MD simulations as shown in ref. [8]. In T. brucei CYP51, egress was observed via the solvent channel more often in the membrane bound form. Moreover, it was distinctly more common for the product, OBT_DM, than for the substrate, OBT, and for the inhibitor VNI. This indicates that the solvent tunnel may act as a route for product to exit into the cytosol.
Fig. 9.
Comparison of the entrance residues of tunnel S of T. brucei CYP51 (PDB ID: 3P99) (A,B) and human CYP51 (PDB ID: 3LD6) (C,D). Representation as in Fig. 6.
Tunnel W was used in T. brucei CYP51 but is not considered as a ligand tunnel because tunnel W leads towards the interface between CYP51 and the cytochrome P450 reductase. Tunnel W is not conserved in T. brucei and human CYP51 (Fig. 10). Tunnel W was not used in human CYP51. The entrance residues of tunnel W in human CYP51 are more polar than those in T. brucei CYP51 (Fig. 10). This can make it more difficult for ligands of CYP51, which are normally hydrophobic ligands, to exit in human CYP51 than in T. brucei CYP51. In standard MD simulations, tunnel W was found to be open in T. brucei CYP51 and closed in human CYP51 [8]. Egress through tunnel W in T. brucei CYP51 tended to occur fastest for VNI, possibly because its greater flexibility facilitated manoeuvring past the heme and through the tunnel.
Fig. 10.
Comparison of the entrance residues of tunnel W of T. brucei CYP51 (PDB ID: 3P99) (A,B) and human CYP51 (PDB ID: 3LD6) (C,D). Representation as in Fig. 6.
3.5. Residues F105 and M460 gate the 2f and S tunnels in T. brucei CYP51
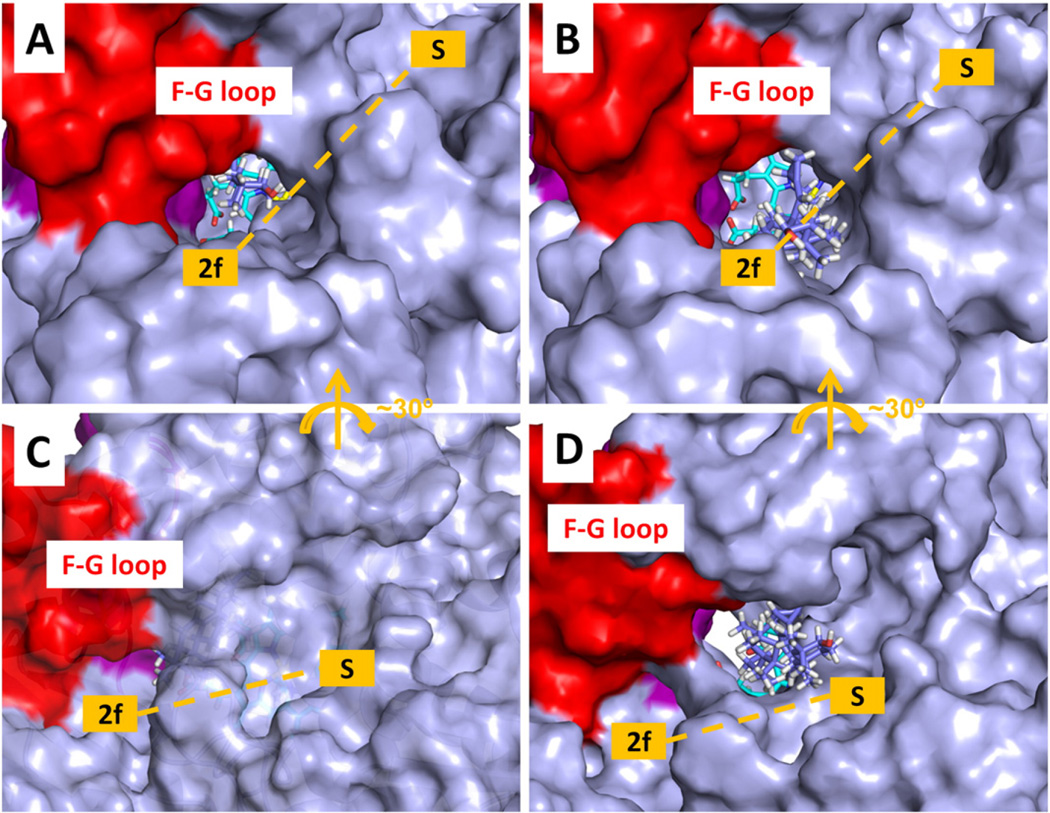
The tunnel through which ligand egress was most often observed in the RAMD simulations of T. brucei CYP51 was tunnel 2 f. Tunnel 2 f was open at the beginning of the RAMD simulations but not wide enough for the large ligands studied to exit. The closest tunnel to 2 f is tunnel S. Tunnel S was initially closed (Fig. 11 A and B). During ligand exit, tunnel 2 f opened wider or tunnel S opened from the closed conformation (Fig. 11 C and D) to allow exit of the ligands.
Fig. 11.
Tunnel opening during egress of the ligand MCP from T. brucei CYP51. The protein is shown with a solvent accessible molecular surface representation. The heme is shown in stick representation (cyan carbons) and the ligand MCP is also in stick representation (violet carbons). The F-G loop is shown in red and the B-C loop in purple. (A) First snapshot of a RAMD trajectory with tunnel 2 f open. (B) Snapshot from the same trajectory at 132 ps showing that tunnel 2 f has opened wider to allow the MCP to exit. (C) First snapshot of another RAMD trajectory with the tunnel S closed to a 1.4 Å radius probe. (D) Snapshot from the same trajectory at 902 ps showing that tunnel S has opened wide enough to allow MCP to exit. The white space below the heme cofactor shows the open tunnel W.
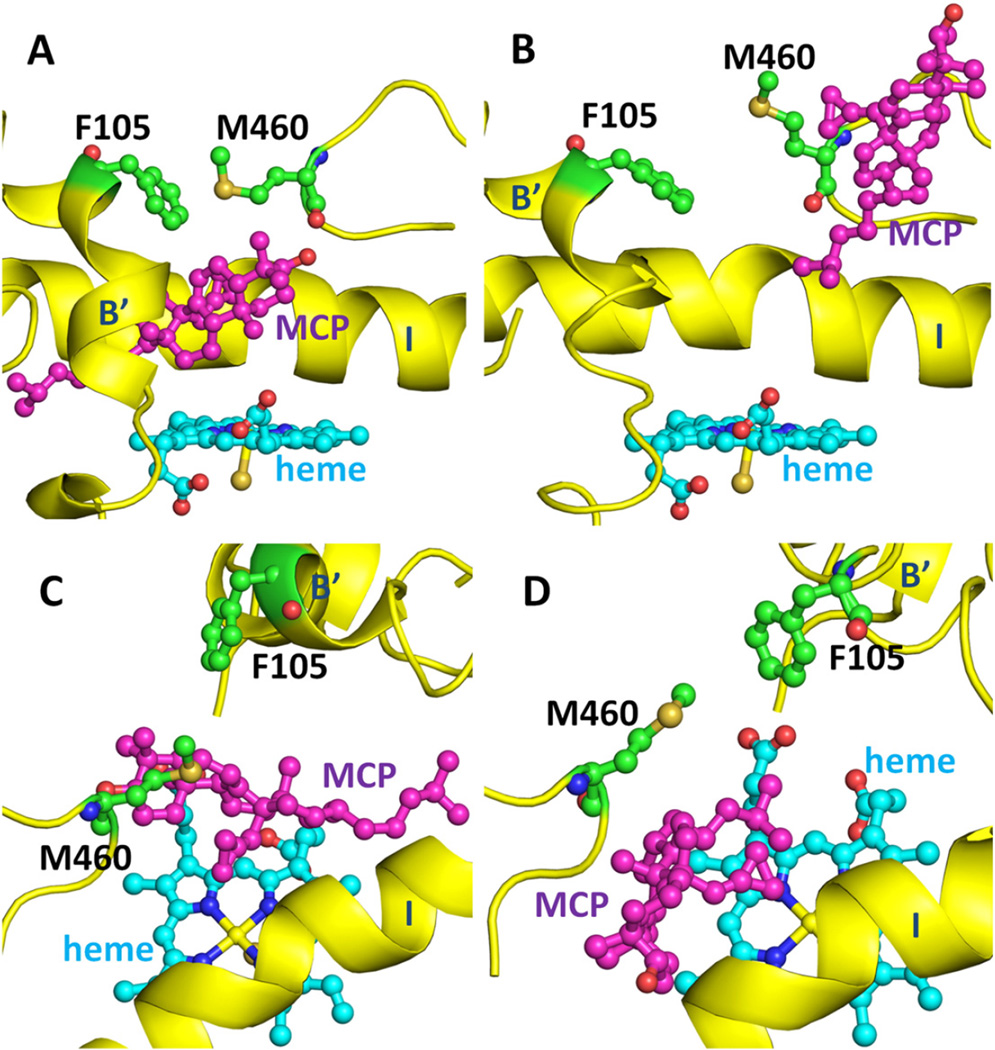
By analyzing the RAMD trajectories, we found two residues (F105 in the B’ helix and M460 in the β4 hairpin) that played an important role in the opening of these two tunnels in T. brucei CYP51 and can be considered the gating residues of these two tunnels. Long distances between these two residues correspond to open tunnels that can accommodate the entrance and exit of the ligands. They may help to keep the ligands in the active site during the catalytic reaction by interacting with each other, see Fig. 12.
Fig. 12.
Reorientation of the gating residues of tunnel 2 f and tunnel S, F105 and M460, in T. brucei CYP51 during egress of the ligand, MCP. The side chain orientations of F105 and M460 are shown before (A) and during (B) ligand exit via tunnel 2 f and before (C) and during (D) ligand exit via tunnel S. The secondary structure is shown in yellow and the heme in cyan. Different views of the corresponding snapshots with a surface representation of the protein are shown in Fig. S4A–D.
F105 is leucine in fungi and animals and isoleucine in T. cruzi and M460 is leucine in fungi [5]. In human CYP51, F105 is substituted by leucine (L134) whereas methionine is conserved at the M460 position (M487). Comparison of the I105F mutant with the wild-type T. cruzi CYP51 [51] showed that the mutation altered substrate preferences and that the catalytic rate of T. cruzi CYP51 for OBT increased 60-fold upon mutation to phenylalanine at position 105 to a rate close to that of T. brucei CYP51. Moreover, the catalytic rate for OBT was close to that of L. infantum and plant Sorgum bicolor CYP51s ((Fig. 6 of ref. (4)), both of which have phenylalanine at position 105, like all Leishmania and plant CYP51 orthologs [51], The fungal C. albicans CYP51 and the human CYP51, which both have leucine at position 105, have catalytic rates for OBT and other sterol substrates that are ca. 5 fold faster than T. brucei CYP51 (Fig. 6 of ref. [4]). Interestingly, the T. brucei F105L mutant shows a broader substrate binding profile than the wild-type, more similar to human and C. albicans CYP51 (GL, unpublished data). Residue 105 lines the active site and these data show its importance for interacting with and orienting ligands, and thereby distinguishing substrates based on their methylation at the C4 position. Our results suggest that residue 105 can further affect catalysis by gating of the active site and affecting ligand passage.
In the RAMD simulations, the distance between F105 and M460 increased sharply upon ligand egress through tunnel 2 f in T. brucei CYP51 whereas the corresponding distance between L134 and M487 in human CYP51 did not change upon ligand egress through tunnel 2 f (Fig. S6). This difference in gating may contribute to the quicker egress of ligands from human CYP51 than from T. brucei CYP51. In our RAMD simulations, the concerted movement of the side chains of F105 and M460 in T. brucei CYP51 can result in either tunnel 2 f or tunnel S opening for ligand passage (Fig. 9 B and D), depending on the directions of the side chain conformational changes. Because tunnel 2 f leads to the membrane and tunnel S leads to the solvent, these two gating residues can determine whether the ligands are released into the membrane or into the solvent.
4. Conclusions
The egress pathways of four ligands (two inhibitors, VNI and MCP, a substrate, OBT, and its product, OBT_DM) from the buried active site of T. brucei CYP51 and of OBT and OBT_DM from that of human CYP51 were studied with RAMD simulations. By simulating ligand egress, we find that the preferred exit route of the ligands is via tunnel 2 f in both soluble and membrane-bound forms of T. brucei CYP51 and in the soluble form of human CYP51. In the RAMD simulations of T. brucei CYP51, tunnels 1, S, and W are also observed to serve as egress routes for the ligands to the solvent but are much less frequently observed than tunnel 2 f. For human CYP51, ligand egress through tunnels S and W was not observed in any of the simulated trajectories. However, tunnel 1 was used more often in human CYP51 than in T. brucei CYP51, suggesting that tunnel 1 may serve as a ligand tunnel to the solvent in human CYP51. The use of tunnel 1 is supported by the crystal structure of Mycobacterium tuberculosis CYP51 which shows one ligand trapped in tunnel 1.
The tunnel entrance residues may contribute to the ligand specificity of T. brucei CYP51 and human CYP51. The substitution of F129 in T. brucei CYP51 by M158 in human CYP51 may facilitate ligand passage through tunnel 1 in human CYP51. In T. brucei CYP51, the binding site residues, F105 and M460, can act as gating residues of tunnel 2 f and tunnel S. They can not only modulate the opening of the two tunnels but also modulate whether the ligands should go to the membrane or the solvent. In human CYP51, F105 is substituted by a leucine which may affect access to tunnel S and reduce gating of ligand passage along tunnel 2 f.
Although the membrane resulted in a more open tunnel 2 f in T. brucei CYP51, the RAMD egress times tended to be longer than in the model of the soluble protein, possibly because the membrane slowed protein and ligand motion. In all the simulations of membrane-bound models of T. brucei CYP51, the ligands preferred tunnel 2 f as the exit tunnel and entered the membrane after leaving the protein. Therefore, in CYP51, the results suggest that the substrates come from the membrane and the products can go back to the membrane after the catalytic reaction, where they are available as substrates for other membrane-bound enzymes in the sterol biosynthetic pathway.
In previous standard MD simulations, we have observed similar flexibility of the T. brucei and human CYP51 enzymes in terms of computed B-factors [8]. Although some differences in tunnel opening were observed in standard simulations, the present RAMD simulations permitted a much more complete sampling of ligand tunnels. Despite the application of a small artificial force, our analysis shows that the differences observed between T. brucei and human CYP51 in RAMD simulations are related to differences in their sequences. The simulations described here thus map out the potential ligand egress routes and their differences between the T. brucei and human CYP51 and between the soluble and membrane-bound T. brucei CYP51. They provide a basis for future detailed calculations of the free energy barriers of the tunnel gates and the free energy profiles for ligand passage through the different tunnels. They could also be complemented by longer simulations to sample the long timescale conformational dynamics of the membrane-bound proteins. Despite its limitations, the combination of standard and RAMD simulations employed here provides new insights that contribute to understanding the ligand access and egress mechanisms of CYP51. The analysis of tunnel entrance residues show differences between T. brucei and human CYP51 which may help to guide the design of parasite-specific inhibitors.
Supplementary Material
Acknowledgements
We gratefully acknowledge the support of the Klaus Tschira Foundation. XY acknowledges the support of the Hartmut Hoffmann-Berling International Graduate School (HBIGS) of Molecular and Cellular Biology, Heidelberg University. VC acknowledges the support of the Max Planck Society. GM acknowledges the support of German Academic Exchange Service (DAAD). GIL acknowledges the support of the USA National Institutes of Health, grant GM067871.
Abbreviations
- CYP
cytochrome P450
- MCP
14α-methylenecyclopropyl-Δ7-24,25-dihydrolanosterol
- MD
molecular dynamics
- OBT
obtusifoliol
- OBT_DM
14α-demethylated obtusifoliol
- PDB
Protein data bank
- POPC
1-palmitoyl-2-oleoyl-sn-glycero-3-phosphocholine
- RAMD
random acceleration molecular dynamics
- VNI
N-[(1R)-1-(2,4-dichlorophenyl)-2-(1 H-imidazol-1-yl)ethyl]-4-(5-phenyl-1,3,4-oxadiazol-2-yl) benzamide
Footnotes
Supplementary data to this article can be found online at http://dx.doi.org/10.1016/j.bbagen.2015.10.015.
Transparency document
The Transparency document associated with this article can be found, in online version.
References
- 1.Lepesheva GI, Park H-W, Hargrove TY, Vanhollebeke B, Wawrzak Z, Harp JM, Sundaramoorthy M, Nes WD, Pays E, Chaudhuri M, Villalta F, Waterman MR. Crystal structures of trypanosoma brucei sterol 14alpha-demethylase and implications for selective treatment of human infections. J. Biol. Chem. 2010;285:1773–1780. doi: 10.1074/jbc.M109.067470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Villalta F, Dobish MC, Nde PN, Kleshchenko YY, Hargrove TY, Johnson CA, Waterman MR, Johnston JN, Lepesheva GI. VNI cures acute and chronic experimental chagas disease. J. Infect. Dis. 2013;208:504–511. doi: 10.1093/infdis/jit042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lepesheva GI, Hargrove TY, Anderson S, Kleshchenko Y, Furtak V, Wawrzak Z, Villalta F, Waterman MR. Structural insights into inhibition of sterol 14alpha-demethylase in the human pathogen Trypanosoma cruzi. J. Biol. Chem. 2010;285:25582–25590. doi: 10.1074/jbc.M110.133215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hargrove TY, Wawrzak Z, Liu J, Nes WD, Waterman MR, Lepesheva GI. Substrate preferences and catalytic parameters determined by structural characteristics of sterol 14alpha-demethylase (CYP51) from leishmania infantum. J. Biol. Chem. 2011;286:26838–26848. doi: 10.1074/jbc.M111.237099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hargrove TY, Wawrzak Z, Liu J, Waterman MR, Nes WD, Lepesheva GI. Structural complex of sterol 14α-demethylase (CYP51) with 14α-methylenecyclopropyl-Delta7-24, 25-dihydrolanosterol. J. Lipid Res. 2012;53:311–320. doi: 10.1194/jlr.M021865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Andriani G, Amata E, Beatty J, Clements Z, Coffey BJ, Courtemanche G, Devine W, Erath J, Juda CE, Wawrzak Z, Wood JT, Lepesheva GI, Rodriguez A, Pollastri MP. Antitrypanosomal lead discovery: identification of a ligand-efficient inhibitor of Trypanosoma cruzi CYP51 and parasite growth. J. Med. Chem. 2013;56:2556–2567. doi: 10.1021/jm400012e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lepesheva GI, Hargrove TY, Kleshchenko Y, Nes WD, Villalta F, Waterman MR. CYP51: a major drug target in the cytochrome P450 superfamily. Lipids. 2008;43:1117–1125. doi: 10.1007/s11745-008-3225-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yu X, Cojocaru V, Mustafa G, Salo-Ahen OMH, Lepesheva GI, Wade RC. Dynamics of CYP51: implications for function and inhibitor design. J. Mol. Recognit. 2015;28:59–73. doi: 10.1002/jmr.2412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Denisov IG, Shih AY, Sligar SG. Structural differences between soluble and membrane bound cytochrome P450s. J. Inorg. Biochem. 2012;108:150–158. doi: 10.1016/j.jinorgbio.2011.11.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cojocaru V, Winn PJ, Wade RC. The ins and outs of cytochrome P450s. Biochim. Biophys. Acta. 2007;1770:390–401. doi: 10.1016/j.bbagen.2006.07.005. [DOI] [PubMed] [Google Scholar]
- 11.Lüdemann SK, Lounnas V, Wade RC. How do substrates enter and products exit the buried active site of cytochrome P450cam? 1. Random expulsion molecular dynamics investigation of ligand access channels and mechanisms. J. Mol. Biol. 2000;303:797–811. doi: 10.1006/jmbi.2000.4154. [DOI] [PubMed] [Google Scholar]
- 12.Lüdemann SK, Lounnas V, Wade RC. How do substrates enter and products exit the buried active site of cytochrome P450cam? 2. Steered molecular dynamics and adiabatic mapping of substrate pathways. J. Mol. Biol. 2000;303:813–830. doi: 10.1006/jmbi.2000.4155. [DOI] [PubMed] [Google Scholar]
- 13.Carlsson P, Burendahl S, Nilsson L. Unbinding of retinoic acid from the retinoic acid random expulsion molecular dynamics. Biophys. J. 2006;91:3151–3161. doi: 10.1529/biophysj.106.082917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wang T, Duan Y. Chromophore channeling in the G-protein coupled receptor rhodopsin. J. Am. Chem. Soc. 2007;129:6970–6971. doi: 10.1021/ja0691977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Vashisth H, Abrams CF. Ligand escape pathways and (Un)binding free energy calculations for the hexameric insulin-phenol complex. Biophys. J. 2008;95:4193–4204. doi: 10.1529/biophysj.108.139675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Pavlova M, Klvana M, Prokop Z, Chaloupkova R, Banas P, Otyepka M, Wade RC, Tsuda M, Nagata Y, Damborsky J. Redesigning dehalogenase access tunnels as a strategy for degrading an anthropogenic substrate. Nat. Chem. Biol. 2009;5:727–733. doi: 10.1038/nchembio.205. [DOI] [PubMed] [Google Scholar]
- 17.Winn PJ, Lüdemann SK, Gauges R, Lounnas V, Wade RC. Comparison of the dynamics of substrate access channels in three cytochrome P450s reveals different opening mechanisms and a novel functional role for a buried arginine. Proc. Natl. Acad. Sci. U. S. A. 2002;99:5361–5366. doi: 10.1073/pnas.082522999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Schleinkofer K, Winn PJ, Lüdemann SK, Wade RC. Do mammalian cytochrome P450s show multiple ligand access pathways and ligand channelling? EMBO Rep. 2005;6:584–589. doi: 10.1038/sj.embor.7400420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yu X, Cojocaru V, Wade RC. Conformational diversity and ligand tunnels of mammalian cytochrome P450s. Biotechnol. Appl. Biochem. 2013;60:134–145. doi: 10.1002/bab.1074. [DOI] [PubMed] [Google Scholar]
- 20.Cojocaru V, Winn PJ, Wade RC. Multiple, ligand-dependent routes from the active site of cytochrome P450 2C9. Curr. Drug Metab. 2012;13:143–154. doi: 10.2174/138920012798918462. [DOI] [PubMed] [Google Scholar]
- 21.Wade RC, Winn PJ, Schlichting I. Sudarko, A survey of active site access channels in cytochromes P450. J. Inorg. Biochem. 2004;98:1175–1182. doi: 10.1016/j.jinorgbio.2004.02.007. [DOI] [PubMed] [Google Scholar]
- 22.Li W, Liu H, Scott EE, Gräter F, Halpert JR, Luo X, Shen J, Jiang H. Possible pathway(s) of testosterone egress from the active site of cytochrome P450 2B1: a steered molecular dynamics simulation. Drug Metab. Dispos. 2005;33:910–919. doi: 10.1124/dmd.105.004200. [DOI] [PubMed] [Google Scholar]
- 23.Li W, Liu H, Luo X, Zhu W, Tang Y. Possible pathway (s) of metyrapone egress from the active site of cytochrome P450 3A4: a molecular dynamics simulation. Drug Metab. Dispos. 2007;35:689–696. doi: 10.1124/dmd.106.014019. [DOI] [PubMed] [Google Scholar]
- 24.Shen Z, Cheng F, Xu Y, Fu J, Xiao W, Shen J, Liu G, Li W, Tang Y. Investigation of indazole unbinding pathways in CYP2E1 by molecular dynamics simulations. PLoS ONE. 2012;7:e33500. doi: 10.1371/journal.pone.0033500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sgrignani J, Magistrato A. Influence of the membrane lipophilic environment on the structure and on the substrate access/egress routes of the human aromatase enzyme. A computational study. J. Chem. Inf. Model. 2012;52:1595–1606. doi: 10.1021/ci300151h. [DOI] [PubMed] [Google Scholar]
- 26.Kingsley LJ, Lill MA. Including ligand-induced protein flexibility into protein tunnel prediction. J. Comput. Chem. 2014:1748–1756. doi: 10.1002/jcc.23680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kingsley LJ, Lill MA. Substrate tunnels in enzymes: Structure-function relationships and computational methodology. Proteins Struct. Funct. Bioinf. 2015:599–611. doi: 10.1002/prot.24772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.De Waal PW, Sunden KF, Furge LL. Molecular dynamics of CYP2D6 polymorphisms in the absence and presence of a mechanism-based inactivator reveals changes in local flexibility and dominant substrate access channels. PLoS ONE. 2014;9:e108607. doi: 10.1371/journal.pone.0108607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lepesheva GI, Nes WD, Zhou W, Hill GC, Waterman MR. CYP51 from trypanosoma brucei is obtusifoliol-specific. Biochemistry. 2004;43:10789–10799. doi: 10.1021/bi048967t. [DOI] [PubMed] [Google Scholar]
- 30.Cojocaru V, Balali-Mood K, Sansom MSP, Wade RC. Structure and dynamics of the membrane-bound cytochrome P450 2C9. PLoS Comput. Biol. 2011;7:e1002152. doi: 10.1371/journal.pcbi.1002152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Strushkevich N, Usanov SA, Park HW. Structural basis of human CYP51 inhibition by antifungal azoles. J. Mol. Biol. 2010;397:1067–1078. doi: 10.1016/j.jmb.2010.01.075. [DOI] [PubMed] [Google Scholar]
- 32.Jójárt B, Martinek TA. Performance of the general amber force field in modeling aqueous POPC membrane bilayers. J. Comput. Chem. 2007;28:2051–2058. doi: 10.1002/jcc.20748. [DOI] [PubMed] [Google Scholar]
- 33.Hornak V, Abel R, Okur A, Strockbine B, Roitberg A, Simmerling C. Comparison of multiple amber force fields and development of improved protein backbone parameters. Proteins. 2006;65:712–725. doi: 10.1002/prot.21123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Jorgensen WL, Chandrasekhar J, Madura JD, Impey RW, Klein ML. Comparison of simple potential functions for simulating liquid water. J. Chem. Phys. 1983;79:926. [Google Scholar]
- 35.Harris DL, Park J-Y, Gruenke L, Waskell L. Theoretical study of the ligand-CYP2B4 complexes: effect of structure on binding free energies and heme spin state. Proteins. 2004;55:895–914. doi: 10.1002/prot.20062. [DOI] [PubMed] [Google Scholar]
- 36.Dupradeau F-Y, Pigache A, Zaffran T, Savineau C, Lelong R, Grivel N, Lelong D, Rosanski W, Cieplak P. The R.E.D. tools: advances in RESP and ESP charge derivation and force field library building. Phys. Chem. Chem. Phys. 2010;12:7821–7839. doi: 10.1039/c0cp00111b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Vanquelef E, Simon S, Marquant G, Garcia E, Klimerak G, Delepine JC, Cieplak P, Dupradeau F-Y. R.E.D. Server: a web service for deriving RESP and ESP charges and building force field libraries for new molecules and molecular fragments. Nucleic Acids Res. 2011;39:W511–W517. doi: 10.1093/nar/gkr288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Frisch MJ, Trucks GW, Schlegel HB, Scuseria GE, Robb MA, Cheeseman JR, Scalmani G, Barone V, Mennucci B, Petersson GA, Nakatsuji H, Caricato M, Li X, Hratchian HP, Izmaylov AF, Bloino J, Zheng G, Sonnenberg JL, Hada M, et al. Gaussian 09. 2009 [Google Scholar]
- 39.Case DA, Darden TA, Cheatham TE, III, Simmerling CL, Wang J, Duke RE, Luo R, Walker RC, Zhang W, Merz KM, Reberts B, Hayik S, Roitberg A, Seabra G, Swails J, Goetz AW, Kolossvary I, Wong KF, Paesani F, et al. Amber 12. San Francisco: University of Califonia; 2012. [Google Scholar]
- 40.Case DA, Darden TA, Cheatham TE, III, Simmerling CL, Wang J, Duke RE, Luo R, Walker RC, Zhang W, Merz KM, Reberts B, Wang B, Hayik S, Roitberg A, Seabra G, Kolossvary I, Wong KM, Paesani F, Vanicek J, et al. Amber 11. San Francisco: University of Califonia; 2010. [Google Scholar]
- 41.Phillips JC, Braun R, Wang W, Gumbart J, Tajkhorshid E, Villa E, Chipot C, Skeel RD, Kalé L, Schulten K. Scalable molecular dynamics with NAMD. J. Comput. Chem. 2005;26:1781–1802. doi: 10.1002/jcc.20289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ryckaert J-P, Ciccotti G, Berendsen HJ. Numerical integration of the cartesian equations of motion of a system with constraints: molecular dynamics of n-alkanes. J. Comput. Phys. 1977;23:327–341. [Google Scholar]
- 43.Humphrey W, Dalke A, Schulten K. VMD: visual molecular dynamics. J. Mol. Graph. 1996;14:33–38. doi: 10.1016/0263-7855(96)00018-5. [DOI] [PubMed] [Google Scholar]
- 44.Schrodinger LLC. The PyMOL Molecular Graphics System. 2010 [Google Scholar]
- 45.Schrodinger LLC. Maestro, version 9.3. 2012 [Google Scholar]
- 46.Monk BC, Tomasiak TM, Keniya MV, Huschmann FU, Tyndall JD, O'Connell JD, Cannon RD, JG MD, Rodriguez A, Finer-Moore JS, Stroud RM. Architecture of a single membrane spanning cytochrome P450 suggests constraints that orient the catalytic domain relative to a bilayer. Proc. Natl. Acad. Sci. 2014;111:3865–3870. doi: 10.1073/pnas.1324245111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wade RC, Motiejunas D, Schleinkofer K, Sudarko WPJ, Banerjee A, Kariakin A, Jung C. Multiple molecular recognition mechanisms. Cytochrome P450–a case study. Biochim. Biophys. Acta. 2005;1754:239–244. doi: 10.1016/j.bbapap.2005.07.044. [DOI] [PubMed] [Google Scholar]
- 48.Berka K, Hendrychová T, Anzenbacher P, Otyepka M. Membrane position of ibuprofen agrees with suggested access path entrance to cytochrome P450 2C9 active site. J. Phys. Chem. A. 2011;115:11248–11255. doi: 10.1021/jp204488j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Hendrychova T, Berka K, Navratilova V, Anzenbacher P, Otyepka M. Dynamics and hydration of the active sites of mammalian cytochromes P450 probed by molecular dynamics simulations. Curr. Drug Metab. 2012;13:177–189. doi: 10.2174/138920012798918408. [DOI] [PubMed] [Google Scholar]
- 50.Eddine AN, von Kries JP, Podust MV, Warrier T, Kaufmann SHE, Podust LM. X-ray structure of 4,4′-dihydroxybenzophenone mimicking sterol substrate in the active site of sterol 14alpha-demethylase (CYP51) J. Biol. Chem. 2008;283:15152–15159. doi: 10.1074/jbc.M801145200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Lepesheva GI, Zaitseva NG, Nes WD, Zhou W, Arase M, Liu J, Hill GC, Waterman MR. CYP51 from Trypanosoma cruzi: a phyla-specific residue in the B’ helix defines substrate preferences of sterol 14alpha-demethylase. J. Biol. Chem. 2006;281:3577–3585. doi: 10.1074/jbc.M510317200. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.