Abstract
We outline structure–function contributions from our laboratories on protein–RNA recognition events that monitor siRNA length, 5′-phosphate and 2-nucleotide 3′ overhangs, as well as the architecture of Argonaute, its externally bound siRNA complex, and Argonaute-based models involving guide-strand-mediated mRNA binding, cleavage, and release.
Small RNAs mediate many gene regulatory events, including posttranscriptional gene silencing in plants, RNA interference (RNAi) in animals, the assembly and function of heterochromatin, as well as microRNA (miRNA) pathways at the genome level (for review, see Hannon 2002; Dykxhoorn et al. 2003; Bartel 2004; Baulcombe 2004; Meister and Tuschl 2004; Verdel et al. 2004; Filipowicz et al. 2005; Hammond 2005; Voinnet 2005; Zamore and Haley 2005; Kim and Nam 2006). These processes are collectively known as RNA silencing, which represents a conserved biological response to expressed double-stranded RNA molecules. Dicer-like enzymes (RNase III family nucleases) recognize and process these double-stranded RNAs (dsRNAs) into small interfering RNAs (siRNAs) (Bernstein et al. 2001), characterized by 21- to 25-nucleotide-long sequences (Hamilton and Baulcombe 1999; Hammond et al. 2000; Parrish et al. 2000; Zamore et al. 2000), composed of duplex (19–23 bp) and 2-nucleotide 3′-overhang segments, with 5′-phosphate and 3′-hydroxyl termini (Elbashir et al. 2001a, b). siRNAs mediate the silencing by guiding a nuclease complex, RNA-induced silencing complex (RISC), to recognize and cleave target mRNAs, with the targeting specificity dictated by base-pairing interaction between guide and target RNA strands (Hammond et al. 2000; Zamore et al. 2000; Elbashir et al. 2001a).
Structural biology approaches provide a unique perspective into the molecular events associated with specific recognition of siRNA by protein domains and the precise processing of mRNA targets mediated by Ago within the RISC (for review, see Collins and Cheng 2005; Filipowicz et al. 2005; Hall 2005; Lingel and Sattler 2005; Parker and Barford 2006; Song and Joshua-Tor 2006). We describe below the structure–function contributions from our laboratories on protein–RNA recognition events that monitor siRNA length, 5′-phosphate and 2-nucleotide 3′-overhangs, as well as the architecture of Argonaute (Ago) proteins, their externally bound siRNA complexes, and Ago-based models involving guide-strand-mediated mRNA binding, cleavage, and release. We conclude by putting our results into perspective by summarizing the structural results from other laboratories on dsRNA processing by Dicer and outlining future challenges associated with attempts to determine the structures of ternary protein–RNA complexes associated with three key steps in the biogenesis of miRNAs.
VIRAL SUPPRESSION THROUGH siRNA LENGTH MEASUREMENT
In plants, one of the major functions of RNA silencing is to act as a defense against invading viruses by sequence-specifically targeting their mRNAs for degradation in response to production of siRNAs from dsRNA replication intermediates (for review, see Baulcombe 2004; Wang and Metzlaff 2005). Many viruses, as a counter-defense against RNA silencing, encode proteins that can specifically inhibit the silencing machinery (for review, see Li and Ding 2001; Silhavy and Burgyan 2004; Voinnet 2005), including an inhibitor encoded by an animal virus (Li et al. 2002). This inhibition could occur through sequestration of the siRNA by the viral suppressor, and a molecular understanding of this process emerged following the successful crystal structure determination of the viral suppressor p19 (Silhavy et al. 2002; Lakatos et al. 2005) bound to siRNA residues from the Traci Tanaka-Hall (Vargason et al. 2003) and our (Ye et al. 2003) laboratories. Studies of viral suppressors are important for our understanding of RNA silencing mechanisms, given a recent result that a cellular miRNA mediates antiviral defense in human cells (Lecellier et al. 2005).
Viral Suppressor p19–siRNA Complex
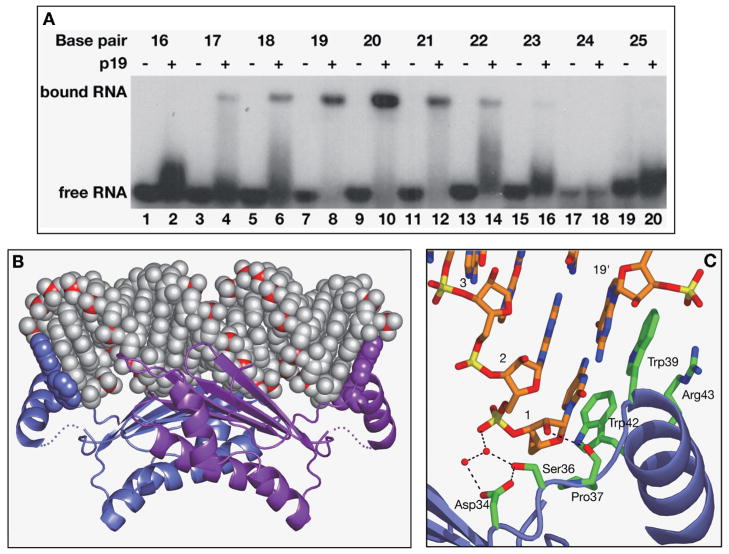
Our efforts first focused on defining the siRNA structural element(s) involved in p19 recognition. Electrophoretic mobility-shift assays (EMSA) established that p19 bound 21-mer siRNAs, but not their corresponding DNA counterparts of the same length and sequence. Furthermore, although the duplex characteristics of siRNA were a prerequisite for p19 recognition, the 2-nucleotide 3′ overhangs could be deleted with little impact on complex formation. A systematic study of p19 recognition of siRNA duplexes ranging in length from 16 bp to 25 bp established that optimal p19 binding spanned duplexes in the 19- to 21-bp length range, with binding attenuated for either shorter or longer lengths. These results established that the viral suppressor p19 measures the length of siRNAs to a resolution of 20 ±1 bp (Fig. 1A) (Ye et al. 2003).
Figure 1.
(A) Electrophoretic mobility-shift assays defining the length dependence of self-complementary siRNA duplexes for complex formation with viral suppressor p19. (B) The crystal structure of the TBSV p19–21-mer siRNA duplex (19-bp and 2-nucleotide 3′-overhang) (Ye et al. 2003). The RNA is shown in a space-filling representation with phosphorus atoms in red. The 2-nucleotide 3′-overhang bases are disordered and not shown. The protein is shown in a ribbon representation except for the pair of bracketing Trp residues at either end, which are shown in a space-filling representation. (C) Stick (RNA) and ribbon (protein) representation of stacking and hydrogen-bonding recognition of one RNA end in the complex. (Reprinted, with permission, from Ye et al. 2003 [Nature Publishing Group].)
The crystal structure of p19 from Tomato Bush Stunt virus (TBSV) in complex with a 21-nucleotide (19-bp) siRNA at 1.85 Å resolution is shown in Figure 1B (Ye et al. 2003). p19 forms a symmetric dimer in the complex where the RNA-binding folds of each monomer align to form an antiparallel eight-stranded β-sheet. The siRNA duplex, which adopts an A-helix conformation, is cradled within the concave face of this β-sheet platform, burying 1300 Å2 of solvent-accessible surface area.
Intermolecular protein–RNA contacts span the entire length of the electrostatically positive-charged surface of the p19 homodimer, thereby interacting with the central minor groove and two adjacent partial major grooves of the siRNA. The observed direct and water-mediated intermolecular contacts are restricted to the backbone phosphates and 2′-hydroxyl groups consistent with sequence-independent siRNA recognition by the p19 protein. The RNA minor-groove-binding face of p19 is rich in serine and threonine residues, thereby forming an unusual hydroxyl group network for hydrogen bonding with 2′-hydroxyl groups of the siRNA. DNA lacks 2′-hydroxyl groups, and this readily explains the ability of p19 to distinguish between siRNA and DNA counterpart duplexes.
The unique feature of p19 is highlighted by the presence of α-helical reading heads attached to the β-sheet-containing RNA-binding fold of each monomer. These α-helical reading heads project from opposite ends of the p19 homodimer, thereby positioning pairs of tryptophans that stack over the terminal base pairs; in essence, measuring and bracketing both ends of the siRNA duplex. The observed stacking of the 3′- and the 5′-terminal bases with W39 and universally conserved W42, respectively, essentially extends the terminal base pairs at either end by an additional step (Fig. 1C). This bracketing is further anchored by the stacking of the long hydrophobic side chain of conserved R43 over W39.
The peptide segments linking the α-helical reading heads to the β-sheet-containing RNA-binding fold are disordered in the crystal. Such structural plasticity could account for the ability of p19 to accommodate 19- to 21-mer siRNAs, its caliper-like action restricted to siRNAs spanning ± 1 bp in length.
Tanaka-Hall and colleagues focused their efforts on p19 from Carnation Italian Ringspot virus (CIRV) and have reported on the crystal structure of the complex of this p19 with a 21-mer siRNA (contains 19 bp) at 2.5 Å resolution (Vargason et al. 2003). There is excellent agreement between the structures of the TBSV (Ye et al. 2003) and CIRV (Vargason et al. 2003) p19–siRNA complexes, with both studies highlighting the principle that p19 acts as a molecular caliper for sequestering siRNAs based on their length.
The structural studies on the CIRV p19–siRNA complex were complemented by in vitro binding assays and in vivo suppression assays that monitored the impact of p19 mutations and siRNA variants on complex formation (Vargason et al. 2003). These studies established the importance of the bracketing residues W39 and W42, since substitution of both tryptophans simultaneously adversely affected complex formation. The binding constant was estimated to be 0.17 nM from EMSA-binding studies for CIRV p19 binding to a 21-mer siRNA containing a 5′-phosphate and a 2-nucleotide overhang at the 3′ end. Removal of the 5′-phosphate resulted in a 23-fold loss in binding, consistent with the 5′-phosphate forming a hydrogen bond with the indole NH of W42.
Viral Suppressor B2–dsRNA Complex
Recently, a crystal structure has also been reported for the complex of viral suppressor B2 from insect Flock House virus (FHV) and dsRNA at 2.6 Å resolution (Chao et al. 2005). FHV B2 suppressor has been shown to bind dsRNA independent of length with 1 nM affinity, but not dsDNA, single-stranded RNA (ssRNA), or DNA–RNA hybrids. The structure of the complex of FHV B2 with an 18-mer dsRNA has established that it does not use the unprecedented caliper-like principle highlighted above for the p19–siRNA complex (Vargason et al. 2003; Ye et al. 2003). Rather, B2 forms a dimer whose four-α-helix bundle topology binds to one face of the RNA duplex, thereby potentially inhibiting both cleavage of dsRNA by Dicer and incorporation of dsRNAs into RISC (Chao et al. 2005).
Viral Suppressor p21
One other viral suppressor, p21 (Chapman et al. 2004), has been characterized structurally in the free state, but not yet in its complex with siRNA. The crystal structure of the viral suppressor p21 from Beet Yellow virus (BSV) solved at 3.3 Å resolution in our laboratory exhibits an octameric ring architecture with a large central cavity of 90 Å in diameter (Ye and Patel 2005). Each monomer within the BSV p21 octamer is composed of amino- and carboxyterminal α-helical domains, which associate with their neighboring counterparts through symmetric head-to-head and tail-to-tail interactions. In vitro binding studies establish that p21 binds both ssRNA and dsRNA, independent of length. There is a putative RNA-binding surface composed of conserved, positive charged amino acids, which line the inner surface of the octameric ring (Ye and Patel 2005).
siRNA 3′-OVERHANG RECOGNITION BY THE PAZ DOMAIN
The PAZ domain is an RNA-binding module found in Ago and some Dicer proteins. Despite initial suggestions regarding the PAZ domain’s potential for mediating protein–protein interactions, it became clear as soon as its structure was determined and binding studies were undertaken that it is an RNA-binding module. Given this background, much effort has focused on elucidating the principles behind protein–RNA recognition involving the PAZ domain.
Architecture of the PAZ Domain
The structure of the PAZ domain has been determined in the free state and is composed of a central five-stranded β-barrel flanked by two α-helices, together with a flap-like segment composed of a β-hairpin and an α-helix (Lingel et al. 2003; Song et al. 2003; Yan et al. 2003). NMR chemical shift and mutation studies have mapped the nucleic-acid-binding region to a hydrophobic cleft containing conserved residues between its β-barrel core and αβ flap elements.
Structure of PAZ–siRNA–mimic-like Complex
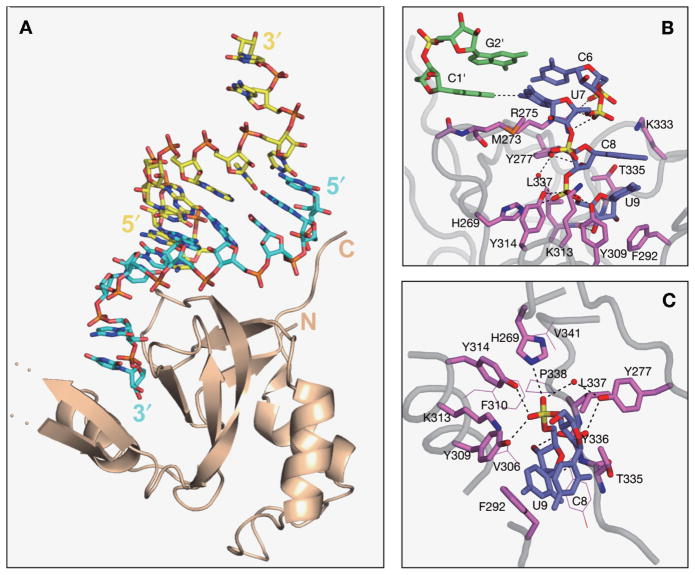
We have solved the crystal structure of the PAZ domain from human Argonaute1 (hAgo1) bound to both ends of a 9-mer siRNA-like duplex. The RNA forms a 7-bp duplex with 2-nucleotide 3′-overhangs at either end, which are targeted by independent non-interacting PAZ modules (Ma et al. 2004). The two strands of the siRNA-like duplex contact specific PAZ domains in a highly asymmetric manner, such that the 3′-overhang-containing strand interacts with the PAZ domain along its entire 9-nucleotide length, whereas the complementary strand’s interactions are restricted to its 5′-terminal residue (Fig. 2A). The bound RNA adopts a 5′ to 3′ stacked helical trajectory, except for a sharp clockwise 100° rotation in the phosphodiester backbone between the duplex and 2-nucleotide 3′-overhang segments (Fig. 2B), resulting in the insertion of the 2-nucleotide overhang into the preformed hydrophobic pocket between the central β-barrel and the αβ flap module.
Figure 2.
(A) The structure of one PAZ domain interacting with an siRNA-like end in the crystal structure of the human Ago1 PAZ-9-mer siRNA duplex (7-bp and 2-nucleotide 3′-overhang) (Ma et al. 2004). The complex is shown in stick (RNA) and ribbon (protein) representation. The RNA strand bound by its 3′-end is colored blue, whereas the strand bound by the 5′-end is colored yellow. (B) A stick representation of the interaction between the PAZ domain with the duplex terminus and 3′-overhang segments. Note the change in the RNA phosphodiester backbone between duplex and overhang segments. (C) Interactions between the 2-nucleotide 3′-overhang of the RNA and the walls of the conserved binding pocket on the PAZ domain. The residues directly contacting the RNA are shown in a stick representation, and other residues contributing to the hydrophobic pocket are shown as thin lines. Key hydrogen-bonding interactions from protein side chains to the internucleotide phosphate and the sugar hydroxyls are highlighted by dashed lines. (Reprinted, with permission, from Ma et al. 2004 [Nature Publishing Group].)
The two overhang nucleotides at the 3′-end retain a stacked A-helical conformation, with the terminal residue buried deep within the pocket. Strikingly, the internucleotide phosphate linking the overhang bases is targeted through hydrogen bonding by the hydroxyls of three tyrosines, one of which is water-mediated, and a histidine, all conserved residues (Fig. 2C) (Ma et al. 2004). The sugar ring of the terminal overhang residue is anchored in place through van der Waals packing, augmented by hydrogen bonds of its 2′- and 3′-OH groups with the amide backbone, while portions of its base and sugar stack over a conserved Phe ring. Mutations of the aromatic and hydrophobic residues that line the binding pocket and contact the terminal overhang base, sugar, and internucleotide linkage, adversely affect binding affinity. In contrast, the penultimate overhang base and sugar make fewer contacts with PAZ residues.
The stacked overhang bases are not involved in hydrogen bonding and face outward within an open pocket, suggesting that both pyrimidines and the larger purines can be equally well accommodated in a sequence-independent manner. The overhang-containing strand tracks the surface of the PAZ domain through electrostatic contacts between its phosphate backbone and five basic arginine and lysine side chains. In contrast, the base edges positioned within either groove of the RNA helix are not contacted by the PAZ domain, consistent with sequence-independent recognition.
The 2-nucleotide overhang bases contribute critically to binding, since removal of one or both overhang nucleotides results in 85-fold and >5000-fold reduction in binding affinity as measured by surface plasmon resonance. The binding pocket is incompatible with bulky 3′-end modifications, but accommodates a 2′-OCH3 with an 18-fold reduction in affinity. The PAZ domain also binds ssRNA, but with 50-fold reduction in binding affinity (Ma et al. 2004).
The above structural studies complemented by binding assays on the hAgo1 PAZ domain bound to a siRNA-like duplex (Ma et al. 2004) establish that PAZ could serve as an siRNA-end-binding module for siRNA transfer in the RNA silencing pathway, and as an anchoring site for the 3′-end of guide RNA within silencing effector complexes. The next challenge will be to solve the structure of Dicer PAZ bound to siRNA and identify similarities and differences with the known structure of the hAgo1 PAZ–siRNA complex.
Structure of PAZ-ssRNA Complex
The above studies on the crystal structure of hAgo1 PAZ–siRNA-like complex have been complemented by NMR structural studies of a uridine nucleotide bound to Drosophila melanogaster Ago1 (DmAgo1) PAZ (Yan et al. 2003) and a 5-mer ssRNA bound to DmAgo2 PAZ (Lingel et al. 2004). The terminal nucleotide binds in the hydrophobic binding pocket without sequence specificity. There are differences in detail between the stacked overhang bases in the crystal structure of the hAgo1 PAZ–siRNA-like complex (Ma et al. 2004) and the partially stacked overhang bases in the DmAgo2–ssRNA complex (Lingel et al. 2004), perhaps suggestive of conformational plasticity within the open binding pocket.
siRNA 5′-PHOSPHATE RECOGNITION BY THE Piwi PROTEIN
The Piwi protein from the archaen Archaeoglobus fulgidus (Af) has played a key role in our understanding of siRNA 5′-phosphate end recognition at the molecular level. The Mid and PIWI domains of the AfPiwi protein also constitute the carboxyterminal half of Ago proteins and hence structure–function studies of AfPiwi–siRNA complexes directly affect our functional understanding of guide-strand-mediated mRNA cleavage by Ago protein within RISC. Indeed, the 5′-phosphate end of siRNA is a prerequisite for both the stability and slicing fidelity during the processing steps within RISC (Rivas et al. 2005).
Architecture of a Bacterial Piwi Protein
The crystal structure of AfPiwi protein in the free state at 1.85 Å resolution established its two-domain architecture, with a substantial interface between its component Mid and PIWI domains (Parker et al. 2004). Strikingly, the PIWI domain adopted an RNase H scaffold, imparting the Piwi protein, as was first outlined for the Ago protein (Song et al. 2004), with potential mRNA cleavage or “slicer” activity. In addition, a basic pocket was identified within the Mid domain that was lined by pairs of conserved lysine and glutamine residues, supplemented by insertion of the carboxyterminal carboxylate, whose charge was neutralized by an accompanying divalent cation (Parker et al. 2004).
Structure of a Bacterial Piwi–siRNA Complex
Biochemical studies in Dm lysates have demonstrated that the 5′-phosphate on the guide RNA is essential for its assembly into RISC (Nykanen et al. 2001). 5′-Phosphorylated single-stranded siRNAs exhibit more potent gene silencing activity than their non-phosphorylated counterparts in HeLa cells (Martinez et al. 2002). In addition, the position of the cleavage site within the mRNA that is paired to the guide RNA in RISC is defined by the 5′-end of the guide RNA strand (Elbashir et al. 2001a,b). A major advance in the field occurred with the reporting of the crystal structures of AfPiwi protein bound to siRNA simultaneously from the David Barford laboratory (Parker et al. 2005) and our laboratory (Ma et al. 2005). These studies provided the structural basis for guide-strand 5′-end recognition and insights into guide-strand-mediated mRNA target cleavage.
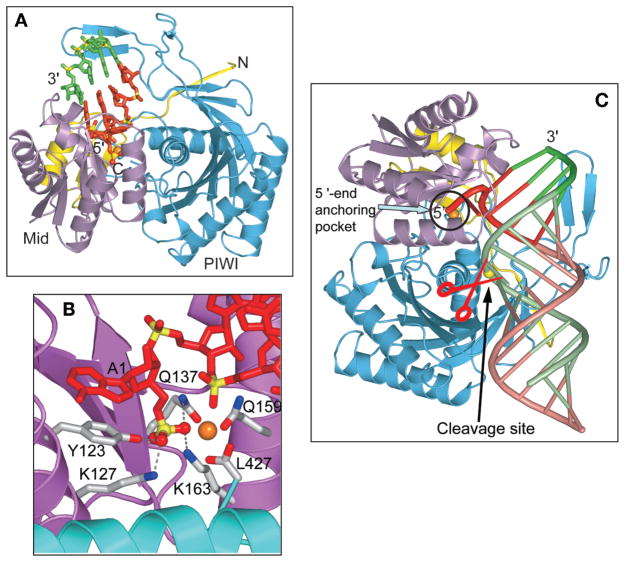
Our group has solved the 2.5 Å crystal structure of AfPiwi protein bound to a 5′-phosphate-containing 21-mer (19-bp) siRNA, capable of self-complementary duplex formation (Ma et al. 2005). We could trace the electron density for the Mid and PIWI domains of the Piwi protein, but we were restricted to a short 4-bp segment of the RNA duplex, positioned adjacent to the 5′-phosphate and its attached unpaired nucleotide (Fig. 3A). The identifiable RNA duplex segment is positioned within a basic channel spanning the Mid–PIWI inter-domain interface of the protein, whereas the 5′-phosphate inserts into the conserved basic pocket composed mainly of residues from the Mid domain and the carboxyterminal carboxylate of the PIWI domain. The 5′-phosphate is anchored within this pocket through a network of hydrogen bonds involving the side chains of Y123, K127, K163, and Q137, and a bound divalent cation (Fig. 3B). The divalent cation is octahedrally coordinated to two nucleic acid phosphate oxygens, three protein-based oxygens, and a water molecule in the complex. The similarities between the free (Parker et al. 2004) and siRNA-bound (Ma et al. 2005; Parker et al. 2005) AfPiwi protein structures is consistent with formation of a pre-formed 5′-phosphate-binding pocket.
Figure 3.
(A) The crystal structure of the A. fulgidus Piwi-21-mer siRNA duplex (19-bp and 2-nucleotide 3′-overhang) (Ma et al. 2005). The Piwi protein, shown in a ribbon representation, consists of a yellow-colored aminoterminal element (1–37), a magenta-colored Mid domain (38–167), and a cyan-colored PIWI domain (168–427). The segment of bound RNA that can be monitored in the structure is shown in a stick representation, with the protein interacting strand containing the 5′-phosphate colored red and its complementary partner strand colored green. (B) The 5′-phosphate-binding site in the complex. The 5′-phosphate is positioned in a basic pocket lined by K127, K163, Q137, and Q159 of the Mid domain and the carboxyterminal carboxylate from the PIWI domain, and a bound divalent cation in orange. Bases A1 and G2 are splayed apart, with unpaired A1 stacked on Y123. (C) Model of the A. fulgidus Piwi protein bound to a 5′-phosphate-containing siRNA duplex complex. The 5′-end nucleotide and 4-bp duplex (colored in red for the guide strand and green for the target strand) observed in the crystal structure of the complex were extended by an A-form duplex (colored tan for the guide strand and light green for the target strand). The 5′-end anchoring pocket and putative catalytic site are circled and labeled by an arrow, respectively, and the phosphate at the cleavage site is marked by a yellow ball. (Reprinted, with permission, from Ma et al. 2005 [Nature Publishing Group].)
We propose that it is the guide RNA strand whose phosphate is anchored within the basic pocket of the Piwi protein. The base (A1 in our case) at the 5′-end of the guide RNA strand is unpaired and stacks over the aromatic ring of invariant Y123 in the complex (Fig. 3B). The two paired RNA strands bind the Piwi protein in a strongly asymmetric manner, such that the 5′-end of the guide strand (colored red in Fig. 3A) interacts extensively with the protein, whereas minimal contacts are made with the mRNA strand (colored green, Fig. 3A).
The wild-type AfPiwi protein binds the siRNA with an apparent Kd of 0.19 μM as monitored from filter-binding studies in our laboratory. Mutations of K127, Q137, and K163 that hydrogen-bond to the 5′-phosphate and Y123 that stacks on the unpaired base adjacent to the 5′-phosphate, either one at a time or in pairs to alanine, result in less than an order of magnitude loss in binding affinity (Ma et al. 2005). The 5′-phosphate stabilization involves multiple hydrogen-bonding interactions with conserved residues that line the binding pocket, and it appears that disruption of either single or pairs of interactions result in a modest decrease in binding affinity.
5′-End Specific Recognition of Guide Strand
A significant portion of the bound 21-mer is disordered in the crystal structure of the complex. We therefore constructed a model in which the trajectory of the 4-bp duplex was extended to form two turns of an A-form helix that spans one face of both the Mid and PIWI domains (Fig. 3C). In this model, the proposed mRNA cleavage site, between positions 10 and 11 as defined from the 5′-end of the guide strand (Elbashir et al. 2001a,b), is positioned opposite the putative catalytic residues associated with the RNAse H fold of the PIWI domain.
We next used sequence alignments to identify the corresponding amino acids that would line the 5′-end-binding pocket of hAgo2, the RISC component that mediates guide-strand-mediated cleavage of target mRNA (Liu et al. 2004; Meister et al. 2004). This allowed us to investigate whether mutation of key residues lining the 5′-end-binding pocket would affect hAgo2 mRNA cleavage activity. Although efficient cleavage of radiolabeled target mRNA was observed for wild-type hAgo2, single alanine mutants of the hAgo2-binding pocket residues K533, Q545, and K570 showed reduced cleavage activity, whereas double alanine mutants showed more severe defects in cleavage capacity (Ma et al. 2005). These results establish that the 5′-end-binding pocket identified in AfPiwi protein is conserved in hAgo2 and is critical for its small RNA-guided cleavage activity.
Given that there are no specific intermolecular contacts directed to the 2′-OH groups of the RNA in the crystal structure of the complex, it appears likely that the AfPiwi protein may also target other types of nucleic acids. Surprisingly, the results of double filter-binding studies establish that the binding is 20-fold tighter for ssDNA than for ssRNA, with binding affinity decreasing on proceeding from DNA–DNA and RNA–DNA to RNA–RNA duplexes (Ma et al. 2005). Related assays also established that 5′-phosphate-containing RNA binds the AfPiwi protein an order of magnitude more tightly than its non-phosphorylated counterpart.
The structure of the complex revealed that the stacked bases 2 to 5 at the 5′-end of the guide RNA are accessible for pairing with their complementary counterparts on the mRNA. Biochemical and bioinformatics analyses of miRNA genes have indicated a high degree of sequence conservation restricted to positions 2–8, indicative of a significant contribution of the 5′-end to activity and specificity (Doench and Sharp 2004; Mallory et al. 2004; Lewis et al. 2005). This concept receives support from the structure of our complex, in which the mRNA initially nucleates with the 5′-end of the protein-bound guide RNA strand (Stark et al. 2003; Haley and Zamore 2004), followed by the remaining segment zippering-up to form the guide RNA–mRNA duplex. Our observation that the first base of the guide RNA strand is not available for pairing with the mRNA in the structure of the complex is consistent with previously reported non-sequence-specific recognition of the first nucleotide within the 5′-end-binding pocket (Stark et al. 2003; Doench and Sharp 2004; Lewis et al. 2005) and that disruption of this pair has no effect on cleavage activity, and under certain conditions can even facilitate target cleavage (Haley and Zamore 2004).
The related crystal structure of the AfPiwi protein bound to a 16-nucleotide siRNA was solved at 2.2 Å resolution (Parker et al. 2005), simultaneously with our contribution (Ma et al. 2005). This group could monitor 8 bp of duplex, together with the 5′-phosphate and its adjacent unstacked base positioned within the binding pocket. In addition, they could also follow the 2-nucleotide overhang at the 3′-end, which was shown to pass through a short exit channel. In essence, both groups have observed similar structures for the complexes and reached similar conclusions related to their functional impact. In summary, the structures of the AfPiwi protein–siRNA complexes (Ma et al. 2005; Parker et al. 2005) have provided the first molecular insights into 5′-end recognition of the guide RNA strand, aspects of the nucleation step associated with guide RNA–mRNA recognition, and the basis for distance-based identification of the mRNA cleavage site. Most importantly, perturbation of the 5′-end-binding pocket affects on mRNA cleavage efficiency (Ma et al. 2005).
ARGONAUTE—THE CATALYTIC ENGINE WITHIN RISC
RISC plays a key role in modulating eukaryotic gene expression through either chromatin remodeling, sequence-specific mRNA cleavage, or translational repression (for review, see Meister and Tuschl 2004; Sontheimer 2005; Tomari and Zamore 2005). RISC-associated Ago proteins (Cerutti et al. 2000; Hammond et al. 2001; Carmell et al. 2002), with their PAZ and PIWI domains, provide both architectural and catalytic functionalities associated with this scaffold serving as a platform for siRNA guide-strand selection and subsequent guide-strand-mediated cleavage of complementary mRNAs (Hutvanger and Zamore 2002; Martinez et al. 2002).
Architecture of Bacterial Argonautes
Our initial understanding of Ago architecture emerged following the report of the 2.25 Å crystal structure of Pyrococcus furiosus Ago (PfAgo) in the free state (Song et al. 2004). In contrast to the AfPiwi protein that is composed of only two domains, the archaeal PfAgo is composed of four domains labeled N, PAZ, Mid, and PIWI, with the PIWI domain shown to adopt a RNase H fold composed of a conserved DD-containing catalytic motif with potential for target RNA cleavage activity. Subsequently, our group reported the 2.9 Å crystal structure of Aquifex aeolicus Ago (AaAgo) and showed that the eubacterial AaAgo adopts a bilobal architecture composed of PAZ-containing (PAZ and N domains and linkers L1 and L2) and PIWI-containing (Mid and PIWI domains) lobes (Fig. 4A) (Yuan et al. 2005).
Figure 4.
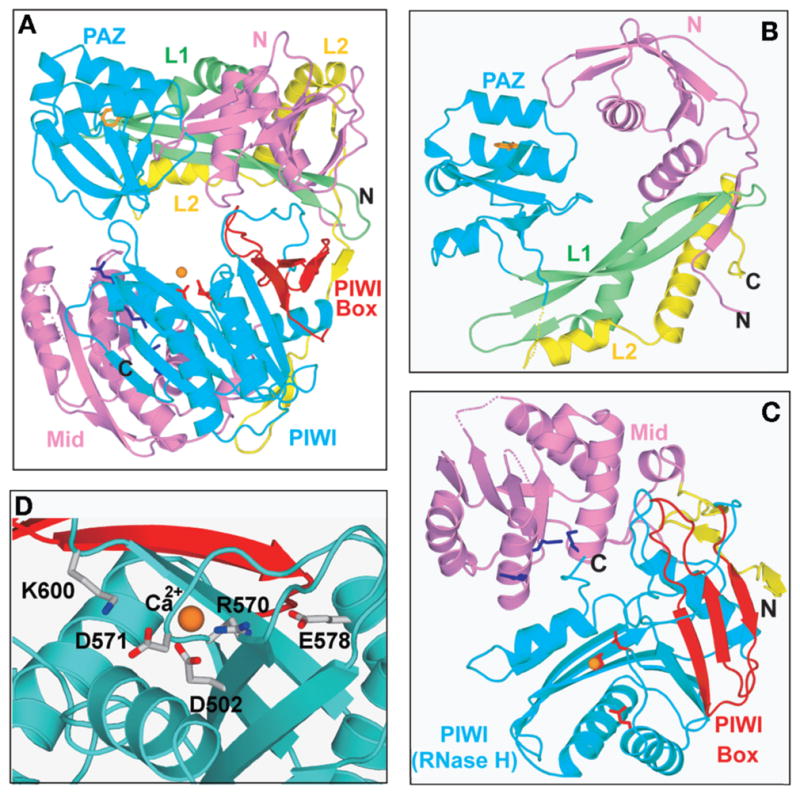
The crystal structure of A. aeolicus Argonaute (Yuan et al. 2005). The protein was crystallized in the presence of rU8, but the RNA was disordered and could not be traced. The view emphasizes the bilobal topology of Ago, with the aminoterminal PAZ-containing lobe on top and the carboxyterminal PIWI-containing lobe on the bottom. The various domains and linkers are numbered and color-coded. (B) Relative arrangements of the magenta-colored N (1–108), green-colored linker L1 (108–165), cyan-colored PAZ (166–262), and segment of yellow-colored linker L2 (263–311) within the PAZ-containing lobe. A Trp ring lining the PAZ-binding pocket is colored orange. (C) Relative arrangements of segment of the yellow-colored linker L2 (312–334), magenta-colored Mid domain (335–488), and cyan-colored PIWI domain (489–706) within the PIWI-containing lobe. Lys residues lining the 5′-phosphate- binding pocket on the Mid domain are colored dark blue. The DDE triad residues lining the catalytic binding pocket on the PIWI domain are colored red. The PIWI box (622–650) is colored red. (D) Relative positioning of invariant catalytic acidic D502, D571, and E578 residues and bound Ca cation on the surface of the RNase H fold of the PIWI domain. Invariant acidic R570 is also positioned in the catalytic pocket, whereas conserved basic K600 is directed toward the catalytic pocket. The Ca cation is also coordinated by D683, which is a His residue in hAgo2. (Reprinted, with permission, from Yuan et al. 2005 [© Elsevier].)
The N, L1, and PAZ domains within the PAZ-containing lobe of AaAgo are placed in a triangular arrangement (Fig. 4B), with each domain interacting with the other two domains. The linker L1 and L2 are in close proximity and, together with the N and PAZ domains, form a compact globular fold. The PAZ domain within the AaAgo context contains the same nucleic-acid-binding pocket as reported previously for isolated PAZ domains (Lingel et al. 2003, 2004; Song et al. 2003; Yan et al. 2003) and its siRNA complex (Ma et al. 2004). The AaAgo PAZ pocket is lined by a similarly spatially positioned cluster of hydrophobic and aromatic amino acids, implying a similar functional role in binding the 3′-end and in orienting the backbone of the guide RNA. One surface of the PAZ-containing lobe, the one directed toward the PIWI-containing lobe, exhibits a pronounced basic electrostatic surface.
The Mid and PIWI domains within the PIWI-containing lobe of AaAgo interact with each other through an extensive interface (Fig. 4C). This lobe contains the 5′-phosphate recognition pocket identified earlier for the AfPiwi protein (Parker et al. 2004) and its siRNA complexes (Parker et al. 2004; Ma et al. 2005). As reported previously for AfPiwi, this pocket in AaAgo is also positioned near the interface between the Mid and PIWI domains and is lined by conserved aromatic and basic residues positioned on the surface of the Mid domain and the inserted carboxyterminal carboxylate from the PIWI domain. The PIWI domain of AaAgo adopts an RNase H fold containing a catalytic DD-containing motif, with spatial characteristics similar to that which emerged from the crystal structures of PfAgo (Song et al. 2004) and AfPiwi (Parker et al. 2004). The RNase-H-like scaffold of the PIWI domain of AaAgo projects highly conserved (D502, D571, and E578) and nonconserved (D683) acidic residues, a basic residue (R570) and a bound Ca2+ cation (Fig. 4D). The spatial organization of the DD-containing motif projecting from the RNase H scaffold of the AaAgo PIWI-containing lobe has similarities with the corresponding DDE-containing RNase H motifs in RNase HII (Lai et al. 2000), as well as related motifs in integrases, reverse transcriptases, and transposases (Yang and Steitz 1995). The AaAgo PIWI-containing lobe also contains a PIWI box (Fig. 4A) (Cerutti et al. 2000), composed of three β-strands, and is positioned close to a pivot point linking the two lobes, and given its surface position, is available for recognition by Dicer (Tahbaz et al. 2004). One surface of the PIWI-containing lobe, the one directed toward the PAZ-containing lobe, exhibits a pronounced basic electrostatic surface, with its most basic segment lining the periphery of the composite 5′-phosphate-binding pocket.
Comparison of the PfAgo (Song et al. 2004) and AaAgo (Yuan et al. 2005) crystal structures highlights some differences in architecture and also perspectives as to relative alignments of individual domains. Thus, the AaAgo PAZ domain adopts a “closed” architecture with an α-helical pair subdomain positioned over the central β-barrel (Yuan et al. 2005), whereas the PfAgo PAZ domain adopts an “open” architecture, with the α-helical pair subdomain flipped out of the central β-barrel (Song et al. 2004). The AaAgo structure has a narrow channel between the N and PAZ domains and a wide channel between the PAZ and PIWI domains (Yuan et al. 2005), with the opposite trend observed in the PfAgo structure (Song et al. 2004). Finally, the AaAgo structure adopts a bilobal architecture defined by PAZ-containing (N and PAZ domains) and PIWI-containing (Mid and PIWI domains) lobes connected by a short hinge segment (Fig. 4A) (Yuan et al. 2005). In contrast, the PfAgo structure adopts an architecture where the PAZ domain is positioned over a crescent-shaped scaffold composed of the remaining three domains (Song et al. 2004). These contrasting architectures identify different linkers and hinge elements as potential contributors to inter-domain flexibility.
Bacterial Argonautes as Site-specific DNA-guided Endoribonucleases
The binding affinity and specificity of AaAgo for nucleic acids was tested in our laboratory using a double filter-binding assay (Wong and Lohman 1993). Unexpectedly, AaAgo bound most tightly to a 21-mer ssDNA (0.01 μM), with a tenfold reduction on binding to the corresponding 21-mer ssRNA. AaAgo also bound more tightly to a DNA–RNA hybrid duplex (0.64 μM) compared to a dsRNA duplex (>10 μM) (Yuan et al. 2005). Our laboratory also measured the same trend in binding affinities with PfAgo and Thermus thermophilus (Tth) Ago (J.-B. Ma et al., unpubl.).
The catalytic cleavage activity of AaAgo for radiolabeled target mRNA was tested in our laboratory as a function of 5′-phosphorylated guide DNA and RNA strands of varying length (18, 21, and 24 nucleotides), divalent ions (Ca, Mg, Mn) and temperature (35°C and 55°C). Cleavage by thermophilic AaAgo was most efficient for guide DNA strands (independent of length), in the presence of Mn as divalent cation, and at elevated temperatures (55°C) (Yuan et al. 2005). These unanticipated findings open new avenues for future investigation of the role of guide DNAs in mediating the functional activities of archaeal and eubacterial Ago proteins.
The number of divalent cations required for Ago-mediated slicing activity (Schwarz et al. 2004) needs further investigation. The catalytic triad residues are coordinated to a divalent ion in the AaAgo structure (Yuan et al. 2005), and Mn can be incorporated into the same site following soaking of this divalent cation into crystals of PfAgo (Rivas et al. 2005). One anticipates that the “slicer” activity of Ago is associated with a catalytic triad involving two coordinated divalent cations, similar to what has been recently observed for RNase H complexes with a DNA/RNA hybrid (Nowotny et al. 2005). Perhaps the second divalent cation accompanies the bound nucleic acid and will only be observable structurally in Ago complexes containing bound nucleic acid.
Although it was previously proposed that the guide strand was selectively loaded onto Ago within RISC (Schwarz et al. 2003), more recent research is consistent with Ago receiving the siRNA duplex (Matranga et al. 2005; Rand et al. 2005), and using either its catalytic potential to cleave the passenger strand or to expel the passenger strand with an RNA helicase action in case the Ago protein has no intrinsic cleavage activity.
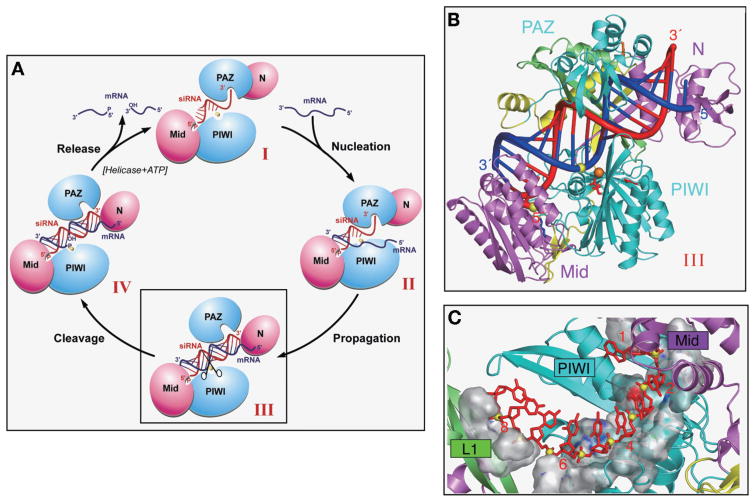
Model of Guide Strand-mediated mRNA Binding, Cleavage, and Release
We have proposed a four-step catalytic cycle model of guide-strand-mediated mRNA binding, cleavage, and release within the context of the Ago scaffold within RISC (Fig. 5A) (Yuan et al. 2005), with similar perspectives into the mechanism of guide-strand-mediated mRNA cleavage also outlined in published reviews on RNA interference (Filipowicz 2005; Tomari and Zamore 2005). Conformer I, which corresponds to guide-strand-bound Ago, involves anchoring of both the 5′-phosphate and 2-nucleotide 3′-overhang ends of the guide strand in the basic pocket of the Mid domain and the aromatic-lined pocket of the PAZ domain, respectively. The bases are restricted to a stacked helical conformation toward the 5′-end due to constraints imposed by the binding channel, and their Watson-Crick edges are available for pairing with the target strand. Conformer II, which corresponds to the nucleation step, involves annealing of the mRNA with the accessible Watson-Crick base edges of the 5′-end of the guide strand, thereby maximizing pairing interactions spanning residues 2–8 of the guide strand. Conformer III, which corresponds to the propagation step, involves zippering-up of the mRNA to form a full-length bound guide strand–mRNA duplex. It is anticipated that steric constraints associated with this conformer will require release of the 2-nucleotide 3′-overhang from its binding pocket within the PAZ domain. Conformer IV, which corresponds to the cleavage step, involves cleavage of the mRNA phosphodiester bond between residues 10 and 11, as measured from the 5′-end of the guide strand, by the precisely positioned catalytic DD-containing residues of the RNase H motif of the PIWI domain. Release of mRNA fragments occurs on transition from conformer IV back to conformer I, perhaps facilitated by ATP-dependent RNA helicases. The catalytic cleavage models (Filipowicz 2005; Tomari and Zamore 2005; Yuan et al. 2005) incorporate concepts that have emerged from structural studies of PAZ–siRNA (Ma et al. 2004) and Piwi–siRNA (Ma et al. 2005; Parker et al. 2005) complexes, as well as ideas that relate to preorganization of the 5′-end (Haley and Zamore 2004; Martinez and Tuschl 2004) and concepts that have emerged from miRNA target predictions (Lewis et al. 2003, 2005; Mallory et al. 2004), together with functional (Doench et al. 2003; Doench and Sharp 2004) and kinetic (Haley and Zamore 2004) studies on the contribution of 3′ bases of small RNAs to catalytic rate.
Figure 5.
(A) Schematic of the catalytic cycle involving guide-RNA-dictated mRNA loading, cleavage, and product release within the context of the Ago scaffold (Yuan et al. 2005). The states represented by conformers I, II, III, and IV are described in the text. The mRNA nucleation step corresponds to the transition from conformer I to II, the mRNA propagation step corresponds to the transition from conformer II to III, the mRNA cleavage step corresponds to the transition from conformer III to IV, and the product release step corresponds to the transition from conformer IV back to conformer I. (B) A view of the model of the AaAgo–DNA/RNA hybrid complex. The orientation of Ago is rotated along the vertical axis relative to the one shown in Fig. 4A. The color coding and labeling of domains and linkers, as well as key amino acids, are the same as listed in Fig. 4A. The hybrid duplex between the guide DNA strand (colored red) and the mRNA strand (colored blue) is shown in a tubular representation, with a thicker diameter for the sugar-phosphate backbone and thinner diameter for the bases. The cleavable phosphate positioned between residues 10 and 11 on the mRNA strand (as counted from the 5′-end of the guide strand) is shown by a yellow ball. (C) The phosphodiester backbone corresponding to positions 2–8 from the 5′-end of the guide strand are positioned within a trough-like segment of the Mid and PIWI domains in the model of the complex. The guide strand is shown in red, with phosphorus atoms as yellow balls. The trough is shown in a surface representation and exhibits surface complementarity with the sugar-phosphate backbone of the 5′-end region of the guide strand. (Reprinted, with permission, from Yuan et al. 2005 [© Elsevier].)
Sterochemically Robust Model of AaAgo Bound to Guide DNA–mRNA
Our modeling efforts have focused on conformer III of the AaAgo complex with a bound duplex composed of a guide DNA and mRNA strands (Yuan et al. 2005), given the findings of the double filter-binding and cleavage assays. The guide DNA–mRNA duplex was successively docked into a stereochemically compatible basic channel within the AaAgo scaffold by anchoring the 5′-phosphate of the guide DNA strand within the 5′-end recognition pocket of the Mid domain, while positioning the scissible mRNA phosphate near the Ca-coordinated DD-containing motif of the RNase H fold of the PIWI domain, with maintenance of the DNA–RNA duplex in a standard helical form. Steric clashes could be relieved by interactive modeling and required repositioning and reorientation of L1 linker and the PAZ domain. A stereochemically robust model of the complex was obtained following molecular dynamics calculations. A view of the model of conformer III is shown in Figure 5B, with the guide DNA and mRNA strands colored red and blue, respectively, positioned between the basic electrostatic surfaces of the mutually facing lobes of the AaAgo bilobal scaffold (Yuan et al. 2005). The phosphodiester backbone corresponding to positions 2–8 from the 5′-end of the guide strand are positioned in a stacked alignment within a trough-like segment of the Mid and PIWI domains (Fig. 5C), with their Watson-Crick edges available for recognition with complementary residues of the mRNA strand.
Structure of AaAgo with Externally Bound siRNA
Considerable effort has gone into attempts at generating complexes of AaAgo with single- and double-stranded nucleic acids containing RNA and/or DNA strands for structural characterization of one or more of the conformers in the proposed catalytic cycle outlined in Figure 5A. These efforts have not succeeded to date but, unexpectedly, have resulted in the crystallographic characterization of AaAgo with an externally bound siRNA. Our group has solved the 3.0 Å crystal structures of 22-mer and 26-mer siRNAs bound to AaAgo, where one 2-nucleotide 3′-overhang of the siRNA inserts into a cavity positioned on the outer surface of the PAZ-containing lobe of the bilobal AaAgo architecture in both complexes (Fig. 6A) (Yuan et al. 2006). The first overhang nucleotide stacks over a tyrosine ring of Y119 (Fig. 6B), whereas the second overhang nucleotide, together with the intervening sugar-phosphate backbone, inserts into a preformed surface cavity (Figs. 6B, C).
Figure 6.
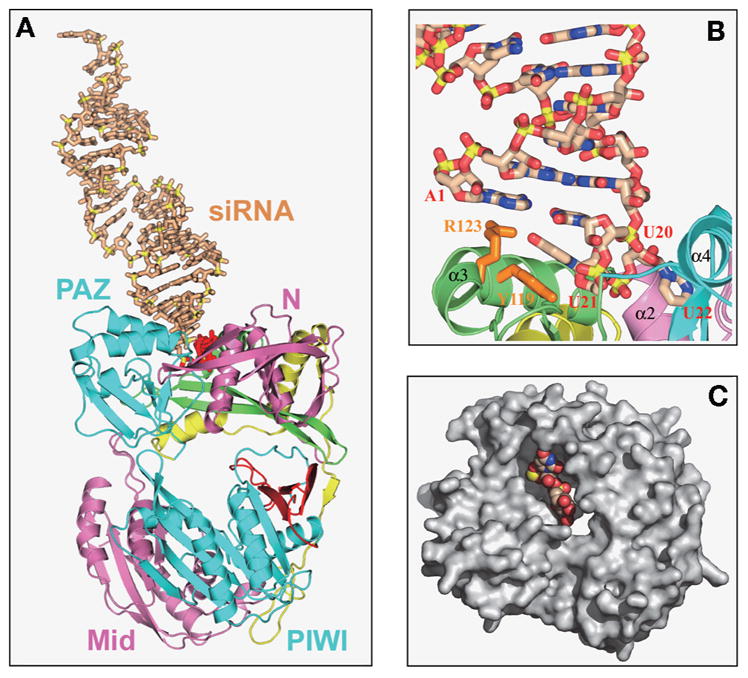
(A) The crystal structure of A. aeolicus Ago bound externally to a 22-mer siRNA (20-bp and 2-nucleotide 3′-overhang) (Yuan et al. 2006). The Ago protein is shown in a ribbon representation with the color coding and labeling of domains and linkers the same as listed in Fig. 4A. The externally bound siRNA, shown in a stick representation, is shown in beige with backbone phosphorus atoms in yellow, except for the 2-nucleotide overhang at the bound end, which is colored red. (B) A view of interactions between the 2-nucleotide overhang at one end of the externally bound 22-mer siRNA and the cavity positioned on the outer surface of the PAZ-containing lobe of AaAgo. The overhang base U21 stacks on the aromatic ring of orange-colored Y119, whereas the overhang base U22 is inserted into a cavity whose walls involve segments of N domain, linker L1, and the PAZ domain. (C) Surface representation highlighting the cavity located on the outward-pointing face of the PAZ-containing lobe of the siRNA-bound AaAgo. The bound 2-nucleotide 3′-overhang is shown in a space-filling representation. (Reprinted, with permission, from Yuan et al. 2006 [© Elsevier].)
Photochemical cross-linking experiments on complexes of 5-iodouridine-labeled siRNA in our laboratory provide support for this externally bound AaAgo–siRNA complex (Yuan et al. 2006). Specifically, cross-linking was only observed when 5-iodouridine was incorporated at the penultimate 3′-overhang position, but not at the 5′-end or in the center of the siRNA duplex, with the extent of cross-linking strongly attenuated for the Y119A mutant. The structure and biochemical results on the externally bound AaAgo–siRNA complex together provide insights into a protein–RNA recognition event that could potentially be associated with the RISC-loading pathway.
Cleavage Specificity of Human Argonautes
There are four human Ago proteins labeled 1 to 4. Functional studies have established that only hAgo2 is capable of guide-strand-mediated cleavage of a complementary RNA target (Hutvagner and Zamore 2002; Martinez et al. 2002; Liu et al. 2004; Meister et al. 2004). The catalytic residues responsible for target cleavage within the PIWI domain of hAgo2 have been identified from mutation studies. Thus, Ala substitution of D597 and D669 resulted in retention of binding but loss of cleavage activity in both in vitro and in vivo experiments (Liu et al. 2004). The catalytic triad contributing to phosphodiester cleavage chemistry in hAgo2 has been identified as a DDH motif based on the results of mutation studies of H807 (Rivas et al. 2005). The catalytic residues are DDR for hAgo1, DDH for hAgo3, and DGR for hAgo4, none of which exhibit mRNA cleavage activity.
Domain-swapping experiments between hAgo2 and hAgo1 in our laboratory have also contributed to our understanding of the unique structural features of hAgo2 required for mRNA cleavage (Yuan et al. 2005). Only chimeral hAgo proteins containing the PIWI domain of hAgo2, either alone or paired with hAgo2 Mid domain, were capable of small RNA-guided cleavage of target RNA. These studies highlight the unique microenvironment restricted to the PIWI domain of hAgo2 and also reinforce earlier observations that the DD-containing motif within the PIWI domain of hAgo2 is the prominent determinant for its unique catalytic activity (Liu et al. 2004; Song et al. 2004; Rivas et al. 2005).
An improved understanding of hAgo2 function will affect our understanding of mechanistic issues related to RNA silencing and in turn aid efforts focused on using this methodology for therapeutic intervention against virus-based diseases, and in the longer run, human genetic diseases.
New Developments within the Ago and Piwi Family
Recent functional studies have identified a new class of short regulatory RNAs that affected RNA silencing. These include Piwi-interacting RNAs (piRNAs) (Aravin et al. 2006; Girard et al. 2006; Grivna et al. 2006; Lau et al. 2006; Watanabe et al. 2006) and repeat-associated small interfering RNAs (rasiRNAs) (Aravin et al. 2003; Saito et al. 2006) that are associated with the Piwi subfamily. The piRNAs are in the 26- to 31-nucleotide range and the rasiRNAs are in the 24- to 29-nucleotide range, with both having a strong preference for U at the 5′-position. These newly identified small RNAs appear to have important implications for sperm developmental regulation and stem cell biology.
DOUBLE-STRANDED RNA PROCESSING BY DICER
We next briefly summarize recent biochemical (Zhang et al. 2004) and structural (Gan et al. 2006; MacRae et al. 2006) efforts that have considerably affected our mechanistic understanding of events associated with dsRNA processing by bacterial RNase III and Dicer (for review, see Carmell and Hannon 2004). Human Dicer is composed of an aminoterminal RNA helicase domain, followed by a domain of unknown function (DUF 283), a PAZ domain, two RNase III domains, and a carboxyterminal RNA-binding domain (RBD).
The Witold Filipowicz group undertook a systematic mutation study of catalytic residues implicated in dsRNA cleavage within Escherichia coli RNase III and human Dicer. These studies have identified a single processing center generated by intramolecular dimerization of its two RNase III domains that is capable of simultaneously asymmetrically cutting partner strands of dsRNA (Zhang et al. 2004). This group also proposed that the PAZ domain in conjunction with the processing center measures the distance from the ends of the dsRNA to the cleavage site, thus generating appropriate-sized siRNAs.
The Xinhua Ji group has solved the crystal structure of a catalytically competent complex of AaRNase III bound to its 2-nucleotide 3′-overhang-containing dsRNA product, which defines the protein–RNA interactions associated with substrate specificity and divalent cation-mediated scissible bond cleavage (Gan et al. 2006). This structure provides direct experimental support at the structural level for a single processing center and the role of induced fit in RNase III protein–RNA recognition and cleavage. The potential role of divalent ions in the catalytic cleavage mechanism has been clarified following comparison between the structures of AaRNase III bound to dsRNA which contains one bound divalent cation at the cleavage site (Gan et al. 2006) and Bacillus halodurans RNase H (BhRNaseH) bound to a DNA/RNA hybrid which contains two bound divalent cations at the cleavage site (Nowotny et al. 2005; Yang et al. 2006).
The Jennifer Doudna laboratory has solved the crystal structure of a catalytically active Giardia Dicer composed of PAZ and two RNase III domains (MacRae et al. 2006). Strikingly, the linkers connecting the individual domains in Dicer adopt folded conformations, such that the overall molecule adopts a hatchet conformation, with the PAZ domain and linkers (a connector helix and a platform domain) forming the handle, while the two RNAse III domains and a linker (the bridging domain) form the blade. This structure, which is of free Dicer, establishes that the PAZ module, which can bind to 2-nucleotide 3′-overhangs at the end of dsRNA, is separated by approximately 65 Å from the catalytic centers of the two ribonuclease III domains, by a flat surface, a length that spans 25 bp of dsRNA. Indeed, dsRNA can be modeled onto this flat surface, thereby suggesting that Dicer is a molecular ruler capable of cleaving dsRNA at a specified distance from its helical end (MacRae et al. 2006). A future challenge will be to structurally characterize either Dicer or its relevant domains with bound dsRNA, in efforts to further understand processing events associated with these RNase III enzymes that play a critical role in RNA silencing. Our laboratory has made progress toward this goal through ongoing crystallographic structural studies on human Dicer PAZ domain and flanking linkers bound to an siRNA-like duplex (J.-B. Ma et al., unpubl.).
microRNA BIOGENESIS
We conclude by outlining an emerging challenge for structural biologists in their efforts at structural characterization of the protein–RNA recognition events associated with the RNA silencing pathway. This challenge involves a molecular level elucidation of the steps associated with the processing of primary microRNAs (pri-miRNAs) into mature miRNA, which in turn are incorporated into RISC (for review, see Du and Zamore 2005; Kim 2005; Kim and Nam 2006). In humans, a protein–RNA complex involving the RNase III nuclease Drosha and DGCR8, a protein containing two dsRBDs, is responsible for converting pri-miRNAs into precursor microRNAs (pre-miRNAs) in the nucleus. The pre-miRNAs are then transported from the nucleus to the cytoplasm by a complex involving exportin-5 (Exp5) and RanGTP. Next, a protein–RNA complex involving the RNase III nuclease Dicer and TRBP, a protein containing three dsRBDs, is responsible for converting pre-miRNAs into mature miRNA duplexes in the cytoplasm. Thus, miRNA biogenesis involves three critical complexes, each involving two proteins and a RNA, whose structural elucidation could greatly advance our understanding of miRNA biogenesis at the molecular level. Because human Drosha and Dicer are large proteins that are both difficult to express and solubilize in E. coli, it will be necessary to identify minimalist domains and their interaction partners, which still retain their essential catalytic activity.
Acknowledgments
The structural research was supported by National Institutes of Health (NIH) grant AI068776 to Dinshaw J. Patel and the functional research by NIH grant GM068476 to Thomas Tuschl. Yi Pei is supported by a Ruth Kirschstein NIH Postdoctoral Fellowship. The authors declare that they have no financial interests. The structural, mutation, and binding research on the TBSV p19–siRNA complex (Keqiong Ye with assistance from Lucy Malinina), hAgo1 PAZ–siRNA complex (Jin-Biao Ma with assistance from Keqiong Ye), A. fulgidus Piwi–siRNA complex (Jin-Biao Ma with assistance from Yu-Ren Yuan), A. aeolicus Argonaute (Yu-Ren Yuan and Jin-Biao Ma) and A. aeolicus Ago externally bound siRNA complex (Yu-Ren Yuan), and the modeling research on the A. aeolicus Ago-DNA/RNA hybrid complex (Vitaly Kuryavyi) was undertaken under the supervision of Dinshaw J. Patel. The functional research associated with target RNA cleavage assays (Yi Pei and Gunter Meister) was undertaken under the supervision of Thomas Tuschl. This contribution was written by Dinshaw J. Patel with input from Thomas Tuschl.
References
- Aravin AA, Lagos-Quintana M, Yalcin A, Zavolan M, Marks D, Snyder B, Gaasterland T, Meyer J, Tuschl T. The small RNA profile during Drosophila melanogaster development. Dev Cell. 2003;5:337. doi: 10.1016/s1534-5807(03)00228-4. [DOI] [PubMed] [Google Scholar]
- Aravin A, Gaidatzis D, Pfeffer S, Lagos-Quintana M, Ladgraf P, Iovino N, Morris P, Brownstein MJ, Kuramochi-Miyagawa S, Nakano T, et al. A novel class of small RNAs bind to MLL1 protein in mouse testis. Nature. 2006;442:203. doi: 10.1038/nature04916. [DOI] [PubMed] [Google Scholar]
- Bartel DP. MicroRNAs: Genomics, biogenesis, mechanism and function. Cell. 2004;116:281. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- Baulcombe D. RNA silencing in plants. Nature. 2004;431:356. doi: 10.1038/nature02874. [DOI] [PubMed] [Google Scholar]
- Bernstein E, Caudy AA, Hammond SM, Hannon GJ. Role for a bidentate ribonuclease in the initiation step of RNA interference. Nature. 2001;409:363. doi: 10.1038/35053110. [DOI] [PubMed] [Google Scholar]
- Carmell MA, Hannon GJ. RNase III enzymes and the initiation of gene silencing. Nat Struct Mol Biol. 2004;11:214. doi: 10.1038/nsmb729. [DOI] [PubMed] [Google Scholar]
- Carmell MA, Xuan Z, Zhang MQ, Hannon GJ. The Argonaute family: Tentacles that reach into RNAi, developmental control, stem cell maintenance, and tumorigenesis. Genes Dev. 2002;16:2733. doi: 10.1101/gad.1026102. [DOI] [PubMed] [Google Scholar]
- Cerutti L, Mian N, Bateman A. Domains in gene silencing and cell differentiation proteins: The novel PAZ domain and redefinition of the Piwi domain. Trends Biochem Sci. 2000;25:481. doi: 10.1016/s0968-0004(00)01641-8. [DOI] [PubMed] [Google Scholar]
- Chao JA, Lee JH, Chapados BR, Debler EW, Schneemann A, Williamson JR. Dual modes of RNA-silencing suppression by Flock House virus protein B2. Nat Struct Mol Bioll. 2005;12:952. doi: 10.1038/nsmb1005. [DOI] [PubMed] [Google Scholar]
- Chapman EJ, Prokhnevsky AI, Gopinath K, Dolja VV, Carrington JC. Viral RNA silencing suppressors inhibit the microRNA pathway at the intermediate step. Genes Dev. 2004;18:1179. doi: 10.1101/gad.1201204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collins RE, Cheng X. Structural domains of RNAi. FEBS Lett. 2005;579:5841. doi: 10.1016/j.febslet.2005.07.072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doench JG, Sharp PA. Specificity of microRNA target selection in translational repression. Genes Dev. 2004;18:504. doi: 10.1101/gad.1184404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doench JG, Petersen CP, Sharp PA. siRNAs can function as miRNAs. Genes Dev. 2003;17:438. doi: 10.1101/gad.1064703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du T, Zamore PD. microPrimer: The biogenesis and function of microRNA. Development. 2005;132:4645. doi: 10.1242/dev.02070. [DOI] [PubMed] [Google Scholar]
- Dykxhoorn DM, Novina CD, Sharp PA. Killing the messenger: Short RNAs that silence gene expression. Nat Rev Mol Cell Biol. 2003;4:457. doi: 10.1038/nrm1129. [DOI] [PubMed] [Google Scholar]
- Elbashir SM, Lendeckel W, Tuschl T. RNA interference is mediated by 21- and 22-nucleotide RNAs. Genes Dev. 2001a;15:188. doi: 10.1101/gad.862301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elbashir SM, Martinez J, Patkaniowska A, Lendeckel W, Tuschl T. Functional anatomy of siRNAs for mediating efficient RNAi in Drosophila melanogaster embryo lysate. EMBO J. 2001b;20:6877. doi: 10.1093/emboj/20.23.6877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Filipowicz W. RNAi: The nuts and bolts of the RISC machine. Cell. 2005;122:17. doi: 10.1016/j.cell.2005.06.023. [DOI] [PubMed] [Google Scholar]
- Filipowicz W, Jaskiewicz L, Kolb FA, Pillai RS. Post-transcriptional gene silencing by siRNAs and miRNAs. Curr Opin Struct Biol. 2005;15:331. doi: 10.1016/j.sbi.2005.05.006. [DOI] [PubMed] [Google Scholar]
- Gan J, Tropea JE, Austin BP, Court DL, Waugh DS, Ji X. Structural insight into the mechanism of double-stranded RNA processing by ribonuclease III. Cell. 2006;124:355. doi: 10.1016/j.cell.2005.11.034. [DOI] [PubMed] [Google Scholar]
- Girard A, Sachidanandam R, Hannon GJ, Carmell MA. A germline-specific class of small RNAs binds mammalian Piwi proteins. Nature. 2006;442:199. doi: 10.1038/nature04917. [DOI] [PubMed] [Google Scholar]
- Grivna ST, Beyret E, Wang Z, Lin H. A novel class of small RNAs in mouse spermatogenic cells. Genes Dev. 2006;20:1709. doi: 10.1101/gad.1434406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haley B, Zamore PD. Kinetic analysis of RNAi enzyme complex. Nat Struct Mol Biol. 2004;11:599. doi: 10.1038/nsmb780. [DOI] [PubMed] [Google Scholar]
- Hall TM. Structure and function of argonaute proteins. Structure. 2005;13:1403. doi: 10.1016/j.str.2005.08.005. [DOI] [PubMed] [Google Scholar]
- Hamilton AJ, Baulcombe DC. A species of small antisense RNA in posttranscriptional gene silencing in plants. Science. 1999;286:950. doi: 10.1126/science.286.5441.950. [DOI] [PubMed] [Google Scholar]
- Hammond SM. Dicing and slicing: The core machinery of the RNA interference pathway. FEBS Lett. 2005;579:5822. doi: 10.1016/j.febslet.2005.08.079. [DOI] [PubMed] [Google Scholar]
- Hammond SM, Bernstein E, Beach D, Hannon GJ. An RNA-directed nuclease mediates post-transcriptional gene silencing in Drosophila cells. Nature. 2000;404:293. doi: 10.1038/35005107. [DOI] [PubMed] [Google Scholar]
- Hammond SM, Boettcher S, Caudy AA, Kobayashi R, Hannon GN. Argonaute2, a link between genetic and biochemical analysis of RNAi. Science. 2001;293:1146. doi: 10.1126/science.1064023. [DOI] [PubMed] [Google Scholar]
- Hannon GJ. RNA interference. Nature. 2002;418:244. doi: 10.1038/418244a. [DOI] [PubMed] [Google Scholar]
- Hutvagner G, Zamore PD. A microRNA in a multiple turnover RNAi enzyme complex. Science. 2002;297:2056. doi: 10.1126/science.1073827. [DOI] [PubMed] [Google Scholar]
- Kim VN. MicroRNA biogenesis: Coordinating cropping and dicing. Nat Rev Mol Cell Biol. 2005;6:376. doi: 10.1038/nrm1644. [DOI] [PubMed] [Google Scholar]
- Kim VN, Nam JW. Genomics of microRNA. Trends Genet. 2006;22:165. doi: 10.1016/j.tig.2006.01.003. [DOI] [PubMed] [Google Scholar]
- Lai L, Yokota H, Hung LW, Kim R, Kim SH. Crystal structure of archael RNase Hii: A homologue of human major RNase H. Structure. 2000;8:897. doi: 10.1016/s0969-2126(00)00179-9. [DOI] [PubMed] [Google Scholar]
- Lakatos L, Csorba T, Pantaleo V, Chapman EJ, Carrington JC, Liu YP, Dolja VV, Calvino LF, Lopz-Moya JJ, Burgyan J. Small RNA binding is a common strategy to suppress RNA silencing by several viral suppressors. EMBO J. 2005;25:2768. doi: 10.1038/sj.emboj.7601164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lau NC, Seto AG, Kim J, Kuramochi-Miyagawa S, Nakano T, Bartel DP, Kingston RE. Characterization of the piRNA complex from rat testes. Science. 2006;313:363. doi: 10.1126/science.1130164. [DOI] [PubMed] [Google Scholar]
- Lecellier CH, Dunoyer P, Arar K, Lehmann-Che J, Eyquem S, Himber C, Saib A, Voinnet O. A cellular microRNA mediates antiviral defense in human cells. Science. 2005;308:557. doi: 10.1126/science.1108784. [DOI] [PubMed] [Google Scholar]
- Lewis BP, Burge CB, Bartel DP. Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell. 2005;120:15. doi: 10.1016/j.cell.2004.12.035. [DOI] [PubMed] [Google Scholar]
- Lewis BP, Shih IH, Jones-Rhodes MW, Bartel DP, Burge CB. Prediction of mammalian microRNA targets. Cell. 2003;115:787. doi: 10.1016/s0092-8674(03)01018-3. [DOI] [PubMed] [Google Scholar]
- Li H, Li WX, Ding SW. Induction and suppression of RNA silencing by an animal virus. Science. 2002;296:1319. doi: 10.1126/science.1070948. [DOI] [PubMed] [Google Scholar]
- Li WX, Ding SW. Viral suppressors of RNA silencing. Curr Opin Biotechnol. 2001;12:150. doi: 10.1016/s0958-1669(00)00190-7. [DOI] [PubMed] [Google Scholar]
- Lingel A, Sattler M. Novel modes of protein-RNA recognition in the RNAi pathway. Curr Opin Struct Biol. 2005;15:107. doi: 10.1016/j.sbi.2005.01.010. [DOI] [PubMed] [Google Scholar]
- Lingel A, Simon B, Izaurralde E, Sattler M. Structure and nucleic acid-binding of the Drosophila Argonaute2 PAZ domain. Nature. 2003;426:465. doi: 10.1038/nature02123. [DOI] [PubMed] [Google Scholar]
- Lingel A, Simon B, Izaurralde E, Sattler M. Nucleic acid 3′-end recognition by the Argonaute2 PAZ domain. Nat Struct Mol Biol. 2004;11:576. doi: 10.1038/nsmb777. [DOI] [PubMed] [Google Scholar]
- Liu J, Carmell MA, Rivas FV, Marsden CG, Thompson JM, Song JJ, Hammond SM, Joshua-Tor L, Hannon GJ. Argonaute2 is the catalytic engine of mammalian RNAi. Science. 2004;305:1437. doi: 10.1126/science.1102513. [DOI] [PubMed] [Google Scholar]
- Ma JB, Ye K, Patel DJ. Structural basis for overhang-specific small interfering RNA recognition by the PAZ domain. Nature. 2004;429:318. doi: 10.1038/nature02519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma JB, Yuan YR, Meister G, Pei Y, Tuschl T, Patel DJ. Structural basis for 5′-end specific recognition of the guide RNA strand by A. fulgidus PIWI protein. Nature. 2005;434:666. doi: 10.1038/nature03514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacRae IJ, Zhou K, Li F, Repic A, Brooks AN, Cande WZ, Adams PD, Doudna JA. Structural basis for double-stranded RNA processing by Dicer. Science. 2006;311:195. doi: 10.1126/science.1121638. [DOI] [PubMed] [Google Scholar]
- Mallory AC, Reinhardt BJ, Jones-Rhoades MW, Tang G, Zamore PD, Barton MK, Bartel DP. MicroRNA control of PHABULOSA in leaf development: Importance of pairing to the microRNA 5′ region. EMBO J. 2004;23:3356. doi: 10.1038/sj.emboj.7600340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez J, Tuschl T. RISC is a 5′ phosphomonoester-producing RNA endonuclease. Genes Dev. 2004;18:975. doi: 10.1101/gad.1187904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez J, Patkaniowska A, Urlaub H, Luhrmann R, Tuschl T. Single-stranded antisense siRNAs guide target RNA cleavage in RNAi. Cell. 2002;110:563. doi: 10.1016/s0092-8674(02)00908-x. [DOI] [PubMed] [Google Scholar]
- Matranga C, Tomari Y, Shin C, Bartel DP, Zamore PD. Passenger-strand cleavage facilitates assembly of siRNA into Ago2-containing RNAi enzyme complexes. Cell. 2005;123:607. doi: 10.1016/j.cell.2005.08.044. [DOI] [PubMed] [Google Scholar]
- Meister G, Tuschl T. Mechanisms of gene silencing by double-stranded RNA. Nature. 2004;431:343. doi: 10.1038/nature02873. [DOI] [PubMed] [Google Scholar]
- Meister G, Landthaler M, Patkaniowska A, Dorsett Y, Teng G, Tuschl T. Human Argonaute2 mediates RNA cleavage targeted by miRNA and siRNA. Mol Cell. 2004;15:185. doi: 10.1016/j.molcel.2004.07.007. [DOI] [PubMed] [Google Scholar]
- Nowotny M, Gaidamakov SA, Crouch RJ, Yang Y. Crystal structures of RNase H bound to an RNA/DNA hybrid: Substrate specificity and metal-dependent catalysis. Cell. 2005;121:1005. doi: 10.1016/j.cell.2005.04.024. [DOI] [PubMed] [Google Scholar]
- Nykanen A, Haley B, Zamore PD. ATP requirements and small interfering RNA structure in the RNA interfering pathway. Cell. 2001;107:309. doi: 10.1016/s0092-8674(01)00547-5. [DOI] [PubMed] [Google Scholar]
- Parker JS, Barford D. Argonaute: A scaffold for the function of short regulatory RNAs. Trends Biochem Sci. 2006;31:622. doi: 10.1016/j.tibs.2006.09.010. [DOI] [PubMed] [Google Scholar]
- Parker JS, Roe SM, Barford D. Crystal structure of a PIWI protein suggests mechanisms for siRNA recognition and slicer activity. EMBO J. 2004;23:4727. doi: 10.1038/sj.emboj.7600488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parker JS, Roe SM, Barford D. Structural insights into mRNA recognition from a PIWI-domain-siRNA-guide complex. Nature. 2005;434:663. doi: 10.1038/nature03462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parrish S, Fleenor J, Xu S, Mello C, Fire A. Functional anatomy of a dsRNA trigger: Differential requirement for the two trigger strands in RNA interference. Mol Cell. 2000;6:1077. doi: 10.1016/s1097-2765(00)00106-4. [DOI] [PubMed] [Google Scholar]
- Rand TA, Petersen S, Du F, Wang X. Argonaute2 cleaves the anti-guide strand of siRNA during RISC activation. Cell. 2005;123:621. doi: 10.1016/j.cell.2005.10.020. [DOI] [PubMed] [Google Scholar]
- Rivas FV, Tolia NH, Song JJ, Aragon JP, Liu J, Hannon GJ, Joshua-Tor L. Purified Argonaute2 and an siRNA form recombinant human RISC. Nat Struct Mol Biol. 2005;12:340. doi: 10.1038/nsmb918. [DOI] [PubMed] [Google Scholar]
- Saito K, Nishida KM, Mori T, Kawamura Y, Miyoshi K, Nagami T, Siomi H, Siomi MC. Specific association of Piwi with rasiRNA derived from retrotransposon and heterochromatic regions in the Drosophila genome. Genes Dev. 2006;20:2214. doi: 10.1101/gad.1454806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwarz DS, Tomari Y, Zamore PD. The RNA-induced silencing complex is a Mg2+-dependent endonuclease. Curr Biol. 2004;14:787. doi: 10.1016/j.cub.2004.03.008. [DOI] [PubMed] [Google Scholar]
- Schwarz DS, Hutvagner G, Du T, Xu Z, Aronin N, Zamore PD. Asymmetry in the assembly of the RNAi enzyme complex. Cell. 2003;115:199. doi: 10.1016/s0092-8674(03)00759-1. [DOI] [PubMed] [Google Scholar]
- Silhavy D, Burgyan J. Effects and side-effects of viral RNA silencing suppressors on short RNAs. Trends Plant Sci. 2004;9:76. doi: 10.1016/j.tplants.2003.12.010. [DOI] [PubMed] [Google Scholar]
- Silhavy D, Molnar A, Lucioli A, Sziiya G, Hornyik C, Tavazza M, Burgyan J. A viral protein suppresses RNA silencing and binds silencing-generated, 21- to 25-nucleotide double-stranded RNAs. EMBO J. 2002;21:3070. doi: 10.1093/emboj/cdf312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song JJ, Joshua-Tor L. Argonaute and RNA-getting into the groove. Curr Opin Struct Biol. 2006;16:5. doi: 10.1016/j.sbi.2006.01.010. [DOI] [PubMed] [Google Scholar]
- Song JJ, Smith SK, Hannon GJ, Joshua-Tor L. Crystal structure of Argonaute and its implications for RISC slicer activity. Science. 2004;305:1434. doi: 10.1126/science.1102514. [DOI] [PubMed] [Google Scholar]
- Song JJ, Liu J, Tolia NH, Schneiderman J, Smith SK, Martienssen RA, Hannon GJ, Joshua-Tor L. The crystal structure of the Argonaute2 PAZ domain reveals an RNA-binding motif in RNAi effector complexes. Nat Struct Biol. 2003;10:1026. doi: 10.1038/nsb1016. [DOI] [PubMed] [Google Scholar]
- Sontheimer EJ. Assembly and function of RNA silencing complexes. Nat Rev Mol Cell Biol. 2005;6:127. doi: 10.1038/nrm1568. [DOI] [PubMed] [Google Scholar]
- Stark A, Brennecke J, Russel RB, Cohen SM. Identification of Drosophila microRNA targets. PLoS Biol. 2003;1:397. doi: 10.1371/journal.pbio.0000060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tahbaz N, Kolb FA, Zhang H, Jaronczyk K, Filipowicz W, Hobman TC. Characterization of the interactions between mammalian PAZ PIWI domain proteins and Dicer. EMBO Rep. 2004;5:1. doi: 10.1038/sj.embor.7400070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomari Y, Zamore PD. Perspective: Machine for RNAi. Genes Dev. 2005;19:517. doi: 10.1101/gad.1284105. [DOI] [PubMed] [Google Scholar]
- Vargason JM, Szittya G, Burgyan J, Tanaka-Hall TM. Size selective recognition of siRNA by an RNA silencing suppressor. Cell. 2003;115:799. doi: 10.1016/s0092-8674(03)00984-x. [DOI] [PubMed] [Google Scholar]
- Verdel A, Jia S, Gerber S, Sugiyama T, Gygi S, Grewal SI, Moazed D. RNAi-mediated targeting of heterochromatin by the RITS complex. Science. 2004;303:672. doi: 10.1126/science.1093686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voinnet O. Induction and suppression of RNA silencing: Insights from viral infections. Nat Rev Genet. 2005;6:206. doi: 10.1038/nrg1555. [DOI] [PubMed] [Google Scholar]
- Wang MB, Metzlaff M. RNA silencing and antiviral defense in plants. Curr Opin Plant Biol. 2005;8:216. doi: 10.1016/j.pbi.2005.01.006. [DOI] [PubMed] [Google Scholar]
- Watanabe T, Takeda A, Tsukiyama T, Mise K, Okuno T, Sasaki H, Minami N, Imai H. Identification and characterization of two novel classes of small RNAs in the mouse germline: Retrotransposon-derived siRNAs in oocytes and germline small RNAs in testes. Genes Dev. 2006;20:1732. doi: 10.1101/gad.1425706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong I, Lohman TM. A double-filter method for nitrocellulose-filter binding: Application to protein-nucleic acid interactions. Proc Natl Acad Sci. 1993;90:5428. doi: 10.1073/pnas.90.12.5428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan KS, Yan S, Farooq A, Han A, Zeng L, Zhou MM. Structure and conserved RNA binding of the PAZ domain. Nature. 2003;426:468. doi: 10.1038/nature02129. [DOI] [PubMed] [Google Scholar]
- Yang W, Steitz TA. Recombining the structures of HIV integrase, RuvC and RNase H. Structure. 1995;3:131. doi: 10.1016/s0969-2126(01)00142-3. [DOI] [PubMed] [Google Scholar]
- Yang W, Lee JY, Nowotny M. Making and breaking nucleic acids: Two-Mg2+-ion dependent catalysis and substrate specificity. Mol Cell. 2006;22:5. doi: 10.1016/j.molcel.2006.03.013. [DOI] [PubMed] [Google Scholar]
- Ye K, Patel DJ. RNA silencing suppressor p21 of beet yellow virus forms an RNA binding octameric ring structure. Structure. 2005;13:1375. doi: 10.1016/j.str.2005.06.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye K, Malinina L, Patel DJ. Recognition of small interfering RNA by a viral suppressor of RNA silencing. Nature. 2003;426:874. doi: 10.1038/nature02213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan YR, Pei Y, Chen HY, Tuschl T, Patel DJ. A potential protein-RNA recognition event along the RISC-loading pathway from the structure of A. aeolicus Argonaute with externally bound siRNA. Structure. 2006;14:1557. doi: 10.1016/j.str.2006.08.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan YR, Pei Y, Ma JB, Kuryavyi Y, Zhadina M, Meister G, Chen HY, Dauter Z, Tuschl T, Patel DJ. Crystal structure of A. aeolicus Argonaute, a site-specific DNA-guided endoribonuclease, provides insights into RISC-mediated mRNA cleavage. Mol Cell. 2005;19:405. doi: 10.1016/j.molcel.2005.07.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zamore PD, Haley B. Ribognome: The big world of small RNAs. Science. 2005;309:1519. doi: 10.1126/science.1111444. [DOI] [PubMed] [Google Scholar]
- Zamore PD, Tuschl T, Sharp PA, Bartel DP. RNAi: Double-stranded RNA directs the ATP-dependent cleavage of mRNA at 21 to 23 nucleotide intervals. Cell. 2000;101:25. doi: 10.1016/S0092-8674(00)80620-0. [DOI] [PubMed] [Google Scholar]
- Zhang H, Kolb FA, Jakiewicz L, Westhof E, Filipowicz W. Single processing center models for human Dicer and bacterial ribonuclease III. Cell. 2004;118:57. doi: 10.1016/j.cell.2004.06.017. [DOI] [PubMed] [Google Scholar]