Abstract
Maintenance of genomic methylation patterns is mediated primarily by DNA methyltransferase-1 (DNMT1). We have solved structures of mouse and human DNMT1 composed of CXXC, tandem bromo-adjacent homology (BAH1/2), and methyltransferase domains bound to DNA-containing unmethylated CpG sites. The CXXC specifically binds to unmethylated CpG dinucleotide and positions the CXXC–BAH1 linker between the DNA and the active site of DNMT1, preventing de novo methylation. In addition, a loop projecting from BAH2 interacts with the target recognition domain (TRD) of the methyltransferase, stabilizing the TRD in a retracted position and preventing it from inserting into the DNA major groove. Our studies identify an autoinhibitory mechanism, in which unmethylated CpG dinucleotides are occluded from the active site to ensure that only hemimethylated CpG dinucleotides undergo methylation.
Maintenance of genomic methylation patterns in mammals (1–3) is required for monoallelic expression of imprinted genes (4), for the transcriptional silencing of retrotransposons (5, 6), and for X chromosome inactivation in females (7). The eukaryotic maintenance DNA methyltransferase DNMT1 is a multimodular protein composed of a replication foci-targeting domain (RFD), a DNA-binding CXXC domain, a pair of bromo-adjacent homology (BAH) domains, and a C-terminal catalytic domain (Fig. 1A). We solved the structures of mouse DNMT1(650–1602) and human DNMT1 (646–1600) bound to duplex DNA–containing unmethylated CG sites. Our structural studies reveal a role for autoinhibition in maintenance DNA methylation, by which occlusion of unmethylated CpG dinucleotides from de novo methylation ensures that only hemimethylated CpG dinucleotides gain access to the active site.
Fig. 1.
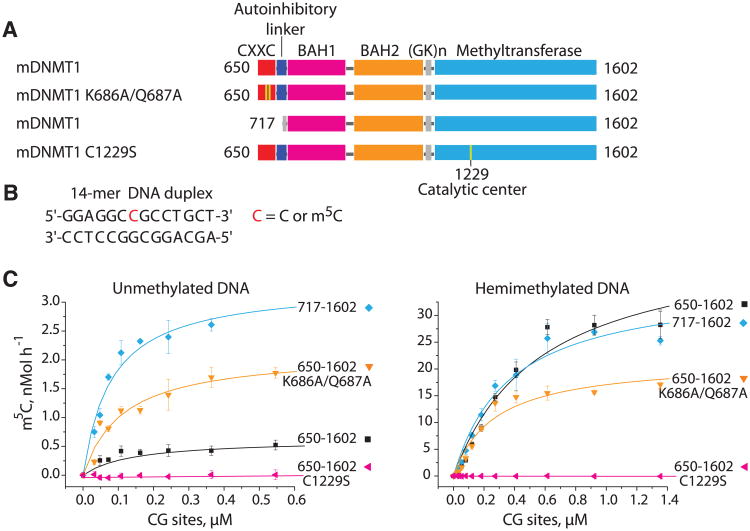
Structural overview of mDNMT1(650–1602)–DNA 19-nucleotide oligomer complex with bound AdoHcy. (A) Color-coded domain architecture and numbering of mDNMT1 sequence. The thin vertical light blue bars indicate binding positions of zinc ions. (B) Ribbon representation of the complex in two orthogonal views. The CXXC, BAH1, BAH2, and methyltransferase domain are colored in red, light purple, orange and light blue, and DNA and zinc ions are colored in light brown and dark purple, respectively; CXXC-BAH1 linker in dark blue, BAH1-BAH2 linker in silver, (GK)n-containing BAH2-methyltransferase linker in black, and bound AdoHcy as in space-filling representation.
Structures of mouse and human DNMT1-DNA complexes
The crystal structure of an enzymatically active mouse DNMT1 (mDNMT1; residues 650 to 1602) in complex with S-adenosyl homocysteine (AdoHcy) and a 19-base pair (bp) DNA duplex was solved at 3.0 Å resolution (table S1). The DNA contained two unmethylated CpG dinucleotides separated by 8 bp. We can trace the CXXC domain (Fig. 1B, red), both BAH1 (Fig. 1B, light purple) and BAH2 (Fig. 1B, orange) domains, and the C-terminal catalytic methyltransferase domain (Fig. 1B, light blue), as well as all 19-bp of the bound DNA (Fig. 1B, light brown) (Fig. 1B and fig. S1). The CXXC and BAH1 domains are at opposite ends of the methyltransferase domain and are connected by a long linker segment (CXXC– BAH1 linker) (Fig. 1B, blue, and fig. S2). The BAH1 and BAH2 domains are separated by an α-helical linker (Fig. 1B, silver), with both BAH domains positioned on the surface remote from the bound DNA. The (GK)n linker segment that connects the BAH2 domain to the catalytic domain is disordered in the complex and is shown by a dashed line in Fig. 1B. The catalytic domain forms the core of the complex and contacts both BAH domains and the DNA. A molecule of AdoHcy (Fig. 1B) is positioned in the active site of the catalytic domain. We observed four Zn2+ cations (Fig. 1B, purple balls) in the structure of the complex, two in Cys4-coordination within the CXXC domain, whereas two others involve coordination of single Zn ions in Cys3His-coordination in BAH1 and in the target recognition domain (TRD) (8) of the methyltransferase. The bound 19-bp DNA adopts an almost ideal B-form duplex in the complex (fig. S3). The 2.5 Å structure of mDNMT1(731–1602) lacking the CXXC domain and the CXXC-BAH1 linker in the free state (figs. S4 and S5 and table S1) is very similar to the structure of the mDNMT1(650– 1602) DNA complex [Cα root mean square deviation (RMSD) of 1.0 Å over 814 aligned residues] and confirms the domain structure and relative orientations of BAH1, BAH2, and the catalytic domain. No structure has yet been reported for DNMT1 constructs bound to hemimethylated DNA.
Mouse DNMT1 and human DNMT1 (hDNMT1) exhibit 85% sequence identity (fig. S6). We have also solved the crystal structure of hDNMT1(646–1600) bound to the same 19-bp DNA and AdoHcy at 3.6 Å resolution (figs. S7 to S9 and table S2). The structures of the mouse and human proteins are very similar, with a Cα RMSD of 1.1 Å over 877 aligned residues, which is suggestive of a common mechanism of action. However, the methyltransferase domain is repositioned relative to the CXXC domain and DNA by a 1-bp translation along the DNA axis. Such differences probably arise from the flexibility of the CXXC-BAH1 linker and could also be due to the different packing environment between the two complexes.
CXXC domain binds unmethylated CpG dinucleotides
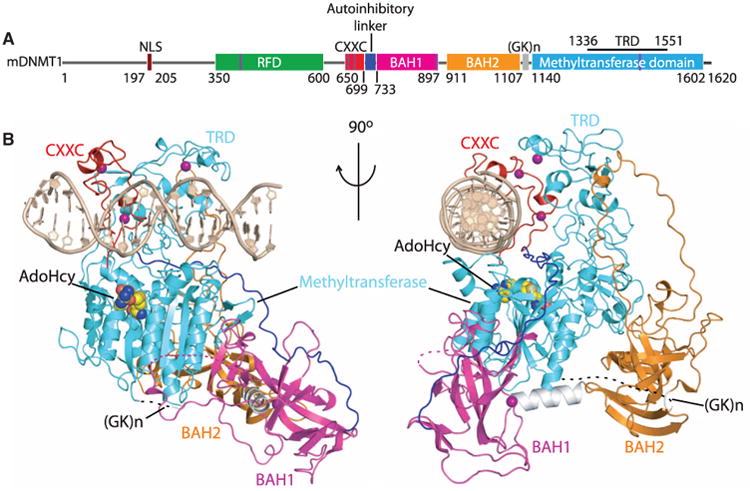
The CXXC domain of mDNMT1 adopts a crescent-like fold similar to that adopted by the MLL1 CXXC domain (9, 10). Two short helical segments position eight Cys residues in two clusters for coordination of a pair of embedded Zn2+ cations (fig. S10, A and B). All sequence-specific contacts with DNA are made via the CXXC domain in the DNMT1-DNA complex. The CXXC domain targets both the major and minor grooves of the DNA over a CpG-containing 4-bp footprint. A loop segment (Arg684-Ser685-Lys686-Gln687) from the CXXC domain penetrates into the major groove (Fig. 2, A and B) and forms base-specific and phosphodiester intermolecular interactions (Fig. 2C), which is reminiscent of the MLL1 CXXC domain interaction with DNA (9, 10).
Fig. 2.
Intermolecular contacts between CXXC domain of mDNMT1(650–1602) and DNA 19-nucleotide oligomer. (A) Ribbon representation of the CXXC domain bound to DNA. The CpG step is rendered in yellow. (B) Surface electrostatic representation of the CXXC domain bound to DNA. (C) Schematic view of intermolecular interactions involving the CXXC domain in the mDNMT1-DNA 19-nucleotide oligomer complex (boxed red rectangle), with intermolecular contacts shown by red arrows. The residue labels are colored according to their respective domains in Fig. 1. (D) Hydrogen-bonding interactions between side chains from the CXXC domain and the guanine base edges of the CpG step in the DNA major groove. The nitrogen, oxygen, and phosphorous atoms are shown in dark blue, red, and yellow, respectively. The bases of the CpG dinucleotide from one strand are shaded. (E) Hydrogen-bonding interactions between backbone carbonyl oxygens from the CXXC domain and cytosine amino groups of the CpG step in the DNA major groove. (F) Hydrogen-bonding interactions between arginine side chains of the CXXC domain and the phosphodiester backbone of the DNA along the DNA minor groove. (G) Potential steric clashes between methylated cytosine modeled on either strand of the CpG step and the CXXC domain of DNMT1 in the structure of the complex. Van der Waals radii are rendered in red for DNMT1 and gray for modeled methylated cytosine.
The guanine bases in the CpG dinucleotide are recognized by side-chain interactions involving Lys686 and Gln687 of the CXXC domain (Fig. 2D), whereas the cytosine bases in the CpG dinucleotide are recognized by backbone interactions involving Ser685 and Lys686 of the CXXC domain (Fig. 2E). DNA recognition is further anchored by salt bridges between arginine side chains of the CXXC domain and the phosphodiester backbone of the DNA [Fig. 2F and supporting online material (SOM) text] (11). The CXXC domain has been reported to specifically bind unmethylated CpG dinucleotides (9, 12–14). Our structural data confirms this conclusion: Methylation of either cytosine in the CpG step would create severe steric clashes with peptide atoms (Fig. 2G).
Methyltransferase domain
The methyltransferase domain of mDNMT1 adopts a class I methyltransferase fold (8) and folds into two subdomains, designated the catalytic core and the TRD (fig. S11A), which are separated by a large cleft that in the M.HhaI-DNA complex is occupied by DNA. The catalytic core of DNMT1 is dominated by a mixed seven-stranded β-sheet that is flanked by three α-helices on either side (Fig. 1A and fig. S11B). At one end, this central β-sheet is further joined by a two-stranded anti-parallel β-sheet from the BAH1 domain (fig. S12C). The TRD subdomain is inserted between the central β-sheet and the last three α-helices of the catalytic core. The majority of the TRD folds into an independent structural unit and is stabilized in part by a Cys3His-coordinated Zn2+ ion (figs. S10D and S11C). In addition, a hairpin-like fold at the start of the TRD forms hydrophobic contacts with the catalytic core and the BAH1 domain (fig. S11D). We observed intermolecular contacts between amino acid side chains (primarily arginine) of the catalytic core, and phosphate groups flanking the (C4pG5)•(C5′pG4′) segment of the unmethylated DNA duplex in the complex (Fig. 2C; fig. S13, A to E; and SOM text).
Comparison of mDNMT1-DNA and M.HhaI-DNA complexes
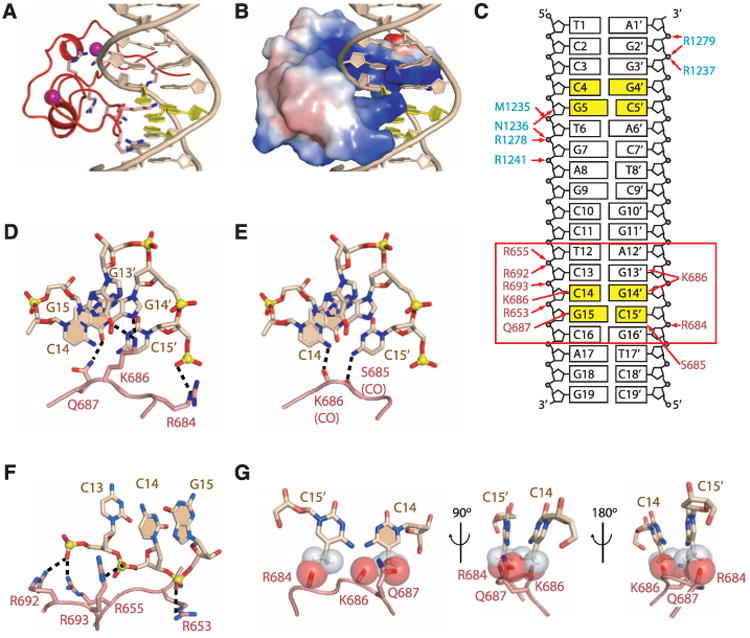
We have compared the structure of the mDNMT1(650–1602)–DNA 19-nucleotide oligomer complex (Fig. 3A, TRD and catalytic core only) with the published structure (15) of the M.Hha1-DNA complex (Fig. 3B). An overlay of these two complexes is shown in stereo in Fig. 3C, after superposition of the catalytic cores of their methyltransferase domains (Cα RMSD = 2.0 Å over 218 aligned residues). Five (I, VI, VIII, IX, and X) out of six conserved sequence motifs of mDNMT1 adopt conformations very similar to those of their counterparts in M.HhaI (fig. S14, A and C to F). Motif IV, which is part of the catalytic loop and contains the catalytic Cys residue, in contrast adopts different conformations in the two complexes (fig. S14B).
Fig. 3.
Comparison of mDNMT1 with M.HhaI in their DNA-bound complexes. (A) The crystal structure of the mDNMT1(650–1602)–DNA 19-nucleotide oligomer complex. The CXXC, BAH1, and BAH2 domains and CXXC-BAH1 linker of mDNMT1 have been removed for clarity. The bound DNA is in light brown, with the TRD and catalytic core in light and dark blue, respectively. (B) The crystal structure of the M.HhaI-DNA complex [PDB: 1MHT (8)]. The bound DNA is in light purple, with the TRD and catalytic core in pale and dark green, respectively. (C) Structural comparison of mDNMT1(650–1602)–DNA 19-nucleotide oligomer complex and M.HhaI-DNA complex in a stereo view looking down the DNA helix axis, after superposition of their methyltransferase domains. AdoHcy is shown in space-filling view. The everted cytosine in the M.HhaI complex is shown in ball-and-stick view in dark purple. (D) Electrostatic surface representation of mDNMT1 CXXC domain and the CXXC-BAH1 linker in the context of the structure of the mDNMT1(650–1602)–DNA 19-nucleotide oligomer complex. The BAH2-TRD loop is highlighted with thicker lines. (E) The proposed model for autoinhibitory mechanism in maintenance DNA methylation. In the autoinhibitory state, the CXXC domain and the auto-inhibitory linker (in red) occlude the active site. In addition, the BAH2-TRD loop (in red) restrains the TRD in a retracted position so that it does not interact with CpG sites on the DNA.
With respect to the methyltransferase domains, the bound DNAs in M.HhaI and mDNMT1 complexes align in the same orientation along the DNA axis, but the unmethylated DNA (Fig. 3, light brown) in the mDNMT1 complex is displaced laterally (by the width of a DNA duplex) so that it is positioned further away from the active site (Fig. 3, A and C). In the M.HhaI complex, the DNA (Fig. 3, light purple) is embedded within the cleft formed by the catalytic core and the TRD domain, with the target cytosine inserted into the active site (Fig. 3, B and C, and fig. S15A). The DNA in the structure of the mDNMT1 complex (Fig. 3, light brown) is anchored by the CXXC domain in a position that is distant from the active site of DNMT1 (fig. S15B). The exclusion of the unmethylated DNA from the active site is the result of an autoinhibitory CXXC-BAH1 linker, which contains a highly acidic segment spanning residues D703 to D711 and is positioned directly between the DNA and the active site (Fig. 3D; the CXXC domain and the CXXC-BAH1 linker are presented in an electrostatic surface representation) (16).
The mDNMT1 and M.HhaI TRD subdomains have little sequence similarity (fig. S11A) and exhibit the greatest structural divergence. They both contain a common 8–amino acid DNA recognition loop, whereas a second 8–amino acid DNA recognition loop in M.HhaI is replaced by a longer 73–amino acid loop in mDNMT1 (fig. S16, A and B). In our structure of the mDNMT1-DNA complex, the TRD subdomain is held in a retracted position away from the DNA through interaction with the BAH2-TRD loop (Figs. 1B and 3D and fig. S12, A and B).
BAH domains
The BAH1 and BAH2 domains of mDNMT1 are connected by an α-helix and are arranged in a dumbbell configuration (Fig. 1B, right). Despite low sequence conservation (fig. S17A), both BAH domains adopt a common fold (fig. S17, B to D) that is similar to the Orc1p BAH domain (fig. S17E) (17, 18). In addition, a Cys3His-coordinated Zn2+ anchors the BAH1 domain to the linker α-helix (fig. S10C).
Both BAH domains are physically associated with the methyltransferase domain (Fig. 1B and fig. S12). In the BAH2 domain, a long loop (BAH2-TRD loop) (Fig. 1B and figs. S12A and S18) projects upward and is anchored near its tip to the TRD of the methyltransferase domain (Fig. 3D and fig. S12A). This interaction is predicted to prevent the interaction of the TRD with DNA (Fig. 1B and fig. S12, A and B). BAH1 and BAH2 domains present a large accessible surface area that is peripheral to the catalytic domain (Fig. 1B) and could serve as a platform for interactions with other proteins.
Enzymatic activities of wild-type, truncated, and mutant DNMT1
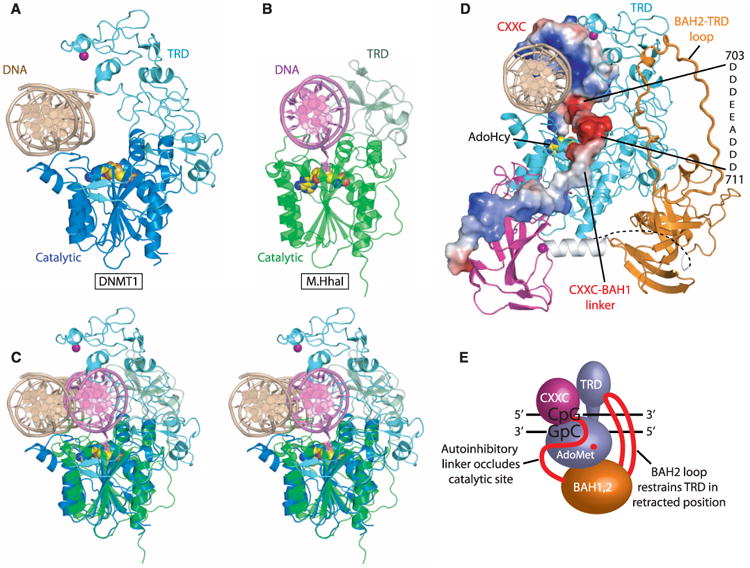
We have used enzymatic methylation assays on full-length, truncated, and mutant DNMT1 (Fig. 4 and fig. S19) to test predictions derived from the structural data. We have used a 14-nucleotide oligomer DNA duplex that contains a single CpG dinucleotide in the hemimethylated or unmethylated state (Fig. 4B). Enzymatic assays have been done on truncated mDNMT1 (Fig. 4C), truncated hDNMT1, commercially purchased full-length hDNMT1 (fig. S19), and on both unmethylated and hemimethylated 14-nucleotide oligomer DNA substrates, with the steady-state Michaelis-Menten kinetic parameters listed in table S3.
Fig. 4.
Efficiency of unmethylated and hemimethylated DNA m5C-methylation by mouse wild-type and mutant DNMT1 proteins. (A) Domain structures of mouse DNMT1 proteins used for activity assay. The Lys686Ala/Gln687Ala and Cys1229Ser mutations are shown as yellow bars. (B) Sequence of 14-nucleotide oligomer DNA duplex used for enzymatic assay. (C) Enzyme kinetics on de novo and maintenance methylation of DNMT1 mutants after removal or mutation of CXXC domain. Rates of 3H-CH3 in nMol per hour transferred to unmethylated and hemimethylated DNA duplexes by 1 nM of mDNMT1(650–1602) wild-type, Lys686Ala/Gln687 Ala and Cys1229Ser mutants and mDNMT1(717–1602) proteins were plotted as a function of CpG site concentration; the steady-state Michaelis-Menten parameters that were estimated from these plots are listed in table S3. Each reaction point was repeated in triplicate; mean and SD values are shown.
We found that the initial rate kcat for un-methylated and hemimethylated substrate was 0.25 ± 0.04 and 18 ± 2 hour−1, respectively, for full-length (1–1616) hDNMT1 (fig. S19 and table S3). The values for truncated (646–1600) hDNMT1 were 0.18 ± 0.02 and 12 ± 1 hour−1. These data confirm that the activities and preference for hemimethylated substrates were comparable for the full-length and truncated proteins.
The cocrystal structure of mDNMT1-DNA complex showed that the CXXC domain and CXXC-BAH1 linker occlude DNA from the active site. We tested mDNMT1 proteins composed of amino acids 717–1602 (CXXC domain and a segment of the CXXC-BAH1 linker deleted) and found that kcat on unmethylated substrates increased from 0.60 ± 0.03 hour−1 for the 650–1602 protein to 3.3 ± 0.2 hour−1 for 717–1602 (Fig. 4C, left, and table S3). In contrast, there is a modest drop in kcat on hemimethylated 14-nucleotide oligomer DNA on proceeding from 650–1602 (kcat = 45 ± 6 hour−1) to 717–1602 (kcat = 36 ± 3 hour−1) mDNMT1 (Fig. 4C, right, and table S3). Thus, the relative preference for hemimethylated over unmethylated 14-nucleotide oligomer DNA drops sevenfold on proceeding from mDNMT1 construct 650–1602, to 717–1602 (table S3).
In addition, residues that contact guanosine bases within the CpG site were mutated to abolish these contacts (mDNMT1 650–1602, K686A/ Q687A double mutant), which was confirmed by gel shift assay (fig. S20). This mutant protein also showed a related increase in kcat (2.1 ± 0.2 hour−1) on unmethylated substrates and a modest decrease in kcat (22 ± 2 hour−1) on hemimethylated subtrates (Fig. 4C and table S3)—thus, the preference for hemimethylated over unmethylated 14-nucelotide oligomer DNA drops sixfold for mDNMT1(650– 1602) on proceeding from wild-type to the CXXC-containing dual mutant (table S3).
These data (a more detailed analysis of the kinetic data are outlined in the SOM text) show that either removal or mutagenesis of the CXXC domain and CXXC-BAH1 linker increases the catalytic activity of mDNMT1 specifically on un-methylated substrates (Fig. 4, A and C), as predicted from the structure of the mDNMT1-DNA cocrystal. There are additional mechanisms that increase the fidelity of maintenance methylation: Removal/ mutation of the CXXC domain yields a six- to sevenfold increase in the rate of de novo methylation, but the truncated protein still retains a preference for hemimethylated DNA (Fig. 4B).
Autoinhibition in maintenance DNA methylation
Our findings allow us to propose a mechanism for autoinhibitory regulation of DNMT1 at unmethylated CpG sites on DNA: Unmethylated DNA is excluded from the active site of mDNMT1 by the binding of the CXXC domain, whereas the presence of the acidic autoinhibitory CXXC-BAH1 linker positioned directly between the DNA and the active site prevents entrance of DNA into the catalytic pocket (Fig. 3D). Further, the BAH2-TRD loop anchors the TRD (Fig. 3D) in a retracted position (Fig. 3A) and prevents it from binding in the DNA major groove.
Maintenance methylation is tightly coupled to DNA replication (19). We propose that unmethylated CpG sites are protected from de novo methylation through binding by the CXXC domain as CpG dinucleotides emerge from the replication complex. This increases the efficiency of maintenance methylation through inhibition of de novo methylation.
Supplementary Material
Acknowledgments
The research was supported by funds from the Abby Rockefeller Mauze Trust and Maloris Foundation to D.J.P. and from NIH to T.H.B. We thank S. Murakami for technical assistance and Z. Wang and H. Li for their assistance with x-ray data collection and structure determination. We would also like to thank the staff of NE-CAT beamlines at the Advanced Photon Source, Argonne National Laboratory, and the X-29 beamline at the Brookhaven National Laboratory, for access and assistance with data collection. The structures of the complexes have been deposited in the Protein Data Bank (PDB).
Footnotes
The accession codes are mDNMT1(650–1602)– DNA complex with bound AdoHcy (3PT6), hDNMT1(646– 1600)–DNA complex with bound AdoHcy (3PTA) and mDNMT1(731–1602) with bound AdoHcy (3PT9).
References
- 1.Law JA, Jacobsen SE. Nat Rev Genet. 2010;11:204. doi: 10.1038/nrg2719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Cheng X, Blumenthal RM. Structure. 2008;16:341. doi: 10.1016/j.str.2008.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Goll MG, Bestor TH. Annu Rev Biochem. 2005;74:481. doi: 10.1146/annurev.biochem.74.010904.153721. [DOI] [PubMed] [Google Scholar]
- 4.Li E, Beard C, Jaenisch R. Nature. 1993;366:362. doi: 10.1038/366362a0. [DOI] [PubMed] [Google Scholar]
- 5.Bourc'his D, Bestor TH. Nature. 2004;431:96. doi: 10.1038/nature02886. [DOI] [PubMed] [Google Scholar]
- 6.Walsh CP, Chaillet JR, Bestor TH. Nat Genet. 1998;20:116. doi: 10.1038/2413. [DOI] [PubMed] [Google Scholar]
- 7.Panning B, Jaenisch R. Cell. 1998;93:305. doi: 10.1016/s0092-8674(00)81155-1. [DOI] [PubMed] [Google Scholar]
- 8.Cheng X, Kumar S, Posfai J, Pflugrath JW, Roberts RJ. Cell. 1993;74:299. doi: 10.1016/0092-8674(93)90421-l. [DOI] [PubMed] [Google Scholar]
- 9.Allen MD, et al. EMBO J. 2006;25:4503. doi: 10.1038/sj.emboj.7601340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cierpicki T, et al. Nat Struct Mol Biol. 2010;17:62. doi: 10.1038/nsmb.1714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Materials and methods are available as supporting material on Science Online.
- 12.Birke M, et al. Nucleic Acids Res. 2002;30:958. doi: 10.1093/nar/30.4.958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lee JH, Voo KS, Skalnik DG. J Biol Chem. 2001;276:44669. doi: 10.1074/jbc.M107179200. [DOI] [PubMed] [Google Scholar]
- 14.Pradhan M, et al. Biochemistry. 2008;47:10000. doi: 10.1021/bi8011725. [DOI] [PubMed] [Google Scholar]
- 15.Klimasauskas S, Kumar S, Roberts RJ, Cheng X. Cell. 1994;76:357. doi: 10.1016/0092-8674(94)90342-5. [DOI] [PubMed] [Google Scholar]
- 16.Single-letter abbreviations for the amino acid residues are as follows: A, Ala; C, Cys; D, Asp; E, Glu; F, Phe; G, Gly; H, His; I, Ile; K, Lys; L, Leu; M, Met; N, Asn; P, Pro; Q, Gln; R, Arg; S, Ser; T, Thr; V, Val; W, Trp; and Y, Tyr. In the mutants, other amino acids were substituted at certain locations; for example, H134R indicates that histidine at position 134 was replaced by arginine.
- 17.Hou Z, Bernstein DA, Fox CA, Keck JL. Proc Natl Acad Sci U S A. 2005;102:8489. doi: 10.1073/pnas.0503525102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhang Z, Hayashi MK, Merkel O, Stillman B, Xu RM. EMBO J. 2002;21:4600. doi: 10.1093/emboj/cdf468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Leonhardt H, Page AW, Weier HU, Bestor TH. Cell. 1992;71:865. doi: 10.1016/0092-8674(92)90561-p. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.