Abstract
In this study we developed and characterized a delivery system for the epigenetic demethylating drug, decitabine, to sensitize temozolomide-resistant human glioblastoma multiforme (GBM) cells to alkylating chemotherapy. A poly(lactic-co-glycolic acid) (PLGA) and polyethylene glycol (PEG) based nano-conjugate was fabricated to encapsulate decitabine and achieved a better therapeutic response in GBM cells. After synthesis, the highly efficient uptake process and intracellular dynamics of this nano-conjugate was monitored by single-molecule fluorescence tools. Our experiments demonstrated that, under an acidic pH due to active glycolysis in cancer cells, the PLGA-PEG nano-vector could release the conjugated decitabine at a faster rate, after which the hydrolyzed lactic acid and glycolic acid would further acidify the intracellular microenvironment, thus providing a “positive feedback” to increase the effective drug concentration and realize growth inhibition. In temozolomide-resistant GBM cells, decitabine can potentiate the cytotoxic DNA alkylation by counteracting cytosine methylation and reactivating tumor suppressor genes, such as p53 and p21. Owing to excellent internalization and endo-lysosomal escape enabled by the PLGA-PEG backbone, the encapsulated decitabine exhibited a better anti-GBM potential than free drug molecules. Hence, the synthesized nano-conjugate and temozolomide could act in synergy to deliver a more potent and long-term anti-proliferation effect against malignant GBM cells.
Keywords: Decitabine, Temozolomide, Glioblastoma Multiforme, Single-molecule Spectroscopy, Drug delivery, Nano-conjugate
INTRODUCTION
Arising from normal glial cells, human GBM constitutes the most prevalent primary brain tumor of adults. Even with gross total resection in surgery and adjuvant therapies, the overall median survival for GBM patients is only 14.6 months and the two-year survival is approximately 30%.1 As a primary choice of chemotherapy against GBM, temozolomide can elicit a drastic anti-tumor effect via inducing efficient alkylation of guanine residues in DNA. However, after short, temporary remission GBM would rapidly relapse and become chemo-resistant due to its infiltration-growth and heterogeneous histological composition: those tumor cells that evade surgical removal and bear drug-resistant potential could lead to a more aggressive phenotype.
Epigenetic aberration extensively participates in cancer initiation and progression, as well as in the etiology of human GBM.2, 3 Through modifications that occur on DNA, histone and chromatin structure, epigenetic mechanism fine-tunes the genetic activity in a time- and tissue-dependent manner without changing the DNA sequence. Among the elucidated epigenetic modifications, DNA methylation – the addition of a methyl group to the 5-carbon of cytosine, is the most investigated. Global DNA hypomethylation and promoter hypermethylation of particular tumor suppressor genes have been characterized as epigenetic hallmarks in a variety of human cancers.4, 5 For GBM, it has been discovered that the abnormal expression of O-6-methylguanine-DNA-methyltransferase (MGMT) gene, mainly regulated by DNA methylation, substantially contributes to the failure of alkylating (i.e. temozolomide) treatment.6 In the general paradigm of epigenetic regulation, hypermethylating a gene promoter has the tendency to switch off or suppress the downstream transcription, but this negative correlation does not seem to be a robust approach for the regulation of MGMT in human GBM.7–9 Instead, a positive correlation between the gene body methylation and MGMT expression was recently noted. Hence pretreating MGMT-expressing cells with decitabine, a cytosine analogue with its 5-carbon substituted by nitrogen, can considerably sensitize tumor cells to temozolomide.10 In the light of these findings, it will be of tremendous value to optimize current epigenetic treatments to advance regular chemotherapies against GBM.
Utilizing DNA demethylating agents, such as 5-azacytidine (AZA) and decitabine (5-aza-2′-deoxycytidine, DAC), in myelodysplastic syndrome to complement traditional treatments has shown encouraging improvement in patient prognosis.11 However, to apply these drugs in solid tumors, especially brain tumors, is quite challenging, although some laboratory experiments and pre-clinical attempts have been conducted.12–15 Several major hurdles need to be overcome. First, unmodified nucleoside drugs have a short circulating time and low solubility before their degradation and clearance from human body.16 Second, a large number of chemo-molecules, including epigenetic drugs, are unable to reach an effective concentration in the central nervous system due to the blood-brain barrier (BBB). Third, the nucleoside transporter-dependent internalization by target cells also impedes an efficient intracellular accumulation of AZA and DAC.17 Moreover, without extra targeting strategy or surface modification, the inability for small-molecule drugs to differentiate cancer cells from normal cells would cause a series of unexpected side effects.
In this study, our objective is to compose a PLGA-PEG-based polymeric nano-vector for decitabine delivery, to sensitize MGMT-expressing GBM cells to temozolomide treatment. Featuring excellent biocompatibility and pH-sensitive biodegradability,18 PLGA-nanoparticle has proven to be a promising chemo-carrier for brain tumors.19–21 In line with existing research, our synthesized nano-conjugate exhibited outstanding delivering efficiency and rapid endo-lysosomal escape in GBM cells. Further, the acidic environment in cancer cells, due to the Warburg effect, can potently accelerate drug release from the PLGA-PEG backbone, which confers the nano-conjugate a targeting preference to impact cancer cells over normal cells. In combination with temozolomide, the free form of decitabine can intrinsically provoke a cytotoxic effect against MGMT-expressing cells, whereas our nano-conjugate formulation further improves the uptake and long-term efficacy of decitabine, which provides a superior option for future in vivo drug delivery.
METHODS AND MATERIALS
Synthesis of PLGA-PEG-OCH3 (Conjugate 1)
The PLGA-PEG di-block was synthesized as described in previous studies with minor modification.22 PLGA 50:50 with average molecular weight 8586 Da (0.31 mM) was dissolved in dry dichloromethane (DCM 10 ml). The carboxylic acid of PLGA was activated by 0.62 mM NHS and 0.375 mM DCC overnight at room temperature. The side product dicyclohexylurea (DCU) was filtered out; activated PLGA-NHS was precipitated by ice cold diethyl ether and dried under vacuum. The activated PLGA (2 g) was then added into methoxy polyethylene glycol (NH2-PEG-OCH3) (0.42 mM) solution in 10 ml dry DCM, and the reaction mixture was stirred at room temperature for 24 hours. The PLGA-PEG conjugate was precipitated out from the reaction mixture by adding diethyl ether and centrifuged at 3,220 g for 10 minutes. The supernatant was discarded and the polymer was washed with a mixture of diethyl ether and hexane 50:50 (3 × 5 ml) to remove excess reagents. Finally, the polymer was dried under vacuum for 24 hours to obtain the conjugated product. The polymer was characterized by 1H NMR spectroscopy. 1H NMR (DMSO, 400 MHz): δH 5.36 – 5.00 (m, PLGA-CH2-), 4.94 – 4.57 (m, PLGA-CH2-), 3.67 – 3.58 (m, PEG-CH2-), 3.38 (s, PEG-OCH3), 1.52 (s, PLGA -CH3-).
Addition of -COOH group to PLGA block of PLGA-PEG (Conjugate 2)
The PLGA block of Conjugate 1 was modified by reacting with succinic anhydride to produce COOH-PLGA-PEG. 1.78 g of Conjugate 1 was dissolved in dry DCM (10 ml) containing 4-Dimethylaminopyridine (DMAP 25% w/v) as a catalyst, and succinic anhydride (39.3 mg) was added. After stirring overnight at 50°C, the DCM was removed in vacuum. The Conjugate 2 was re-dissolve in dry DCM and precipitated in ice cold diethyl ether, dialyzed against water for 4 hours using MWCO 3,500 and lyophilized. The polymer was characterized by 1H NMR spectroscopy. 1H NMR (DMSO, 400 MHz): δH 5.41 – 5.15 (m, PLGA-CH2-), 5.02 – 4.59 (m, PLGA-CH2-), 3.71 – 3.60 (m, PEG-CH2-), 3.38 (s, PEG-OCH3), 5.74 (s, SA-O-CH2), 1.57 (s, PLGA -CH3-).
Synthesis of DAC-PLGA-PEG (Conjugate 3)
Conjugate 2 (0.08 mM) was dissolved in dry DCM containing 0.23 mM DPTS and cooled at 0°C. For the reaction mixture, a solution of decitabine (0.09 mM) dissolved in dry N,N-dimethylformamide (DMF) was added with continuous stirring. Then 0.23 mM DCC in pyridine was added slowly to the solution and the reaction was carried out at 4°C for 72 hours under nitrogen atmosphere. The progress of reaction was monitored by TLC with CHCl3/CH3OH (1:4, v/v) as eluent (product Rf = 0.68). The DCM and pyridine were removed under reduced pressure and the product was re-dissolved in DCM and precipitated in ice cold diethyl ether. Untreated drug and other impurities were removed followed by dialysis. The dialysis was precipitated in ice cold diethyl ether/hexane 50:50 mixture, and was freeze-dried to keep at −20°C. The free drug and drug conjugate were characterized by UV and 1H NMR spectroscopy. 1H NMR of DAC (DMSO, 400 MHz): δH 8.56 (s, 1H, H-6), 7.51 (s, 2H, NH2), 6.15 (t, 1H), 5.25 (s, 1H, OH-2′), 5.00 (s, 1H, OH), 4.2(s, 1H, H), 3.72 (1H, H), 3.6 – 3.7 (m, 1H, H), 3.7 – 3.5 (m, 2H, H) 23 and 1H NMR of DAC-PLGA-PEG (Conjugate 3) (400 MHz): δ (ppm) = 8.56 (s, 1H, H-6), 7.52–7.49 (s, 2H, NH2), 6.15 (t, 1H), 5.26 (s, SA-O-CH2), 5.25 (m, PLGA-CH2-), 4.91–4.83 (m, PLGA-CH2-) 4.2(s, 1H, H), 3.76 (1H, H), 3.6 – 3.7 (m, 1H, H), 3.7 – 3.5 (m, 2H, H), 3.49 (m, PEG-CH2-), 3.22 (s, PEG-OCH3), 1.46 (s, PLGA -CH3-).
Synthesis of PLGA-PEG-FITC (Conjugate 4)
The di-block polymer PLGA-PEG-NH2 (600 mg) was prepared and dissolved in 20 ml DCM. FITC (22.3 mg) was dissolved in 250 μl of mixture of DCM/pyridine and was added into the co-polymer solution. The reaction was allowed to run at 4°C for 24 hours in dark. The resulting conjugate was precipitated by ice cold diethyl ether (125 ml) and the product was centrifuged at 3,220 g for 30 minutes. The supernatant was discarded and the polymeric product was washed thoroughly by diethyl ether (5 ml × 3) and dried under vacuum overnight. The amount of fluorophore conjugated was determined using the fluorescence spectrometer at an excitation wavelength of 485 and emission at 530 nm.
Preparation of nano-micelles for biological tests
The DAC-PLGA-PEG (DC) and fluorescent DAC-PLGA-PEG (FDC) nano-micelles were prepared by film dispersion method. The DC conjugates with and without FDC conjugates were dissolved in chloroform at room temperature. The solvent was removed under reduced pressure to form a dry drug containing polymer film. This film was hydrated with 10 mM HEPES buffer (pH 7.4) at room temperature for 30 minutes. The nano-micelles were extruded through a 200-nm polycarbonate membrane. The concentration of drug is determined by UV spectrophotometer for DC and by fluorescent spectrometer for FDC with 485 nm as excitation and 530 nm as emission.
Characterization of nano-micelles
The size and morphology of final products were determined by transmission electron microscope (TEM, Philips, CM1000) with 100-kV acceleration voltage. Nano-conjugate were diluted to 0.5 mg/ml with HEPES buffer (pH 7.4) and stained with 0.8 % uranyl acetate. The zeta potential of nano-micelles was measured with a ZetaPlus analyzer (Brookhaven). Gel permeation chromatography (GPC) was performed using an Agilent 1200 series Wyatt Dawn Heleos-II system equipped with UV, LSD & RID detector. N-Dimethylformamide was used as the mobile phase at flow rate of 0.3 ml/min through OHpak SB-803 HQ [8.0 × 300 (mm): inner diameter × length] column at 40°C. GPC calibration curve was constructed using polystyrene standards.
Cell culture and cytotoxicity assay
Human GBM cell lines SF767 and U87 were cultured in Iscove’s Modified Dulbecco’s Medium (IMDM) supplemented with 10% fetal bovine serum, 1% antibiotics and 1% glutamate. MTT assay was used to test the short-term cell viability with different treatments. In a 96-well plate assay, 3,000 cells were seeded in each well and grown for 24 hours. Then the culture medium was replaced with fresh medium containing predetermined agents. After 72 hours, the medium was quickly removed and cells were rinsed with clean 1 × PBS. Cells were incubated with 100 ml fresh medium containing 10 ml thiazolyl blue tetrazolium bromide solution (MTT, 5 mg/ml in PBS as stock) for 3–4 hours and the precipitated formazan was dissolved in 100 ml acidic isopropanol prior to analysis by microplate reader at an absorbance of 570 nm. For long-term proliferation assay, 5,000 cells were seeded to a T25 flask at day 0, and the cell number under each specific condition was counted for 8 days.
DNA methylation assay
During long-term treatment (8 days), genomic DNA was extracted from cells to determine the global methylation level at Day 4 and Day 8. DNA extraction and purification were performed with DNeasy Blood & Tissue Kit (Qiagen). Global methylation was quantified by MethylFlash™ Methylated DNA Quantification Kit (Epigentek). 100 ng DNA was input for immunoassay and the generated colorimetric signal was measured by using a VersamaxTM absorbance microplate reader (Molecular Device). The percentage of DNA methylation was calculated by normalizing the optical intensity as to the positive control DNA.
Single-molecule fluorescence detection
Single-molecule fluorescence experiments (FCS, PCH and FLIM) were performed with a Microtime200 scanning confocal time-resolved system (Picoquant GmbH). A 465-nm picosecond pulsed laser was applied to excite the FITC tag on nano-conjugate. The excitation beam was delivered to the sample stage through an apochromatic water immersion objective (60×, N.A. = 1.2) and the emitted fluorescence was collected by the same objective, from which the emission was separated by a dual band dichroic (z467/638rpc, Chroma). A 50-μm pinhole was used to reject the off-focus photons, and the signal beam was further filtered by a band-pass filter (520/40 nm, Chroma) before reaching the single photon avalanche photodiode detector (SPAD) (SPCM-AQR, PerkinElmer Inc.).
Fluorescence correlation spectroscopy (FCS)
Autocorrelation function G(τ) in FCS is the measure of similarity between observations as a function of the time lag (τ).24, 25 For fluorescent molecules diffusing in-and-out a femtoliter-level detection volume enable by confocal instrumentation, the autocorrelation function can be obtained as:
| (1) |
where <F> is the average fluorescence intensity, and δF(t) = F(t) − <F(t)>. For molecules diffusing freely in a 3D Gaussian space, Equation 1 can be re-written as:
| (2) |
where , w0 and z0 are the lateral and axial radii of the detection profile, respectively. τD is the diffusion time, which reflects the average dwelling time of the fluorescent molecules in the detection volume and can be correlated to the diffusion coefficient (D) as: .
Photon counting histogram (PCH)
The sensitivity of FCS mainly depends on the molecular size rather than the molecular weight of diffusors, which requires a minimum of 1.6-fold change in hydrodynamic radius or at least 4.1-fold change in molecular weight to distinguish two species of spherical shape (Figure S6B).26 PCH provides another inherent characteristic – the molecular brightness (ε), which reflects on the stoichiometry of fluorescent molecules when molecular oligomerization cannot bring about a significant difference in diffusion.27, 28 Defined as the average number of photons detected per molecule per integration interval (e.g. cpsm, counts per second per molecule) and can be calculated as follows:
| (3) |
where εi is the brightness for species i, fi the fractional intensity and σ2 the variance. PCH analysis can simultaneously determine the average number and brightness of molecule within a 3D Gaussian volume. The gamma factor (γ) is an excitation-dependent value and assigned as 0.3536 for single photon system. The algorithm was integrated into an ImageJ plugin and kindly provided by Stowers Institute for Medical Research.
Fluorescence lifetime imaging microscopy (FLIM)
Fluorescence lifetime (τ) is defined as the average time the fluorophore stays at the excited state prior to emitting the first photon and can be expressed as:
| (4) |
where Γ is the emissive rate and knr the non-radiative decay rate. Fluorescence lifetime is independent of the molecular concentration and excitation intensity, but it is quite sensitive to the surrounding physicochemical factors such as pH. In the time-domain lifetime measurement, the pixel-by-pixel emitted photons were recorded using time-correlated single photon counting (TCSPC) module and stored in time-tagged time-resolved (TTTR) format (Time Harp200, PicoQuant GmbH), based on which the lifetime can be calculated as the amplitude of detected photons decays to 1/e: F(t) = F0e−t/τ. In FLIM images, a TCSPC histogram is generated for each pixel to reconstruct a color-coded lifetime image.
Drug release kinetics
The DAC-PLGA-PEG nano-conjugate was suspended in 500 μl of PBS (pH 5 or 7.4) and dialyzed (MWCO 3.0 kDa) against 1.5 ml of medium. Then the tubes were placed in an orbital shaker setting at 120 rpm and 37°C. At predetermined time (1h, 2h, 4h, 8h, 12h, 24h, 48h, 72h), 10 μl aliquot was withdrawn, and replaced with fresh buffer. The released DAC concentration was quantified by UV-Vis spectroscopy at a characteristic wavelength of DAC, λmax 244 nm (pH 5) and 220 nm (pH 7.4) (Figure S5).29 The release of DAC from DAC-PLGA-PEG micelle was expressed as a percentage of the drug released and plotted as a function of time.
Quantitative RT-PCR
RNA extraction and reverse transcription were conducted using RNeasy Mini Kit (Qiagen) and iScript™ cDNA Synthesis Kit (Bio-Rad) respectively, according to manufacturers’ instructions. Before cDNA synthesis, genomic DNA was removed by DNase I (Life Technologies). PCR amplification was performed in a StepOnePlus™ system with SYBR® Green PCR Master Mix (Applied Biosystems). The GAPDH gene was used as internal control and ΔΔCT method was applied to compare gene expression.
Flow cytometry
For cell cycle analysis, cells were fixed with 70% ethanol and stained with 20 μg/ml propidium iodide solution containing 0.1% Triton X-100 and 200 μg/ml RNase A, according to standard protocol.30 Flow cytometry was performed in a FC500 MPL system (Beckman Coulter). 10,000 cells were collected for each condition, and the cell cycle histograms were analyzed with Watson pragmatic model in Flowjo software.
Statistical analysis
In significance test: student t-test was used for two sample comparison, and ANOVA test was used for multiple sample comparison. All analysis was performed with OriginPro software.
RESULTS AND DISCUSSION
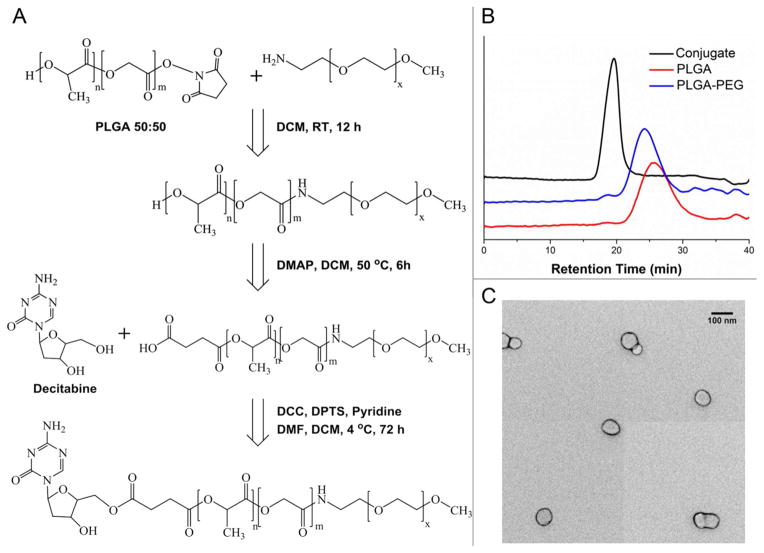
Decitabine-containing nano-conjugate was prepared with a three-step synthetic route (Figure 1A). First, N-hydroxysuccinimide (NHS)-activated PLGA reacted with PEG to form the co-polymer PLGA-PEG. Then the terminal hydroxyl group of PLGA-PEG was converted to carboxylic group by reaction with succinic acid, and the succinylated PLGA-PEG co-polymer was allowed to react with the 5′-OH of decitabine in the presence of N,N′-dicyclohexylcarbodiimide (DCC) and 4-(Dimethylamino) pyridinium-4-toluene sulfonate (DPTS), to result in the formation of DAC-PLGA-PEG conjugate. The final product was confirmed by 1H NMR spectroscopy with all the characteristic peaks and integration values as indicated in Figures S1–3. In addition, it has been reported that carbodiimide derivatives and DPTS are useful esterification reagents with prominent reaction yield.31 The number-averaged molecular weight of the DAC-PLGA-PEG co-polymer was found to be 9,975, which was calculated by using the ratio among the peak areas at 5.25 ppm (-CH protons of PLA), 4.85 ppm (-CH protons of PGA), 3.49 ppm (-CH2 protons of PEG) and 8.5 ppm (-NH2 of decitabine). The decitabine content in DAC-PLGA-PEG conjugate was estimated to be 2.29% (by weight) with UV-Vis spectroscopy, and the difference between PLGA, PLGA-PEG and DAC-PLGA-PEG was further confirmed by GPC. The incremental reduction in the retention time of products along the synthesis clearly indicates an increase in the molecular size following each attaching reaction (Figure 1B).
Figure 1.
Synthesis of DAC-PLGA-PEG nano-conjugate. (A) Brief schematic for chemical conjugation. (B) The successful attachment of each component was validated by the decreased retention time in GPC measurement. (C) TEM image of the formed nano-micelles.
The solution of micelle-like nano-conjugate of DAC-PLGA-PEG was prepared with Bangham method which was initially used for liposome production.32, 33 DAC-PLGA-PEG conjugates were dispersed in organic solvent. Then, the organic solvent was removed under reduced pressure. Finally, the dry polymeric film deposited on the flask wall was hydrated by adding neutral buffer solution under agitation at room temperature. This method is widespread and easy to handle; however, dispersed polymer in aqueous buffer yields a population of heterogeneous particles both in size and shape. Thus the nano-micelles were further extruded through polycarbonate filters to obtain a more uniform solution.34 The morphology of nano-micelles was then analyzed by TEM (Figure 1C), from which the spherical geometry of the particle with an average diameter of ~55 nm was determined. In addition, with surface PEGylation, the originally negative PLGA-based nano-micelles were tune to have a neutral (slightly negative) zeta potential (Figure S4C). The physical characteristics of polymeric nanoparticles such as size, shape and surface charge are critical factors for their stability, in vivo pharmacokinetics and bio-availability. In vivo nano-vectors must first evade the reticuloendothelial system (RES) which can opsonize foreign nanomaterials and subsequently induce their degradation via phagocytosis by macrophages.35 PLGA-nanoparticles below 100 nm with hydrophilic surface modification such as PEG could greatly avoid opsonization and minimize the clearance by RES. This would prolong the circulation time of nanoparticles and allow the delivery of more payloads to tumor lesions.36 The other rationale for us to add PEGylation onto this nano-conjugate lies in that positively charged surface particles bind easily to a variety of cells in the body, including vascular endothelial cells, before reaching the diseased loci. Therefore it would be preferential to maintain the nano-vectors as either neutral or slightly anionic for a better stabilization and less non-specific drug loss.37, 38
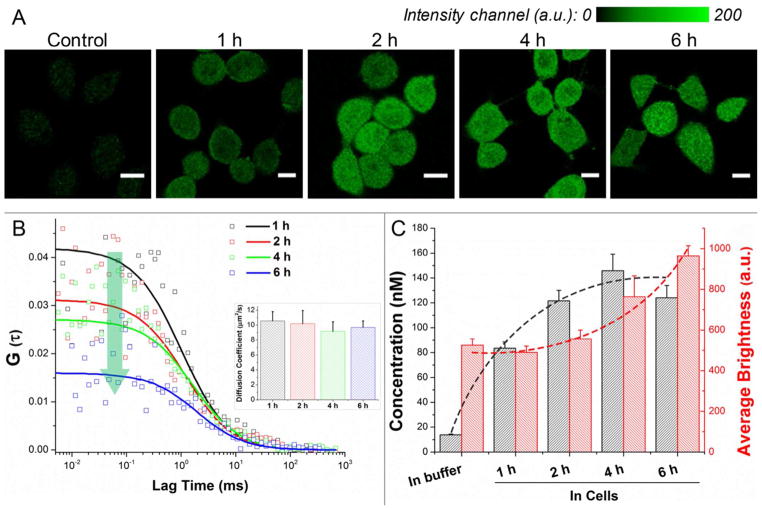
Exploration of drug release kinetics and diffusion by single-molecule spectroscopy in living single cells enables us to record the spatiotemporal distribution of this nano-conjugate in real time with the resolution down to microsecond and in femtoliter volume.39–41 By utilizing ultrashort pulsed laser and SPAD system, the fluorescence fluctuation in an extremely small volume (~0.6 femtoliter under 465 nm excitation) can be extracted and interpreted with FCS and PCH as presented in Figure 2.42 In order for the fluorescence fluctuation to be detected, the concentration of fluorescent molecules should be in the pM to nM range to avoid signal deficiency or saturation. For live cell experiments, DAC-PLGA-PEG and PLGA-PEG-FITC were mixed in a 2:1 ratio to render the formed nano-micelles fluorescent, which did not affect the basic physicochemical properties of PLGA-PEG nanoparticles (Figure S4). The nano-micelles in buffer solution (determined by FCS) were incubated (initial concentration of 13.8 nM) with SF767 cells for up to 6 hours (Figure 2A–B). The intracellular concentration of nano-micelles could reach 83.5 ± 5.0 nM within the first hour, quantitatively confirming the high uptake rate and delivery efficiency of this PLGA-PEG-based nano-vector. After 4 hours, the intracellular concentration peaked at 145.8 ± 13.4 nM as shown in Figure 2C. Notably, the average diffusion coefficient of nano-micelles in living cells did not experience drastic change at different time points (Figure 2B, inset), which indicates that no significant aggregation or abrupt particle disintegration occurred within 6 hours. From PCH, the average brightness of nano-micelles displayed a gradual increase (Figure 2C, red dash line). We therefore speculated that some PLGA fragments containing FITC molecules hydrolyzed from early-taken micelles would integrate into newly internalized micelles due to physical adsorption or hydrophobic force. From the fluorescence images, the PLGA-PEG-based nano-micelles exhibit a uniform distribution inside the whole cell without compartmental entrapping (Figures 2A and S6A), indicative of its elusion from the endo-lysosomal system. The results from FCS and PCH suggest that the maximum cell internalization of the PLGA-PEG nano-conjugate could occur within 1 hour for GBM cells, followed by a gradual hydrolysis process.
Figure 2.
Single-molecule characterization of cell uptake and intracellular behavior of the nano-micelles. (A) Confocal fluorescence images of cells incubated with fluorescent nano-micelles. (B) Autocorrelation curves from single living cells for FCS measurement. By fitting with an appropriate model, the diffusion coefficient (inset panel), (C) the average number/concentration and brightness of molecules in the detection volume can be determined (n > 20, mean + S.E.M.).
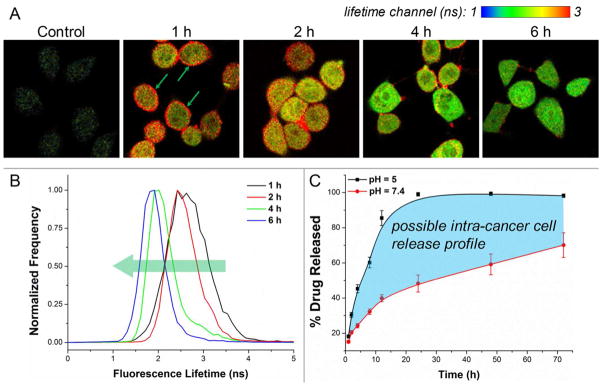
Upon the hydrolysis of PLGA backbone, the intracellular accumulation of lactic acid and glycolic acid was expected, and these two products would further acidify the internal environment to facilitate a drug release cascade. To some extent, the further acidified pH could not only accelerate the hydrolysis rate of PLGA, but also slow down the triazine ring-opening decomposition of decitabine,29 thus supporting a better efficacy of our nano-conjugate in against cancer cells. The fluorescence lifetime of FITC is a sensitive measure of its surrounding pH, and an acidic environment can cause a significant reduction in the fluorescence lifetime of FITC.43 Hence, FLIM was applied to indirectly assess the process of acidification and drug release inside cells. In FLIM, color-coded lifetime is visualized based on a pixel-by-pixel TCSPC calculation. Figure 3A shows the time-resolved FLIM images at different incubation times. It is clear, especially from the 1 hour-incubation image, that SF767 GBM cells boast an acidic internal environment since FITC on the membrane exhibits a significant longer lifetime (green arrows) before internalization. Along with incubation, the significantly shortened fluorescence lifetime of FITC inside cells was noted in FLIM images, which also can be distinguished from the globally shifted lifetime histograms (Figure 3B). To verify the influence of pH on PLGA hydrolysis, we performed the in vitro hydrolysis assay to study the drug release kinetics of our nano-conjugate at pH 5.0 and 7.4, respectively. As shown in Figure 3C, the hydrolysis rate was significantly faster at acidic pH than in neutral environment. Approximately 85% of the conjugated decitabine was released at pH 5.0 within the first 12 hours, whereas only ~40% was released at pH 7.4. This validates the susceptibility of the ester bond-mediated PLGA-conjugation to acid; however, a more comprehensive release study is required for future in vivo applications due to the lack of analytical methods for simultaneously detecting decitabine and its degradation.44, 45
Figure 3.
The influence of pH on nano-conjugate hydrolysis and drug release. (A) Time-resolved FLIM images of the FITC molecules integrated into the nano-micelles are presented. The reduction of FITC lifetime inside cells implicates a further acidification of intracellular pH. (B) Lifetime histograms from the above images are shown. (C) The intra-cancer cell drug release was projected based on in vitro drug release kinetics (n = 3, phosphate buffers).
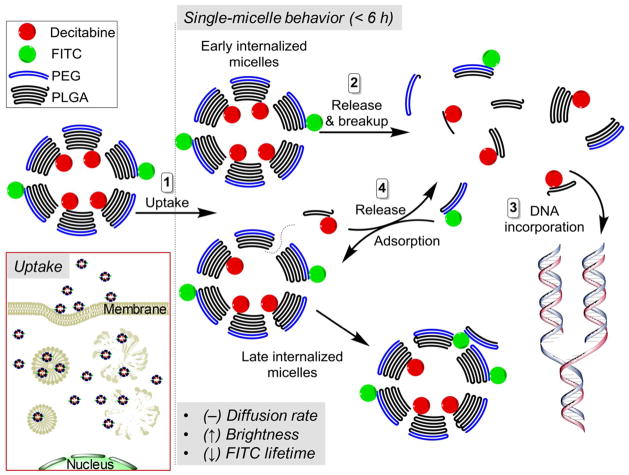
Based on the results from our single-molecule experiments, the intracellular behavioral mode of DAC-PLGA-PEG was proposed (Figure 4). Having a size of 50–60 nm, the formed nano-micelles can rapidly enrich in GBM cells within 1 – 2 hours and the delivery of decitabine here, to a great extent, is thus independent of nucleoside transporters. Instead, the initial linear uptake trend and the later saturated uptake trend, revealed by FCS measurements (Figure 2C, black dash line), evidently suggest that the cell internalization of sub-100 nm PLGA-nanoparticles is the outcome of a cooperation between fluid-phase pinocytosis and clathrin-mediated endocytosis,46 which is different from the recently proposed mechanism for large-size PLGA-nanoparticles (i.e. via extracellular release of the drug payload to contacting cells).47 The rapid endo-lysosomal escape is another advantage for PLGA-based delivery since a further anionic-to-cationic reversal of the particle surface charge facilitates the nano-micelles to interact with endo-lysosomal membrane and achieve the escape within 10 minutes.48 Moreover, the internalized nano-micelle can well maintain its integrity within a considerable time (at least 4 – 6 hours), and its hydrolysis is subject to acidic pH in cancer cells. Taken together, our single-molecule profiling has for the first time quantitated the intracellular dynamics of a PLGA-based delivery system. Further, under in vivo condition, considering the “enhanced permeation and retention” (EPR) effect of tumor microenvironment, the long-circulating and BBB-penetrating PLGA-PEG nano-micelles are expected to be an ideal nano-vector to target brain tumors.
Figure 4.
Proposed single-molecule behavior of PLGA-PEG-based nano-micelles in GBM cells.
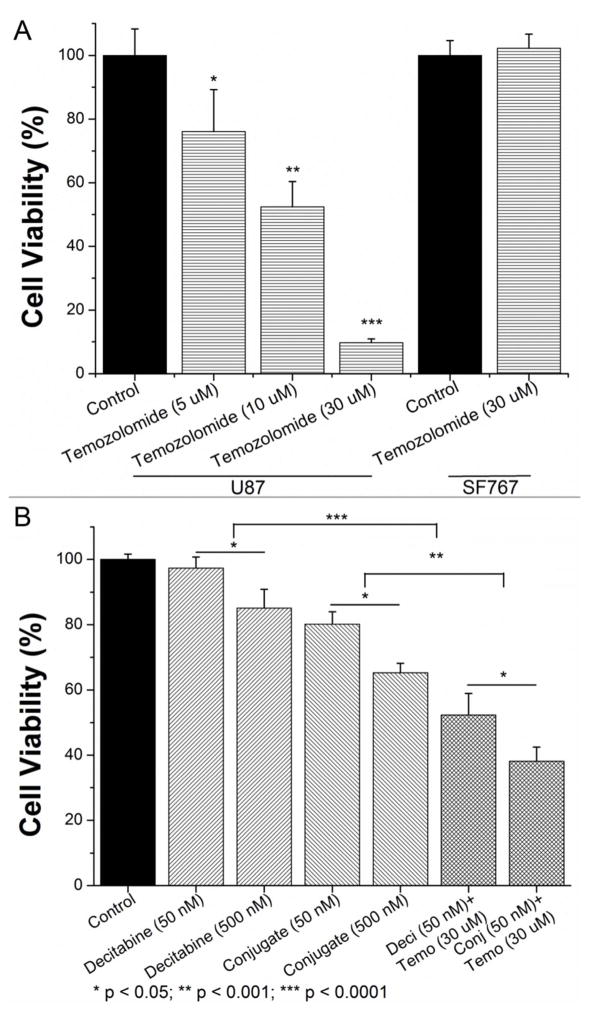
To explore the therapeutic potential of this nano-conjugate, we applied MTT assay to evaluate the cell viability under different conditions. First of all, the MGMT-mediated resistance to temozolomide treatment was assessed in different cell lines. MGMT-positive SF767 cells and MGMT-negative U87 cells were treated with temozolomide at a concentration of up to 30 μM for 72 hours. As expected, U87 cells were quite sensitive to temozolomide treatment and a dosage of 30 μM can fully suppress the cell proliferation (IC50 was estimated to be around 20 μM49). In contrast, SF767 cells did not show any significant response to 30 μM temozolomide (Figure 5A). In the following combinatorial treatments, SF767 cells were first sensitized with decitabine or our synthesized nano-conjugate with equivalent drug content (50 nM) for 24 hours, followed by addition of 30 μM temozolomide for another 48 hours. Notably, both decitabine and the nano-conjugate exhibited a concentration-dependent cytotoxicity against MGMT-expressing SF767 cells because of the non-specific effect, but the decitabine encapsulated in nano-micelles was more effective than the free drug possibly due to the better internalization efficiency. After sensitization with 50 nM decitabine as in free form or in nano-conjugate form, 30 μM of temozolomide could successfully suppress the cell viability by 47% and 62%, respectively (Figure 5B). This set of experiments thereby suggests a synergistic effect between decitabine/nano-conjugate and temozolomide.
Figure 5.
Therapeutic potential of combinatorial treatments. (A) Distinct temozolomide-resistance for MGMT-expressing (SF767) and MGMT-negative (U87) GBM cells was assessed. (B) The synergistic effect between decitabine and temozolomide was tested in SF767 cells. In the meantime, the difference in potency of decitabine in free form and in conjugate form was demonstrated (n = 6, mean + standard deviation).
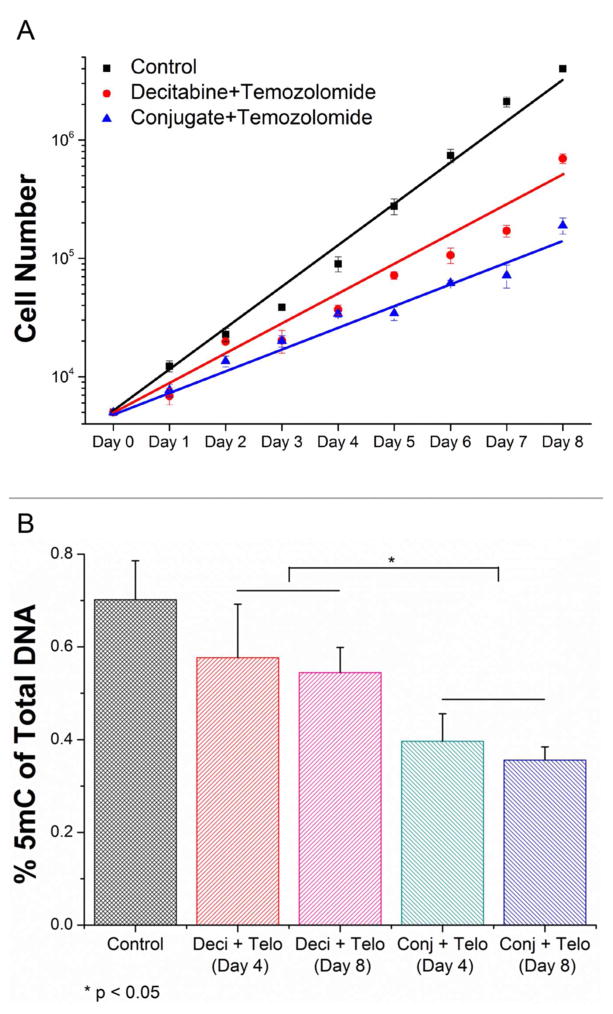
Next, we assessed the long-term effects of the combinatorial treatments to support the superior therapeutic efficacy of decitabine in the nano-conjugate form. A crucial pitfall for conventional PLGA-based delivery system comes from its undesired burst release of drug, so here we opted to chemically conjugate decitabine to the PLGA backbone instead of to physically load the free drug. This design merits a better estimation for drug dosage and also a sustainable release. In this regard, after MTT assay we extended the monitoring time for cell response, in terms of proliferation rate and DNA methylation change, to 8 days. It is clear from Figure 6A that the proliferation rate of SF767 cells under the conjugate and temozolomide treatment is significantly lower (by ~70%) than the rate under decitabine and temozolomide treatment in a long run. We also noted that a larger extent of reduction in 5-methylcytosine (5mC) could be achieved and maintained by decitabine nano-conjugate (Figure 6B), validating the drug effects on DNA methyltransferases and 5mC content.
Figure 6.
Long-term effect of combinatorial treatments. (A) SF767 cell proliferation was monitored for 8 days upon indicated treatments (for decitabine and conjugate, 50 nM of equal drug content was applied; for temozolomide, 30 μM was given; n = 3 replicates). (B) During the whole course, the quantity of DNA methylation (i.e. percentage of 5mC) was assessed at three time points (pre-treatment, middle, and post-treatment; n = 3, mean + standard deviation).
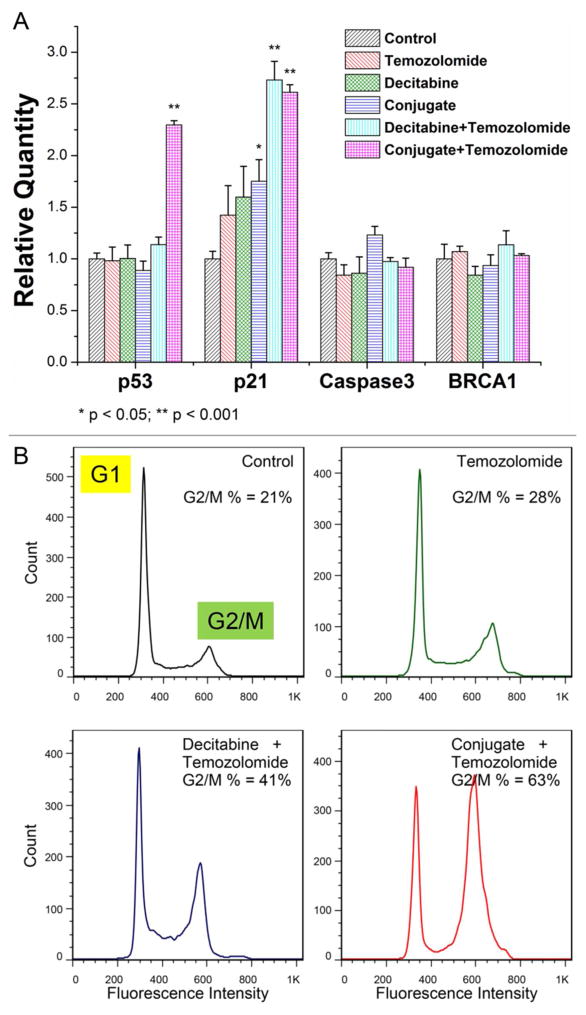
Since both decitabine and temozolomide function through inducing DNA alkylation but on different nucleosides, the next immediate query is to test whether their synergistic effect against MGMT-expressing GBM cells stems from triggering more extensive DNA damages. Quantitative RT-PCR was thus applied to check four critical genes responding to DNA damage and apoptosis initiation: p53, p21, Caspase3 and BRCA1. Consistent with the MTT assay results, temozolomide alone cannot evoke any significant changes in the transcription of these genes in SF767 cells, whereas the combinatorial treatment (conjugate and temozolomide) radically increased both transcripts of p53 and p21 (Figure 7A). However, we did not observe significant changes in Caspase3 and BRCA1, which verified the decitabine-induced Caspase-independent apoptotic response and impaired BRCA1-pathway in human gliomas.50, 51 The induction of p53 and p21 could in turn retard cell cycle progression and cell proliferation. Examined by flow cytometry, a much larger proportion of cells were found to be arrested in the G2/M-phase when subjected to combinatorial treatment (Figure 7B), which is in agreement with the previous research on decitabine pharmacology.52–54 Collectively, our data suggest that the main mechanism for decitabine to potentiate temozolomide treatment hinges on amplifying the effect of DNA damage, though we cannot rule out the possibility that the observed increase in p53 and p21 might also be attributed to the induced demethylation of their promoters by decitabine.
Figure 7.
Decitabine nano-conjugate better sensitizes MGMT-expressing GBM cells to temozolomide by inducing DNA damage and G2/M arrest in cell cycle progression. (A) Transcriptional levels of four DNA damage responsive genes were determined by quantitative RT-PCR after 72 hours treatment (n = 4, mean + standard deviation). (B) Cell cycle progression was evaluated by flow cytometry.
CONCLUSION
Taking advantage of the synergistic effect between decitabine and temozolomide to target human GBM, we address a major problem regarding drug delivery that has a promising significance in enabling future in vivo application of the decitabine involved chemotherapies. A PLGA-PEG-constituted nano-conjugate was herein devised to encapsulate decitabine, and the formed nano-micelles achieved a highly efficient cell uptake. The pH-dependent hydrolysis of PLGA renders this nano-conjugate more fragile in acidic microenvironment, which would aid in a more potent drug release and cytotoxic effect in cancer cells, specifically. Our experiments also indicate that the complementary effect of decitabine to temozolomide stems from inducing more significant DNA damage and triggering the p53/p21-dependent apoptotic pathway in MGMT-expressing GBM cells. Future efforts should be placed in understanding the in vivo pharmacokinetics and pharmacodynamics of this nano-vector, as well as its therapeutic potential in treating other solid tumors.
Acknowledgments
We thank Christopher J. Collins and Vivek Badwaik (Purdue University) for their assistance in NMR facility and HEC Pakistan training grant for funding Dr. Asia Naz. We want to thank Dr. Rebecca Packer (Colorado State University) for providing human GBM cell lines.
Footnotes
SUPPORTING INFORMATION
Supplementary results and materials as descried in the text. This material is available free of charge via the internet at http://pubs.acs.org/.
References
- 1.Das S, Marsden PA. Angiogenesis in glioblastoma. The New England journal of medicine. 2013;369(16):1561–3. doi: 10.1056/NEJMcibr1309402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Nagarajan RP, Costello JF. Epigenetic mechanisms in glioblastoma multiforme. Seminars in cancer biology. 2009;19(3):188–97. doi: 10.1016/j.semcancer.2009.02.005. [DOI] [PubMed] [Google Scholar]
- 3.Sharma S, Kelly TK, Jones PA. Epigenetics in cancer. Carcinogenesis. 2010;31(1):27–36. doi: 10.1093/carcin/bgp220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Esteller M. Epigenetics in cancer. The New England journal of medicine. 2008;358(11):1148–59. doi: 10.1056/NEJMra072067. [DOI] [PubMed] [Google Scholar]
- 5.Heyn H, Esteller M. DNA methylation profiling in the clinic: applications and challenges. Nature reviews Genetics. 2012;13(10):679–92. doi: 10.1038/nrg3270. [DOI] [PubMed] [Google Scholar]
- 6.Esteller M, Garcia-Foncillas J, Andion E, Goodman SN, Hidalgo OF, Vanaclocha V, Baylin SB, Herman JG. Inactivation of the DNA-repair gene MGMT and the clinical response of gliomas to alkylating agents. The New England journal of medicine. 2000;343(19):1350–4. doi: 10.1056/NEJM200011093431901. [DOI] [PubMed] [Google Scholar]
- 7.Brell M, Tortosa A, Verger E, Gil JM, Vinolas N, Villa S, Acebes JJ, Caral L, Pujol T, Ferrer I, Ribalta T, Graus F. Prognostic significance of O6-methylguanine-DNA methyltransferase determined by promoter hypermethylation and immunohistochemical expression in anaplastic gliomas. Clinical cancer research : an official journal of the American Association for Cancer Research. 2005;11(14):5167–74. doi: 10.1158/1078-0432.CCR-05-0230. [DOI] [PubMed] [Google Scholar]
- 8.Christmann M, Nagel G, Horn S, Krahn U, Wiewrodt D, Sommer C, Kaina B. MGMT activity, promoter methylation and immunohistochemistry of pretreatment and recurrent malignant gliomas: a comparative study on astrocytoma and glioblastoma. International journal of cancer Journal international du cancer. 2010;127(9):2106–18. doi: 10.1002/ijc.25229. [DOI] [PubMed] [Google Scholar]
- 9.Kreth S, Thon N, Eigenbrod S, Lutz J, Ledderose C, Egensperger R, Tonn JC, Kretzschmar HA, Hinske LC, Kreth FW. O-methylguanine-DNA methyltransferase (MGMT) mRNA expression predicts outcome in malignant glioma independent of MGMT promoter methylation. PloS one. 2011;6(2):e17156. doi: 10.1371/journal.pone.0017156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Moen EL, Stark AL, Zhang W, Dolan ME, Godley LA. The role of gene body cytosine modifications in MGMT expression and sensitivity to temozolomide. Molecular cancer therapeutics. 2014;13(5):1334–44. doi: 10.1158/1535-7163.MCT-13-0924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yang XJ, Lay F, Han H, Jones PA. Targeting DNA methylation for epigenetic therapy. Trends Pharmacol Sci. 2010;31(11):536–546. doi: 10.1016/j.tips.2010.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Eramo A, Pallini R, Lotti F, Sette G, Patti M, Bartucci M, Ricci-Vitiani L, Signore M, Stassi G, Larocca LM, Crino L, Peschle C, De Maria R. Inhibition of DNA methylation sensitizes glioblastoma for tumor necrosis factor-related apoptosis-inducing ligand-mediated destruction. Cancer research. 2005;65(24):11469–77. doi: 10.1158/0008-5472.CAN-05-1724. [DOI] [PubMed] [Google Scholar]
- 13.Cowan LA, Talwar S, Yang AS. Will DNA methylation inhibitors work in solid tumors? A review of the clinical experience with azacitidine and decitabine in solid tumors. Epigenomics. 2010;2(1):71–86. doi: 10.2217/epi.09.44. [DOI] [PubMed] [Google Scholar]
- 14.Nie J, Liu L, Li X, Han W. Decitabine, a new star in epigenetic therapy: the clinical application and biological mechanism in solid tumors. Cancer letters. 2014;354(1):12–20. doi: 10.1016/j.canlet.2014.08.010. [DOI] [PubMed] [Google Scholar]
- 15.Turcan S, Fabius AW, Borodovsky A, Pedraza A, Brennan C, Huse J, Viale A, Riggins GJ, Chan TA. Efficient induction of differentiation and growth inhibition in IDH1 mutant glioma cells by the DNMT Inhibitor Decitabine. Oncotarget. 2013;4(10):1729–36. doi: 10.18632/oncotarget.1412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Coade SB, Pearson JD. Metabolism of adenine nucleotides in human blood. Circulation research. 1989;65(3):531–7. doi: 10.1161/01.res.65.3.531. [DOI] [PubMed] [Google Scholar]
- 17.Damaraju VL, Mowles D, Yao S, Ng A, Young JD, Cass CE, Tong Z. Role of Human Nucleoside Transporters in the Uptake and Cytotoxicity of Azacitidine and Decitabine. Nucleos Nucleot Nucl. 2012;31(1–3):236–255. doi: 10.1080/15257770.2011.652330. [DOI] [PubMed] [Google Scholar]
- 18.Danhier F, Ansorena E, Silva JM, Coco R, Le Breton A, Preat V. PLGA-based nanoparticles: An overview of biomedical applications. J Control Release. 2012;161(2):505–522. doi: 10.1016/j.jconrel.2012.01.043. [DOI] [PubMed] [Google Scholar]
- 19.Gelperina S, Maksimenko O, Khalansky A, Vanchugova L, Shipulo E, Abbasova K, Berdiev R, Wohlfart S, Chepurnova N, Kreuter J. Drug delivery to the brain using surfactant-coated poly(lactide-co-glycolide) nanoparticles: Influence of the formulation parameters. Eur J Pharm Biopharm. 2010;74(2):157–163. doi: 10.1016/j.ejpb.2009.09.003. [DOI] [PubMed] [Google Scholar]
- 20.Wohlfart S, Khalansky AS, Gelperina S, Maksimenko O, Bernreuther C, Glatzel M, Kreuter J. Efficient Chemotherapy of Rat Glioblastoma Using Doxorubicin-Loaded PLGA Nanoparticles with Different Stabilizers. PloS one. 2011;6(5) doi: 10.1371/journal.pone.0019121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jain DS, Athawale RB, Bajaj AN, Shrikhande SS, Goel PN, Nikam Y, Gude RP. Unraveling the cytotoxic potential of Temozolomide loaded into PLGA nanoparticles. Daru. 2014;22(1):18. doi: 10.1186/2008-2231-22-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cheng J, Teply BA, Sherifi I, Sung J, Luther G, Gu FX, Levy-Nissenbaum E, Radovic-Moreno AF, Langer R, Farokhzad OC. Formulation of functionalized PLGA-PEG nanoparticles for in vivo targeted drug delivery. Biomaterials. 2007;28(5):869–876. doi: 10.1016/j.biomaterials.2006.09.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Redkar S, Joshi-Hangal R. Compositions and formulations of decitabine polymorphs and methods of use thereof. Google Patents. 2006 [Google Scholar]
- 24.Magde D, Webb WW, Elson E. Thermodynamic Fluctuations in a Reacting System - Measurement by Fluorescence Correlation Spectroscopy. Phys Rev Lett. 1972;29(11):705. [Google Scholar]
- 25.Lakowicz JR. Principles of fluorescence spectroscopy. 3. Springer; New York: 2006. p. xxvi.p. 954. [Google Scholar]
- 26.Meseth U, Wohland T, Rigler R, Vogel H. Resolution of fluorescence correlation measurements. Biophysical journal. 1999;76(3):1619–31. doi: 10.1016/S0006-3495(99)77321-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chen Y, Muller JD, So PT, Gratton E. The photon counting histogram in fluorescence fluctuation spectroscopy. Biophysical journal. 1999;77(1):553–67. doi: 10.1016/S0006-3495(99)76912-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chen Y, Muller JD, Ruan Q, Gratton E. Molecular brightness characterization of EGFP in vivo by fluorescence fluctuation spectroscopy. Biophysical journal. 2002;82(1 Pt 1):133–44. doi: 10.1016/S0006-3495(02)75380-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Rogstad DK, Herring JL, Theruvathu JA, Burdzy A, Perry CC, Neidigh JW, Sowers LC. Chemical decomposition of 5-aza-2′-deoxycytidine (Decitabine): kinetic analyses and identification of products by NMR, HPLC, and mass spectrometry. Chemical research in toxicology. 2009;22(6):1194–204. doi: 10.1021/tx900131u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Juan G, Traganos F, Darzynkiewicz Z. Methods to identify mitotic cells by flow cytometry. Method Cell Biol. 2001;63:343. doi: 10.1016/s0091-679x(01)63019-x. [DOI] [PubMed] [Google Scholar]
- 31.Aryal S, Hu CMJ, Zhang LF. Combinatorial Drug Conjugation Enables Nanoparticle Dual-Drug Delivery. Small. 2010;6(13):1442–1448. doi: 10.1002/smll.201000631. [DOI] [PubMed] [Google Scholar]
- 32.Yu B, Lee RJ, Lee LJ. Microfluidic methods for production of liposomes. Methods in enzymology. 2009;465:129–41. doi: 10.1016/S0076-6879(09)65007-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zhang L, Pornpattananangku D, Hu CM, Huang CM. Development of nanoparticles for antimicrobial drug delivery. Current medicinal chemistry. 2010;17(6):585–94. doi: 10.2174/092986710790416290. [DOI] [PubMed] [Google Scholar]
- 34.Laouini A, Jaafar-Maalej C, Limayem-Blouza I, Sfar S, Charcosset C, Fessi H. Preparation, characterization and applications of liposomes: state of the art. Journal of Colloid Science and Biotechnology. 2012;1(2):147–168. [Google Scholar]
- 35.Owens DE, Peppas NA. Opsonization, biodistribution, and pharmacokinetics of polymeric nanoparticles. Int J Pharmaceut. 2006;307(1):93–102. doi: 10.1016/j.ijpharm.2005.10.010. [DOI] [PubMed] [Google Scholar]
- 36.Acharya S, Sahoo SK. PLGA nanoparticles containing various anticancer agents and tumour delivery by EPR effect. Adv Drug Deliver Rev. 2011;63(3):170–183. doi: 10.1016/j.addr.2010.10.008. [DOI] [PubMed] [Google Scholar]
- 37.Sourabhan S, Kaladhar K, Sharma CP. Method to enhance the encapsulation of biologically active molecules in PLGA nanoparticles. Trends Biomater Artif Organs. 2009;22(3):211–5. [Google Scholar]
- 38.Yallapu MM, Gupta BK, Jaggi M, Chauhan SC. Fabrication of curcumin encapsulated PLGA nanoparticles for improved therapeutic effects in metastatic cancer cells. J Colloid Interf Sci. 2010;351(1):19–29. doi: 10.1016/j.jcis.2010.05.022. [DOI] [PubMed] [Google Scholar]
- 39.Slaughter BD, Schwartz JW, Li R. Mapping dynamic protein interactions in MAP kinase signaling using live-cell fluorescence fluctuation spectroscopy and imaging. Proceedings of the National Academy of Sciences of the United States of America. 2007;104(51):20320–5. doi: 10.1073/pnas.0710336105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Chen J, Irudayaraj J. Quantitative investigation of compartmentalized dynamics of ErbB2 targeting gold nanorods in live cells by single molecule spectroscopy. ACS nano. 2009;3(12):4071–9. doi: 10.1021/nn900743v. [DOI] [PubMed] [Google Scholar]
- 41.Cui Y, Cho IH, Chowdhury B, Irudayaraj J. Real-time dynamics of methyl-CpG-binding domain protein 3 and its role in DNA demethylation by fluorescence correlation spectroscopy. Epigenetics : official journal of the DNA Methylation Society. 2013;8(10):1089–100. doi: 10.4161/epi.25958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kim SA, Heinze KG, Schwille P. Fluorescence correlation spectroscopy in living cells. Nature methods. 2007;4(11):963–73. doi: 10.1038/nmeth1104. [DOI] [PubMed] [Google Scholar]
- 43.French T, So PT, Weaver DJ, Jr, Coelho-Sampaio T, Gratton E, Voss EW, Jr, Carrero J. Two-photon fluorescence lifetime imaging microscopy of macrophage-mediated antigen processing. Journal of microscopy. 1997;185(Pt 3):339–53. doi: 10.1046/j.1365-2818.1997.d01-632.x. [DOI] [PubMed] [Google Scholar]
- 44.Stresemann C, Lyko F. Modes of action of the DNA methyltransferase inhibitors azacytidine and decitabine. International Journal of Cancer. 2008;123(1):8–13. doi: 10.1002/ijc.23607. [DOI] [PubMed] [Google Scholar]
- 45.Neupane YR, Srivastava M, Ahmad N, Soni K, Kohli K. Stability indicating RP-HPLC method for the estimation of Decitabine in bulk drug and lipid based Nanoparticles. Int J Pharm Sci Res. 2014;7:294. [Google Scholar]
- 46.Panyam J, Labhasetwar V. Dynamics of endocytosis and exocytosis of poly(D,L-lactide-co-glycolide) nanoparticles in vascular smooth muscle cells. Pharm Res. 2003;20(2):212–20. doi: 10.1023/a:1022219003551. [DOI] [PubMed] [Google Scholar]
- 47.Xu PS, Gullotti E, Tong L, Highley CB, Errabelli DR, Hasan T, Cheng JX, Kohane DS, Yeo Y. Intracellular Drug Delivery by Poly(lactic-co-glycolic acid) Nanoparticles, Revisited. Mol Pharmaceut. 2009;6(1):190–201. doi: 10.1021/mp800137z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Panyam J, Zhou WZ, Prabha S, Sahoo SK, Labhasetwar V. Rapid endo-lysosomal escape of poly(DL-lactide-co-glycolide) nanoparticles: implications for drug and gene delivery. Faseb J. 2002;16(10):1217–26. doi: 10.1096/fj.02-0088com. [DOI] [PubMed] [Google Scholar]
- 49.Yoshino A, Ogino A, Yachi K, Ohta T, Fukushima T, Watanabe T, Katayama Y, Okamoto Y, Naruse N, Sano E, Tsumoto K. Gene expression profiling predicts response to temozolomide in malignant gliomas. Int J Oncol. 2010;36(6):1367–1377. doi: 10.3892/ijo_00000621. [DOI] [PubMed] [Google Scholar]
- 50.Deng T, Zhang Y. Possible involvement of activation of P53/P21 and demethylation of RUNX 3 in the cytotoxicity against Lovo cells induced by 5-Aza-2′-deoxycytidine. Life Sci. 2009;84(9–10):311–320. doi: 10.1016/j.lfs.2008.12.015. [DOI] [PubMed] [Google Scholar]
- 51.Granzotto A, Bencokova Z, Vogin G, Devic C, Joubert A, Balosso J, Foray N. DNA Double-Strand Breaks Repair and Signaling of Human Gliomas and Normal Brain Cells in Response to Radiation: Potential Impact of the ATM- and BRCA1- Dependent Pathways, Brain Tumors - Current and Emerging Therapeutic Strategies. InTech; 2011. p. 432. [Google Scholar]
- 52.Amatori S, Papalini F, Lazzarini R, Donati B, Bagaloni I, Rippo MR, Procopio A, Pelicci PG, Catalano A, Fanelli M. Decitabine, differently from DNMT1 silencing, exerts its antiproliferative activity through p21 upregulation in malignant pleural mesothelioma (MPM) cells. Lung Cancer. 2009;66(2):184–190. doi: 10.1016/j.lungcan.2009.01.015. [DOI] [PubMed] [Google Scholar]
- 53.Hollenbach PW, Nguyen AN, Brady H, Williams M, Ning Y, Richard N, Krushel L, Aukerman SL, Heise C, MacBeth KJ. A comparison of azacitidine and decitabine activities in acute myeloid leukemia cell lines. PloS one. 2010;5(2):e9001. doi: 10.1371/journal.pone.0009001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Shin DY, Kang HS, Kim GY, Kim WJ, Yoo YH, Choi YH. Decitabine, a DNA methyltransferases inhibitor, induces cell cycle arrest at G2/M phase through p53-independent pathway in human cancer cells. Biomed Pharmacother. 2013;67(4):305–311. doi: 10.1016/j.biopha.2013.01.004. [DOI] [PubMed] [Google Scholar]