Abstract
Although various functions of RNA are carried out in conjunction with proteins, some catalytic RNAs, or ribozymes, which contribute to a range of cellular processes, require little or no assistance from proteins. Furthermore, the discovery of metabolite-sensing riboswitches and other types of RNA sensors has revealed RNA-based mechanisms that cells use to regulate gene expression in response to internal and external changes. Structural studies have shown how these RNAs can carry out a range of functions. In addition, the contribution of ribozymes and riboswitches to gene expression is being revealed as far more widespread than was previously appreciated. These findings have implications for understanding how cellular functions might have evolved from RNA-based origins.
More than 40 years of extensive research has revealed that RNAs participate in a range of cellular functions. Some RNAs, despite being composed of only four chemically similar nucleotides, can fold into distinct three-dimensional architectures. In many cases, these constitute simple scaffolds that provide binding sites for proteins that function together with the RNAs. However, the discovery of the first catalytic RNAs — later named RNA enzymes, or ribozymes — in the 1980s demonstrated that RNAs can also have important functional roles in their own right1,2. The list of naturally occuring ribozymes is short, and additions over the years have been rare, with each new discovery eliciting considerable excitement in the field.
Studies of molecular evolution suggest that RNA molecules significantly influenced the development of modern organisms by mediating the genetic flow from DNA to proteins, as well as through their own contribution to catalytic functions. The ‘RNA world’ hypothesis implies that RNA molecules appeared before DNA and proteins3. Nevertheless, even with a head start, RNA catalysts do not prevail in the modern world. Moreover, most ubiquitous ribozymes require proteins for efficient catalysis in vivo, and most true protein-devoid catalytic RNAs have been found in only a few viral-like sources, suggesting that the contribution of protein-free RNAs to the functions of modern cells has been limited.
The discovery of riboswitches4–6 and other RNA-based sensors reignited interest in the roles of protein-devoid RNA-based elements. These RNA sensors can be broadly defined as mRNA regions that are capable of modulating gene expression in response to internal and external inputs, without the initial participation of proteins. Unlike ribozymes, many of these sensors direct gene expression purely through changes in RNA conformation. For instance, riboswitches alter their conformations in response to small metabolites. However, the identification of the bacterial ribozyme glmS, which specifically binds a metabolite and cleaves the mRNA encoding the protein that controls the metabolism of that metabolite, has established a closer link between riboswitches and ribozymes7. Recent findings of novel riboswitches, ribozymes and other RNA-based regulatory elements further highlight the essential contribution of such RNAs in gene expression regulation.
In this Review, we mainly focus on naturally occurring RNAs that have distinct three-dimensional structures and that can function without the help of proteins. We first present an overview of ribozymes, riboswitches and other related RNA sensors, and highlight their impact on gene regulation. We then analyse their structure–function relationships, dissecting the features that are essential for gene expression control and other cellular processes. Finally, we compare the functions of protein-free RNAs with those of proteins and RNA–protein complexes, providing a perspective on the evolution of the role of RNAs in the gene regulatory processes of modern organisms.
Ribozymes
Although naturally occurring ribozymes, excluding the ribosome, all catalyse the same reaction of RNA strand scission and ligation, they can be divided into two groups according to their main function: cleaving ribozymes, which include self-cleaving RNAs and trans-cleaving ribonuclease P (RNase P); and splicing ribozymes, which are large RNAs involved in the excision of introns from precursor RNAs (pre-RNAs) (TABLE 1).
Table 1.
Major classes and distribution of ribozymes and RNA switches
| Class | Group | Members | Main functions | Natural ligand | Size (nucleotides) | Distribution |
|---|---|---|---|---|---|---|
| Cleaving ribozymes | ||||||
| Self-cleaving | Satellite RNAs | Hammerhead | Gene control (?); processing of multimeric transcripts during rolling-circle replication | 65 | Viroids, plant viral satellite RNA, eukaryotes (plants, crickets, amphibians, schistosomes) | |
| Hairpin | 75 | Plant viral satellite RNA | ||||
| VS | 155 | Satellite RNA of Neurospora spp. mitochondria | ||||
| HDV | 85 | Human satellite virus | ||||
| mRNAs | CoTC | Transcription termination (?) | GTP | 190 | Primates | |
| CPEB3 | Splicing regulation (?) | 70 | Mammals | |||
| glmS | Gene control | GlcN6P | 170 | Gram+ bacteria | ||
| Trans-cleaving | RNase P | tRNA processing | 140–500 | Prokaryotes, eukaryotes | ||
| Splicing ribozymes | ||||||
| Group I | Self-splicing | Guanosine | 200–1500 | Organelles (fungi, plants, protists), bacteria, bacteriophages, mitochondria (animals) | ||
| Group II | Self-splicing | 300–3000 | Organelles (fungi, plants, protists), bacteria, archaea | |||
| RNA switches | ||||||
| Thermosensors | Gene control | Variable | Phages, bacteria, eukaryotes | |||
| sRNAs | Gene control | Hfq | >85 | Bacteria | ||
| T-boxes | Gene control | tRNA | 190 | Mostly Gram+ bacteria | ||
| Metabolites | Coenzymes | TPP | Gene control | TPP | 100 | Bacteria, archaea, eukaryotes (fungi, plants) |
| FMN | Gene control | FMN | 120 | Bacteria | ||
| AdoCbl | Gene control | AdoCbl | 200 | Bacteria | ||
| SAM-I | Gene control | SAM | 105 | Mostly Gram+ bacteria | ||
| SAM-II | Gene control | SAM | 60 | α- and β-proteobacteria | ||
| SAM-III (SMK) | Gene control | SAM | 80 | Gram– bacteria | ||
| Amino acids | Lysine | Gene control | Lysine | 175 | γ-proteobacteria, Thermotogales, Firmicutes | |
| Glycine (I+II) | Gene control | Glycine | 110 | Bacteria | ||
| Nucleobases | Guanine | Gene control | Guanine, hypoxanthine | 70 | Gram+ bacteria | |
| Adenine | Gene control | Adenine | 70 | Bacteria | ||
| preQ1 | Gene control | preQ1 | 35 | Bacteria | ||
| Magnesium | mgtA | Gene control | Mg2+ | 70 | Gram– bacteria | |
AdoCbl, adenosylcobalamin; CoTC, co-transcriptional cleavage; FMN, flavin mononucleotide; GlcN6P, glucosamine-6-phosphate; Gram−, Gram-negative; Gram+, Gram-positive; HDV, hepatitus δ virus; Hfq, host factor Q, interacting with some sRNAs; preQ1, pre-quenosine-1; SAM, S-adenosylmethionine; sRNA, small RNA; TPP, thiamine pyrophosphate; VS,Varkud satellite. ? indicates an unconfirmed function.
Cleaving ribozymes
Self-cleaving ribozymes range from ~40–200 nucleotides in length, and have various secondary structures and three-dimensional folds (FIG. 1). For the purpose of this Review, this group can be tentatively split according to their function in either RNA replication or mRNA cleavage.
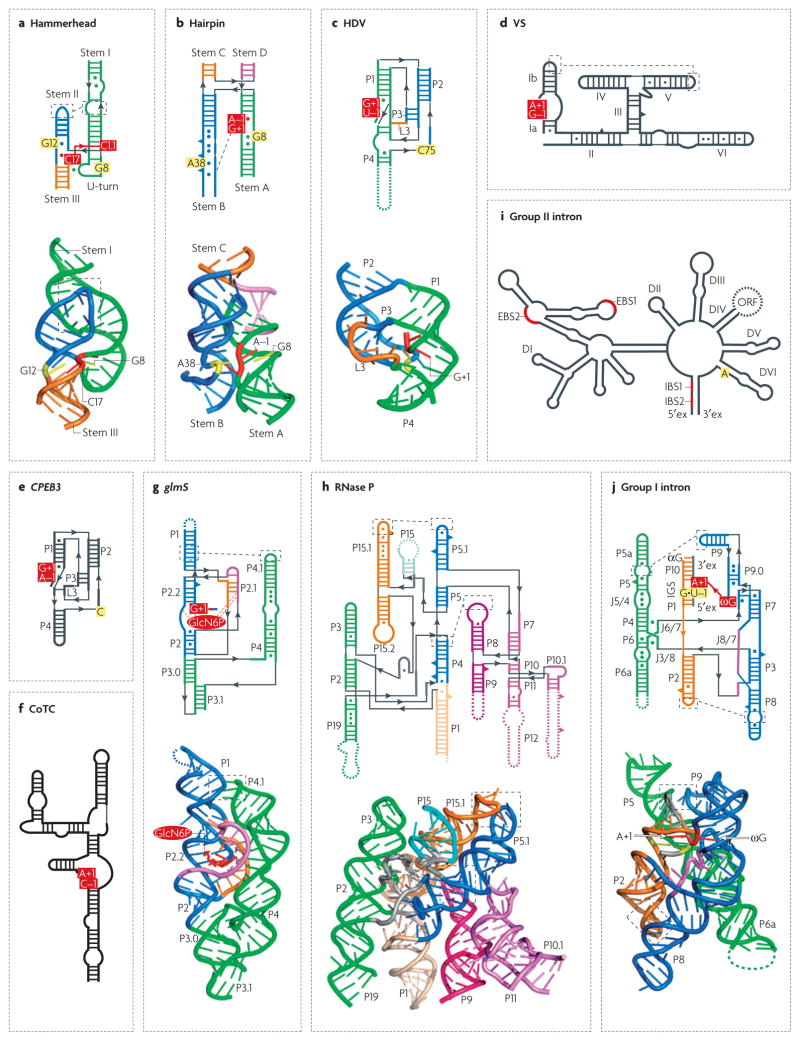
Figure 1. Domain organization and secondary and three-dimensional structures of ribozymes.
Secondary structures are depicted in thick lines and are connected by thin black lines with arrows. Watson–Crick and non-canonical base pairs are shown as solid lines and circles, respectively. Bulged-out nucleotides are represented as triangles. Ribozymes with known three-dimensional structures are coloured according to secondary structure elements and domains. Three-dimensional structures, if available, are shown in a ribbon-and-stick representation below the secondary structures. The two nucleotides that lie adjacent to the scissile phosphate are indicated by red boxes. Nucleotides that are implicated in catalysis (panels a–f and i) or that are essential for molecular recognition (panel j) are indicated by yellow boxes. Black dashed squares and lines highlight important tertiary interactions. Coloured dashed lines indicate elements that are missing in the structure or that are substituted by non-natural sequences. a | Hammerhead ribozyme76. b | Hairpin ribozyme78. c | Hepatitis δ virus ribozyme74. d | Varkud satellite (VS) ribozyme12. e | CPEB3 ribozyme13. f | Human CoTC ribozyme14. g | Bacillus antracis glmS ribozyme; glucosamine-6-phosphate (GlcN6P) is represented as a red oval, with its interactions with RNA indicated by dashed lines69. h | Bacillus subtilis RNase P, B-type73. i | Domain organization of the group II intron, with IBS and EBS designating intron and exon binding sequences, respectively21,22. The yellow-coloured A designates a conserved unpaired adenosine that participates in splicing. j | Asoarcus spp. BH72 group I intron in the state that precedes the second step of splicing79. The internal guide sequence (IGS) aligns the 5′ and 3′ exons (ex), which are shown in grey. ωG and αG designate the 3′-terminal guanosine nucleotide of the intron and the external guanosine that is linked to the intron after the first step of splicing, respectively. Secondary structures in panels a, c, d, e, f, g, h and j are modified with permission from REF. 76 © (2006) Cell Press, REF. 117 © (2007) Cell Press, REF. 118 © (1995) National Academy of Sciences (USA), REF. 13 © (2006) American Association for the Advancement of Science, Nature REF. 14 © (2004) Macmillan Publishers Ltd, REF. 75 © (2006) American Association for the Advancement of Science and REF. 69 © (2007) Current Biology Ltd, REF. 119 © (2006) Elsevier Sciences and REF. 120 © (2005) Elsevier Sciences, respectively.
The ribozymes of the first subgroup, which include the hammerhead8, hairpin9, hepatitis δ virus (HDV)10,11 and Varkud satellite (VS)12 ribozymes (FIG. 1a–d), are predominantly found in satellite RNAs of plant origin. These RNAs are the product of a rolling-circle replication mechanism involving ribozyme processing of long multimeric RNAs into short monomers that, following cyclization, participate in the next round of replication.
The second subgroup includes the recently discovered, more diverse ribozymes that reside within eukaryotic pre-mRNAs (the CPEB3 (REF. 13) and co-transcriptional cleavage (CoTC)14 ribozymes) and a bacterial mRNA (the glmS ribozyme)7. The CPEB3 ribozyme (FIG. 1e) lies within the second intron of the mammalian CPEB3 gene, which encodes cytoplasmic polyadenylation element binding protein 3 (REF. 13). The secondary structure and cleavage-site organization of this catalytic RNA closely resemble those of the HDV ribozyme (FIG. 1c). Although the exact function of the CPEB3 ribozyme is unclear, it might function in the regulation of CPEB 3 biosynthesis by interfering with mRNA splicing and facilitating mRNA degradation and/or the production of truncated protein forms.
The CoTC element (FIG. 1f) has been identified downstream of the protein-coding sequences and poly(A) sites of primate β-globin genes, suggesting that CoTC has a role in transcription termination and RNA processing14. Indeed, CoTC integrity was found to be crucial for efficient termination of transcription of these genes by RNA polymerase II15, and CoTC RNA can be targeted by several proteins that have been implicated in RNA processing and degradation (A. Akoulitchev, personal communication). Auto-catalytic cleavage by the CoTC ribozyme requires the presence of GTP or its derivatives. Because the rate of CoTC self-cleavage is low, the biological significance of this ribozyme remains to be validated.
The glmS ribozyme (FIG. 1g), which was identified in the 5′ region of the bacterial glmS gene, also requires a specific cofactor for self-cleavage. This gene encodes an amidotransferase7, which generates glucosamine-6- phosphate (GlcN6P), a molecule that is used in cell-wall biosynthesis. When sufficient GlcN6P is present, the ribozyme uses GlcN6P as a coenzyme in the self-cleaving reaction that is thought to cause degradation of the glmS mRNA, thereby turning off the gene.
Almost all self-cleaving ribozymes break the RNA backbone through the same reversible phosphodiester-cleavage reaction (FIG. 2a). Nevertheless, they have different catalytic pockets and use different cleavage mechanisms, which are still being debated16. The CoTC ribozyme catalyses a different reaction14, producing 3′-hydroxyl (3′-OH) and 5′-phosphate (5′-P) ends, and apparently carries out autolytic cleavage in the same way as RNase P.
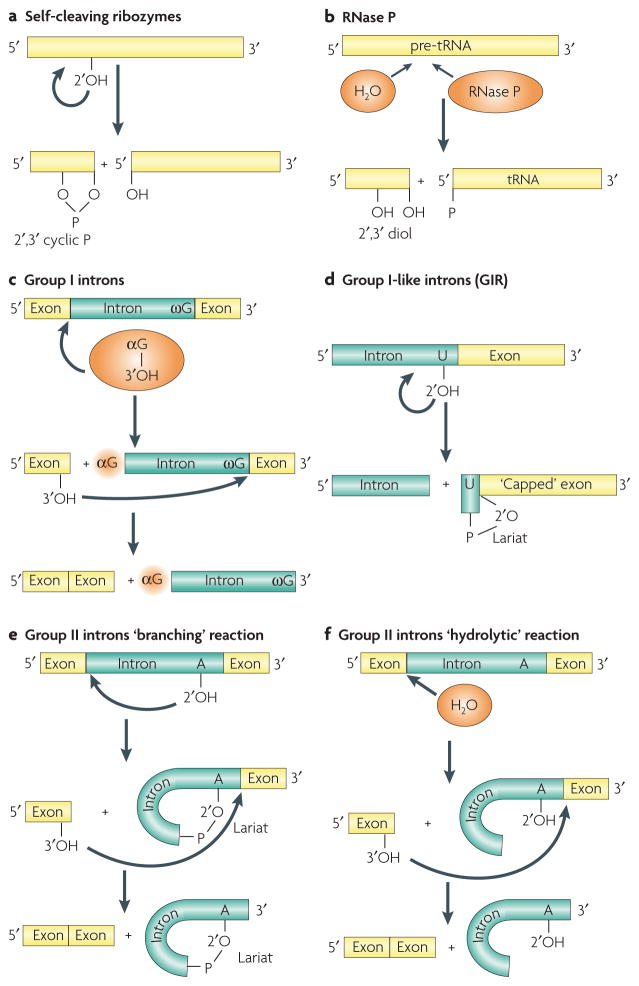
Figure 2. Reactions catalysed by ribozymes.
a | The typical reaction of self-cleaving ribozymes, initiated by 2′-hydroxyl (OH) attack, and yielding 2′,3′-cyclic phosphate (P) and 5′-OH termini. b | The catalytic cleavage of pre-tRNA by RNase P (REF. 2). A water molecule serves as a nucleophile, and the reaction yields 2′,3′-diol and 5′-P termini. c | Self-splicing by group I introns1. The reaction is initiated by nucleophilic attack by the 3′-OH of external guanosine (αG) at the 5′ splice site. This results in covalent linkage of αG to the 5′ end of the intron and release of the 3′-OH of the 5′ exon. In the second step, the 3′-OH attacks the 3′ splice site located immediately after the conserved guanosine (ωG), resulting in excision of the intron with αG at the 5′ end and release of the ligated exons. d | The ‘capping’ reaction of the Didium iridis GIR1 ribozyme is similar to the first step of the ‘branching’ reaction of group II introns25. The reaction joins nucleotides by a 2′,5′-phosphodiester linkage, thereby forming a 3-nucleotide ‘lariat’ that might be a protective 5′ cap of the mRNA. e | The self-splicing of group II introns by a branching reaction22. In the first step, the 5′ splice site is attacked by the 2′-OH of a conserved unpaired adenosine located in domain VI, resulting in formation of a 2′,5′-phosphodiester linkage. In the next step, the free 3′-OH group of the 5′ exon attacks the 3′ splice site, liberating the circular intron lariat and ligated exons. f | Alternative ‘hydrolytic’ self-splicing of group II introns22. The reaction involves a water molecule as a nucleophile and produces a linear intron.
RNase P is the only naturally occurring ribozyme identified so far that performs a multiple-turnover RNA cleavage reaction in trans, involving multiple substrate molecules2. This ubiquitous ribozyme removes extra sequences from the 5′ ends of pre-tRNAs and some other RNAs, by a different mechanism from the reaction catalysed by self-cleaving ribozymes (FIG. 2b). RNase P contains distinct protein subunits that are essential for its in vivo function, but protein-free RNase P from both bacteria2 and eukaryotes17 exhibits detectable activity in vitro. Despite differences in sequence, length and secondary structure, RNAse P RNAs contain five universally conserved regions that constitute the structural core of the RNA18. These regions include the P4 helix, the P10–P12 and the P2–P15.2 regions (FIG. 1h).
RNase P is structurally and evolutionarily related to RNase MRP, which is found only in eukaryotes. Both RNases contain similar RNA components and share several proteins19. Nevertheless, each RNase has its own substrate preferences. In contrast to RNase P, RNase MRP is implicated in 5.8S pre-ribosomal RNA (pre-rRNA) processing and in RNA cleavage during mitochondrial DNA synthesis, but not in pre-tRNA processing.
Splicing ribozymes
Typical nuclear mRNA splicing requires a complex machinery that involves the assembly of RNA–protein complexes. Splicing ribozymes can perform the precise excision of an intron and the covalent linkage of the boundary exons, with or without assistance from specific protein factor(s). This group encompasses two classes of heterogeneous self-splicing introns (groups I and II), which can be found in many tRNA, mRNA and rRNA precursors (TABLE 1).
Most large self-splicing introns fold into two types of multidomain secondary structure that define their division into groups I and II20,21 (FIG. 1i,j). In each group, helical elements that form the catalytic core of the molecule are relatively conserved, whereas the peripheral regions vary, allowing classification into several subgroups. Smaller introns that have a group II-like domain VI and a streamlined domain I, but lack domains II–V, establish a separate family21. Self-splicing occurs in vitro for most group I and for several group II introns; however, a protein machinery is required for efficient splicing in vivo22. Introns can also encode RNA maturases, homing endonucleases and reverse transcriptases (with only group II introns), which help introns to splice and to spread within genomes.
In contrast to the cleaving ribozymes, the splicing ribozymes perform two consecutive reactions: cleavage and ligation of RNA. Sequence specificity and the locations of splice sites are defined by the interactions between the 5′ region of the intron (the internal guide sequence, or IGS) and two exons (domains P1 and P10) in group I introns (FIG. 1j), and by two or three pairs of interactions between intron binding sites (IBSs) and exon binding sites (EB Ss) in group II introns22 (FIG. 1i). The first step of self-splicing requires an exogenous nucleophile, and is therefore similar to the RNase P-mediated reaction. However, instead of water, group I and II introns use external guanosine (αG) and internal adenosine as nucleophiles, respectively (FIG. 2c,e). Growing evidence suggests that some group II introns that lack the crucial adenosine undergo an alternative hydrolytic pathway23 (FIG. 2f). Remarkably, the active sites of group II introns can accommodate a diversity of nucleophiles (2′-OH group, 3′-OH group and water) and substrates (DNA and RNA) and, in addition to splicing, can catalyse several other reactions, which are either adaptations of the basic splicing activity or distinct reactions such as terminal transferase activity24. Somewhat unexpectedly, the group I-like ribozyme GIR1 from the slime mould Didymium iridis forms a 2′,5′-lariat, similar to that of the group II introns25 (FIG. 2d). This lariat structure apparently serves as a cap, which protects the intron-encoded endonuclease mRNA against degradation.
RNA switches
The term riboswitch is usually applied to metabolite-sensing RNA switches, which share important characteristics with other RNA sensors that are responsive to cations, temperature and regulatory RNA molecules (TABLE 1). Indeed, all these mRNA-based control systems, identified in many prokaryotes and some eukaryotes, can sense various stimuli and transduce them to the gene expression apparatus, with proteins being recruited only at a later stage. By contrast, in classical transcriptional attenuation control, ribosomes or specialized proteins are directly involved in the initial step of regulation by helping to sense metabolite levels.
Temperature sensing
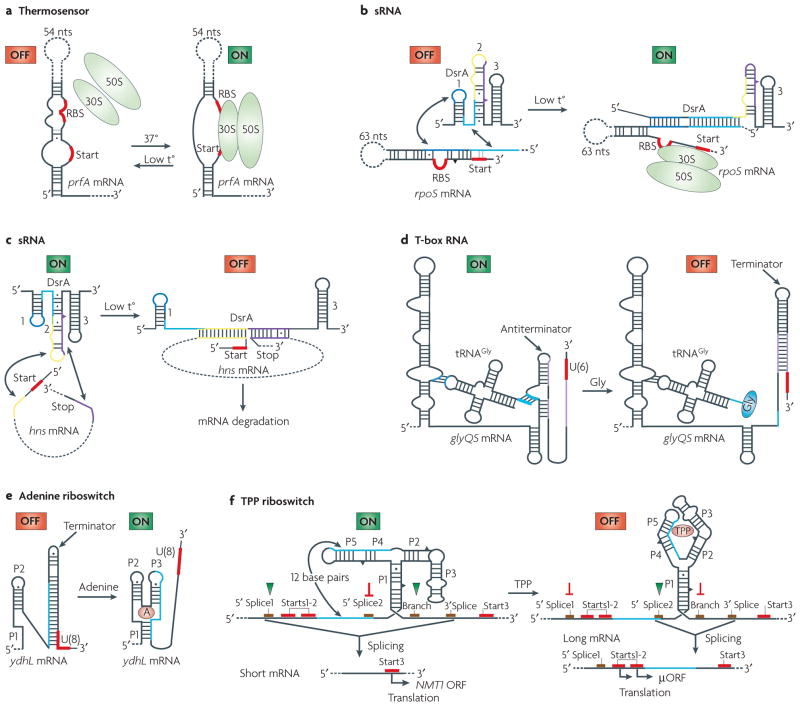
RNA thermosensors26,27 are the simplest RNA switches, yet they control adaptation to an important physical parameter that affects multiple cellular processes. Because RNA secondary structure is highly influenced by temperature, a thermosensor can be as primitive as a stem–loop RNA structure that bears a ribosome binding site (RBS) and an initiation codon28 (FIG. 3a). At low temperatures, this RNA-based thermometer adapts a conformation that masks the RBS and prevents ribosome binding. At elevated temperatures, secondary structure elements melt locally, thereby allowing ribosome binding to the exposed RBS and translation of the mRNA. Intrinsic thermometers are well documented in two instances: during pathogenic invasion and during heat-shock responses. When the pathogen Listeria monocytogenes enters an animal host, it encounters a warmer environment, and virulence-associated genes that are normally silent at low temperatures become activated. This occurs because of a thermosensor located within the 5′ untranslated region (5′-UTR) of the prfA mRNA28 (FIG. 3a). A simple RNA thermometer, the repression of heat-shock gene expression (ROSE) element, which was identified in the hspA 5′-UTR, also upregulates a heat-shock gene at high temperatures in the bacterium Bradyrhizobium japonicum29. More complex RNA structures, including large trans-acting RNAs, can participate in temperature sensing, typically during heat and cold shocks, in various organisms including bacteriophages26, bacteria27,30 and eukaryotes31.
Figure 3. Gene regulation by RNA switches.
RNA regions that are involved in gene expression switching are shown in the same colour. a | Translation activation of virulence genes in the pathogen Listeria monocytogenes28. An increase in temperature melts the secondary structure around the ribosome binding site (RBS) and start codon, allowing ribosome binding and translation initiation. b | Upregulation of Escherichia coli σs-factor gene by the DsrA antisense short RNA (sRNA). DsrA RNA121 pairs with the translational operator of the rpoS gene122 using two sequences (coloured blue and light blue) located within helices 1 and 2 (REFs 33,34,121). This base pairing exposes translation initiation signals for ribosome binding and increases mRNA stability101. c | Downregulation of transcription regulator HNS by DsrA sRNA. DsrA RNA, transcribed in response to low temperature, pairs with 5′ and 3′ regions of hns mRNA and causes faster turnover of the mRNA101, possibly by RNase E degradation123. d | Transcription termination of the Bacillus subtilis glycyl-tRNA synthetase gene by aminoacylated tRNAGly (REF. 124). Non-aminoacylated tRNAGly interacts with the T-box region of mRNA using an anticodon and an acceptor helix125, and promotes the formation of the anti-terminator stem–loop structure. The aminoacylated tRNA cannot contact mRNA using the acceptor stem, thus allowing the formation of the transcription terminator. e | Transcription activation of the purine efflux pump by the adenine riboswitch45. In the absence of adenine, transcription of B. subtilis ydhL mRNA is aborted as a result of formation of a transcription terminator. Adenine binding stabilizes the metabolite-sensing domain and prevents the formation of the terminator. f | Thiamine pyrophosphate (TPP)-riboswitch-mediated alternative splicing of mRNA in Neurospora crassa43. In the absence of TPP, the mRNA adopts a structure that occludes the 5′ splice site by base pairing with the P4–P5 region of the riboswitch. Pre-mRNA splicing from 5′ splice site 1 leads to production of a short mRNA and expression of the NMT1 gene. TPP binding causes a structural change in the RNA, opening the 5′ splice site 2 and occluding the branch site. Therefore, splicing is inhibited (not shown) and, were it to proceed, would result in the formation of a long mRNA. The alternatively spliced and non-spliced mRNAs both carry a short ORF (μORF) that begins from initiation codons 1–2 and competes with translation of the main ORF, thereby repressing NMT1 expression. Key splicing determinants are activated (indicated by green arrows) and inhibited (indicated by red lines) during different occupancy states of the TPP-sensing domain. Panels a, b and c, d and f are modified with permission from REF. 28 © (2002) Cell Press, REF. 126 © (2000) National Academy of Sciences (USA), REF. 124 © (2005) Academic Press and Nature REF. 43 © (2007) Macmillan Publishers Ltd.
RNA controls RNA
A similar strategy of either sequestering or exposing an RBS is utilized in gene expression control by some regulatory antisense short RNAs (sRNAs) in bacteria. These riboregulators are transcribed in response to various changes in the environment, and can in turn regulate genes by base pairing with target mRNAs in the regions overlapping or adjacent to the RBS. The sRNA–mRNA interactions can occur within a single region, as for the replication control of plasmid R1 by CopA RNA32 and the control of biosynthesis of the RNA polymerase σs-factor by DsrA RNA33,34, or within two regions that are distant or close to each other, as for DsrA- and OxyS-mediated control of transcription factor production33,35,36 (FIG. 3b–c). Note that many sRNAs require pairing with the RNA chaperone protein Hfq (host factor Q) for their activity37,38. In addition to translation, RNAs can also direct transcription of genes by targeting transcription termination signals. The most spectacular example of such regulation involves tRNA. Non-aminoacylated tRNATyr (REF. 39), tRNAGly (REF. 40) and others41 can selectively activate transcription of the cognate aminoacyl-tRNA synthetase gene via specific interaction of the anticodon and acceptor stem with the so-called T-box region located in the 5′-UTR (FIG. 3d). This binding promotes formation of the anti-terminator stem and disruption of the terminator hairpin, thereby producing the ‘on’ state of the gene. However, aminoacylated tRNA does not bind mRNA using its aminoacylated acceptor stem; therefore, the terminator stem forms, rather than the anti-terminator, causing transcriptional abortion.
Regulation by metabolites
Metabolite-binding riboswitches, numerous examples of which are found typically embedded in the 5′-UTRs of bacterial metabolic genes, represent genetic elements that are reminiscent of T-boxes (TABLE 1). Riboswitches typically consist of evolutionarily conserved ~35–200-nucleotide sensing or aptamer domains — which specifically bind metabolites when concentrations exceed a threshold — and expression platforms that form or carry gene regulatory signals42. Riboswitches can adapt alternative metabolite-free and metabolite-bound conformations, which control gene expression as on–off switches. In bacteria, this is achieved by the formation of structures that form and disrupt either transcription terminators or hairpins that carry translation initiation signals. In fungi, a thiamine pyrophosphate (TPP)-specific riboswitch located within an intron was found to be involved in the regulation of splicing43,44. Regulation generally depends on alternative base pairing of the small region involved in the formation of helix P1, which locks the sensing domain in the metabolite-bound conformation. Although most riboswitches are integrated into negative-feedback regulation loops, some activate gene expression; for example, an adenine-specific riboswitch achieves this by disrupting a transcriptional terminator45 (FIG. 3e).
To date, metabolite-sensing riboswitches were found to direct expression of the bacterial genes that are implicated in the biosynthesis and transport of various metabolites, including co-enzymes4–6,46–52, amino acids53,54 and nucleobases45,55,56 (FIG. 4). A sugar derivative is also sensed by the glmS ribozyme7. Discovery of the magnesium sensor that controls the magnesium transporter MgtA of Salmonella enterica57 suggests that riboswitches are involved in cellular processes that until now were not thought to be subject to riboregulation. The ligands of several conserved RNA elements are still to be identifed, and these RNA elements might represent novel riboswitch classes46,58. Remarkably, riboswitches can discriminate between highly similar compounds, such as S-adenosylhomocysteine (SAH) and S-adenosylmethionine (SAM), which differ by only a single methyl group47,49,52 (FIG. 4c). Recent studies have identified several metabolite-like antimicrobial compounds that function by targeting riboswitches. Among them are the TPP analogue pyrithiamine pyrophosphate (PTPP), which differs from TPP by the central ring59 (FIG. 4b), two analogues of lysine, L-aminoethylcysteine (AEC) and DL-4-oxalysine, which contain carbon substitutions in the lysine side chain60 (FIG. 4i), and the FMN analogue roseoflavin61 (FIG. 4f). Interestingly, high specificity for a particular ligand can be conferred by different sequences, and some metabolites such as SAM interact with several types of sensing domain46–49,52 (FIG. 4c–e), suggesting that riboswitches have taken on similar functions by following different evolutionary paths.
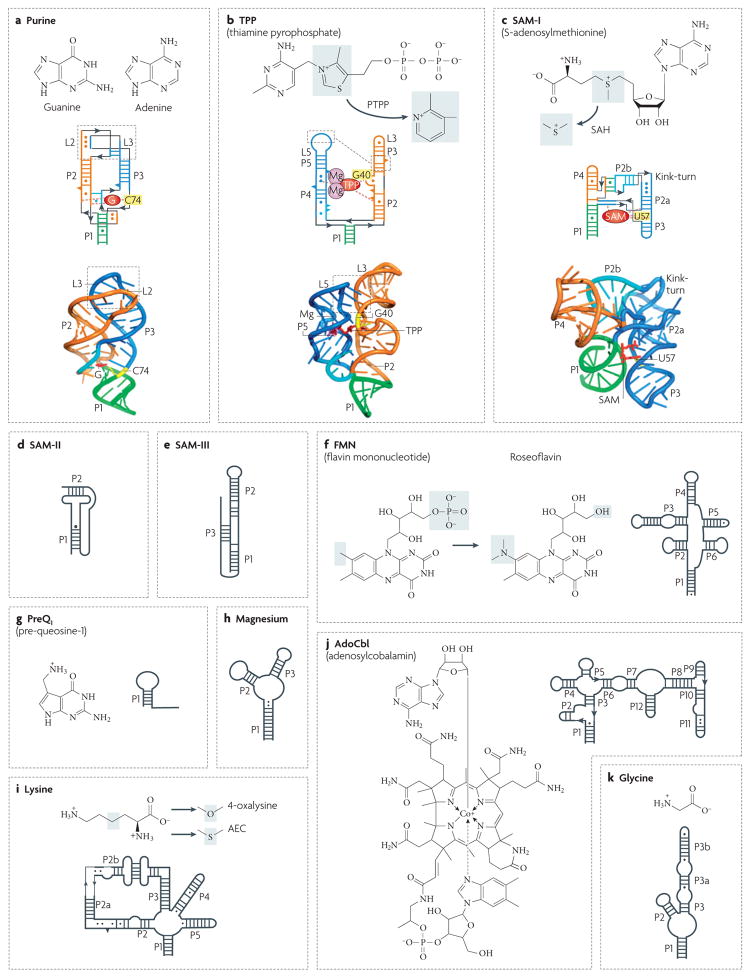
Figure 4. Secondary and tertiary structures of riboswitches.
Secondary structures are depicted by thick lines and are connected by black lines with arrows. Watson–Crick and non-canonical base pairs are shown as solid lines and circles, respectively. Natural riboswitch ligands and riboswitch-binding antibiotics are shown next to the secondary structures. Grey shadings indicate areas in which the ligands undergo changes. a | The Bacillus subtilis xpt gene guanine riboswitch bound to guanine (represented as a red G)66. The discriminatory nucleotide C74 is coloured yellow. b | The TPP riboswitch from the Escherichia coli thiM gene bound with TPP (in red)65. A pair of hydrated Mg2+ cations is shown in magenta. G40 interacting with the pyrimidine moiety of TPP is shown in yellow. The antibiotic pyrithiamine pyrophosphate (PTPP) differs from TPP by the central ring. c | The class I SAM Thermoanaerobacter tengcongensis riboswitch in complex with SAM (in red)64. U57 interacting with the purine moiety of SAM is shown in yellow. The grey area highlights a methyl group that is missing in S-adenosylhomocysteine (SAH). d | A class II SAM riboswitch from the Agrobacterium tumefaciens metA gene46. e | An SMK (class III SAM) riboswitch from the Streptococcus gordonii metK gene48. Helix P3 is formed by Shine–Dalgarno and anti-Shine–Dalgarno sequences. f | The FMN riboswitch from the B. subtilis ribD gene50. g | The preQ1 riboswitch from the B. subtilis queC gene56. h | The magnesium riboswitch from the Salmonella enterica mgtA gene57. i | The lysine riboswitch from the B. subtilis lysC gene54. j | The AdoCbl riboswitch from the E. coli btuB gene5. k | The glycine type II riboswitch from Vibrio cholerae gcvT gene53. Secondary structures in panels c, d, e, f, g, h, i, j and k modified with permission from Nature REF. 64 © (2006) Macmillan Publishers Ltd, REF. 46 © (2005) BioMed Central Ltd, Nature Structural & Molecular Biology REF. 48 © (2006) Macmillan Publishers Ltd, REF. 50 © (2002) National Academy of Sciences (USA), Nature Structural & Molecular Biology REF. 56 © (2007) Macmillan Publishers Ltd, REF. 57 © (2006) Cell Press, Nature Biotechnology REF. 61 © (2006) Macmillan Publishers Ltd, REF. 5 © (2002) Current Biology Ltd and REF. 53 © (2004) American Association for the Advancement of Science, respectively.
Structure–function relationships
The chemical simplicity of RNA molecules raises the question of whether diverse RNA-based genetic elements utilize a limited number of architectural components and folding rules. The recently solved three-dimensional structures of several riboswitches62–67 and the majority of ribozyme types68–80 have shed light on this question.
Architectures that have an impact on function
The structures of ribozymes and riboswitches highlight the concept of modularity, a principle that is also used by many other RNAs to generate and maintain specific architectures81. All RNAs, ranging from simple thermometers (FIG. 3) to complex ribozymes (FIG. 1), can be viewed as hierarchical assemblies that are composed of helices as the major building blocks. The helices associate into bundles by end-to-end stacking and edge-to-edge docking, and are connected together by junctional regions and tertiary interactions. Many oligonucleotide elements that are present in secondary structures as non-paired sequences can form compact and often helix-like topologies. A comparison of riboswitch and small ribozyme structures reveals that the key elements defining these architectures are junctions composed of three (FIGS 1a;4a,b) or more (FIGS 1b;4c) helical segments that converge together. In large ribozymes68,73,80, the TPP riboswitch63,65,67 and the hairpin77 ribozyme, these junctions comprise structural elements that are necessary for the correct orientation of the helical domains, in some cases demonstrating a remarkable convergence in terms of global architecture82. This feature is also observed for the SAM-I riboswitch64 and the P4–P6 and P3–P7 domains of group I introns68,71,72.
To reinforce junctional conformations, spatial constraints are provided by diverse and complex tertiary interactions, which most often involve loop–loop62,66 (FIGS 1a,h;4a), loop–helix63,65,67,76 (FIGS 1g;4b) and helix– helix interlocking connections between peripheral regions. In the absence of crucial tertiary loop–loop contacts, the hammerhead ribozyme shows a 100-fold loss of activity83,84, and purine riboswitches do not interact with their ligands62. Ribozymes, riboswitches and other RNAs share several conserved three-dimensional nucleotide combinations called structural motifs. These include the T-loop, a 5-bp hairpin that is closed by a special non-canonical pair found in the TPP riboswitch63,65,67 and RNase P85, and the kink-turn, a sharp bend in the backbone of the RNA strand found in the SAM-I riboswitch64 (FIG. 4c) and the Tetrahymena thermophila group I intron72.
Folding of ribozymes and riboswitches
Not surprisingly, splicing reactions and larger substrates require longer ribozymes with intricate three-dimensional structures, whereas smaller self-cleaving ribozymes and riboswitches adopt simpler folds. It is therefore notable that ongoing in vitro studies indicate that large ribozymes self-assemble into almost-active conformations in the presence of magnesium86, and subsequently undergo smaller substrate-induced conformational changes. By contrast, riboswitches require their ligands for efficient folding, thus demonstrating a typical induced-fit binding mechanism. The name ‘riboswitch’ implies a switch between two states in response to the presence of metabolites; however, more than half of the putative riboswitches that have been identified are predicted to function through transcriptional attenuation, when RNA polymerase must make a choice between mRNA elongation and transcription termination soon after the sensor domain of the riboswitch has been synthesized. After that event, the energy barrier for re-folding of the domain would be too great to overcome, suggesting that, instead of switching between conformations, a riboswitch makes a ‘once-in-a-lifetime’ choice between ‘on’ and ‘off ‘ conformations87.
Because of the high speed of RNA polymerase, some riboswitches such as the FMN riboswitch88 might not have sufficient time for metabolite–RNA interactions to reach thermodynamic equilibrium before the regulatory decision is made; therefore, they might require higher concentrations of metabolites (relative to the apparent dissociation constants) to affect transcription4,50. Other riboswitches might need to attain thermodynamic equilibrium with ligands to make a choice between continuation or termination of transcription. However, the adenine-sensing riboswitch utilizes kinetic or equilibrium control depending on the speed of transcription and external variables such as temperature89. Interestingly, co-transcriptional folding seems to dictate the formation of the tuning-fork-like architecture that is observed for several riboswitches62,64–67. Such a mechanism, in which two helical segments trap the metabolite between them, thereby stabilizing the transient helix P1 and preventing it from adopting an alternative base-pairing alignment, might be superior to single-site recognition, especially for bulky and extended metabolites.
An interesting situation is seen in the context of phage and bacterial mRNAs that contain self-splicing introns. In bacteria, transcription and translation are coupled, and translating ribosomes can prevent the formation of an elaborate intron structure that requires the splice sites to be in close proximity. Therefore, efficient splicing requires that RNA folding is coordinated with transcription and translation90.
Catalysis and binding: similar trends?
A direct comparison between molecular recognition mechanisms used by ribozymes and riboswitches is difficult to undertake owing to the fact that ribozymes target a specific sugar-phosphate backbone position whereas riboswitches recognize various small molecules. An analysis of available structures also indicates that ribozymes and riboswitches exhibit great structural diversity despite superficial similarities in their global architectures. For example, a comparison of RNAs with similar scaffolds, such as the three-way helical junctions that are shared by the hammerhead ribozyme, purine and TPP riboswitches (FIGS 1a;4a,b), shows that they are used for conceptually different goals (BOX 1). In the hammerhead ribozyme76, an open catalytic pocket formed by a three-way junction functions to precisely position a scissile phosphate. By contrast, the three-way junction of the purine riboswitches62,66 consists of several stacked layers of base triples, and nucleobase ligands such as adenine, guanine or the guanine analogue hypoxanthine are sandwiched between layers so that ~98% of the surface area is shielded from solvent62. In the TPP riboswitch63,65,67, the junction adopts a simpler fold and the ligand is recognized outside of the junctional region by two helical segments (FIG. 4b). The same principle of bridging two helical segments by the ligand is exploited by the SAM-I riboswitch64 (FIG. 4c). Despite different ligand-binding properties, the binding event in purine, TPP and SAM-I riboswitches leads to the same result: stabilization of the overall junctional region, which in turn contributes to the stabilization of the transient helix P1, which is involved in the gene expression switch.
Box 1. The architecture of binding and catalytic pockets.
The functioning of ribozymes and riboswitches depends on the precise formation of catalytic domains and binding pockets, respectively. How do these RNA molecules achieve an exceptionally high specificity for recognition and catalysis despite a limited arsenal of functional groups, and are there common principles that underlie ligand–substrate recognition?
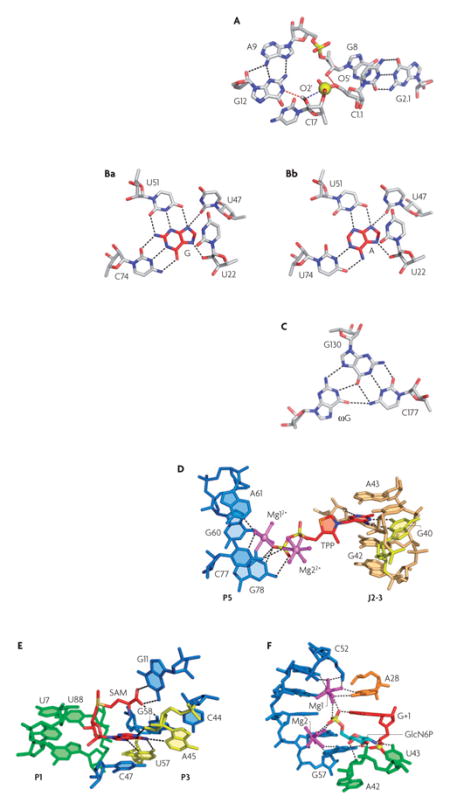
In the hammerhead ribozyme76, the scissile phosphate (indicated by a yellow sphere in panel A) is recognized in an ‘open’ pocket. Nucleotides C17 and C1.1, which are neighbours to the scissile phosphate, are splayed apart. C1.1 forms hydrogen bonds (indicated by black dashed lines) with G2.1, whereas C17 is intercalated into the core of the junction. The nucleophilic 2′-hydroxyl (OH) of C17 is positioned in line with the scissile phosphate and the 5′-oxygen of the leaving group (indicated by a blue dashed line coming from the phosphate group). The conserved G8 and G12 are located nearby to facilitate acid–base catalysis. Red dashed lines indicate hydrogen bonds that are potentially active for the catalysis.
Purine riboswitches contain a ‘closed’ ligand-binding pocket that is located between several layers of triples that constitute the three-way junction62,66. The bound guanine (G) and adenine (A) are surrounded by uridines along their periphery66 (shown in panels Ba and Bb, respectively). Specific recognition of the ligands is achieved by Watson–Crick pairing with a discriminatory nucleotide at position 74 (either cytosine or uridine in guanine and adenine riboswitches, respectively45,55,62,66,115).
Similarly to purine riboswitches, the guanosine-binding pocket of group I introns68,71,72 is built by several stacked triples, and the 3′ splice site (ωG) is bound by extensive stacking and the hydrogen bond interactions of its nucleobase. However, ωG is not fully enveloped and is likely to be inserted or removed without a pronounced conformational change in the pocket. ωG interacts with a guanosine residue that is engaged in a G130–C177 base pair, forming a nucleotide triple (shown in panel C). Specific recognition of guanosine also has an essential role in the selection of the 5′ splice site by the ribozyme68. In this case, a conserved wobble pair is formed between the terminal nucleotide of the 5′ exon, U-1 and a G within the intron’s internal guide sequence (IGS) (FIG. 1j). Such pairing exposes the exocyclic amine and 2′-OHs of the G for readout by a wobble receptor element, containing two non-canonical A•A pairs, in J5–4.
In the thiamine pyrophosphate (TPP) riboswitch63,65,67 (shown in panel D), the ligand is bound in an extended conformation by two helical segments that lie outside of the junctional region65. The pyrimidine-like ring pairs with G40 (coloured yellow) and is stacked between G42 and A43, which constitute part of a T-loop motif (coloured orange). Two Mg2+ ions (the coordinated water molecules are shown in magenta) are bound to the pyrophosphate moiety, which interacts with the RNA through direct contact of phosphate oxygens with the base edges of C77 and G78. In addition, Mg12+ makes direct contacts with G60 and G78, and also makes water-mediated contacts with several other nucleotides.
The ligand also bridges two helical segments in the SAM-I riboswitch (shown in panel E)64. However, unlike TPP, the bound S-adenosylmethionine (SAM) adopts a compact conformation. The purine moiety of SAM specifically binds U57 and A45 (coloured yellow), and is sandwiched between a methionine moiety and C47. Mainchain atoms of methionine interact with the (G58–C44)•G11 base triple. SAM forms van der Waals interactions with U7 and U88, positioning P1 close to P3. These multiple interactions zip up helices P1 and P3, and stabilize the P1 helix and the overall four-way junctional fold, with the former constituting the integral part of the gene expression switch.
The preformed open glucosamine-6-phosphate (GlcN6P)-binding pocket of the glmS ribozyme (shown in panel F)69,75 consists of nucleotides from the P2 loop (FIG. 1g) that form a double pseudoknot conformation, which is observed in the structures of the hepatitis δ virus (HDV)70 and Diels–Alder ribozymes116. Like the HDV ribozyme, the double pseudoknot in the glmS ribozyme forms an active-site cleft, with the scissile phosphate located at the bottom. GlcN6P is locked in place by stacking with the G+1 nucleotide and multiple direct and magnesium-mediated interactions involving ring and phosphate moieties. The reaction mechanism might include deprotonation of the 2′-OH of A-1 by guanine (not shown), and protonation of the 5′-O leaving group by GlcN6P, consistent with the observation that GlcN6P is essential for catalysis. Note that Mg2+ does not directly participate in catalysis.
Interesting parallels are observed when riboswitch structures are compared with hairpin ribozyme and group I intron structures. Formation of the catalytic pocket in the hairpin ribozyme77 via docking of two helical domains might be considered to be reminiscent of TPP and SAM-I riboswitches (FIG. 1b). In group I introns, the guanosine-binding pocket, formed by nucleotides at the junction of J6–J7 and P7 and occupied by ωG (the 3′ splice site)68,71,72, is surrounded by several stacked base triples (FIG. 1j), reminiscent of the purine-binding site in purine riboswitches. Unlike other ribozymes and riboswitches, the glmS ribozyme contains pre-formed catalytic and ligand-binding sites and, in contrast to riboswitches, binds its ligand GlcN6P in an open, solvent-accessible pocket69,75 (BOX 1; FIG. 1g).
Regulation of gene expression
Given that RNase P has a universal catalytic function in both prokaryotes and eukaryotes, there has been a long-standing appreciation of the important role that ribozymes have in cells. The discovery of riboswitches and novel ribozymes, however, points to a different trend for protein-devoid RNAs; namely, a direct involvement in a range of gene-control mechanisms in both prokaryotes and eukaryotes.
The role of ribozymes in gene expression
Self-splicing introns continually modulate the genomic organization of various organisms by mediating the mobility of genetic material within the genome and by spreading between species. How important are these ribozymes for the regulation of gene expression? Self-splicing introns can interrupt genes, but the ability of introns to self-excise from mRNA potentially renders them neutral to the host91. Nevertheless, in the roaA gene of the Euglena gracilis chloroplast, self-splicing introns cause alternative splicing of pre-mRNAs to produce two distinct transcripts92, highlighting one way in which these RNAs can affect gene expression. Splicing might also be an important mechanism for posttranscriptional regulation. For example, the addition of a 3′ CC A sequence, which is required for amino-acylation, to a plastid tRNA is less efficient when intron removal is blocked93. Importantly, in addition to homing, which occurs when introns spread into similar genes, self-splicing introns can also invade ectopic or non-homologous sites22, and these new integration sites might not necessarily be neutral for gene expression.
Other self-cleaving ribozymes, such as the CPEB3 and CoTC ribozymes, which seem to be involved in splicing13 and transcription14, respectively, might have a direct impact on gene expression. These ribozymes have been identified in only some mammalian species, and further study is needed to determine whether similar RNA elements exist, providing a more generally applicable mechanism for their function. By contrast, small self-cleaving ribozymes (hammerhead, hairpin, VS and HDV ribozymes) have specialized functions in replication, and are therefore unlikely to influence expression of the host genes specifically and directly.
A few more catalytically active self-cleaving sequences have been found in humans13 and plants94. Homology searches in various genomes have also identified several hammerhead-like motifs, the activities of which have not been demonstrated95,96. Potentially, cleavages produced by the catalytically active elements might provide specific entry sites for distinct exonucleases, with direct implications for transcriptional termination, intron degradation and mRNA turnover. Future research should reveal whether these ribozymes are an integral part of gene expression mechanisms or are involved in specific mechanisms of gene expression regulation. So far, a regulatory function has been clearly shown for only the glmS ribozyme, which cleaves off the 5′ mRNA region and, probably through mRNA degradation, downregulates expression of the amidotransferase gene. Unexpectedly, the pre-rRNA-processing enzyme RNase MRP also participates in mRNA degradation by targeting specific mRNAs that are involved in cell-cycle regulation97, suggesting that RNase MRP might be involved in gene expression control.
The complexity of riboswitch-dependent regulation in bacteria
The number of genes controlled by metabolite-sensing riboswitches (~2% in some bacteria)55 is comparable with the number of genes regulated by metabolite-sensing proteins. Indeed, a bacterial genome can contain riboswitches that are specific to different metabolites, as well as multiple riboswitches of the same type, each regulating a different operon. Moreover, two sensing domains or even two complete switches can lie adjacent to each other, resulting in a more complex mechanism of gene expression regulation53,98. For example, tandem glycine-specific aptamers bind glycine cooperatively and accomplish regulation through a single expression platform, thus providing a greater dynamic response to small changes in glycine concentration53. Another adjoining arrangement constitutes two complete riboswitches that independently sense different metabolites, such as SAM and adenosylcobalamin (AdoCbl), and function as a two-input Boolean NOR logic gate, wherein binding of either ligand causes repression98. Tandem riboswitches can also be composed of two complete riboswitches of the same type. Such composite arrangements, exhibited by some TPP riboswitches98,99, candidate riboswitches98 and T-box RNAs98,100, probably enable a greater dynamic range for gene control and greater responsiveness to changes in metabolite concentration98.
Remarkably, depending on the expression platform, riboswitches of the same type are capable of either repression or activation of gene expression, greatly increasing their regulatory potential. A similar trend is seen in regulation by bacterial sRNAs, although this involves an increased level of sophistication. The DsrA sRNA can pair with two different mRNAs, hns and rpoS, down- and upregulating their translation, respectively33,34,101 (FIG. 3b,c). To add further complexity, rpoS mRNA can, in turn, be repressed by another sRNA, OxyS, possibly by titration of Hfq37.
Riboswitches as mediators of eukaryotic gene expression
To date, virtually all riboswitch-like riboregulators have been found in bacterial species. Because of differences in transcription, translation and mRNA structure, eukaryotes rely on post-transcriptional gene-control mechanisms that differ to some extent from their prokaryotic counterparts. Nevertheless, functional TPP riboswitches have been identified in the 5′-UTR introns of fungal genes44 and the 3′-UTRs of plant genes involved in thiamine biosynthesis102, suggesting a regulatory role in splicing and mRNA processing, respectively. Indeed, a recent study has shown that fungal TPP riboswitches can either downregulate or upregulate gene expression using an elegant alternative splicing mechanism43 (FIG. 3f). Computer searches have also identified TPP riboswitch-like motifs in two other eukaryotic species and in archaea, and SAM-II, preQ1 and AdoCbl riboswitch-like sequences have been found in eukaryotes103, although some of them such as preQ1 and AdoCbl cannot be functional riboswitches. Furthermore, a novel candidate riboswitch that senses arginine has been recently suggested to control the arginase gene in fungi104. These examples highlight the plasticity of riboswitch-mediated genetic control and point out the need for experimental studies that might reveal the distinct features of eukaryotic versus prokaryotic regulatory mechanisms.
RNAs versus proteins
As they are composed of many more variable building blocks, proteins are generally considered to be more versatile than RNAs, so why did nature also implement RNA-based catalysts and regulatory elements? In fact, the versatility of RNA might have been underestimated. Studies of ribozymes and RNA sensors have shown that, despite having a much simpler composition, RNA molecules have a number of features that are typical of protein molecules. Like their protein counterparts, ribozymes and especially riboswitches interact with small-molecule cofactors, and some of these regulatory and catalytic RNAs show allosterically controlled activity. Similarly to proteins, RNAs demonstrate high affinity and specificity towards their ligands and can recognize a wide range of molecules, from simple glycine to bulky AdoCbl. Also in common with proteins, some RNA regulators, such as tandem riboswitches, have a modular organization that seems to be crucial for cooperative and other types of complex control.
In addition to their similarities with proteins, RNAs might have functional advantages over proteins in some situations. Direct sensing of metabolites and other molecules by mRNA provides a significant advantage for cells, which can save energy and resources by eliminating the need for special regulatory proteins. Another important feature of RNA regulators that are positioned in cis is a synchronized response to changing surroundings. In contrast to the long-lived trans-acting protein factors, regulatory elements that are embedded within an mRNA exclusively control that mRNA, providing precise regulation of gene expression.
Both RNA and protein enzymes use binding interactions and energy to facilitate catalysis105. Remarkably, despite a limited arsenal of functional groups and a simple composition, some ribozymes turn out to be efficient catalysts. The cleavage-rate constants for the HDV106 and hammerhead ribozymes (~1 sec−1 and 15 sec−1, respectively)107 approach the corresponding values for the protein enzyme ribonuclease A (160 sec−1 and 69 sec−1 for UpG and CpC substrates, respectively)108. Nevertheless, many ribozymes have much slower rate constants (~1 min−1)109, with these values being sufficient for typical ribozyme reactions involving single cleavage and ligation in cis. The cleavage and ligation reactions can be carried out by specific host endoribonucleases and ligases. However, virus-like RNAs, the sources of many ribozymes, could improve their chances for survival by encoding cleavage and ligation activity within their sequences, thereby eliminating a dependence on specialized host enzymes.
Finally, cells undoubtedly benefit from RNA-dependent gene expression control under stress conditions. Relatively small sRNAs can efficiently and precisely regulate protein synthesis by base pairing with target mRNAs, thereby preventing a waste of resources for protein production.
Despite the benefits described above, some features of RNA-based regulatory systems can be limiting. For instance, because RNA typically degrades more quickly than proteins in vivo, RNA regulators that work in trans, such as sRNAs, are probably not well suited to functioning under conditions in which control is required over an extended period of time. Current research suggests that many RNA catalysts and regulators might be considered as relics of a pre-protein era, either perfectly adapted to particular functions or still subject to evolutionary pressure for replacement by proteins. Indeed, functions that are carried out by certain ribozymes and riboswitches have been taken over by protein-based systems in other species110.
Descendants of the RNA world?
If RNAs constituted the predominant species in the early biological world as suggested by the RNA world hypothesis, are modern RNA modules direct descendants of those primordial molecules? Given the key participation of RNA in fundamental cellular processes such as protein synthesis, it is tempting to consider a positive answer, especially as it is assumed that proteins, at first, largely assisted functional RNAs. Therefore, the existing ribonucleoprotein enzymes, including the ribosome, RNase P and the spliceosome, could be considered as remnants of the RNA world, although their origins cannot be unambiguously traced owing to the many missing links in their evolutionary history. A similar problem is encountered in the evolutionary analysis of modern small ribozymes. The complexity and narrow distribution of VS, HDV and hairpin ribozymes argues against their descent from the RNA world. However, the distribution of the hammerhead ribozyme in highly divergent organisms, suggesting an ancient origin, might be misleading: this ribozyme might instead have arisen independently multiple times111. The evolutionary relationships between similar ribozymes in different species might be even more intricate than initially thought, given the similarity between the HDV and CPEB3 ribozymes, which, along with the limited distribution of the HDV ribozyme, suggests a human origin for this viral ribozyme13.
Knowledge of the origin and evolution of self-splicing introns, although uncertain, is important for understanding the origins of spliceosomal introns112. Indeed, terminal structures of group II self-splicing introns, which contain the parts that carry out the splicing reaction, show a remarkable similarity to the structures of the spliceosomal small nuclear RNAs (snRNAs)22. This prompted speculation that self-splicing introns are ancestors of spliceosomal introns. Growing evidence suggests that group II introns, which are present in many bacteria, are among the most ancient genetic entities, and probably moved into the evolving eukaryotic genome from the α-proteobacterial progenitor of mitochondria, later giving rise to the spliceosomal introns and splicing machinery112.
Several aspects of riboswitches are suggestive of their RNA world origin. Riboswitch scaffolds have probably stayed unaltered over long periods of time, because riboswitches recognize coenzymes and nucleobases that have remained unchanged for billions of years113. These nucleotide-like molecules could initially have been tethered to RNAs and used as prosthetic groups for catalytic reactions. With the emergence of proteins and their take-over of catalytic functions, the ancient cofactor-dependent ribozymes could have lost their catalytic ability and instead evolved to create RNA switches42. In addition, the presence of some riboswitches in diverse organisms supports their ancient origin, although even this criterion is not absolute because of possible lateral gene transfer.
Perspectives and future challenges
Modern genomic and biochemical techniques provide unique opportunities to identify novel roles for RNAs that function independently of proteins, as shown by the recent discoveries of riboswitches and several ribozymes that are encoded by cellular genomes. Some of these studies, such as the discovery of riboswitches4–6, have taken advantage of well-established biological observations, suggesting the existence of metabolite-dependent gene expression control. Other examples, such as the metabolite-dependent glmS and CPEB3 ribozymes, were discovered serendipitously during the characterization of a potential riboswitch7 or by careful labour-intensive experimentation involving in vitro selection to identify self-cleaving RNAs13. Many of these exciting findings, such as the identification of glmS7, CoTC13 and CPEB314 RNAs, have highlighted our limited understanding of key biological processes including the termination of mRNA transcription, and mRNA processing and degradation.
A priority for future studies is to characterize the self-cleavage activities of recently identified genome-encoded ribozymes13,94, as well as candidate ribozyme and RNA sensor sequences46,58,95,96,103, that might reveal new aspects of genetic regulation by RNA elements. As a pivotal example, we cite the recent and long-awaited dissection of the functional role of eukaryotic riboswitches, which participate in gene expression control by alternative splicing of their pre-mRNAs43. This outstanding work will encourage experimentation on archaeal and other eukaryotic riboswitches, especially those that are found in the 3′ region of mRNAs and are potentially involved in novel types of regulation. These biochemical and genetic studies should be supplemented by searches for novel RNA sensors and ribozymes using new computer-based approaches that can identify candidate RNA sequences with low sequence homology114.
Despite considerable progress in studies of ribozymes and riboswitches at the atomic level, future research must resolve many uncertainties in functional mechanisms, especially for some classical (VS ribozyme, group II introns and RNase P) and recently identified (CoTC and CPEB3) ribozymes, for which structures are either not yet available or are restricted to a subset of functional states. This need also applies to many riboswitches for which three-dimensional structures await determination. Comprehensive research in this area would enhance our mechanistic understanding of the biological function of these molecules and more complex RNA-containing cellular machineries such as the spliceosome, the ribosome and telomerase.
Acknowledgments
This work was supported by the US National Institutes of Health grant GM073618.
Glossary
- Satellite RNAs
Subviral agents whose multiplication in a host cell depends on coinfection with a helper virus
- Rolling-circle replication
A process of replication of some circular genomes, whereby one strand is replicated first and the second strand is replicated after completion of the first one
- RNA maturase
A protein enzyme, intronspecific, that acts as a cofactor to facilitate splicing
- Homing endonucleases
dsDNA-specific deoxyribonucleases that recognize large asymmetrical DNA sequences, which are not stringently defined, and which mobilize these DNA elements by facilitating their integration into new genomic sites. Because these sites lack introns and inteins (protein introns), this form of mobility is termed ‘homing’. Homing endonucleases are encoded by intervening sequences embedded in either introns or inteins
- Reverse transcriptase
An RNA-dependent DNA polymerase that transcribes ssRNA into dsDNA
- Transcriptional attenuation
A regulatory mechanism whereby gene expression is controlled through the formation of alternative structures in the mRNA sequence that inhibit or facilitate the progression of transcription
- Aptamer
Derived from the Latin aptus, meaning ‘to fit’, aptamers are oligonucleotide or peptide molecules that bind specific target molecules. The term is usually applied to nucleic acid molecules that are created following selection from a large random sequence pool, as well as to natural metabolitesensing domains in riboswitches, which possess similar recognition properties to artificially generated aptamers
- Boolean NOR logic gate
Boolean logic, named after George Boole, is a complete system for logical operations. A logic gate performs a logical operation on one or more logical inputs and produces a single logical output. The logical NOR, or joint denial, is a logical operator, meaning that the output is true if none of the inputs are true. Consequently, if one or both inputs are true, then the output is not true
- Prosthetic group
A non-protein component of a conjugated protein that is usually essential for the protein’s function. Prosthetic groups are also called coenzymes and cofactors
- Lateral transfer
A process of transferring genetic material from one organism to another without reproduction. Also called horizontal gene transfer
- Telomerase
An RNA-containing reverse transcriptase that adds DNA sequence repeats (telomeric repeats) to the 3′ end of DNA strands in eukaryotic chromosomes using its RNA component as a synthesis template
Footnotes
Competing interests statement
The authors declare no competing financial interests.
References
- 1.Cech TR, Zaug AJ, Grabowski PJ. In vitro splicing of the ribosomal RNA precursor of Tetrahymena: involvement of a guanosine nucleotide in the excision of the intervening sequence. Cell. 1981;27:487–496. doi: 10.1016/0092-8674(81)90390-1. [DOI] [PubMed] [Google Scholar]
- 2.Guerrier-Takada C, Gardiner K, Marsh T, Pace N, Altman S. The RNA moiety of ribonuclease P is the catalytic subunit of the enzyme. Cell. 1983;35:849–857. doi: 10.1016/0092-8674(83)90117-4. [DOI] [PubMed] [Google Scholar]
- 3.Gilbert W. Origin of life: the RNA World. Nature. 1986;319:618. [Google Scholar]
- 4.Mironov AS, et al. Sensing small molecules by nascent RNA: a mechanism to control transcription in bacteria. Cell. 2002;111:747–756. doi: 10.1016/s0092-8674(02)01134-0. References 4–6 describe the discovery of metabolite-binding riboswitches. [DOI] [PubMed] [Google Scholar]
- 5.Nahvi A, et al. Genetic control by a metabolite binding mRNA. Chem Biol. 2002;9:1043–1049. doi: 10.1016/s1074-5521(02)00224-7. [DOI] [PubMed] [Google Scholar]
- 6.Winkler W, Nahvi A, Breaker RR. Thiamine derivatives bind messenger RNAs directly to regulate bacterial gene expression. Nature. 2002;419:952–956. doi: 10.1038/nature01145. [DOI] [PubMed] [Google Scholar]
- 7.Winkler WC, Nahvi A, Roth A, Collins JA, Breaker RR. Control of gene expression by a natural metabolite-responsive ribozyme. Nature. 2004;428:281–286. doi: 10.1038/nature02362. This work showed that a riboswitch can function as a ribozyme. [DOI] [PubMed] [Google Scholar]
- 8.Forster AC, Symons RH. Self-cleavage of plus and minus RNAs of a virusoid and a structural model for the active sites. Cell. 1987;49:211–220. doi: 10.1016/0092-8674(87)90562-9. [DOI] [PubMed] [Google Scholar]
- 9.Buzayan JM, Gerlach WL, Bruening G. Non-enzymatic cleavage and ligation of RNAs complementary to a plant virus satellite RNA. Nature. 1986;323:349–353. [Google Scholar]
- 10.Sharmeen L, Kuo MY, Dinter-Gottlieb G, Taylor J. Antigenomic RNA of human hepatitis δ virus can undergo self-cleavage. J Virol. 1988;62:2674–2679. doi: 10.1128/jvi.62.8.2674-2679.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wu HN, et al. Human hepatitis δ virus RNA subfragments contain an autocleavage activity. Proc Natl Acad Sci USA. 1989;86:1831–1835. doi: 10.1073/pnas.86.6.1831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Saville BJ, Collins RA. A site-specific selfcleavage reaction performed by a novel RNA in Neurospora mitochondria. Cell. 1990;61:685–696. doi: 10.1016/0092-8674(90)90480-3. [DOI] [PubMed] [Google Scholar]
- 13.Salehi-Ashtiani K, Luptak A, Litovchick A, Szostak JW. A genomewide search for ribozymes reveals an HDV-like sequence in the human CPEB3 gene. Science. 2006;313:1788–1792. doi: 10.1126/science.1129308. The identification of several functional ribozymes in the human genome. [DOI] [PubMed] [Google Scholar]
- 14.Teixeira A, et al. Autocatalytic RNA cleavage in the human β-globin pre-mRNA promotes transcription termination. Nature. 2004;432:526–530. doi: 10.1038/nature03032. [DOI] [PubMed] [Google Scholar]
- 15.West S, Gromak N, Proudfoot NJ. Human 5′ → 3′ exonuclease XRN2 promotes transcription termination at co-transcriptional cleavage sites. Nature. 2004;432:522–525. doi: 10.1038/nature03035. [DOI] [PubMed] [Google Scholar]
- 16.Fedor MJ, Williamson JR. The catalytic diversity of RNAs. Nature Reviews Mol Cell Biol. 2005;6:399–412. doi: 10.1038/nrm1647. [DOI] [PubMed] [Google Scholar]
- 17.Kikovska E, Svard SG, Kirsebom LA. Eukaryotic RNase P RNA mediates cleavage in the absence of protein. Proc Natl Acad Sci USA. 2007;104:2062–2067. doi: 10.1073/pnas.0607326104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chen JL, Pace NR. Identification of the universally conserved core of ribonuclease P RNA. RNA. 1997;3:557–560. [PMC free article] [PubMed] [Google Scholar]
- 19.Lindahl L, Fretz S, Epps N, Zengel JM. Functional equivalence of hairpins in the RNA subunits of RNase MRP and RNase P in Saccharomyces cerevisiae. RNA. 2000;6:653–658. doi: 10.1017/s1355838200992574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Michel F, Dujon B. Conservation of RNA secondary structures in two intron families including mitochondrial-, chloroplast- and nuclear-encoded members. EMBO J. 1983;2:33–38. doi: 10.1002/j.1460-2075.1983.tb01376.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Michel F, Umesono K, Ozeki H. Comparative and functional anatomy of group II catalytic introns — a review. Gene. 1989;82:5–30. doi: 10.1016/0378-1119(89)90026-7. [DOI] [PubMed] [Google Scholar]
- 22.Pyle AM, Lambowitz AM. In: The RNA World. Gesteland RF, Cech TR, Atkins JF, editors. Cold Spring Harbor Laboratory Press; Cold Spring Harbor: 2006. pp. 469–506. [Google Scholar]
- 23.Vogel J, Borner T. Lariat formation and a hydrolytic pathway in plant chloroplast group II intron splicing. EMBO J. 2002;21:3794–3803. doi: 10.1093/emboj/cdf359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Muller MW, Stocker P, Hetzer M, Schweyen RJ. Fate of the junction phosphate in alternating forward and reverse self-splicing reactions of group II intron RNA. J Mol Biol. 1991;222:145–154. doi: 10.1016/0022-2836(91)90201-g. [DOI] [PubMed] [Google Scholar]
- 25.Nielsen H, Westhof E, Johansen S. An mRNA is capped by a 2′, 5′ lariat catalyzed by a group I-like ribozyme. Science. 2005;309:1584–1587. doi: 10.1126/science.1113645. [DOI] [PubMed] [Google Scholar]
- 26.Altuvia S, Kornitzer D, Teff D, Oppenheim AB. Alternative mRNA structures of the cIII gene of bacteriophage λ determine the rate of its translation initiation. J Mol Biol. 1989;210:265–280. doi: 10.1016/0022-2836(89)90329-x. [DOI] [PubMed] [Google Scholar]
- 27.Morita MT, et al. Translational induction of heat shock transcription factor sigma32: evidence for a built-in RNA thermosensor. Genes Dev. 1999;13:655–665. doi: 10.1101/gad.13.6.655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Johansson J, et al. An RNA thermosensor controls expression of virulence genes in Listeria monocytogenes. Cell. 2002;110:551–561. doi: 10.1016/s0092-8674(02)00905-4. [DOI] [PubMed] [Google Scholar]
- 29.Chowdhury S, Maris C, Allain FH, Narberhaus F. Molecular basis for temperature sensing by an RNA thermometer. EMBO J. 2006;25:2487–2497. doi: 10.1038/sj.emboj.7601128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lybecker MC, Samuels DS. Temperature-induced regulation of RpoS by a small RNA in Borrelia burgdorferi. Mol Microbiol. 2007;64:1075–1089. doi: 10.1111/j.1365-2958.2007.05716.x. [DOI] [PubMed] [Google Scholar]
- 31.Shamovsky I, Ivannikov M, Kandel ES, Gershon D, Nudler E. RNA-mediated response to heat shock in mammalian cells. Nature. 2006;440:556–560. doi: 10.1038/nature04518. [DOI] [PubMed] [Google Scholar]
- 32.Kolb FA, et al. Progression of a loop–loop complex to a four-way junction is crucial for the activity of a regulatory antisense RNA. EMBO J. 2000;19:5905–5915. doi: 10.1093/emboj/19.21.5905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lease RA, Cusick ME, Belfort M. Riboregulation in Escherichia coli: DsrA RNA acts by RNA:RNA interactions at multiple loci. Proc Natl Acad Sci USA. 1998;95:12456–12461. doi: 10.1073/pnas.95.21.12456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Majdalani N, Cunning C, Sledjeski D, Elliott T, Gottesman S. DsrA RNA regulates translation of RpoS message by an anti-antisense mechanism, independent of its action as an antisilencer of transcription. Proc Natl Acad Sci USA. 1998;95:12462–12467. doi: 10.1073/pnas.95.21.12462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Altuvia S, Weinstein-Fischer D, Zhang A, Postow L, Storz G. A small, stable RNA induced by oxidative stress: role as a pleiotropic regulator and antimutator. Cell. 1997;90:43–53. doi: 10.1016/s0092-8674(00)80312-8. [DOI] [PubMed] [Google Scholar]
- 36.Altuvia S, Zhang A, Argaman L, Tiwari A, Storz G. The Escherichia coli OxyS regulatory RNA represses fhlA translation by blocking ribosome binding. EMBO J. 1998;17:6069–6075. doi: 10.1093/emboj/17.20.6069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zhang A, et al. The OxyS regulatory RNA represses rpoS translation and binds the Hfq (HF-I) protein. EMBO J. 1998;17:6061–6068. doi: 10.1093/emboj/17.20.6061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zhang A, Wassarman KM, Ortega J, Steven AC, Storz G. The Sm-like Hfq protein increases OxyS RNA interaction with target mRNAs. Mol Cell. 2002;9:11–22. doi: 10.1016/s1097-2765(01)00437-3. [DOI] [PubMed] [Google Scholar]
- 39.Grundy FJ, Henkin TM. tRNA as a positive regulator of transcription antitermination in B. subtilis. Cell. 1993;74:475–482. doi: 10.1016/0092-8674(93)80049-k. [DOI] [PubMed] [Google Scholar]
- 40.Grundy FJ, Winkler WC, Henkin TM. tRNA-mediated transcription antitermination in vitro: codon–anticodon pairing independent of the ribosome. Proc Natl Acad Sci USA. 2002;99:11121–11126. doi: 10.1073/pnas.162366799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Gagnon Y, et al. Clustering and co-transcription of the Bacillus subtilis genes encoding the aminoacyl-tRNA synthetases specific for glutamate and for cysteine and the first enzyme for cysteine biosynthesis. J Biol Chem. 1994;269:7473–7482. [PubMed] [Google Scholar]
- 42.Breaker RR. In: The RNA World. Gesteland RF, Cech TR, Atkins JF, editors. Cold Spring Harbor Laboratory Press; Cold Spring Harbor: 2006. pp. 89–108. [Google Scholar]
- 43.Cheah MT, Wachter A, Sudarsan N, Breaker RR. Control of alternative RNA splicing and gene expression by eukaryotic riboswitches. Nature. 2007;447:391–393. doi: 10.1038/nature05769. References 43 and 44 established the involvement of eukaryotic riboswitches in the control of alternative splicing. [DOI] [PubMed] [Google Scholar]
- 44.Kubodera T, et al. Thiamine-regulated gene expression of Aspergillus oryzae thiA requires splicing of the intron containing a riboswitch-like domain in the 5′-UTR. FEBS Lett. 2003;555:516–520. doi: 10.1016/s0014-5793(03)01335-8. [DOI] [PubMed] [Google Scholar]
- 45.Mandal M, Breaker RR. Adenine riboswitches and gene activation by disruption of a transcription terminator. Nature Struct Mol Biol. 2004;11:29–35. doi: 10.1038/nsmb710. [DOI] [PubMed] [Google Scholar]
- 46.Corbino KA, et al. Evidence for a second class of S-adenosylmethionine riboswitches and other regulatory RNA motifs in α-proteobacteria. Genome Biol. 2005;6:R70. doi: 10.1186/gb-2005-6-8-r70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Epshtein V, Mironov AS, Nudler E. The riboswitch-mediated control of sulfur metabolism in bacteria. Proc Natl Acad Sci USA. 2003;100:5052–5056. doi: 10.1073/pnas.0531307100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Fuchs RT, Grundy FJ, Henkin TM. The SMK box is a new SAM-binding RNA for translational regulation of SAM synthetase. Nature Struct Mol Biol. 2006;13:226–233. doi: 10.1038/nsmb1059. [DOI] [PubMed] [Google Scholar]
- 49.McDaniel BA, Grundy FJ, Artsimovitch I, Henkin TM. Transcription termination control of the S box system: direct measurement of S-adenosylmethionine by the leader RNA. Proc Natl Acad Sci USA. 2003;100:3083–3088. doi: 10.1073/pnas.0630422100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Winkler WC, Cohen-Chalamish S, Breaker RR. An mRNA structure that controls gene expression by binding FMN. Proc Natl Acad Sci USA. 2002;99:15908–15913. doi: 10.1073/pnas.212628899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Nou X, Kadner RJ. Adenosylcobalamin inhibits ribosome binding to btuB RNA. Proc Natl Acad Sci USA. 2000;97:7190–7195. doi: 10.1073/pnas.130013897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Winkler WC, Nahvi A, Sudarsan N, Barrick JE, Breaker RR. An mRNA structure that controls gene expression by binding S-adenosylmethionine. Nature Struct Biol. 2003;10:701–707. doi: 10.1038/nsb967. [DOI] [PubMed] [Google Scholar]
- 53.Mandal M, et al. A glycine-dependent riboswitch that uses cooperative binding to control gene expression. Science. 2004;306:275–279. doi: 10.1126/science.1100829. [DOI] [PubMed] [Google Scholar]
- 54.Sudarsan N, Wickiser JK, Nakamura S, Ebert MS, Breaker RR. An mRNA structure in bacteria that controls gene expression by binding lysine. Genes Dev. 2003;17:2688–2697. doi: 10.1101/gad.1140003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Mandal M, Boese B, Barrick JE, Winkler WC, Breaker RR. Riboswitches control fundamental biochemical pathways in Bacillus subtilis and other bacteria. Cell. 2003;113:577–586. doi: 10.1016/s0092-8674(03)00391-x. [DOI] [PubMed] [Google Scholar]
- 56.Roth A, et al. A riboswitch selective for the queuosine precursor preQ1 contains an unusually small aptamer domain. Nature Struct Mol Biol. 2007;14:308–317. doi: 10.1038/nsmb1224. [DOI] [PubMed] [Google Scholar]
- 57.Cromie MJ, Shi Y, Latifi T, Groisman EA. An RNA sensor for intracellular Mg2+ Cell. 2006;125:71–84. doi: 10.1016/j.cell.2006.01.043. [DOI] [PubMed] [Google Scholar]
- 58.Barrick JE, et al. New RNA motifs suggest an expanded scope for riboswitches in bacterial genetic control. Proc Natl Acad Sci USA. 2004;101:6421–6426. doi: 10.1073/pnas.0308014101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Sudarsan N, Cohen-Chalamish S, Nakamura S, Emilsson GM, Breaker RR. Thiamine pyrophosphate riboswitches are targets for the antimicrobial compound pyrithiamine. Chem Biol. 2005;12:1325–1335. doi: 10.1016/j.chembiol.2005.10.007. [DOI] [PubMed] [Google Scholar]
- 60.Blount KF, Wang JX, Lim J, Sudarsan N, Breaker RR. Antibacterial lysine analogs that target lysine riboswitches. Nature Chem Biol. 2007;3:44–49. doi: 10.1038/nchembio842. [DOI] [PubMed] [Google Scholar]
- 61.Blount KF, Breaker RR. Riboswitches as antibacterial drug targets. Nature Biotechnol. 2006;24:1558–1564. doi: 10.1038/nbt1268. [DOI] [PubMed] [Google Scholar]
- 62.Batey RT, Gilbert SD, Montange RK. Structure of a natural guanine-responsive riboswitch complexed with the metabolite hypoxanthine. Nature. 2004;432:411–415. doi: 10.1038/nature03037. References 62 and 66 describe the first threedimensional structures of riboswitches. [DOI] [PubMed] [Google Scholar]
- 63.Edwards TE, Ferre-D’Amare AR. Crystal structures of the thi-box riboswitch bound to thiamine pyrophosphate analogs reveal adaptive RNA–small molecule recognition. Structure. 2006;14:1459–1468. doi: 10.1016/j.str.2006.07.008. [DOI] [PubMed] [Google Scholar]
- 64.Montange RK, Batey RT. Structure of the S-adenosylmethionine riboswitch regulatory mRNA element. Nature. 2006;441:1172–1175. doi: 10.1038/nature04819. [DOI] [PubMed] [Google Scholar]
- 65.Serganov A, Polonskaia A, Phan AT, Breaker RR, Patel DJ. Structural basis for gene regulation by a thiamine pyrophosphate-sensing riboswitch. Nature. 2006;441:1167–1171. doi: 10.1038/nature04740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Serganov A, et al. Structural basis for discriminative regulation of gene expression by adenine- and guanine-sensing mRNAs. Chem Biol. 2004;11:1729–1741. doi: 10.1016/j.chembiol.2004.11.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Thore S, Leibundgut M, Ban N. Structure of the eukaryotic thiamine pyrophosphate riboswitch with its regulatory ligand. Science. 2006;312:1208–1211. doi: 10.1126/science.1128451. [DOI] [PubMed] [Google Scholar]
- 68.Adams PL, Stahley MR, Kosek AB, Wang J, Strobel SA. Crystal structure of a self-splicing group I intron with both exons. Nature. 2004;430:45–50. doi: 10.1038/nature02642. References 68, 71, 72 and 79 provide insights into the group I intron self-splicing mechanism at the atomic level. [DOI] [PubMed] [Google Scholar]
- 69.Cochrane JC, Lipchock SV, Strobel SA. Structural investigation of the GlmS ribozyme bound to its catalytic cofactor. Chem Biol. 2007;14:97–105. doi: 10.1016/j.chembiol.2006.12.005. References 69 and 75 describe the crystal structures of the glmS ribozyme. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Ferre-D’Amare AR, Zhou K, Doudna JA. Crystal structure of a hepatitis δ virus ribozyme. Nature. 1998;395:567–574. doi: 10.1038/26912. [DOI] [PubMed] [Google Scholar]
- 71.Golden BL, Kim H, Chase E. Crystal structure of a phage Twort group I ribozyme-product complex. Nature Struct Mol Biol. 2005;12:82–89. doi: 10.1038/nsmb868. [DOI] [PubMed] [Google Scholar]
- 72.Guo F, Gooding AR, Cech TR. Structure of the Tetrahymena ribozyme: base triple sandwich and metal ion at the active site. Mol Cell. 2004;16:351–362. doi: 10.1016/j.molcel.2004.10.003. [DOI] [PubMed] [Google Scholar]
- 73.Kazantsev AV, et al. Crystal structure of a bacterial ribonuclease P RNA. Proc Natl Acad Sci USA. 2005;102:13392–13397. doi: 10.1073/pnas.0506662102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Ke A, Zhou K, Ding F, Cate JH, Doudna JA. A conformational switch controls hepatitis δ virus ribozyme catalysis. Nature. 2004;429:201–205. doi: 10.1038/nature02522. [DOI] [PubMed] [Google Scholar]
- 75.Klein DJ, Ferre-D’Amare AR. Structural basis of glmS ribozyme activation by glucosamine-6-phosphate. Science. 2006;313:1752–1756. doi: 10.1126/science.1129666. [DOI] [PubMed] [Google Scholar]
- 76.Martick M, Scott WG. Tertiary contacts distant from the active site prime a ribozyme for catalysis. Cell. 2006;126:309–320. doi: 10.1016/j.cell.2006.06.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Rupert PB, Ferre-D’Amare AR. Crystal structure of a hairpin ribozyme-inhibitor complex with implications for catalysis. Nature. 2001;410:780–786. doi: 10.1038/35071009. [DOI] [PubMed] [Google Scholar]
- 78.Rupert PB, Massey AP, Sigurdsson ST, Ferre-D’Amare AR. Transition state stabilization by a catalytic RNA. Science. 2002;298:1421–1424. doi: 10.1126/science.1076093. [DOI] [PubMed] [Google Scholar]
- 79.Stahley MR, Strobel SA. Structural evidence for a two-metal-ion mechanism of group I intron splicing. Science. 2005;309:1587–1590. doi: 10.1126/science.1114994. [DOI] [PubMed] [Google Scholar]
- 80.Torres-Larios A, Swinger KK, Krasilnikov AS, Pan T, Mondragon A. Crystal structure of the RNA component of bacterial ribonuclease P. Nature. 2005;437:584–587. doi: 10.1038/nature04074. [DOI] [PubMed] [Google Scholar]
- 81.Westhof E, Masquida B, Jaeger L. RNA tectonics: towards RNA design. Fold Des. 1996;1:R78–88. doi: 10.1016/S1359-0278(96)00037-5. [DOI] [PubMed] [Google Scholar]
- 82.Lescoute A, Westhof E. Topology of three-way junctions in folded RNAs. RNA. 2006;12:83–93. doi: 10.1261/rna.2208106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.De la Pena M, Gago S, Flores R. Peripheral regions of natural hammerhead ribozymes greatly increase their self-cleavage activity. EMBO J. 2003;22:5561–5570. doi: 10.1093/emboj/cdg530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Khvorova A, Lescoute A, Westhof E, Jayasena SD. Sequence elements outside the hammerhead ribozyme catalytic core enable intracellular activity. Nature Struct Biol. 2003;10:708–712. doi: 10.1038/nsb959. [DOI] [PubMed] [Google Scholar]
- 85.Krasilnikov AS, Yang X, Pan T, Mondragon A. Crystal structure of the specificity domain of ribonuclease P. Nature. 2003;421:760–764. doi: 10.1038/nature01386. [DOI] [PubMed] [Google Scholar]
- 86.Waldsich C, Pyle AM. A folding control element for tertiary collapse of a group II intron ribozyme. Nature Struct Mol Biol. 2007;14:37–44. doi: 10.1038/nsmb1181. [DOI] [PubMed] [Google Scholar]
- 87.Nudler E. Flipping riboswitches. Cell. 2006;126:19–22. doi: 10.1016/j.cell.2006.06.024. [DOI] [PubMed] [Google Scholar]
- 88.Wickiser JK, Winkler WC, Breaker RR, Crothers DM. The speed of RNA transcription and metabolite binding kinetics operate an FMN riboswitch. Mol Cell. 2005;18:49–60. doi: 10.1016/j.molcel.2005.02.032. [DOI] [PubMed] [Google Scholar]
- 89.Wickiser JK, Cheah MT, Breaker RR, Crothers DM. The kinetics of ligand binding by an adenine-sensing riboswitch. Biochemistry. 2005;44:13404–13414. doi: 10.1021/bi051008u. [DOI] [PubMed] [Google Scholar]
- 90.Semrad K, Schroeder R. A ribosomal function is necessary for efficient splicing of the T4 phage thymidylate synthase intron in vivo. Genes Dev. 1998;12:1327–1337. doi: 10.1101/gad.12.9.1327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Ferat JL, Le Gouar M, Michel F. A group II intron has invaded the genus Azotobacter and is inserted within the termination codon of the essential groEL gene. Mol Microbiol. 2003;49:1407–1423. doi: 10.1046/j.1365-2958.2003.03649.x. [DOI] [PubMed] [Google Scholar]
- 92.Jenkins KP, Hong L, Hallick RB. Alternative splicing of the Euglena gracilis chloroplast roaA transcript. RNA. 1995;1:624–633. [PMC free article] [PubMed] [Google Scholar]
- 93.Vogel J, Hess WR. Complete 5′ and 3′ end maturation of group II intron-containing tRNA precursors. RNA. 2001;7:285–292. doi: 10.1017/s1355838201001960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Przybilski R, et al. Functional hammerhead ribozymes naturally encoded in the genome of Arabidopsis thaliana. Plant Cell. 2005;17:1877–1885. doi: 10.1105/tpc.105.032730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Ferbeyre G, Bourdeau V, Pageau M, Miramontes P, Cedergren R. Distribution of hammerhead and hammerhead-like RNA motifs through the GenBank. Genome Res. 2000;10:1011–1019. doi: 10.1101/gr.10.7.1011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Graf S, Przybilski R, Steger G, Hammann C. A database search for hammerhead ribozyme motifs. Biochem Soc Trans. 2005;33:477–478. doi: 10.1042/BST0330477. [DOI] [PubMed] [Google Scholar]
- 97.Gill T, Cai T, Aulds J, Wierzbicki S, Schmitt ME. RNase MRP cleaves the CLB2 mRNA to promote cell cycle progression: novel method of mRNA degradation. Mol Cell Biol. 2004;24:945–953. doi: 10.1128/MCB.24.3.945-953.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Sudarsan N, et al. Tandem riboswitch architectures exhibit complex gene control functions. Science. 2006;314:300–304. doi: 10.1126/science.1130716. [DOI] [PubMed] [Google Scholar]
- 99.Rodionov DA, Dubchak I, Arkin A, Alm E, Gelfand MS. Reconstruction of regulatory and metabolic pathways in metal-reducing δ-proteobacteria. Genome Biol. 2004;5:R90. doi: 10.1186/gb-2004-5-11-r90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Putzer H, Gendron N, Grunberg-Manago M. Co-ordinate expression of the two threonyl-tRNA synthetase genes in Bacillus subtilis: control by transcriptional antitermination involving a conserved regulatory sequence. EMBO J. 1992;11:3117–3127. doi: 10.1002/j.1460-2075.1992.tb05384.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Lease RA, Belfort M. A trans-acting RNA as a control switch in Escherichia coli: DsrA modulates function by forming alternative structures. Proc Natl Acad Sci USA. 2000;97:9919–9924. doi: 10.1073/pnas.170281497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Sudarsan N, Barrick JE, Breaker RR. Metabolite-binding RNA domains are present in the genes of eukaryotes. RNA. 2003;9:644–647. doi: 10.1261/rna.5090103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Griffiths-Jones S, et al. Rfam: annotating non-coding RNAs in complete genomes. Nucleic Acids Res. 2005;33:D121–D124. doi: 10.1093/nar/gki081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Borsuk P, et al. L-arginine influences the structure and function of arginase mRNA in Aspergillus nidulans. Biol Chem. 2007;388:135–144. doi: 10.1515/BC.2007.015. [DOI] [PubMed] [Google Scholar]
- 105.Hertel KJ, Peracchi A, Uhlenbeck OC, Herschlag D. Use of intrinsic binding energy for catalysis by an RNA enzyme. Proc Natl Acad Sci USA. 1997;94:8497–8502. doi: 10.1073/pnas.94.16.8497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Tanner NK, et al. A three-dimensional model of hepatitis δ virus ribozyme based on biochemical and mutational analyses. Curr Biol. 1994;4:488–498. doi: 10.1016/s0960-9822(00)00109-3. [DOI] [PubMed] [Google Scholar]
- 107.Canny MD, et al. Fast cleavage kinetics of a natural hammerhead ribozyme. J Amer Chem Soc. 2004;126:10848–10849. doi: 10.1021/ja046848v. [DOI] [PubMed] [Google Scholar]
- 108.Richards FM, Wyckoff HW. In: The Enzymes. Boyer PD, editor. Academic; New York: 1971. pp. 647–806. [Google Scholar]
- 109.Nahas MK, et al. Observation of internal cleavage and ligation reactions of a ribozyme. Nature Struct Mol Biol. 2004;11:1107–1113. doi: 10.1038/nsmb842. [DOI] [PubMed] [Google Scholar]
- 110.Rodionov DA, Vitreschak AG, Mironov AA, Gelfand MS. Comparative genomics of the methionine metabolism in Gram-positive bacteria: a variety of regulatory systems. Nucleic Acids Res. 2004;32:3340–3353. doi: 10.1093/nar/gkh659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Salehi-Ashtiani K, Szostak JW. In vitro evolution suggests multiple origins for the hammerhead ribozyme. Nature. 2001;414:82–84. doi: 10.1038/35102081. [DOI] [PubMed] [Google Scholar]
- 112.Koonin EV. The origin of introns and their role in eukaryogenesis: a compromise solution to the introns-early versus introns-late debate? Biol Direct. 2006;1:22. doi: 10.1186/1745-6150-1-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.White HB., 3rd Coenzymes as fossils of an earlier metabolic state. J Mol Evol. 1976;7:101–104. doi: 10.1007/BF01732468. [DOI] [PubMed] [Google Scholar]
- 114.Hammann C, Westhof E. Searching genomes for ribozymes and riboswitches. Genome Biol. 2007;8:210. doi: 10.1186/gb-2007-8-4-210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Noeske J, et al. An intermolecular base triple as the basis of ligand specificity and affinity in the guanine-and adenine-sensing riboswitch RNAs. Proc Natl Acad Sci USA. 2005;102:1372–1377. doi: 10.1073/pnas.0406347102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Serganov A, et al. Structural basis for Diels–Alder ribozyme-catalyzed carbon–carbon bond formation. Nature Struct Mol Biol. 2005;12:218–224. doi: 10.1038/nsmb906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Ke A, Ding F, Batchelor JD, Doudna JA. Structural roles of monovalent cations in the HDV ribozyme. Structure. 2007;15:281–287. doi: 10.1016/j.str.2007.01.017. [DOI] [PubMed] [Google Scholar]
- 118.Beattie TL, Olive JE, Collins RA. A secondary structure model for the self-cleaving region of Neurospora VS RNA. Proc Natl Acad Sci USA. 1995;92:4686–4690. doi: 10.1073/pnas.92.10.4686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Torres-Larios A, Swinger KK, Pan T, Mondragon A. Structure of ribonuclease P — a universal ribozyme. Curr Opin Struct Biol. 2006;16:327–335. doi: 10.1016/j.sbi.2006.04.002. [DOI] [PubMed] [Google Scholar]
- 120.Woodson SA. Structure and assembly of group I introns. Curr Opin Struct Biol. 2005;15:324–330. doi: 10.1016/j.sbi.2005.05.007. [DOI] [PubMed] [Google Scholar]
- 121.Rolle K, Zywicki M, Wyszko E, Barciszewska MZ, Barciszewski J. Evaluation of the dynamic structure of DsrA RNA from E. coli and its functional consequences. J Biochem (Tokyo) 2006;139:431–438. doi: 10.1093/jb/mvj045. [DOI] [PubMed] [Google Scholar]
- 122.Brown L, Elliott T. Mutations that increase expression of the rpoS gene and decrease its dependence on hfq function in Salmonella typhimurium. J Bacteriol. 1997;179:656–662. doi: 10.1128/jb.179.3.656-662.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Masse E, Escorcia FE, Gottesman S. Coupled degradation of a small regulatory RNA and its mRNA targets in Escherichia coli. Genes Dev. 2003;17:2374–2383. doi: 10.1101/gad.1127103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Yousef MR, Grundy FJ, Henkin TM. Structural transitions induced by the interaction between tRNAGly and the Bacillus subtilis glyQS T box leader RNA. J Mol Biol. 2005;349:273–287. doi: 10.1016/j.jmb.2005.03.061. [DOI] [PubMed] [Google Scholar]
- 125.Yousef MR, Grundy FJ, Henkin TM. tRNA requirements for glyQS antitermination: a new twist on tRNA. RNA. 2003;9:1148–1156. doi: 10.1261/rna.5540203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Altuvia S, Wagner EGH. Switching on and off with RNA. Proc Natl Acad Sci USA. 2000;97:9824–9826. doi: 10.1073/pnas.97.18.9824. [DOI] [PMC free article] [PubMed] [Google Scholar]