Abstract
Obesity is associated with disturbed lipid metabolism and low-grade inflammation in tissues. The aim of this study was to investigate the association between FA metabolism and adipose tissue (AT) inflammation in the Kuopio Obesity Surgery study. We investigated the association of surgery-induced weight loss and FA desaturase (FADS)1/2 genotypes with serum and AT FA profile and with AT inflammation, measured as interleukin (IL)-1β and NFκB pathway gene expression, in order to find potential gene-environment interactions. We demonstrated an association between serum levels of saturated and polyunsaturated n-6 FAs, and estimated enzyme activities of FADS1/2 genes with IL-1β expression in AT both at baseline and at follow-up. Variation in the FADS1/2 genes associated with IL-1β and NFκB pathway gene expression in SAT after weight reduction, but not at baseline. In addition, the FA composition in subcutaneous and visceral fat correlated with serum FAs, and the associations between serum PUFAs and estimated D6D enzyme activity with AT inflammation were also replicated with corresponding AT FAs and AT inflammation. We conclude that the polymorphism in FADS1/2 genes associates with FA metabolism and AT inflammation, leading to an interaction between weight loss and FADS1/2 genes in the regulation of AT inflammation.
Keywords: fatty acid, fatty acid/desaturases, fatty acid/metabolism, gene expression, genetics, obesity, fatty acid desaturase 1
Low-grade inflammation is associated with metabolic diseases, such as type 2 diabetes, cardiovascular diseases, cancer, and nonalcoholic fatty liver disease (1, 2). It is linked to energy excess in diet, i.e., obesity, and also to the quality of dietary fats, particularly to diets rich in saturated FAs (SFAs) (2, 3). Several studies demonstrate that SFAs promote inflammation through toll-like receptor 4 (TLR4) and activation of the, so called, inflammasome complex, which is responsible for the regulation of interleukin (IL)-1β production (1, 4, 5). In turn, an increased production of IL-1β has a major role in adipose tissue (AT) inflammation by stimulating macrophage infiltration into AT (6, 7).
The role of PUFAs in AT inflammation is more complex. Overall, n-3 PUFAs are considered to be anti-inflammatory (5). However, n-6 PUFAs are related to pro-inflammatory events, most likely due to the fact that arachidonic acid (AA), produced from linoleic acid (LA), is a major substrate for eicosanoids that regulate inflammatory responses (5). The main FA desaturase (FADS) enzymes regulating PUFA metabolism are delta-5 desaturase (D5D) and delta-6 desaturase (D6D), encoded by the FADS1 and FADS2 genes, respectively (8). Genetic variations in the FADS genes and isoforms of these genes have been associated with plasma PUFA concentrations and pro-inflammatory markers, diabetes, and metabolic syndrome, indicating that activity of these enzymes may modify the effect of dietary fat on tissue inflammation (3, 9, 10).
To investigate the association between FA metabolism and tissue inflammation, we measured the serum and AT FA profile in different lipid fractions and IL-1β gene expression in subcutaneous AT (SAT), visceral AT (VAT), liver, and peripheral blood mononuclear cells (PBMCs) after a standardized 4–5 week very low calorie (VLC) diet period in 89 obese individuals undergoing obesity surgery. Furthermore, the effect of surgery-induced weight loss and genotypes of the FADS1 and FADS2 on serum and AT FA profile and AT inflammation, measured as IL-1β gene expression and using TruSeq Targeted RNA Expression NFκB pathway gene panel, was investigated in order to find potential gene-environment interactions.
MATERIAL AND METHODS
Study population
The Kuopio Obesity Surgery (KOBS) study is an ongoing study, with 89 individuals (age 46.3 ± 8.8 years, BMI 44.9 ± 6.3 kg/m2) currently included in it (Table 1) (11). Clinical parameters were assessed prior to the gastric bypass surgery (at baseline, n = 89) and at 1 year after surgery (follow-up, n = 64). Biopsies from liver, SAT and VAT, and PBMCs were collected from the participants at the time of surgery. In addition, SAT and liver biopsies were taken 1 year after the surgery (11 individuals available for follow-up liver biopsy). Blood samples were drawn after a 12 h overnight fast, both at baseline and follow-up. All participants were guided to follow a VLC diet for 4–5 weeks before the surgery with the aim to lose weight (10%) before the operation. A time point after the VLC diet is considered as baseline. For the purposes of this study, it is essential to note that diet was standardized with the aim to reach a caloric intake of 800–1,000 kcal using primarily commercial products for the VLC diet. Weight loss in this study during the 1 year follow-up was −24 ± 9% (Table 1). The study protocol was approved by the Ethics Committee of Northern Savo Hospital District and carried out in accordance with the Helsinki Declaration. Informed written consent was obtained from all participants.
TABLE 1.
Clinical characteristics at baseline and at 1 year follow-up in the KOBS study
| Baseline (n = 89) | 1 Year (n = 64) | P | |
| Age (years) | 46.31 ± 8.77 | 46.85 ± 8.60 | — |
| BMI (kg/m2) | 44.87 ± 6.32 | 34.45 ± 5.67 | 9 × 10−30 |
| Total cholesterol (mmol/l) | 4.48 ± 1.08 | 4.45 ± 1.02 | 3 × 10−01 |
| HDL-cholesterol (mmol/l) | 1.10 ± 0.30 | 1.42 ± 0.39 | 1 × 10−16 |
| LDL-cholesterol (mmol/l) | 2.68 ± 0.96 | 2.50 ± 0.90 | 9 × 10−03 |
| TGs (mmol/l) | 1.58 ± 0.70 | 1.18 ± 0.48 | 3 × 10−09 |
| Fasting glucose (mmol/l) | 6.58 ± 1.65 | 5.51 ± 0.90 | 5 × 10−07 |
| Fasting insulin (mU/l) | 21.91 ± 23.08 | 11.81 ± 11.71 | 2 × 10−08 |
Values are presented as mean ± SD, paired samples t-test.
Biochemical measurements
Plasma glucose, insulin, and serum lipid (total cholesterol, HDL-cholesterol, and TGs) levels were measured from fasting venous blood samples. Plasma glucose was measured by enzymatic hexokinase photometric assay (Konelab Systems reagents; Thermo Fischer Scientific, Vantaa, Finland). Insulin was determined by immunoassay (ADVIA Centaur Insulin IRI, number 02230141; Siemens Medical Solutions Diagnostics, Tarrytown, NY). An enzymatic colorimetric method (Wako NEFA C test kit; Wako Chemicals, Neuss, Germany) was used for serum FFA measurement.
AT gene expression
All samples for gene expression analysis were immediately frozen in liquid nitrogen. Total RNA from AT was extracted using Tri-Reagent (Applied Biosystems, Foster City, CA) and reverse-transcribed using the High Capacity cDNA Reverse Transcription kit (Applied Biosystems) according to the manufacturer’s protocol. Quantitative real-time PCR was carried out with the Applied Biosystems 7500 real-time PCR system using KAPA SYBR FAST qPCR Universal Master Mix (Kapa Biosystems, Woburn, MA). The primer sequences for IL-1β are forward 5′-CAGCTACGAATCTCCGACCA and reverse 5′-TCCATGGCCACAACAACTGA and for RPLP0, which was used as an endogenous control gene, forward 5′-GGCGACCTGGAAGTCCAACT and reverse 5′-CCATCAGCACCACAGCCTTC.
TruSeq targeted RNA expression
A fixed NFκB pathway gene panel (Illumina, San Diego, CA) was used for measuring gene expression levels in human SAT (n = 86) before and 1 year after surgery using the MiSeq system (Illumina) according to instructions provided by the manufacturer. Total RNA (50 ng) from SAT was reverse-transcribed using ProtoScript II reverse transcriptase (New England BioLabs). Oligo pool targeted regions of interest were hybridized to cDNA. Next, hybridized cDNA was extended by DNA polymerase followed by ligation using DNA ligase. The extension-ligation products were amplified with PCR and AMPure XP beads (Beckman Coulter) and used to clean up the PCR products. Equal volumes of the products were pooled together and quantitated with DNA 1000 chip (Agilent Technologies, Wilmington, DE). Finally, the pooled sample was diluted and denatured and sequenced with MiSeq.
Assessment of FA composition
FA composition in serum and AT was analyzed according to previously described methods (12, 13). In short, the serum samples were extracted with chloroform-methanol (2:1) and the different lipid fractions, cholesteryl ester, TG, and phospholipid, were separated by solid phase extraction with an aminopropyl column. The AT samples were pulverized with liquid nitrogen and approximately 40 mg of AT was extracted with chloroform-methanol (2:1), as described above. Serum and AT FAs were transmethylated with 14% boron trifluoride in methanol and were analyzed by a 7890A gas chromatograph (Agilent Technologies) equipped with a 25 m FFAP column. Cholesteryl nonadecanoate (Nu Chek Prep, Inc., Elysian, MA), trinonadecanoin, and phosphatidylcholine dinonadecanoyl (Larodan Fine Chemicals, Malmö, Sweden) served as internal standards. Enzyme activities in different lipid fractions were estimated as product-to-precursor ratios of individual FAs as follows: stearoyl-CoA desaturase (SCD) = 16:1 n-7/16:0 or 18:1 n-9/18:0, elongase = 18:0/16:0 or 18:1 n-7/16:1 n-7, D5D = 20:4 n-6/20:3 n-6, D6D = 18:3 n-6/18:2 n-6, and de novo lipogenesis = 16:0/18:2 n-6. The terms D5D and D6D are used for the estimated enzyme activities in serum and AT, and FADS1 and FADS2 in italic type are used for the genes.
Genotype analyses
The variants, rs174547 (FADS1) and rs174616 (FADS2), were genotyped from 154 DNA samples of the KOBS study using the TaqMan SNP genotyping assay (Applied Biosystems) according to the manufacturer’s protocol. These SNPs were chosen for the current analysis because of their previous association with PUFA levels and desaturase activities (14–17).
Statistical analysis
Statistical analyses were conducted with SPSS software (version 19; SPSS Inc., Chicago, IL). Paired samples t-test was used to compare the baseline and follow-up data and ANOVA was used for the comparison between genotype groups. The correlation between variables was analyzed using Spearman’s nonparametric correlation. The Hardy-Weinberg equilibrium and genotype distribution among study groups were analyzed with the χ2-test. Genetic associations with continuous variables were analyzed with general linear models univariate ANOVA and Bonferroni’s multiple correction was used when there was a significant association in order to find differences between genotype groups. For the TruSeq targeted RNA expression (TREx) analysis, the expression levels for each gene per sample in the NFκB gene panel were normalized based on the total number of aligned reads of the corresponding sample and the results are shown as percentage of total transcript reads. The results of the TREx analysis were analyzed for the deviation of Spearman’s correlations between the expression of individual genes and respective FA parameters from the null hypothesis (correlation coefficient = 0) using one sample t-test. Paired samples (Spearman’s correlation coefficient for individual genes at baseline and follow-up) t-test was used to compare the baseline and follow-up Spearman correlation coefficients. Descriptive statistics are presented as mean ± SD. False discovery rate-adjusted P value (P < 0.05) using the Benjamini and Hochberg method was considered statistically significant in the TREx analysis.
RESULTS
Characteristics
BMI, levels of serum total TGs, LDL-cholesterol, fasting plasma glucose, and insulin decreased, and the level of HDL-cholesterol increased, in response to surgery (P < 0.05, Table 1), as previously published in the same cohort (18, 19).
IL-1β expression is decreased after surgery-induced weight loss in several human tissues
First, to set up the model in which we investigated the relationship between tissue inflammation and FA metabolism, we measured IL-1β mRNA expression in AT, liver, and PBMCs before and after surgery. The expression was decreased in SAT (P = 0.035) and PBMCs (P = 2.25 × 10−8) in response to surgery (Table 2). In the liver, there was a tendency for reduced IL-1β expression after weight loss (P = 0.343). However, the difference was not significant, probably due to the low number of liver samples in the follow-up (n = 11).
TABLE 2.
IL-1β mRNA expression in different tissues at baseline and follow-up in the KOBS study
| IL-1β Expression (AU) | Baseline | Follow-up | P |
| PBMC (n = 56) | 0.00423 ± 0.0026 | 0.00209 ± 0.0016 | 2 × 10−8 |
| AT (n = 65) | 0.00209 ± 0.0031 | 0.00123 ± 0.0014 | 0.035 |
| Liver (n = 11) | 0.00143 ± 0.0018 | 0.00089 ± 0.0005 | 0.343 |
Values are presented as mean ± SD, paired samples t-test. AU, arbitrary unit.
Serum FA profile associates with IL-1β expression in AT
In the analysis of serum and AT FA composition in different lipid fractions, we concentrated on the TG fraction due to the fact that the FA profile in this fraction reflects both the diet and endogenous FA metabolism (20). Furthermore, the strongest correlations between serum FA composition and tissue IL-1β expression were observed in the TG fraction. Correlations between tissue IL-1β expression and FA composition in serum cholesteryl ester, phospholipid, and AT TG fractions are shown in supplementary Table 1.
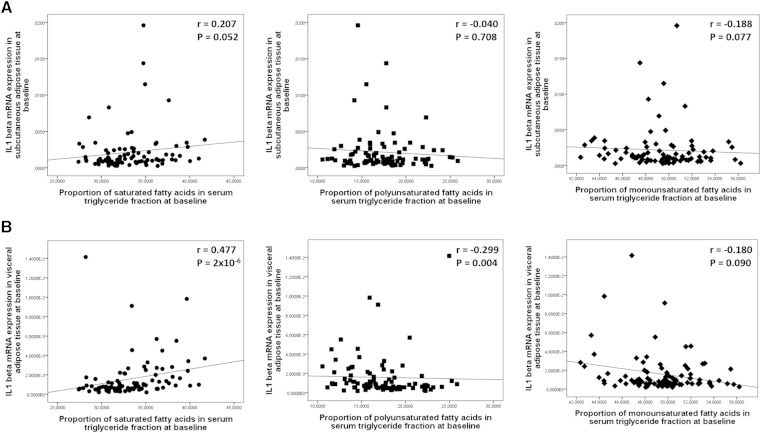
There was a strong positive correlation between the proportion of serum SFA in the TG fraction and IL-1β expression in VAT (Fig. 1B). For individual SFAs in the TG fraction, positive correlations of myristic acid and stearic acid were higher with IL-1β expression in VAT than with IL-1β expression in SAT (Table 3). Total PUFA proportion in the TG fraction correlated negatively with IL-1β expression in VAT, but not in SAT (Fig. 1A, B). This could be explained by negative correlation of LA, a substrate for D6D activity, with IL-1β expression in VAT. On the other hand, the correlation of γ-linolenic acid (GLA), a product of D6D activity, was positive with IL-1β expression both in SAT and in VAT (P = 0.012 and P = 0.001, respectively). In fact, GLA was the only one of the individual FAs that correlated significantly with IL-1β expression in SAT at the follow-up (Table 3). The proportion of serum MUFAs in the TG fraction correlated negatively with IL-1β expression in SAT, but not in VAT (Fig. 1A, B). Among the MUFAs in the TG fraction (Table 3), oleic acid correlated negatively with IL-1β expression in SAT and VAT (P < 0.05), and eicosenoic acid correlated positively with IL-1β expression in SAT at baseline (P < 0.01). There were no correlations between serum FAs in different lipid fractions and IL-1β expression in the liver at baseline (supplementary Table 1) or PBMCs (not shown). Taken together, the correlations of serum FA composition in the TG fraction with IL-1β expression in tissues was stronger in VAT than in SAT, and there were few correlations between serum FA composition in different lipid fractions and IL-1β expression in liver or PBMCs (supplementary Table 1).
Fig. 1.
Scatterplots demonstrating correlations of IL-1β gene expression in SAT (n = 89) (A) and VAT (n = 90) (B) with the proportions (as mole percent) of SFAs (black circles), PUFAs (black squares), and MUFAs (black diamonds) in the serum TG fraction at baseline of the KOBS study.
TABLE 3.
Spearman’s correlation coefficients of IL-1β expression in SAT and VAT with the serum FA profile of the TG fraction in the KOBS study
| FA (mol%) | Baseline | Follow-up | |
| SAT (n = 89) | VAT (n = 90) | SAT (n = 64) | |
| SFA | |||
| Myristic acid (14:0) | 0.301b | 0.453b | 0.161 |
| Palmitic acid (16:0) | 0.129 | 0.430b | 0.117 |
| Stearic acid (18:0) | 0.377b | 0.435b | 0.209 |
| MUFA | |||
| Palmitoleic acid (16:1n-7) | 0.026 | 0.025 | 0.152 |
| Oleic acid (18:1n-9) | −0.223a | −0.234a | −0.068 |
| Vaccenic acid (18:1n-7) | −0.108 | −0.093 | −0.026 |
| Eicosenoic acid (20:1n-9+11) | 0.324b | 0.202 | −0.088 |
| PUFA | |||
| Total n-6 FA | −0.072 | −0.314b | −0.140 |
| LA (18:2n-6) | −0.038 | −0.305b | −0.159 |
| GLA (18:3n-6) | 0.267a | 0.344b | 0.284a |
| Dihomo-γ-linolenic acid (20:3n-6) | 0.133 | 0.158 | 0.173 |
| AA (20:4n-6) | −0.185 | −0.151 | 0.001 |
| Total n-3 FA | 0.011 | −0.160 | −0.025 |
| α-Linolenic acid (18:3n-3) | 0.148 | 0.026 | −0.094 |
| Eicosapentaenoic acid (20:5n-3) | 0.004 | −0.038 | 0.004 |
| Docosahexaenoic acid (22:6n-3) | −0.052 | −0.148 | −0.040 |
P < 0.05.
P < 0.01.
Estimated D6D enzyme activity in serum TG fraction correlates with IL-1β expression in SAT
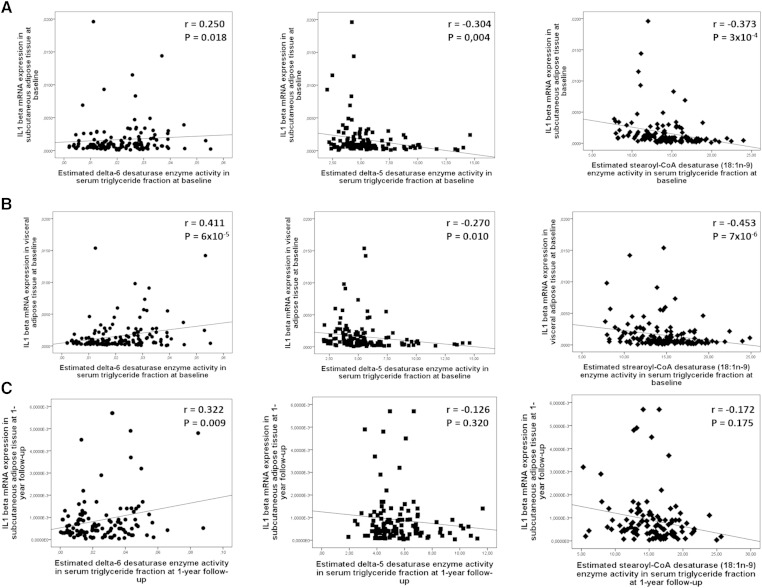
Next we investigated whether the correlations between serum FA composition and AT inflammation could be linked to changes with the activity indexes of the enzymes regulating endogenous FA metabolism. In general, the correlations of the enzyme activity indexes in the TG fraction with IL-1β expression at baseline were similar in both AT depots (Fig. 2). The only exception was the de novo lipogenesis enzyme activity, which correlated positively with IL-1β expression in VAT, but not in SAT (supplementary Table 2). Of the other desaturase enzyme activity indexes, the correlations of D5D and SCD activity on 18 carbon FAs in the TG fraction with IL-1β expression were negative at baseline, but not at follow-up (Fig. 2A–C). The estimated D6D enzyme activity in the TG fraction was the only index that correlated positively with IL-1β expression both in VAT and SAT at baseline, and with IL-1β expression in SAT at follow-up (Fig. 2A, C). There were no correlations of SCD activity on 16 carbon FAs with IL-1β expression (supplementary Table 2). In addition, IL-1β expression in both AT depots correlated positively with elongase enzyme activity on SFAs (18:0/16:0), but there was no correlation with the elongase activity on MUFAs (18:1n-7/16:1n-7, supplementary Table 2). The correlations of enzyme activity indexes with IL-1β expression in liver and PBMCs are shown in supplementary Table 2.
Fig. 2.
Scatterplots demonstrating correlations of IL-1β gene expression in SAT (n = 89) (A), VAT (n = 90) (B) with estimated enzyme activities at baseline, and in SAT (n = 64) (C) at follow-up of the KOBS study. Black circles, D6D (18:3n-6/18:2n-6); black squares, D5D (20:4n-6/20:3n-6); black diamonds, SCD 18:1n-9 (18:1n-9/18:0).
Genetic variations in FADS1 and FADS2 genes associate with D5D and D6D activities and IL-1β expression in AT
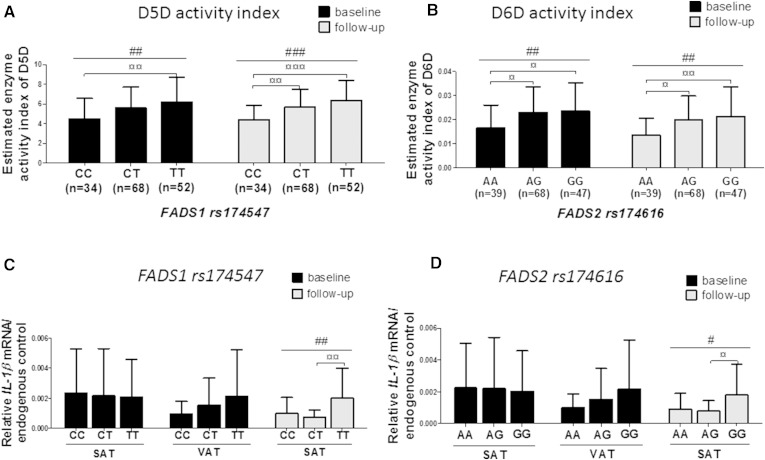
Because the estimated D5D and D6D enzyme activities in the TG fraction correlated with AT inflammation, we next investigated whether genetic variations in FADS1 and FADS2 genes associate with serum FA profile, as published before (14–17), and with IL-1β expression in AT. The SNPs, rs174616 (FADS2) and rs174547 (FADS1), were in Hardy-Weinberg equilibrium (0.277, 0.591), and in linkage disequilibrium (r2 = 0.33, D’= 0.70), as known due the existence of these genes in the same gene cluster (17). There were no associations of these SNPs with anthropometric measures, glucose and insulin concentrations, and with concentrations of serum TGs, or total, LDL-, or HDL-cholesterol at baseline or follow-up, despite a known association with serum lipids in the genome-wide association studies (14, 17). The lack of associations with serum lipids may be explained by a reduced statistical power compared with genome-wide association studies. However, we could clearly demonstrate an association of the FADS1 and FADS2 SNPs with estimated D5D and D6D enzyme activity indexes at baseline and follow-up (P < 0.006, Fig. 3A, B). The estimated D6D enzyme activity in the serum TG fraction increased after weight loss (0.022 ± 0.011 vs. 0.025 ± 0.016, P < 0.001), and associated with FADS2 genotype so that subjects with major allele (G) were associated with increased enzyme activity of D6D after weight loss (0.024 ± 0.012 vs. 0.032 ± 0.018, P < 0.001). In addition, the change of D6D enzyme activity was associated both with FADS2 (P = 0.001) and FADS1 (P = 4.0 × 10−4) genotypes. The estimated D5D enzyme activity was not changed after weight loss (5.540 ± 2.338 vs. 5.673 ± 1.946, P = 0.482).
Fig. 3.
Enzyme activities of D5D (A) and D6D (B), and IL-1β expression in SAT and VAT (C, D) according to the FADS1 and FADS2 genotypes in the KOBS study. #P < 0.05, ##P < 0.01, ###P < 0.001 (univariate general linear model), ¤P < 0.05, ¤¤P < 0.01, and ¤¤¤P < 0.001 (additive genetic model in univariate general linear model analysis with Bonferroni’s correction).
The most important finding of the genetic analysis was that both variants, rs174547 (FADS1) and rs174616 (FADS2), were associated with IL-1β expression in SAT after weight reduction at 1 year follow-up, but not at baseline, indicating that these genes are able to regulate AT inflammation after weight loss-induced reduction in AT inflammation (P = 0.004 and P = 0.031, respectively, Fig. 3C, D). This association was also significant after adjustment with the change of BMI (P = 0.003, P = 0.032, respectively). Individuals having major alleles (T or G) of the FADS1/2 polymorphisms that associate with increased D5D and D6D enzyme activities after weight loss (Fig. 3A, B) continued to also have higher IL-1β expression after weight loss, whereas subjects with minor alleles (C or A) tended to decrease IL-1β expression after weight loss (P = 0.135) (Fig. 3C, D).
Analysis of the TREx NFκB pathway gene panel
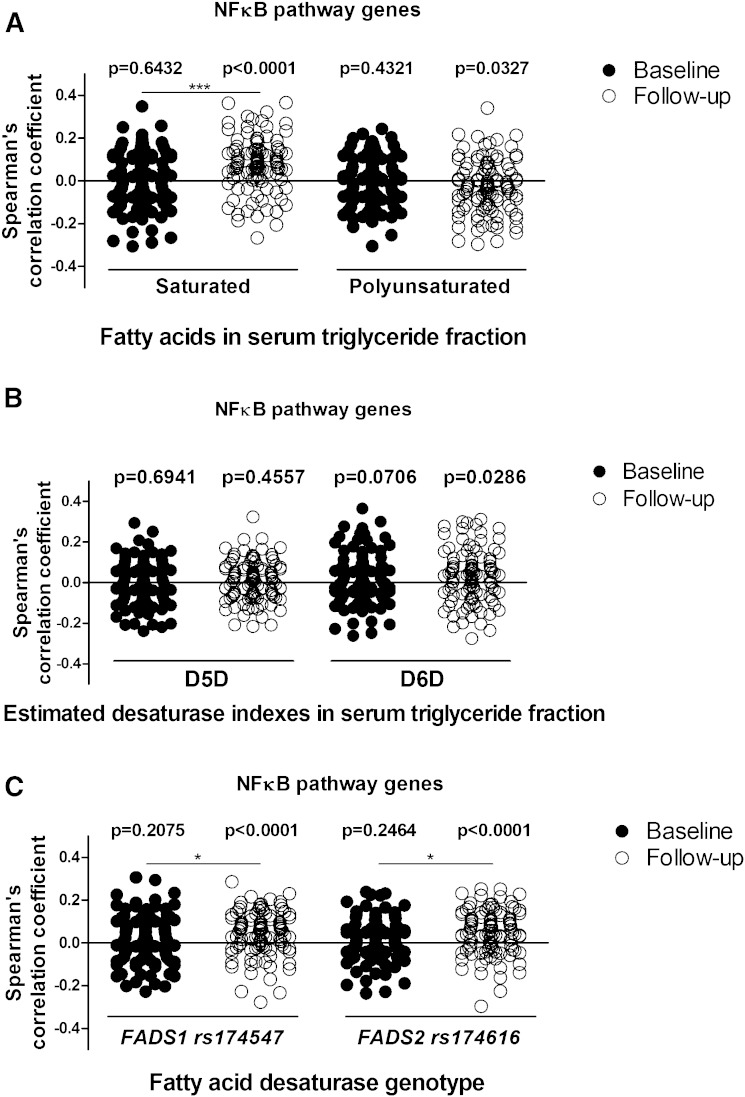
Because our results were based only on the expression of a single gene, IL-1β, we proceeded to the analysis of the NFκB pathway gene panel. Gene expression and correlation results are shown in supplementary Tables 3 and 4. We could verify that inflammatory genes had, on average, a positive correlation with serum SFAs of the TG fraction (average Spearman’s correlation 0.077 ± 0.125) (Fig. 4A) and the estimated D6D enzyme activity index (0.028 ± 0.130) (Fig. 4B), and, on average, a negative correlation with serum PUFAs of the TG fraction (−0.027 ± 0.125) (Fig. 4A) at follow-up, but not at baseline. Importantly, the effect of FADS1 and FADS2 genotypes on inflammatory genes (0.044 ± 0.103 and 0.049 ± 0.104, respectively) was observed at follow-up, but not at baseline (Fig. 4C), similarly as observed with an association between the genotypes and IL-1β gene expression (Fig. 3C, D). Finally, the average correlation of NFκB pathway genes with SFAs of the TG fraction and FADS1/2 genotypes at follow-up differed significantly when compared with baseline (P = 7 × 10−6, P = 0.0125, and P = 0.0153, respectively; Fig. 4A, C).
Fig. 4.
Spearman’s correlation coefficient values of TREx NFκB pathway gene expression in SAT with SFAs and PUFAs (A), estimated D5D and D6D enzyme activities (B), and genetic variations in the FADS1 (rs174547) and FADS2 (rs174616) genes (C) in the KOBS study. One sample t-test was used to analyze the deviation of correlation coefficients from 0 at baseline or follow-up. Paired t-test was used to compare correlation coefficients for genes at baseline and follow-up. *P < 0.05, ***P < 0.0001.
FA composition in the TG fraction of AT
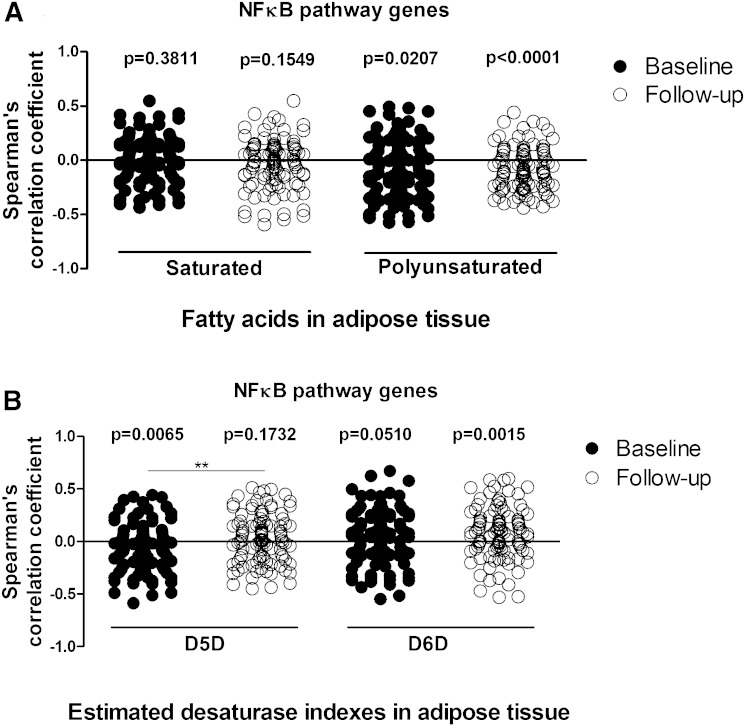
FA composition and correlation results in serum and AT TG fractions are shown in supplementary Table 5. The most abundant FAs in the TG fraction of serum and AT depots were 16:0, 18:1n-9, and 18:2n-6, as also shown before (21). Our results showed strong correlations between the FA composition in the TG fraction in serum and both AT depots (supplementary Table 5). We could replicate the association of serum PUFAs and estimated D6D enzyme activity with AT inflammation, measured either with IL-1β mRNA expression or using the NFκB gene panel, also when we measured AT FAs instead of serum FAs. Similarly to serum analyses, the estimated D6D enzyme activity in the TG fraction of SAT correlated positively with IL-1β expression in SAT at follow-up (supplementary Table 2). Importantly, the results also demonstrated that NFκB pathway-related inflammatory genes in SAT correlated, on average, negatively with PUFA composition in the TG fraction of SAT, both at baseline and at follow-up, similarly as observed with serum FA analyses. Furthermore, expression of NFκB pathway genes correlated negatively with the estimated D5D enzyme activity in the TG fraction of SAT at baseline and positively with estimated D6D enzyme activity in the TG fraction of SAT at follow-up (Fig. 5). Finally, subjects with FADS2 GG genotype tended to have higher estimated D6D enzyme activity in the TG fraction of SAT (P = 0.096) and VAT (P = 0.095), as was shown with serum analysis. Overall, the observations in AT FAs verified the results with serum FAs that the association of lower levels of PUFAs and higher levels of estimated D6D enzyme activity with AT inflammation is revealed after weight loss.
Fig. 5.
Spearman’s correlation coefficient values of TREx NFκB pathway gene expression in SAT with SFAs and PUFAs in AT (A) and estimated D5D and D6D enzyme activities (B) in the KOBS study. One sample t-test was used to analyze the deviation of correlation coefficients from 0 at baseline or follow-up. Paired t-test was used to compare correlation coefficients for genes at baseline and follow-up. **P < 0.01.
DISCUSSION
We investigated the association between FA metabolism and tissue inflammation after standardized VLC diet and after a substantial weight loss induced by obesity surgery. As expected, we observed a decrease in IL-1β expression in several tissues in response to weight loss (Fig. 1) (18, 19). However, the association between serum FA composition in the TG fraction and IL-1β expression were observed both after the standardized diet and after the surgery. This suggests that there is an association between endogenous FA metabolism and IL-1β expression in AT. In fact, estimated desaturase enzyme activities, both in serum and SAT, associated with IL-1β expression in AT, and FADS1 and FADS2 genotypes associated with IL-1β expression and expression of genes in the NFκB pathway after follow-up in AT (Figs. 3, 4). All these findings suggest that the genetic variation in the FADS1/2 genes and the corresponding desaturase enzyme activities are involved in endogenous FA metabolism and AT low-grade inflammation.
The regulation of PUFA-derived pro- and anti-inflammatory eicosanoid levels by D5D and D6D activities are of great interest in the context of AT low-grade inflammation (15). D6D and D5D have been shown to have opposite effects, as shown by an inverse correlation of D5D and a direct correlation of D6D activities with the risk of diabetes and insulin resistance (IR) (22). Recently, it has been shown that a genetic variation in FADS2 gene, encoding for D6D, was associated with decreased risk of type 2 diabetes (23). Interestingly, the most consistent findings of our study related to the FADS2 gene and corresponding D6D enzyme activity in the TG fraction, calculated as the ratio of serum 18:3 n-6/18:2 n-6. First, we could show that both serum and SAT D6D activities associate with IL-1β expression in several tissues (Fig. 2, supplementary Table 2). Second, subjects who had TT or GG genotypes of FADS1/2 polymorphism, which associated with higher D5D and D6D enzyme activities, also associated with higher IL-1β expression in SAT after weight loss, when compared with other genotypes. However, IL-1β expression at baseline was not influenced by FADS1 and FADS2 genetic variations. This suggests a sustained AT inflammation in subjects with TT or GG genotypes of FADS1 and FADS2 genes despite the fact that IL-1β expression decreased after weight reduction. This could be explained, in part, by the fact that the change of BMI was smaller in subjects with GG genotype (P = 0.03) when compared with AA genotype of the FADS2, indicating a possible decreased effect of weight loss on AT low-grade inflammation. It should be noted that D6D, as well as D5D, activities are regulated by insulin (24, 25). Therefore, the effect of the genotype on IL-1β expression may have been revealed in our study after the improvement of hyperinsulinemia in response to weight loss. Overall, these findings confirm the significance of the FADS1 and FADS2 SNPs in the regulation of diabetes risk and tissue inflammation (22). Furthermore, our results indicate a gene-environment interaction in the regulation of AT IL-1β expression by FADSs. As a mechanism, we suggest that genetic variation in FADS2 that increases the activity of D6D resulting in increased levels of serum AA interacts with the inflammatory/anti-inflammatory effects of the diet. AA, as a precursor for pro-inflammatory eicosanoids, could be further metabolized to produce pro-inflammatory leukotrienes, which are known to activate NFκB to produce IL-1β (26).
IL-1β is considered to be a marker of low-grade inflammation, which contributes to development of IR and type 2 diabetes (6, 7). Low-grade inflammation in VAT is more strongly correlated with the metabolic disorders than inflammation in SAT (6, 27), and IL-1β expression is shown to be more abundant in VAT than in SAT in obese individuals (7). In line with this, our results showed a more clear relationship between serum FA profile in the TG fraction and IL-1β expression in VAT than in SAT, suggesting that the interaction between FA metabolism and IL-1β expression is more prominent in VAT. For example, serum total SFAs and individual SFAs (myristic, palmitic, and stearic) in the TG fraction correlated positively with IL-1β expression in VAT, but not in SAT. This is further supported by our results, which showed a negative correlation of IL-1β expression and SCD activity on 18 carbon FAs and by a positive correlation with elongase (18:0/16:0) activity. Decreased SCD activity is associated with increased SFA accumulation and development of inflammation (15), and elongase (18:0/16:0) is responsible for the production of SFAs by the elongation of palmitic acid to stearic acid (28). Previous studies have shown that SFAs stimulate IL-1β-induced AT inflammation through activation of the TLR4/NFκB pathway and inflammasome complex (29, 30). Recently, it has also been shown that MUFAs can attenuate IL-1β-mediated AT inflammation through AMPK activation (29). In line with this, our results also showed a negative correlation between oleic acid and IL-1β expression in both adipose depots at baseline of the KOBS study.
In fact, there are several possible mechanisms that link serum FA composition to the regulation of IL-1β gene expression in AT. Serum FAs reflect AT FAs (21) and, thus, the link between serum FAs and IL-1β gene expression regulation in AT may, in fact, be showing the association between AT FAs and AT inflammation. Alternatively, it is known that FAs have hormone-like effects and are able to regulate gene expression (31) and, thus, serum FAs could also regulate gene expression in tissues. However, the exact mechanisms by which n-6 PUFAs and FADS2 genotype coregulate AT IL-1β expression needs to be elucidated. We propose that the lack of significant correlations between IL-1β expression and FA composition in AT could be explained, in part, by the smaller sample size with AT samples. However, we could clearly demonstrate a correlation between serum and AT FA composition in the TG fraction (supplementary Table 5), demonstrating that serum samples, with some exceptions, can be used as markers of AT FAs. This is in line with previous studies demonstrating that plasma TGs are an important source of endogenous FA and are in constant interaction with AT FAs (32). We postulate that serum FA composition can be used as a marker of endogenous FA metabolism in AT in our study.
We could also verify the observation that the association of lower proportions of serum and AT PUFAs, D6D index, and FADS1/2 genotypes on AT inflammation is revealed after weight loss using an NFκB-related gene expression panel. Because of limited sample size, our power was not high enough to identify individual genes related to serum and AT FA profile in the TG fraction when using false discovery rate-corrected P values (supplementary Table 3). However, as a group, we could verify that NFκB-related genes had, on average, a more positive correlation with serum SFAs and FADS1/2 genotypes at follow-up when compared with baseline. Moreover, we could show, both in serum and in AT, a negative correlation between NFκB-related genes and PUFAs and a positive correlation between NFκB-related genes and estimated D6D enzyme activity at follow-up. Specifically, the finding that an association between AT inflammation and FADS1/2 genotypes was observed only after weight loss suggests an interaction between obesity (or obese state in general) and genotype (Fig. 4C). We suggest that the most likely explanation is that the effect of the genotype is masked by obesity-related AT inflammation and related comorbidities (e.g., hyperglycemia, hyperinsulinemia) at baseline. Furthermore, we suggest that the association is revealed after weight loss and concurrent improvement in IR. On the other hand, this implies that individuals with major alleles of the FADS1 and FADS2 genotypes with the higher estimated D6D enzyme activity may not benefit as much from weight loss with regard to AT inflammation (Fig. 3C, D).
There are some limitations in our study. First, the participants of the study were obese, even after weight reduction, and thus the results cannot be generalized to individuals with normal body weight. On the other hand, low-grade inflammation is closely related to obesity and, thus, it is difficult to investigate in lean individuals. We acknowledge the calculated enzyme activity indexes for FADSs are estimates of the actual enzyme activity. However, they have been widely used as surrogate markers for FA metabolism (33, 34). Finally, we clearly observed that for genetic association studies, our sample size was small because we did not observe the association of the FADS1/2 SNPs with serum lipids (14, 17). However, we still had enough power to detect the direct effect on respective enzyme activities.
We conclude that the genetic variation in the FADS1/2 genes associates with FA metabolism and AT inflammation, leading to an interaction between weight loss and FADS1/2 genes in the regulation of AT inflammation. This supports the hypothesis that genetic regulation of endogenous FA metabolism interacts with obesity in the regulation of tissue inflammation.
Supplementary Material
Acknowledgments
The authors thank Erja Kinnunen, Päivi Turunen, Tiina Sistonen, and Matti Laitinen for their technical assistance.
Footnotes
Abbreviations:
- AA
- arachidonic acid
- AT
- adipose tissue
- D5D
- delta-5 desaturase
- D6D
- delta-6 desaturase
- FADS
- FA desaturase
- GLA
- γ-linolenic acid
- IL
- interleukin
- IR
- insulin resistance
- KOBS
- Kuopio Obesity Surgery
- LA
- linoleic acid
- NFκB
- nuclear factor κ B
- PBMC
- peripheral blood mononuclear cell
- SAT
- subcutaneous adipose tissue
- SCD
- stearoyl-CoA desaturase
- SFA
- saturated FA
- TLR4
- toll-like receptor 4
- TREx
- TruSeq targeted RNA expression
- VAT
- visceral adipose tissue
- VLC
- very low calorie
This study was financially supported by grants from the Academy of Finland (contract numbers 120979 and 138006), the Finnish Diabetes Research Foundation, the Finnish Cultural Foundation and Northern Savo Regional Fund, and the Kuopio University Hospital State Research Funding (EVO and VTR).
The online version of this article (available at http://www.jlr.org) contains a supplement.
REFERENCES
- 1.Hellmann J., Zhang M. J., Tang Y., Rane M., Bhatnagar A., and Spite M.. 2013. Increased saturated fatty acids in obesity alter resolution of inflammation in part by stimulating prostaglandin production. J. Immunol. 191: 1383–1392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Schwab U., Lauritzen L., Tholstrup T., Haldorssoni T., Riserus U., Uusitupa M., and Becker W.. 2014. Effect of the amount and type of dietary fat on cardiometabolic risk factors and risk of developing type 2 diabetes, cardiovascular diseases, and cancer: a systematic review. Food Nutr. Res. 58: doi:10.3402/fnr.v58.25145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lee H., and Park W. J.. 2014. Unsaturated fatty acids, desaturases, and human health. J. Med. Food. 17: 189–197. [DOI] [PubMed] [Google Scholar]
- 4.Chait A., and Kim F.. 2010. Saturated fatty acids and inflammation: who pays the toll? Arterioscler. Thromb. Vasc. Biol. 30: 692–693. [DOI] [PubMed] [Google Scholar]
- 5.Teng K. T., Chang C. Y., Chang L. F., and Nesaretnam K.. 2014. Modulation of obesity-induced inflammation by dietary fats: mechanisms and clinical evidence. Nutr. J. 13: 12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Esser N., L’homme L., De Roover A., Kohnen L., Scheen A. J., Moutschen M., Piette J., Legrand-Poels S., and Paquot N.. 2013. Obesity phenotype is related to NLRP3 inflammasome activity and immunological profile of visceral adipose tissue. Diabetologia. 56: 2487–2497. [DOI] [PubMed] [Google Scholar]
- 7.Nov O., Shapiro H., Ovadia H., Tarnovscki T., Dvir I., Shemesh E., Kovsan J., Shelef I., Carmi Y., Voronov E., et al. . 2013. Interleukin-1beta regulates fat-liver crosstalk in obesity by auto-paracrine modulation of adipose tissue inflammation and expandability. PLoS One. 8: e53626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Martinelli N., Girelli D., Malerba G., Guarini P., Illig T., Trabetti E., Sandri M., Friso S., Pizzolo F., Schaeffer L., et al. . 2008. FADS genotypes and desaturase activity estimated by the ratio of arachidonic acid to linoleic acid are associated with inflammation and coronary artery disease. Am. J. Clin. Nutr. 88: 941–949. [DOI] [PubMed] [Google Scholar]
- 9.Ameur A., Enroth S., Johansson A., Zaboli G., Igl W., Johansson A. C., Rivas M. A., Daly M. J., Schmitz G., Hicks A. A., et al. . 2012. Genetic adaptation of fatty-acid metabolism: a human-specific haplotype increasing the biosynthesis of long-chain omega-3 and omega-6 fatty acids. Am. J. Hum. Genet. 90: 809–820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Aslibekyan S., Jensen M. K., Campos H., Linkletter C. D., Loucks E. B., Ordovas J. M., Deka R., Rimm E. B., and Baylin A.. 2012. Fatty acid desaturase gene variants, cardiovascular risk factors, and myocardial infarction in the Costa Rica study. Front. Genet. 3: 72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Pihlajamäki J., Kuulasmaa T., Kaminska D., Simonen M., Kärjä V., Grönlund S., Käkelä P., Pääkkönen M., Kainulainen S., Punnonen K., et al. . 2012. Serum interleukin 1 receptor antagonist as an independent marker of non-alcoholic steatohepatitis in humans. J. Hepatol. 56: 663–670. [DOI] [PubMed] [Google Scholar]
- 12.Venäläinen T., Schwab U., Ågren J., de Mello V., Lindi V., Eloranta A. M., Kiiskinen S., Laaksonen D., and Lakka T. A.. 2014. Cross-sectional associations of food consumption with plasma fatty acid composition and estimated desaturase activities in Finnish children. Lipids. 49: 467–479. [DOI] [PubMed] [Google Scholar]
- 13.Agren J. J., Julkunen A., and Penttila I.. 1992. Rapid separation of serum lipids for fatty acid analysis by a single aminopropyl column. J. Lipid Res. 33: 1871–1876. [PubMed] [Google Scholar]
- 14.Hellstrand S., Sonestedt E., Ericson U., Gullberg B., Wirfalt E., Hedblad B., and Orho-Melander M.. 2012. Intake levels of dietary long-chain PUFAs modify the association between genetic variation in FADS and LDL-C. J. Lipid Res. 53: 1183–1189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Merino D. M., Ma D. W., and Mutch D. M.. 2010. Genetic variation in lipid desaturases and its impact on the development of human disease. Lipids Health Dis. 9: 63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Merino D. M., Johnston H., Clarke S., Roke K., Nielsen D., Badawi A., El-Sohemy A., Ma D. W., and Mutch D. M.. 2011. Polymorphisms in FADS1 and FADS2 alter desaturase activity in young Caucasian and Asian adults. Mol. Genet. Metab. 103: 171–178. [DOI] [PubMed] [Google Scholar]
- 17.Bokor S., Dumont J., Spinneker A., Gonzalez-Gross M., Nova E., Widhalm K., Moschonis G., Stehle P., Amouyel P., De Henauw S., et al.; HELENA Study Group . 2010. Single nucleotide polymorphisms in the FADS gene cluster are associated with delta-5 and delta-6 desaturase activities estimated by serum fatty acid ratios. J. Lipid Res. 51: 2325–2333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Simonen M., Mannisto V., Leppanen J., Kaminska D., Karja V., Venesmaa S., Kakela P., Kuusisto J., Gylling H., Laakso M., et al. . 2013. Desmosterol in human nonalcoholic steatohepatitis. Hepatology. 58: 976–982. [DOI] [PubMed] [Google Scholar]
- 19.Männistö V. T., Simonen M., Soininen P., Tiainen M., Kangas A. J., Kaminska D., Venesmaa S., Käkelä P., Kärjä V., Gylling H., et al. . 2014. Lipoprotein subclass metabolism in nonalcoholic steatohepatitis. J. Lipid Res. 55: 2676–2684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Perassolo M. S., Almeida J. C., Pra R. L., Mello V. D., Maia A. L., Moulin C. C., Camargo J. L., Zelmanovitz T., Azevedo M. J., and Gross J. L.. 2003. Fatty acid composition of serum lipid fractions in type 2 diabetic patients with microalbuminuria. Diabetes Care. 26: 613–618. [DOI] [PubMed] [Google Scholar]
- 21.Hellmuth C., Demmelmair H., Schmitt I., Peissner W., Bluher M., and Koletzko B.. 2013. Association between plasma nonesterified fatty acids species and adipose tissue fatty acid composition. PLoS One. 8: e74927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kröger J., and Schulze M. B.. 2012. Recent insights into the relation of Delta5 desaturase and Delta6 desaturase activity to the development of type 2 diabetes. Curr. Opin. Lipidol. 23: 4–10. [DOI] [PubMed] [Google Scholar]
- 23.Yao M., Li J., Xie T., He T., Fang L., Shi Y., Hou L., Lian K., Wang R., and Jiang L.. 2015. Polymorphisms of rs174616 in the FADS1-FADS2 gene cluster is associated with a reduced risk of type 2 diabetes mellitus in northern Han Chinese people. Diabetes Res. Clin. Pract. 109: 206–212. [DOI] [PubMed] [Google Scholar]
- 24.Vessby B., Gustafsson I. B., Tengblad S., Boberg M., and Andersson A.. 2002. Desaturation and elongation of fatty acids and insulin action. Ann. N. Y. Acad. Sci. 967: 183–195. [DOI] [PubMed] [Google Scholar]
- 25.Brenner R. R. 2003. Hormonal modulation of delta6 and delta5 desaturases: case of diabetes. Prostaglandins Leukot. Essent. Fatty Acids. 68: 151–162. [DOI] [PubMed] [Google Scholar]
- 26.Patterson E., Wall R., Fitzgerald G. F., Ross R. P., and Stanton C.. 2012. Health implications of high dietary omega-6 polyunsaturated Fatty acids. J. Nutr. Metab. 2012: 539426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mathis D. 2013. Immunological goings-on in visceral adipose tissue. Cell Metab. 17: 851–859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Matsuzaka T., Shimano H., Yahagi N., Kato T., Atsumi A., Yamamoto T., Inoue N., Ishikawa M., Okada S., Ishigaki N., et al. . 2007. Crucial role of a long-chain fatty acid elongase, Elovl6, in obesity-induced insulin resistance. Nat. Med. 13: 1193–1202. [DOI] [PubMed] [Google Scholar]
- 29.Joffe Y. T., Collins M., and Goedecke J. H.. 2013. The relationship between dietary fatty acids and inflammatory genes on the obese phenotype and serum lipids. Nutrients. 5: 1672–1705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Finucane O. M., Lyons C. L., Murphy A. M., Reynolds C. M., Klinger R., Healy N. P., Cooke A. A., Coll R. C., McAllan L., Nilaweera K. N., et al. . 2015. Monounsaturated fatty acid-enriched high-fat diets impede adipose NLRP3 inflammasome-mediated IL-1beta secretion and insulin resistance despite obesity. Diabetes. 64: 2116–2128. [DOI] [PubMed] [Google Scholar]
- 31.Garaulet M., Hernandez-Morante J. J., Lujan J., Tebar F. J., and Zamora S.. 2006. Relationship between fat cell size and number and fatty acid composition in adipose tissue from different fat depots in overweight/obese humans. Int. J. Obes. (Lond). 30: 899–905. [DOI] [PubMed] [Google Scholar]
- 32.Garaulet M., Perez-Llamas F., Perez-Ayala M., Martinez P., de Medina F. S., Tebar F. J., and Zamora S.. 2001. Site-specific differences in the fatty acid composition of abdominal adipose tissue in an obese population from a Mediterranean area: relation with dietary fatty acids, plasma lipid profile, serum insulin, and central obesity. Am. J. Clin. Nutr. 74: 585–591. [DOI] [PubMed] [Google Scholar]
- 33.Matthan N. R., Ooi E. M., Van Horn L., Neuhouser M. L., Woodman R., and Lichtenstein A. H.. 2014. Plasma phospholipid fatty acid biomarkers of dietary fat quality and endogenous metabolism predict coronary heart disease risk: a nested case-control study within the Women’s Health Initiative observational study. J. Am. Heart Assoc. 3: doi:10.1161/JAHA.113.000764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Warensjö E., Sundström J., Vessby B., Cederholm T., and Risérus U.. 2008. Markers of dietary fat quality and fatty acid desaturation as predictors of total and cardiovascular mortality: a population-based prospective study. Am. J. Clin. Nutr. 88: 203–209. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.